Abstract
Down syndrome, caused by trisomy 21, is characterized by a variety of medical conditions including intellectual impairments, cardiovascular defects, blood cell disorders and pre-mature aging phenotypes. Several somatic stem cell populations are dysfunctional in Down syndrome and their deficiencies may contribute to multiple Down syndrome phenotypes. Down syndrome is associated with muscle weakness but skeletal muscle stem cells or satellite cells in Down syndrome have not been investigated. We find that a failure in satellite cell expansion impairs muscle regeneration in the Ts65Dn mouse model of Down syndrome. Ts65Dn satellite cells accumulate DNA damage and over express Usp16, a histone de-ubiquitinating enzyme that regulates the DNA damage response. Impairment of satellite cell function, which further declines as Ts65Dn mice age, underscores stem cell deficiencies as an important contributor to Down syndrome pathologies.
Introduction
Trisomy 21, responsible for Down syndrome, is the most common autosomal aneuploidy and the most frequent genetic cause of intellectual disability1–3. Cognitive disabilities, growth defects, muscle weakness, facial abnormalities, cardiac malformations, early-onset Alzheimer’s disease and premature aging manifest in Down syndrome with variable penetrance4,5. Although the cellular and molecular mechanisms driving these different phenotypes are incompletely understood, altered stem cell function is a potential common link. For example, expansion and differentiation defects in neuronal stem cells impair neurogenesis in the developing brain and adult brain of individuals with Down syndrome6–8. Hematopoietic stem cells accumulate DNA damage, prematurely senesce and fail to expand in mouse models of Down syndrome9,10. Thus, stem cell defects in Down syndrome likely contribute to cognitive impairments, blood cell disorders, and pre-mature aging phenotypes in Down syndrome10–13.
Satellite cells, required for muscle regeneration14–17, are typically quiescent and fuse into the multinucleated myotubes of skeletal muscle to maintain the tissue or in response to injury18,19. Following muscle injury, satellite cells exit quiescence, proliferate and then differentiate to repair muscle while a small number of cells self-renewal to maintain the quiescent satellite cell population18. While satellite cell dysfunction contributes to a variety of diseases including muscular dystrophy, cancer cachexia and age-induced muscle wasting20–24, whether Down syndrome trisomy affects satellite cells and contributes to Down syndrome muscle phenotypes is unknown. Since skeletal muscle dysfunction associated with Down syndrome includes muscle weakness, early onset age-induced atrophy and overall diminished mobility, Down syndrome trisomy may impact satellite cell function25–29.
Here we analyze Ts65Dn mice, an established mouse model of Down syndrome, that are trisomic for ~55% of the orthologous protein coding genes on human chromosome 21 and recapitulate many phenotypes observed in individuals with Down syndrome30,31. While pre-injury satellite cell numbers are normal, muscle regeneration is impaired in Ts65Dn mice because of a reduction in satellite cell expansion, arising from an inability of Ts65Dn satellite cells to complete their first cell division upon exit from quiescence. An accumulation of DNA damage and elevated levels of Usp16, a de-ubiquitinating enzyme whose gene is on chromosome 21, accompany the defects in Ts65Dn satellite cell division. The impairment of satellite cell function in Ts65Dn mice provides further evidence that stem cell dysfunction is a common contributor to multiple Down syndrome phenotypes.
