Abstract
Multidrug-resistant lineages of Acinetobacter baumannii (MDRAB) are important nosocomial pathogens. As tigecycline remains active against most MDRAB we sought to investigate whether tigecycline resistance impacts biological fitness. The effects of treatment-emergent tigecycline resistance were investigated in vitro and in vivo using two pre- (AB210; W6976) and post-therapy (AB211; W7282) clinical pairs, recovered from individual patients, where tigecycline resistance was associated with up-regulated efflux activity. All isolates belonged to the same epidemic UK lineage. Significant differences were observed in end-point survival proportions between AB210 and AB211, but not between W6976 and W7282, using the Galleria mellonella infection model. Isolate AB211 outcompeted AB210 in vivo, in contrast to isolate W7282, which was outcompeted by its pre-therapy counterpart, W6972. Whole-genome sequencing of isolates W6976 and W7282 revealed a mutation in the adeABC regulatory gene, adeS in W7282; resulting in a Ser-8 → Arg substitution. Previous whole-genome comparison of AB210 and AB211 also identified a non-synonymous mutation in adeS, among several other lesions in genes involved in biofilm formation and DNA mismatch repair; consistent with the phenotypic differences described here. In conclusion, the differing effects on the wider phenotype were not predictable from the antibiograms or clonal lineage, despite a common mechanism of tigecycline resistance.
Introduction
Acinetobacter baumannii is a Gram-negative opportunistic pathogen that has emerged in the last decade as one of the most problematic causes of healthcare-associated infection1,2. Once established the organism is extremely difficult to eradicate from the environment; it is capable of withstanding desiccation and the action of many disinfectants. Most strains also exhibit multidrug resistance (MDR), with many of the major epidemic clones that have disseminated worldwide retaining susceptibility to only polymyxins and tigecycline (TGC)3,4. Resistance to even these agents has now been described and in the case of TGC can occur due to up-regulation of efflux pumps of the resistance-nodulation-division (RND) family5,6, among other mechanisms.
Although a vast amount is known about mechanisms of antimicrobial resistance there is relatively little information on many of the basic processes contributing to the pathogenicity of A. baumannii. Amongst the processes that A. baumannii may use to establish human infection, studies performed in vitro suggest that it can readily forms biofilms, is able to adhere to and invade host cells7,8 and modulates the host immune response through an interaction with toll-like receptors (TLR) 2 and 49,10. In terms of its ability to thrive within the human host many strains exhibit serum resistance11 and produce siderophores capable of scavenging iron from host proteins12,13. Other potential virulence factors include lipopolysaccharide14,15 production of exopolysaccharide and a capsule16.
The seemingly endless capacity of A. baumannii to develop resistance raises the question of whether this is costly to the organism. Knowledge of the biological cost of a MDR phenotype is important to gain a fuller understanding of the threat such isolates pose to human health.
The availability of susceptible and resistant pairs of clinical isolates obtained from the same patient offers an opportunity to study links between acquired resistance and virulence. These isolates have developed resistance over the course of a human infection, exposed to both the host immune system and antimicrobial chemotherapy. Previously we reported the in vivo emergence of TGC resistance in two MDR A. baumannii (MDRAB) isolates obtained from separate patients, in both cases in association with up-regulation of the AdeABC efflux system (Table 1)5. Data mining the whole-genome sequences of one of the pre-therapy (AB210; TGC-susceptible) and post-therapy (AB211; TGC-resistant) pair suggested that efflux-mediated TGC resistance might be associated with significant phenotypic differences14. In this study we assessed the impact of in vivo TGC exposure on the relative fitness and pathogenic potential of these MDRAB isolates using a range of in vitro and in vivo assays.
Table 1.
Characteristics of A. baumannii isolates used.
| Isolate | Origin | PFGE-assigned clone | MIC (mg/L) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CTX | IM | MEM | TOB | AMK | COL | TGC | |||
| AB210 | Pre-therapy; clinical | OXA-23 clone 1 | >64 | >256 | >32 | >32 | >32 | >64 | ≤0.5 | 0.5 |
| AB211 | Post-therapy; clinical | OXA-23 clone 1 | >64 | >256 | >32 | >32 | 2 | 4 | ≤0.5 | 16 |
| W6976 | Pre-therapy; clinical | OXA-23 clone 1 | >64 | >256 | >32 | >32 | >32 | >64 | 1 | 0.5 |
| W7282 | Post-therapy; clinical | OXA-23 clone 1 | >64 | 256 | 32 | 32 | >32 | >64 | 1 | 8 |
Results
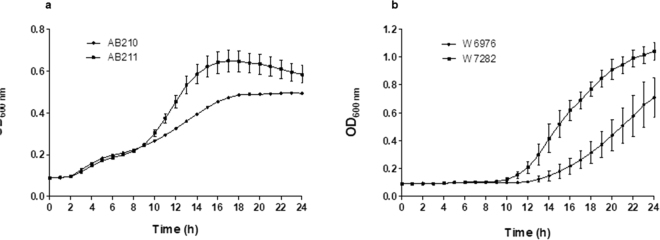
Experiments conducted to compare the in vitro growth rates of each isolate revealed differences in their ability to grow under standard and ‘stressed’ laboratory conditions. Under most conditions, AB210 performed better in numerical terms than AB211, its TGC-resistant counterpart, but not to a statistically significant level. The exception was under iron limitation when AB211 was able to grow faster and to a higher optical density than AB210 in LB broth supplemented with 200 µM 2.2′ dipyridyl (Fig. 1a). Reproducible differences were also observed in the growth rates of the W6976 and W7282 pair. The post-therapy, TGC-resistant isolate was better able to grow at low pH (pH 4.5) (Fig. 1b) suggesting it might be better adapted for survival in acidic environments or acidic compartments of the host.