Results
Impaired satellite cell function and muscle regeneration in Ts65Dn mice
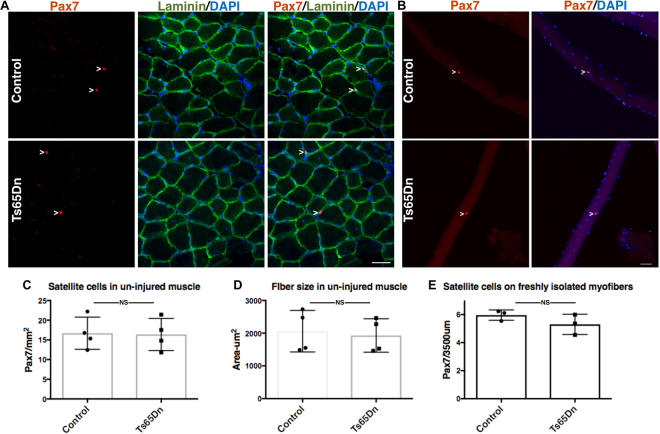
Satellite cell number and myofiber size were analyzed in sections of un-injured tibialis anterior (TA) muscle from 5 mo old wild type mice and Ts65Dn mice by scoring for Pax7 immunoreactive satellite cells15 and by determining the myofiber cross-sectional area using laminin immunoreactivity to identify the myofiber basement membrane, respectively (Fig. 1A). No differences in either the numbers of Pax7+ satellite cells (Fig. 1A,C) or in the average myofiber cross-sectional area were observed between wild type TA muscles and Ts65Dn TA muscles (Fig. 1A,D). To confirm that satellite cell numbers between Ts65Dn muscles and wild type muscles were similar, Pax7+ satellite cell numbers were quantified on individual myofibers isolated from the extensor digitorum longus (EDL) muscle (Fig. 1B,E). Thus, no differences in average myofiber size or differences in the number of Pax7 expressing satellite cells were observed when comparing 5 mo old adult wild type muscles and Ts65Dn muscles.
Figure 1.
Satellite cell number and myofiber size are normal in un-injured Ts65Dn muscle. (A) Un-injured TA muscle sections stained with anti-Pax7 antibody to label satellite cells (red) and laminin (green) to label the basal lamina. Blue is DAPI. White carets mark satellite cells. (B) Myofibers isolated from EDL muscle were fixed immediately and stained with anti-Pax7 antibody to identify satellite cells. Blue is DAPI. White carets mark satellite cells. (C–E) Quantification of Pax7+ satellite cell number and average fiber size in Ts65Dn muscle compared to wild type (n = 3 or 4). Statistical significance was determined using Student’s t test using. P-value < 0.05 were considered significant. NS indicates not significant Scale bars are 40 μm.
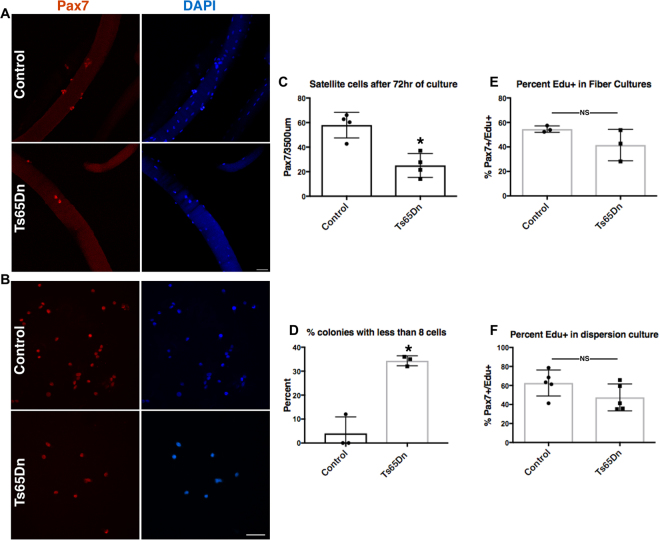
We compared the function of Ts65Dn satellite cells to wild type satellite cells by culturing satellite cells on individual myofibers isolated from EDL muscles32,33. Myofiber-associated satellite cells cultures were maintained for 72 h with the first satellite cell division occurring between 24–36 h and subsequent divisions occurring every 10–12 h thereafter34. Cultures were treated with EdU (5-ethynyl-2′-deoxyuridine) 2 h prior to collection to label cells in S-phase. A 57% reduction in Pax7 immunoreactive cell numbers occurred on myofibers cultured from Ts65Dn mice compared to myofibers from wild type mice (Fig. 2A,C). Clonal analysis of isolated Pax7+ satellite cells, cultured for 72 h, confirmed reduced expansion of Ts65Dn satellite cells, where 35% of satellite cell colonies in Ts65Dn cultures had 8 cells or less compared to only 5% of wild type colonies with 8 or fewer cells per colony (Fig. 2B,D). Although colony size was dramatically different between satellite cells isolated from Ts65Dn mice and wild type mice, the percent of EdU labeled cells did not differ between Ts65Dn and wild type cultures in either myofiber-associated satellite cell cultures or dispersed satellite cell cultures (Fig. 2E,F). Therefore, the deficient Ts65Dn Pax7+ satellite cell expansion is unlikely to arise from an overall slower cell cycle for Ts65Dn satellite cells compared to wild type satellite cells.