Figure 1.
Growth curves of pre-and post-therapy isolates: (a) AB210 and AB211 in LB broth supplemented with 200 µM of the iron chelating agent, 2,2′-dipyridyl; (b) W6976 and W7282 in LB broth pH 4.5. Experiments were performed in triplicate. Error bars represent standard deviation.
The ability of isolates to grow in the presence of bile was also assessed. All four organisms were bile-tolerant (growth in the presence of ≥10% bovine bile) although AB210 and AB211 were inhibited at a concentration of 16% w/v while W6976 and W7282 were not.
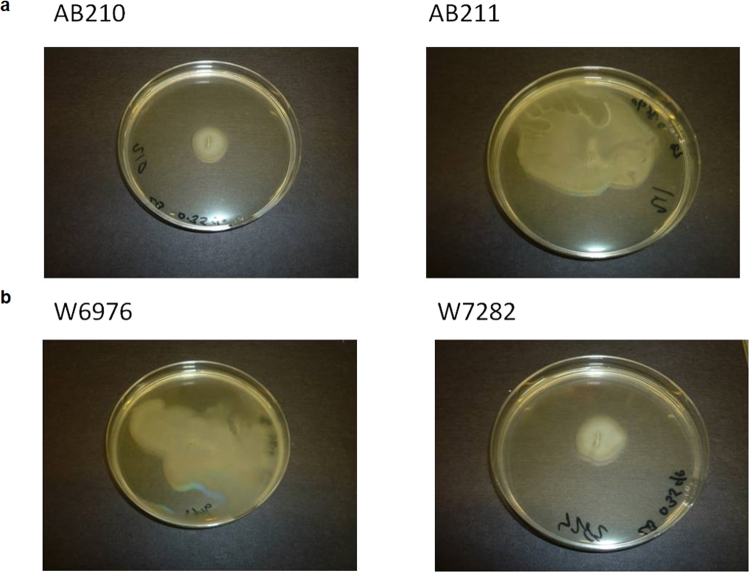
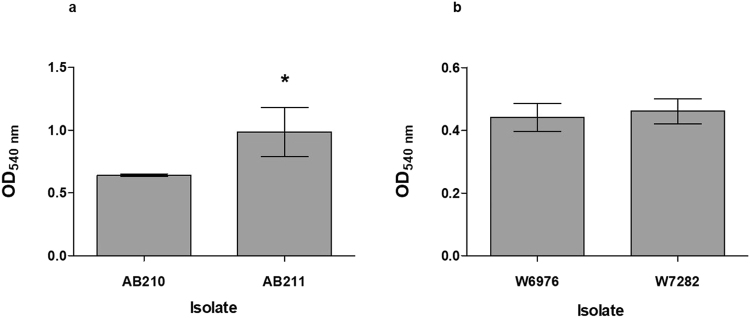
Pre-therapy isolate W6976 appeared to be motile whereas its post-therapy counterpart, W7282, was not. The opposite phenotypes were observed for AB210/AB211 (Fig. 2). When adherence to the wells of polystyrene microtitre plates was compared using a crystal violet assay, the motile, post-therapy isolate AB211 was better able to form a biofilm than AB210 (p = 0.0377), but no differences were found between W6976 and W7282 (p = 0.6024) (Fig. 3).
Figure 2.
Results of the surface motility assay performed on low percentage (0.35%) LB agar plates: (a) AB210 (left) and AB211 (right); (b) W6976 (left) and W7282 (right).
Figure 3.
Biofilm production in the pre-and post-therapy isolates: (a) AB210 and AB211; (b) W6976 and W7282, assessed using a microtitre plate-based crystal violet assay. Experiments were performed in triplicate. Error bars represent standard deviation.
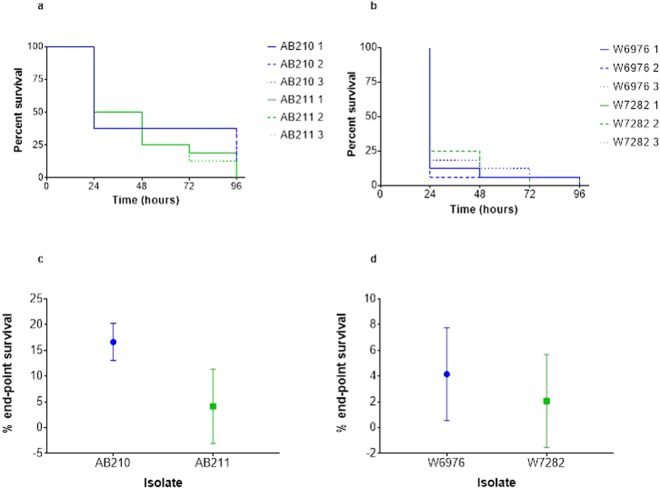
Initial experiments demonstrated that all four A. baumannii isolates were pathogenic to G. mellonella. Analysis of survival curves revealed no reproducible differences in the killing kinetics of AB210 compared with AB211 or between W6976 and W7282 (Fig. 4a and b). However, analysis of end-point survival proportions indicated that AB211 was more virulent than AB210 (p = 0.0382), which was not the case for W7282 when compared with W6976 (p = 0.2593) (Fig. 4c and d). Growth of the isolates in vivo when in direct competition in the same animal was also investigated. After 24 hours incubation at 37 °C, post-therapy isolate AB211 was recovered from infected insects at a median 5-fold higher CFU/larvae compared with pre-therapy isolate AB210, suggesting that it could outcompete its TGC-susceptible counterpart in vivo. In contrast, pre-therapy isolate W6976 out-competed post-therapy isolate W7282 by a median ratio of 3:1 (Fig. 5).
Figure 4.
Survival curves and end-point survival proportions displaying G. mellonella killing: (a) AB210 (blue) and AB211 (green); (b) W6976 (blue) and W7282 (green); (c) average end-point survival percentages (AB210 and AB211); (d) average end-point survival percentages (W6976 and W7282). Data obtained from three biological replicates. Error bars represent standard deviation.
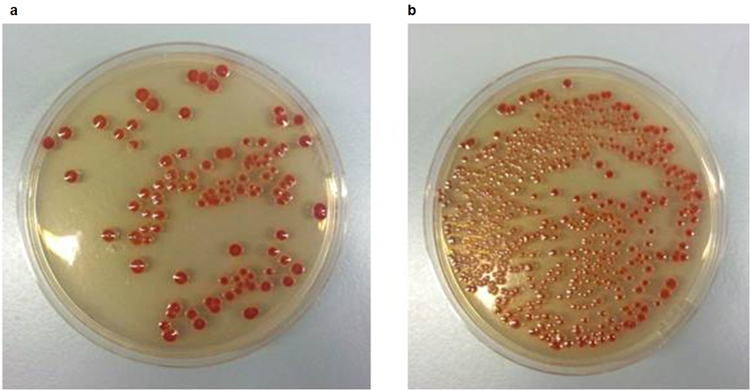
Figure 5.
Images of plates from an in vivo competition assay (AB210 and AB211): (a) AB210 on CHROMagar Acinetobacter with KPC supplement containing 25 mg/L vancomycin and 25 mg/L kanamycin; (b) mixed population of AB210 and AB211 on CHROMagar Acinetobacter with KPC supplement containing 25 mg/L vancomycin.
Hypermutation has been shown to promote both the development of antimicrobial resistance, attenuated virulence, colonization and bacterial persistence in chronic infections15,17. AB211 was found to be a hypermutator as evidenced by the presence of significantly more colonies within the zone of inhibition around a fosfomycin disc compared with AB210 (p = 0.05). No such differences were observed between isolates W6976 and W7282 (p = 0.33) (Table 2).
Table 2.
Hypermutator phenotype screening assay using 200 μg fosfomycin discs.
| Isolate | Number of colonies within zone of inhibition | Median | Mann-Whitney p value (one-tailed) | ||
|---|---|---|---|---|---|
| 1st | 2nd | 3rd | |||
| AB210 | 4 | 0 | 3 | 3 | 0.05 |
| AB211 | 23 | 24 | 27 | 24 | |
| W6976 | 5 | 5 | 4 | 5 | 0.33 |
| W7282 | 6 | 0 | 0 | 0 | |
Whole-genome sequencing of W6976 (BioSample accession: SAMN02471603) and W7282 (BioSample accession: SAMN02471607) produced >40 million and >66 million nucleotide bases with a peak depth of eight- and 14-fold coverage, respectively, which gave estimated genome sizes of 3.91 and 3.95 Mb. Automated gene prediction of isolate W7282 detected 3825 putative coding sequences (CDSs), of which 3478 (91%) were homologous to the A. baumannii ACICU reference sequence (GenBank accession: NC_010611)18. Over 98% of the reads from W6976 mapped to the assembled W7282 genome (GenBank assembly accession: GCA_000248275.2).