Figure 2.
Impaired expansion of Ts65Dn satellite cells in vitro. (A) Pax7+ cells (red) on isolated EDL myofibers after 72 h of culture. Blue is DAPI. (B) Images of satellite cell colonies after 72 h of culture, immunoreactive for Pax7 (red) and DAPI (blue). (C) Quantification of satellite cell number normalized to myofiber length on myofibers cultured for 72 h (n = 4). (D) Quantification of colony size represented as percent of colonies with less than 8 cells after 72 h of culture (n = 3). (E) Quantification of EdU+ satellite cells on 72 h myofiber cultures where EdU was added to media 2 h prior to collection (n = 3). (F) Quantification of EdU+ cells in dispersion cultures after 72 h of culture and EdU given 2 h prior to collection (n = 5). Statistical significance was tested using Student’s t test. Asterisks indicates significant difference and P-value < 0.05. NS indicates not significant. Scale bars are 40 μm.
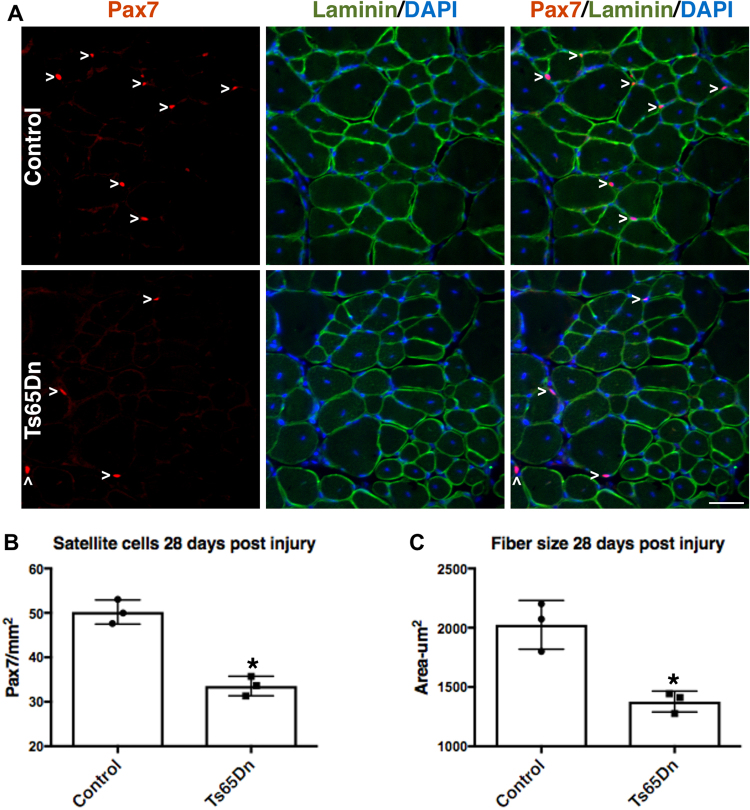
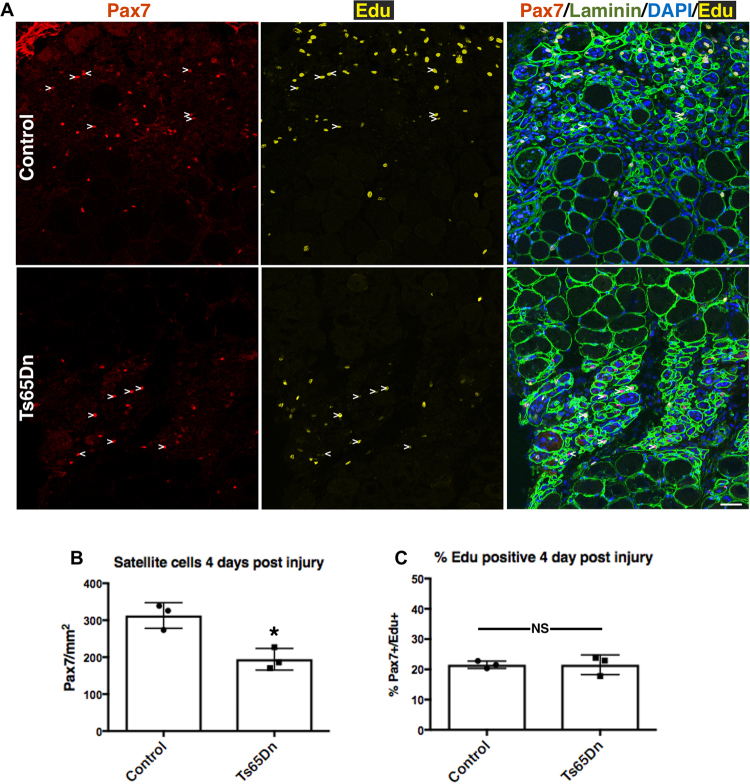
If the deficit in Ts65Dn satellite cell expansion is cell intrinsic and not an aberrancy arising from cell culture, then regeneration of Ts65Dn skeletal muscle should be impaired. Injection of the TA muscle with BaCl2 elicits a severe muscle injury with subsequent satellite cell expansion that peaks at 4 days post BaCl2 injury and resolves by 28 days post injury when satellite cells return to quiescence16,35,36. TA muscles from BaCl2-injected 5 mo old Ts65Dn mice and 5 mo old wild type mice were collected 28 days post BaCl2 injury and quantified for satellite cell numbers and myofiber size. The numbers of Pax7+ satellite cells were reduced by 1.5-fold in Ts65Dn muscle compared to wild type muscle (Fig. 3A,B), with a concomitant reduction in regenerated myofiber size (marked by the presence of central nuclei) in Ts65Dn muscle compared to wild type TA muscles (Fig. 3A,C). To examine satellite cell expansion in vivo, we injured the TA muscles of 5 mo old wild type and Ts65Dn mice using BaCl2 and injected EdU 2 h