Five putative SNPs were detected between the pre- and post- therapy isolates W6976 and W7282, although only one was non-synonymous. This SNP was located in the sensor histidine kinase gene, adeS and was predicted to result in a Ser-8 → Arg substitution. This mutation was not detected by Sanger sequencing when the nucleotide sequence of adeRS was originally investigated5. A larger number of SNPs (n = 18) were found between AB210 and AB211, 8 of which were non-synonymous. Of note, two mutations in AB211 (one nonsense; one non-synonymous) were found in genes encoding putative GGDEF domain containing proteins14. These molecules are thought to influence adhesion and biofilm formation via the second messenger cyclic-di-GMP, which regulates the shift from sessile to planktonic states19. The presence of these mutations in AB211 appears consistent with its motile phenotype and enhanced ability to form a biofilm compared with AB210. A deletion of the N-terminal region of mutS, a gene involved in DNA mismatch repair, was also found in AB21114.
Discussion
The most pressing problem with MDR bacteria is a lack of drugs for effective treatment, though they also pose challenges for those involved in controlling their spread and quantifying the wider implications for public health. TGC and polymyxins are increasingly being used as treatments of last resort, especially for A. baumannii infections. With the threat of resistance to these agents now on the horizon it is timely to investigate how this might impact on the pathogenic potential of MDRAB.
Resistance is clearly advantageous to the organism in the presence of the drug, but the longer-term consequences and biological costs of maintaining a resistant phenotype in the absence of the selecting agent are less clear. Development of resistance via the acquisition of chromosomal mutations has often been associated with significant fitness costs in vitro20–22, but there are fewer studies that have examined this in vivo23. The effects of MDR due to the acquisition of additional genetic material via plasmids or transposons may be even more varied and will depend on whether virulence determinants are linked to the resistance genes24 and if there are metabolic costs associated with the maintenance of large MDR plasmids25,26.
We investigated the biological cost of TGC resistance selected for in vivo during on-label use of the drug for the treatment of MDRAB infections. In each case TGC resistance was likely due to up-regulation of the AdeABC efflux system. Links between antimicrobial resistance, RND pump activity and virulence have been reported for several organisms (e.g. Escherichia coli; Salmonella enterica27; Pseudomonas aeruginosa). Recently, several interesting studies have investigated the complexities of RND efflux pump regulatory systems and the associated impact on antimicrobial susceptibility and virulence in A. baumannii, both in vitro and in vivo28–30. However, to our knowledge this is one of the first studies to investigate these phenomena in A. baumannii using in vivo-selected, tigecycline susceptible and resistant clinical pairs recovered from individual patients.
A. baumannii can be found in the gastrointestinal tract of infected patients suggesting that gut colonisation may be important in the establishment and progression of infection31. In enteric bacteria such as E. coli and S. enterica RND transporters are thought to contribute to the ability of these organisms to survive in the gut as they can protect from the antimicrobial effects of bile salts. All the A. baumannii isolates were bile-tolerant, with no differences between isolates despite the changes in the expression of adeABC.
There were substantial phenotypic differences between the isolates that could clearly impact upon their pathogenic potential. TGC-resistant isolate AB211 was slower growing than AB210 under most laboratory conditions except under iron limitation. An enhanced ability to sequester this essential micronutrient may impart a competitive advantage on AB211 over AB210 in vivo where bacteria often encounter iron-limited environments. This is further supported by the competition experiments in G. mellonella where AB211 was recovered at a median 5-fold higher ratio than AB210. The ability to out-compete its susceptible counterpart in the absence of TGC exposure in vivo could impact on the transmissibility of AB211 in the nosocomial setting in which patients are heavily colonised. Overall, AB211 appeared to display more of a ‘persister phenotype’ with a low metabolic potential and better ability to form biofilms, which could in turn make it harder to eradicate from the environment.
The W6976/W7282 pair behaved quite differently from AB210/AB211. In the G. mellonella competition assay the TGC-susceptible isolate, W6976 was more successful, out-competing the resistant isolate by a median factor of 3:1. However, resistant isolate W7282 was better able to grow under acidic conditions (pH 4.5); the mechanism for this and any relation to RND pump activity is unclear although this may make it better adapted for survival in acidic environments, which would include parts of the gastrointestinal tract and within macrophages.
The acquisition of a resistance mechanism able to influence the normal physiology of the bacterial membrane, either by modulating the transfer of molecules across it (efflux changes) or by reducing its permeability (porin lesions) might usually be considered disadvantageous to the organism. This is supported by a study that investigated the effects of in vivo-selected imipenem resistance on the virulence of Enterobacter cloacae32. This study examined isolates that were either fully susceptible, had reduced susceptibility to imipenem due to enhanced activity of the AcrAB-TolC RND pump (AcrB exhibits 50% homology to AdeB in A. baumannii) or frank resistance when the pump lesion was combined with porin loss. In contrast to our observations on the virulence of TGC-resistant MDRAB in G. mellonella, all the imipenem-resistant E. cloacae isolates exhibited reduced growth and virulence in a Caenorhabditis elegans model. Interestingly, isolates which displayed only reduced susceptibility to imipenem due to up-regulation of the RND pump were more virulent in this model. We chose not to use the C. elegans model as small quantities of ethanol must be added to the media for it to be used in the study of A. baumannii pathogenesis33, ethanol exposure has a number of pleotropic effects on A. baumannii, including overexpression of adeB, which would have complicated our analysis34.
Multidrug resistance in the A. baumannii isolates used in this study was not known to be associated with porin loss. However, the effects of this have been investigated in a pan-drug resistant A. baumannii isolate lacking the outer membrane proteins CarO and OprD, although any contribution of efflux systems was not investigated. This isolate was found to exhibit impaired growth in vitro and had reduced cytotoxicity towards respiratory epithelial cells35, suggesting a significant cost. Nevertheless, these properties seemed to have negligible effect on the ability of the strain to cause an outbreak of infections in critically-ill patients36.
Comparison of the genome sequences of all four isolates revealed insights into possible reasons for the differences observed between the two pairs. Firstly, a greater number of SNPs were found between AB210 and AB211 (n = 18) than between W6976 and W7282 (n = 5). Indeed, the only obvious functional difference between W6976 and W7282 was the mutation encoding the substitution in the AdeS protein predicted to result in up-regulation of AdeABC. Although AB211 also harboured a mutation in adeS there were in addition several large deletions in the genome of this isolate compared with its counterpart. Of interest was a deletion of a portion of the mutS gene in AB211, a gene involved in DNA mismatch repair37.
The role of MutS in Acinetobacter genome plasticity has until recently largely been studied in the Acinetobacter baylii isolate, ADP-1. However, a recent study used a derivative of AB210 (AB201M) to evolve tigecycline resistance in a continuous culture system. Intriguingly, the researchers found that almost all the successful lineages became hypermutators because of the interruption of mutS by a mobile element. Moreover, adeS was found to be the most mutated gene30. Similarly, the post-therapy isolate AB211 exhibited a ‘hypermutator’ phenotype as evidenced by its increased capacity to develop spontaneous resistance to fosfomycin. It is possible that the mutS lesion was in fact the initial event that facilitated the selection and maintenance of the adeS mutation identified previously in AB211. Non-functional MutS could also be the reason for the ability of AB211 to out-compete AB210 in vivo. Any in vivo fitness cost associated with dysregulated RND activity alone, as suggested by the competition studies between W6976 and W7282, could be negated by the adaptive advantages of the mutS mutation.
The loss of TGC for the treatment of MDRAB leaves only polymyxins as viable treatment options. The prevalence of frank resistance to polymyxins in Gram-negative organisms varies widely by geographical region though is most common in the Enterobacteriaceae and A. baumannii38. Of most concern is the recent emergence, evolution and dissemination of plasmid-mediated phosphoethanolamine transferases in recent years. In a recent study, the introduction of the mcr-1 gene into both laboratory and clinical strains of ESKAPE (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter species) pathogens resulted in colistin resistance in A. baumannii among other species and consistent phosphoethanolamine modification of lipid A even in cases where only moderate increases in colistin MICs were observed39. However, to date there have been no reports of A. baumannii clinical isolates harbouring mcr genes. Literature evidence, along with the data presented here, highlight the need for a better understanding of the potential risks to public health posed by MDRAB beyond the antibiogram and clonal lineage.
Methods
Identification and characterisation of A. baumannii isolates
AB210, AB211, W6976 and W7282 were recovered from clinical samples using standard microbiology procedures and were partially characterised previously5,14 (Table 1).
Comparative growth in vitro
In vitro growth rates were determined in LB broth using a microtitre plate-based growth kinetics assay24. Five microlitres of a 0.5 McFarland suspension were inoculated into 145 µL of growth medium in microtitre plates (Cornig, Amsterdam, Netherlands), which were incubated in a VersaMax Microplate Reader (Molecular Devices, Sunnyvale, CA, US) for 24 hours at 37 °C. The OD600 of the cultures was measured every 15 minutes with plates shaken for 10 seconds before each measurement. Growth under physiological stress (low pH, high salt concentration, nutrient and iron depletion) was investigated by growing the isolates in: (i) LB pH 4.5; (ii) LB supplemented with 200 mM NaCl; (iii) in one-third strength LB diluted with saline; and (iv) LB supplemented with 200 µM 2,2′-dipyridyl (an iron chelator); (v) normal, full strength LB. Relative fitness was calculated as Td parent/Td derivative. Differences in mean relative fitness were assessed using t test statistics.
Bile tolerance assays
Sensitivity to bovine bile (Sigma-Aldrich, Poole, United Kingdom) was assessed in vitro using a microtitre plate-based assay. Fifty microlitres of 0.5 McFarland suspensions prepared in LB broth from overnight cultures were added to 150 μL of LB broth ± varying concentrations of bile (0–16% [w/v]). Plates were incubated at 37 °C without shaking overnight and examined for turbidity as a measure of microbial growth. Assay endpoints were confirmed by the addition of 20 µl of alamarBlue reagent (Life Technologies, Paisley, UK).
Motility assays
Surface motility was assessed on low percentage agar plates. Five microlitres of overnight LB broth cultures were stab-inoculated into the centre of LB agar (0.3%) plates and incubated overnight at 37 °C. The ability of the organism to spread across the surface of the plate after overnight incubation was taken as evidence of motility40.