prior to tissue collection at 4 days post injury, the time point when satellite cells reach peak numbers36. Satellite cell expansion in vivo is impaired in Ts56Dn mice compared to wild type at 4d post-injury where reductions in Pax7 immunoreactive satellite cells are significant (Fig. 4A,B). No significant difference in the percentage of EdU positive Pax7+ satellite cells was observed between Ts65Dn and wild type TA muscles at 4d post injury (Fig. 4A,C). The impaired Pax7+ satellite cell expansion is likely responsible for the observed reductions in myofiber size in Ts65Dn mice compared to wild type mice following an induced muscle injury.
Figure 3.
Impaired muscle regeneration in adult Ts65Dn mice. (A) TA muscle collected from 5mo old mice, 28 days post injury, immunoreactive for Pax7 (red) to identify satellite cells and immunoreactive for laminin (green) to identify the basal lamina. DAPI is blue. Carets mark satellite cells. (B–C) Quantification shows satellite cell number (B) and average fiber cross sectional area (C) in regenerated muscle tissue of wild type and Ts65Dn mice (n = 3). Statistical significance was tested using Student’s t test. Asterisks indicates significant difference and P-value < 0.05. Scale bar is 40 μm.
Figure 4.
Reduced number of Ts65Dn satellite cells at 4 days post injury. (A) Images of TA muscle collected 4 days post injury were visualized for Pax7 immunoreactivity (red) to label satellite cells, laminin immunoreactiviey to mark myofibers (green) and EdU (yellow). EdU was injected 2 h prior to collection. Blue is DAPI. White carets mark EdU+ satellite cells. (B) Quantification of satellite cell numbers at 4 days post injury in Ts65Dn muscle and wild type muscle (n = 3). (C) Quantification showing the percentage of Pax7+/EdU+ cells in wild type TA muscle and Ts65Dn TA muscle at 4 day post injury (n = 3). Statistical significance was tested using Student’s t test. Asterisks indicates significant difference and P-value < 0.05. NS indicates not significant. Scale bar is 40 μm.