Biofilm formation
Biofilm formation was assessed using a modified version of the methods described by King et al.41. Overnight LB broth cultures were used to inoculate fresh LB broths which were incubated at 37 °C with shaking until the OD600 reached 1.0. Fifty microlitres of these cultures were added to 50 µL LB broth in wells of a sterile, polystyrene microtitre plates and incubated overnight at 37 °C. After incubation, 100 µL of 1% crystal violet was added to the wells and the plate was incubated for 30 minutes at room temperature. Plates were washed three times using 200 µl of sterile distilled water, after which 200 µL of 95% ethanol was added to each well. One hundred and fifty microlitres of the ethanol from each well was transferred to a clean microtitre plate and the OD540 was measured.
A. baumannii – Galleria mellonellavirulence assays
The virulence of A. baumannii isolates was assessed by their ability to kill G. mellonella (wax moth) larvae using methods described by Peleg et al.42 Larvae were obtained from Livefood UK Limited (Rooks Bridge, Somerset, UK) and stored in the dark at 15 °C in wood shavings.
Briefly, an overnight LB broth culture of A. baumannii was washed twice in sterile saline and serially diluted. Using a 25 µL Hamilton syringe (Cole-Parmer, London, UK), 16 caterpillars were injected via a left proleg with 10 µL of diluted culture containing 104 CFU. Un-inoculated and mock-inoculated (injected with 10 µL sterile saline) caterpillars were used as controls. Larvae were incubated in petri dishes lined with filter paper at 37 °C for 96 hours and scored daily for survival. Those insects that did not respond to touch with a pipette tip were considered dead. Experiments were performed on three separate occasions employing different batches of larvae. Survival curves were produced and analysed using the log-rank test. End-point survival proportions were compared using the one-tailed unpaired t test with Welch’s correction.
In vivocompetition assay
A selective culture medium, CHROMagar Acinetobacter (CHROMagar, Paris, France)43, was used to recover A. baumannii from infected G. mellonella larvae used in competition assays. This medium was supplemented with KPC supplement and vancomycin (25 mg/L) to prevent the growth of carbapenem-susceptible Gram-negative bacteria44 and Gram-positive bacteria, respectively. The ability of this media (CAA-KPC) to suppress growth of normal caterpillar flora was confirmed using un-infected larvae. Caterpillars were mechanically disrupted in 1 mL sterile PBS and 100 μL of the resulting suspension were plated on to supplemented CHROMagar Acinetobacter plates and ISO agar plates. For the recovery and enumeration of A. baumannii from larvae infected with AB211 or W7282 the CAA-KPC media was further supplemented with either kanamycin or TGC as detailed below.
The in vivo competition assay was performed using a starting ratio of 1:1 (AB210: AB211 or W6976: W7282) as determined by viable counts on ISO agar. After 24 hours incubation at 37 °C, the caterpillars were disrupted in 1 mL sterile PBS and serial dilutions were plated on CAA-KPC agar with and without either 25 mg/L kanamycin or 2 mg/L TGC. Ratios were calculated from viable counts on the selective media after 24 hours incubation at 37 °C.
Hypermutation studies
All A. baumannii isolates were screened for a mutator phenotype using a modified version of the fosfomycin disc test method45. Briefly, 104 CFU/ml from an overnight ISO broth culture was used to inoculate ISO agar plates. A disc containing 200 μg fosfomycin was placed on to the plates, which were then incubated for 24 hours at 37 °C. The numbers of colonies growing within the zone of inhibition around the fosfomycin disc were then counted. Differences in median number of colonies were assessed using Mann-Whitney statistics.
Whole-genome DNA sequencing of isolates W6976 and W7282
Isolates W6976 and W7282 were grown on nutrient agar and genomic DNA sequenced using a 454 GS FLX pyrosequencer (Roche, Branford, CT, USA). Draft genomes were assembled de novo from flowgram data using Newbler v. 2.5 (Roche). The resulting contigs were annotated using the automated annotation pipeline on the xBASE server46. Single nucleotide polymorphisms (SNPs) detected in W6976 relative to W7282 were confirmed by PCR and Sanger sequencing of the resulting amplicons using an ABI 3730xl DNA analyser (Applied Biosystems, Warrington, UK).
Acknowledgements
This study was funded in part by a research grant from Pfizer. Both authors would like to acknowledge Mark Pallen, Nick Loman and Jacqueline Chan for advice received on the interpretation and analysis of sequencing data. The results of this study were presented in part at the 5th British Society for Antimicrobial Chemotherapy’s Antibiotic Resistance Mechanisms Workshop, Birmingham, UK.