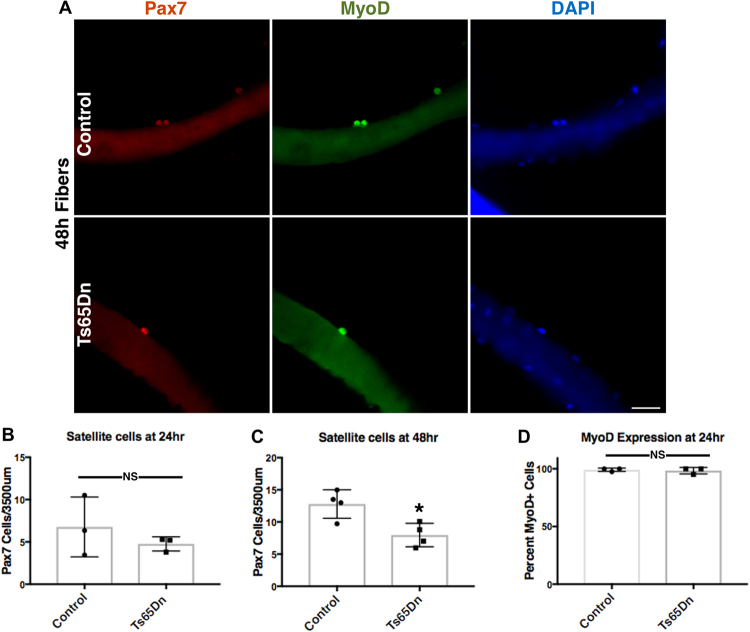
When satellite cells exit quiescence, Myf5 protein and MyoD protein are rapidly induced via inhibition of MyoD mRNA decay, permitting cell growth and mitosis between 24 h and 36 h after exit from G037,38. Following the initial division, subsequent cell cycles are rapid with satellite cells dividing every 8 h–12 h. Isolation of myofibers and their associated satellite cells models in vivo injury, permitting analysis of cell cycle kinetics in culture39–41. No differences in the number of Pax7+ cells were observed 24 h post-isolation in myofiber-associated satellite cells from 5 mo old Ts65Dn mice compared to wild type mice (Fig. 5B). Moreover, the initial numbers of Pax7+ satellite cells on freshly isolated myofibers from Ts65Dn mice (5.30 Pax7+ satellite cells/myofiber length) were indistinguishable from Pax7+ satellite cell numbers on freshly isolated wild type myofibers (5.96 Pax7+ satellite cells/myofiber length). After 48 h of culture, Ts65Dn myofiber-associated Pax7+ satellite cell number are reduced compared to wild type (Fig. 5A,C), where most satellite cells on wild type myofibers appear as doublets, indicating a recent initial cell division (Fig. 5A). In contrast, the majority of myofiber-associated Pax7+ satellite cells on Ts65Dn myofibers have not yet completed their initial division after 48 h of culture (Fig. 5A). MyoD protein, required for S-phase entry, appears 3 h to 6 h post satellite cell activation and can be used to indicate G0 exit38. No differences in the percentage of MyoD+/Pax7+ satellite cells were observed in either wild type myofiber cultures or Ts65Dn myofiber cultures at 24 h post-isolation (Fig. 5A,D). Since both wild type satellite cells and Ts65Dn satellite cells exit G0 by 24 h in culture but the number of satellite cells on Ts65Dn myofibers fails to increase by 48 h of culture, the failure of Ts65Dn Pax7+ satellite cell numbers to increase most likely arises from a delayed or incomplete initial cell division following G0 exit.
Figure 5.
Ts65Dn satellite cells have an impaired first division following exit from quiescence. (A) Images of isolated myofibers grown in culture for 48 h and visualized for Pax7 immunoreactivity (red) and MyoD immunoreactivity (green). Blue is DAPI. Wild type satellite cells are seen as neighbor doublets while Ts65Dn satellite cells are predominately individual cells. (B) Quantification of satellite cell number normalized to myofiber length after 24 h for Ts65Dn myofibers and wild type myofibers (n = 3). (C) Quantification after 48 h of culture for shows a satellites cell on Ts65Dn myofibers compared to wild type myofibers (n = 4). (D) Quantification of MyoD positive satellite cells on myofibers cultured for 24 h from Ts65Dn mice and wild type mice (n = 3). Statistical significance was tested using Student’s t test. Asterisks indicates significant difference and P-value < 0.05. NS indicates not significant. Scale bar is 40 μm.
Ts65Dn satellite cells accumulate DNA damage and further functionally decline with age
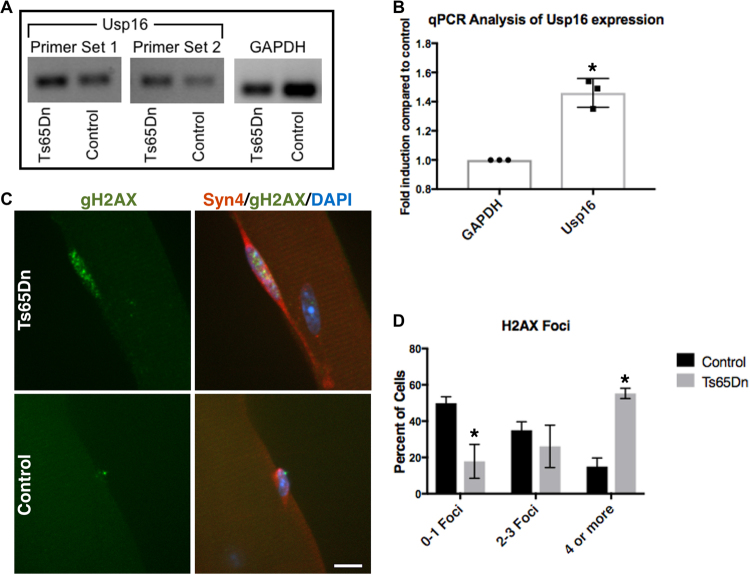
Usp16, encoded on human chromosome 21, regulates gene expression, DNA damage repair, and cell cycle progression by controlling the ubiquitination state of histone 2 A (H2A)42–44. Usp16 expression is elevated 1.5-fold in Ts65Dn satellite cells compared to wild type satellite cells (Fig. 6A,B), as predicted by the three Usp16 alleles present in Ts65Dn mice. Since elevated Usp16 expression contributes to somatic stem cell dysfunction in Down syndrome10 and Usp16 represses DNA damage responses44,45, we asked if Ts65Dn satellite cells accumulate more DNA damage that wild type satellite cells. Individual myofibers were isolated from 6 mo old wild type mice as well as Ts65Dn mice and myofiber-associated satellite cells were cultured for 24 h. Staining for γH2AX, a variant of H2A that is rapidly phosphorylated at sites of double stranded DNA breaks44–46, revealed a four-fold increase of γH2AX foci in Ts65Dn satellite cells compared to wild type satellite cells (Fig. 6C,D). The increased DNA damage in Ts65Dn satellite cells may underlie their inability to effectively complete their first division upon exit from G0.
Figure 6.
Increased Usp16 expression and DNA damage in Ts65Dn satellite cells. (A) Semi-quantitative RT-PCR analysis for Usp16 amplification products from cDNA of Ts65Dn satellite cells compared to wild type satellite cells. The images shown were cropped to save space (full-length gel included in Supplementary Information). (B) Quantitative real-time PCR for Usp16 expression in Ts65Dn satellite cells compared to wild type satellite cells (n = 3). (C) Images for isolated myofibers cultured for 24 h immunoreactive for γH2AX (green, DNA damage) and immunoreactive for Syndecan-4 (red, satellite cells). Blue is DAPI. (D) Quantification of DNA damage foci identified by γH2AX labeling in Ts65Dn satellite cells and wild type satellite cells (n = 3). Statistical significance was tested using Student’s t test. Asterisks indicates significant difference and P-value < 0.05. Scale bar is 10 μm.