Author Contributions
M.H. and D.W.W. undertook the laboratory work and contributed equally in writing of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gordon NC, Wareham DW. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int. J. Antimicrob. Agents. 2010;35:219–26. doi: 10.1016/j.ijantimicag.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 2.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–82. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vila J, Pachón J. Acinetobacter baumannii resistant to everything: what should we do? Clin. Microbiol. Infect. 2011;17:955–6. doi: 10.1111/j.1469-0691.2011.03566.x. [DOI] [PubMed] [Google Scholar]
- 4.Durante-Mangoni E, Zarrilli R. Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 2011;6:407–22. doi: 10.2217/fmb.11.23. [DOI] [PubMed] [Google Scholar]
- 5.Hornsey M, et al. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemoth. 2010;65:1589–1593. doi: 10.1093/jac/dkq218. [DOI] [PubMed] [Google Scholar]
- 6.Peleg AY, Adams J, Paterson DL. Tigecycline Efflux as a Mechanism for Nonsusceptibility in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:2065–9. doi: 10.1128/AAC.01198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, et al. Adherence of Acinetobacter baumannii strains to human bronchial epithelial cells. Res Microbiol. 2006;157:360–366. doi: 10.1016/j.resmic.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Choi C, Lee J, Lee Y, Park T, Lee J. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. Bmc Microbiol. 2008;8:1–11. doi: 10.1186/1471-2180-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erridge M-N, Morgan Y, Poxton IR. Acinetobacter baumannii lipopolysaccharides are potent stimulators of human monocyte activation via Toll-like receptor 4 signalling. J Med Microbiol. 2007;56:165–171. doi: 10.1099/jmm.0.46823-0. [DOI] [PubMed] [Google Scholar]
- 10.March C, et al. Dissection of Host Cell Signal Transduction during Acinetobacter baumannii – Triggered Inflammatory Response. Plos One. 2010;5:e10033. doi: 10.1371/journal.pone.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SW, et al. Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol. Lett. 2009;301:224–31. doi: 10.1111/j.1574-6968.2009.01820.x. [DOI] [PubMed] [Google Scholar]
- 12.Eijkelkamp B, Hassan K, Paulsen I, Brown M. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. Bmc Genomics. 2011;12:1–14. doi: 10.1186/1471-2164-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goel VK, Kapil A. Monoclonal antibodies against the iron regulated outer membrane Proteins of Acinetobacter baumannii are bactericidal. BMC Microbiol. 2001;1:16. doi: 10.1186/1471-2180-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornsey M, et al. Whole-genome comparison of two Acinetobacter baumannii isolates from a single patient, where resistance developed during tigecycline therapy. J Antimicrob Chemoth. 2011;66:1499–1503. doi: 10.1093/jac/dkr168. [DOI] [PubMed] [Google Scholar]
- 15.Oliver A. Mutators in cystic fibrosis chronic lung infection: Prevalence, mechanisms, and consequences for antimicrobial therapy. Int. J. Med. Microbiol. 2010;300:563–72. doi: 10.1016/j.ijmm.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Choi A, Slamti L, Avci F, Pier G, Maira-Litrán T. The pgaABCD Locus of Acinetobacter baumannii Encodes the Production of Poly-β-1-6-N-Acetylglucosamine, Which Is Critical for Biofilm Formation. J Bacteriol. 2009;191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindgren P, Higgins P, Seifert H, Cars O. Prevalence of hypermutators among clinical Acinetobacter baumannii isolates. J Antimicrob Chemoth. 2016;71:661–665. doi: 10.1093/jac/dkv378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iacono M, et al. Whole-Genome Pyrosequencing of an Epidemic Multidrug-Resistant Acinetobacter baumannii Strain Belonging to the European Clone II Group. Antimicrobial Agents and Chemotherapy. 2008;52:2616–2625. doi: 10.1128/AAC.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotter P, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Sander P, et al. Fitness cost of chromosomal drug resistance-conferring mutations. Antimicrob. Agents Chemother. 2002;46:1204–11. doi: 10.1128/AAC.46.5.1204-1211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagel S, Hüllen V, Wiedemann B, Heisig P. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob. Agents Chemother. 1999;43:868–75. doi: 10.1128/aac.43.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersson D. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr Opin Microbiol. 2006;9:461–465. doi: 10.1016/j.mib.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez P, et al. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 2002;50:657–64. doi: 10.1093/jac/dkf185. [DOI] [PubMed] [Google Scholar]
- 24.Petersen A, Aarestrup FM, Olsen JE. The in vitro fitness cost of antimicrobial resistance in Escherichia coli varies with the growth conditions. FEMS Microbiol. Lett. 2009;299:53–9. doi: 10.1111/j.1574-6968.2009.01734.x. [DOI] [PubMed] [Google Scholar]