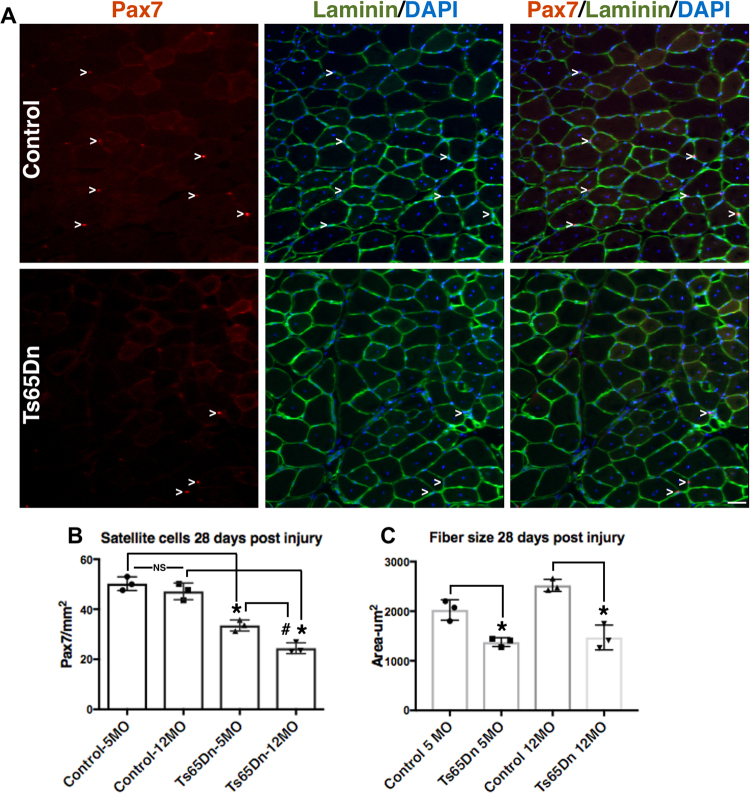
If DNA damage in Ts65Dn satellite cells accumulates over time, Ts65Dn satellite cell function maybe more severely impaired by age than wild type satellite cells. In wild type mice, age-related impairment of muscle regeneration does not appear until 18–20 mo of age and progressively worsens by 28–32 mo of age20,21,47. Muscle regeneration is impaired in 5 mo old and 12 mo old Ts56Dn mice compared to wild type mice (Fig. 7A–C). The average size of regenerated myofibers was reduced in Ts65Dn TA muscle compared to regenerated wild type TA muscles at 5 mo of age and 12 mo of age (Fig. 7A,C). However, myofiber size in injured TA muscles did not differ between 5 mo old and 12 mo old Ts65Dn mice (Fig. 7C). The numbers of Pax7+ satellite cells in Ts65Dn regenerated muscle declines from 5 mo to 12 mo of age and is significantly reduced compared to the numbers of wild type Pax7+ satellite cells in injured muscles at both 5 mo and 12 mo of age (Fig. 7A,B). The numbers of Pax7+ satellite cells in un-injured 12 mo old wild type TA muscles was not significantly different than those in 12 mo old Ts65Dn TA muscles (17.95 ± 2.28 Pax7+ satellite cells/mm2 in wild type vs. 15.55 ± 1.14 Pax7+ satellite cells/mm2 in Ts65Dn). The continuing decline in satellite cell numbers following regeneration in Ts65Dn muscle that occurs with age is consistent with an advancing decline in satellite cell function caused by accumulation of DNA damage.
Figure 7.
Further impairment of satellite cell function in 12 month old Ts65Dn tissue. (A) Satellite cells from TA muscles collected 28 days post injury from 12 mo old mice were quantified by Pax7 immunoreactivity (red) to mark satellite cells and laminin immunoreactivity (green) to identify the basal lamina. DAPI = blue. Carets mark satellite cells. (B) Satellite cell numbers quantified at 28 days post injury muscle for Ts65Dn regenerated muscle compared to wild type muscle at 5 mo of age and 12 mo of age. (C) Myofiber cross sectional area was measured in Ts65Dn regenerated muscle and wild type regenerated muscle at 5 mo of age and 12 mo of age. Statistical significance was tested using Student’s t test. Asterisks or pound sign indicates significant difference and P-value < 0.05. NS indicates not significant. Scale bar is 40 μm.
Discussion
Here we report that reduced satellite cell expansion impairs skeletal muscle regeneration in Ts65Dn mice. Satellite cell expansion defects in Ts65Dn mice arise from an impaired initial cell division following exit from quiescence. However, there are no differences in either cell cycle rates or the time to exit quiescence between Ts65Dn satellite cells and wild type satellite cells. Ts65Dn satellite cells over express the de-ubiquitinating enzyme Usp16, accumulate double stranded DNA breaks, and lose regenerative capability when compared to wild type satellite cells. Therefore, Ts65Dn satellite cell dysfunction may contribute to multiple Down syndrome muscle phenotypes providing additional evidence that somatic stem cell deficiencies contribute to Down syndrome phenotypes.
Down syndrome phenotypes associated with skeletal muscle and mobility include muscle weakness, low muscle tone, and fragile joints48–51. Satellite cells maintain skeletal muscle, are essential for skeletal muscle repair19,52, and thus, decreases in muscle strength in individuals with Down syndrome may arise from deficits in satellite cell function48–50. Although individuals with Down syndrome benefit from physical training, and can increase strength with resistance exercise, it is unclear if their exercise response is blunted27,29,53. Since satellite cells respond to exercise54–56, determining if satellite cell dysfunction contributes to muscle weakness or impairs the exercise response in individuals with Down syndrome may be clinically relevant.