- 25.Marciano D, Karkouti O, Palzkill T. A Fitness Cost Associated With the Antibiotic Resistance Enzyme SME-1 β-Lactamase. Genetics. 2007;176:2381–2392. doi: 10.1534/genetics.106.069443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michon A, et al. Plasmidic qnrA3 Enhances Escherichia coli Fitness in Absence of Antibiotic Exposure. Plos One. 2011;6:e24552. doi: 10.1371/journal.pone.0024552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webber M, et al. The Global Consequence of Disruption of the AcrAB-TolC Efflux Pump in Salmonella enterica Includes Reduced Expression of SPI-1 and Other Attributes Required To Infect the Host. J Bacteriol. 2009;191:4276–4285. doi: 10.1128/JB.00363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richmond GE, et al. The Acinetobacter baumannii Two-Component System AdeRS Regulates Genes Required for Multidrug Efflux, Biofilm Formation, and Virulence in a Strain-Specific Manner. MBio. 2016;7:e00430–16. doi: 10.1128/mBio.00430-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh, M. H., Choi, C. H. & Lee, J. C. The effect of ISAba1-mediated adeN gene disruption on Acinetobacter baumannii pathogenesis. Virulence 1–3 10.1080/21505594.2017.1339859 (2017). [DOI] [PMC free article] [PubMed]
- 30.Hammerstrom TG, Beabout K, Clements TP, Saxer G, Shamoo Y. Acinetobacter baumannii Repeatedly Evolves a Hypermutator Phenotype in Response to Tigecycline That Effectively Surveys Evolutionary Trajectories to Resistance. PLoS ONE. 2015;10:e0140489. doi: 10.1371/journal.pone.0140489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thom K, et al. Patients with Acinetobacter baumannii bloodstream infections are colonized in the gastrointestinal tract with identical strains. Am J Infect Control. 2010;38:751–753. doi: 10.1016/j.ajic.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavigne J-PP, et al. Membrane permeability, a pivotal function involved in antibiotic resistance and virulence in Enterobacter aerogenes clinical isolates. Clin. Microbiol. Infect. 2012;18:539–45. doi: 10.1111/j.1469-0691.2011.03607.x. [DOI] [PubMed] [Google Scholar]
- 33.Smith MG, Des Etages SG, Snyder M. Microbial synergy via an ethanol-triggered pathway. Mol. Cell. Biol. 2004;24:3874–84. doi: 10.1128/MCB.24.9.3874-3884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camarena L, Bruno V, Euskirchen G, Poggio S, Snyder M. Molecular Mechanisms of Ethanol-Induced Pathogenesis Revealed by RNA-Sequencing. Plos Pathog. 2010;6:e1000834. doi: 10.1371/journal.ppat.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernández-Cuenca F, et al. Attenuated virulence of a slow-growing pandrug-resistant Acinetobacter baumannii is associated with decreased expression of genes encoding the porins CarO and OprD-like. Int J Antimicrob Ag. 2011;38:548–549. doi: 10.1016/j.ijantimicag.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Valencia R, et al. Nosocomial Outbreak of Infection With Pan–Drug‐Resistant Acinetobacter baumannii in a Tertiary Care University Hospital. Infect Cont Hosp Ep. 2009;30:257–263. doi: 10.1086/595977. [DOI] [PubMed] [Google Scholar]
- 37.Young D, Ornston N. Functions of the Mismatch Repair Gene mutS from Acinetobacter sp. Strain ADP1. J Bacteriol. 2001;183:6822–6831. doi: 10.1128/JB.183.23.6822-6831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srinivas P, Rivard K. Polymyxin Resistance in Gram-negative Pathogens. Curr Infect Dis Rep. 2017;19:38. doi: 10.1007/s11908-017-0596-3. [DOI] [PubMed] [Google Scholar]
- 39.Liu, Y.-Y. Y. et al. Structural Modification of Lipopolysaccharide Conferred by mcr-1 in Gram-Negative ESKAPE Pathogens. Antimicrob. Agents Chemother. 61, (2017). [DOI] [PMC free article] [PubMed]
- 40.Rashid H, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc National Acad Sci. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King LB, Swiatlo E, Swiatlo A, McDaniel LS. Serum resistance and biofilm formation in clinical isolates of Acinetobacter baumannii. FEMS Immunol. Med. Microbiol. 2009;55:414–21. doi: 10.1111/j.1574-695X.2009.00538.x. [DOI] [PubMed] [Google Scholar]
- 42.Peleg A, et al. Galleria mellonella as a Model System To Study Acinetobacter baumannii Pathogenesis and Therapeutics. Antimicrob Agents Ch. 2009;53:2605–2609. doi: 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordon & Wareham Evaluation of CHROMagar Acinetobacter for Detection of Enteric Carriage of Multidrug-Resistant Acinetobacter baumannii in Samples from Critically Ill Patients. J Clin Microbiol. 2009;47:2249–2251. doi: 10.1128/JCM.00634-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wareham & Gordon Modifications to CHROMagar Acinetobacter for improved selective growth of multi-drug resistant Acinetobacter baumannii. J Clin Pathol. 2011;64:164–167. doi: 10.1136/jcp.2010.083469. [DOI] [PubMed] [Google Scholar]
- 45.Ellington M, Livermore D, Pitt T, Hall L, Woodford N. Mutators among CTX-M β-lactamase-producing Escherichia coli and risk for the emergence of fosfomycin resistance. J Antimicrob Chemoth. 2006;58:848–852. doi: 10.1093/jac/dkl315. [DOI] [PubMed] [Google Scholar]
- 46.Chaudhuri R, et al. x BASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 2008;36:D543–D546. doi: 10.1093/nar/gkm928. [DOI] [PMC free article] [PubMed] [Google Scholar]