Our data add skeletal muscle stem cells to a growing list of dysfunctional stem cells and dysfunctional progenitor cells in Down syndrome that include HSCs, neuronal stem cells, mammary epithelial cells and neural crest progenitors8,10,12,57,58. Common functional stem cell defects associated with Down syndrome include impaired expansion and impaired self-renewal10,11, which may contribute to accelerated aging phenotypes10,12,13,59. Declines in Ts65Dn satellite cell function, which are apparent in 5 mo old Ts65Dn mice and are exacerbated in 1 year old Ts65Dn mice do not occur in1 year old wild type mice. The declines in Ts65Dn satellite cell function correlate with sarcopenia appearing at younger ages with more pronounced atrophy in Ts65Dn mice than in age-matched wild type mice28. Bolstering somatic stem cell function in individuals with Down syndrome may slow the accelerated aging and thus, is a potential therapeutic target.
What molecular mechanisms drive Down syndrome stem cell and satellite cell dysfunction? In hematopoietic stem cells, Usp16 over expression prematurely induces senescence via p16Ink4a expression10. In Ts65Dn satellite cells, Usp16 expression is increased but we found no evidence of increased p16Ink4a expression (data not shown). However, the presence of γH2AX foci and increased Usp16 expression in Ts65Dn satellite cells could be responsible for increased DNA damage and decreased DNA damage repair60. Ubiquintation of histone 2A at sites of DNA damage recruits DNA damage repair proteins, while Usp16 and other de-ubiquintases restore ubiquitin levels to normal approximately 24 h after damage60–62. Thus, the increased Usp16 expression in TS65Dn satellite cells could disrupt DNA repair, induce DNA damage, and impair satellite cell expansion. Over expression of Usp16 is likely not solely responsible for the increased DNA damage as Down syndrome cells have an increased susceptibility to genotoxic stress63. Moreover, increased oxidative stress in Down syndrome cells leads to increased DNA damage and to increased apoptosis64–67. While we could not detect apoptotic cells on Ts65Dn myofiber cultures using TUNEL staining (data not shown), it is possible that a small number of satellite cells are undergoing apoptosis. Although, our EdU analysis did not detect differences in cell cycle rates for Ts65Dn satellite cells compared to wild type cells, cell cycle deficiencies have been reported in other Ts65Dn adult stem cell populations10,11. Thus, we cannot rule out cell cycle defects not detectable in our assay. Our data suggest that the primary mechanism responsible for reducing Ts65DN satellite cell numbers is a delay in the initial cell division, reducing final cell numbers. The cause of DNA damage in Ts65Dn satellite cells and how DNA damage leads to reductions in satellite cell numbers remains unknown. Whether the reduced satellite cell numbers arise from cell autonomous changes has not been addressed and cell non-autonomous effects may contribute to satellite cell dysfunction. Furthermore, the Ts65Dn muscle regeneration defects are not likely caused by satellite cell defects alone. For example, immune deficiency observed in Down syndrome68,69 could contribute to satellite cell dysfunction, as immune cells are required for skeletal muscle regeneration70–73. In conclusion, the data presented here identify potential causes of Down syndrome associated skeletal muscle weakness. In addition, satellite cells are another somatic stem cell that function aberrantly in a mouse model of Down syndrome and our data link DNA damage to Down syndrome stem cell dysfunction.
Methods
Mice
Mice were bred and housed according to National Institutes of Health (NIH) guidelines for the ethical treatment of animals in a pathogen-free facility at the University of Colorado at Boulder. University of Colorado Institutional Animal Care and Use Committee (IACUC) approved all animal protocols and procedures. Ts65Dn mice were purchased from Jackson Labs and Ts65Dn trisomic female mice (Jackson Labs Stock No. 005252) were crossed with wild type disomic F1 hybrid male mice (Jackson Laboratory Stock No. 001875). The male F1 hybrid mice are offspring of a cross between C57BL/6JEiJ females (B6Ei) and C3H/HeSnJ males (C3Sn) and commonly used in crosses as a background for deleterious mutations. For all experiments, wild type mice are disomic sex-matched littermates from the crosses described above.
Mouse Injuries and EdU injections
Mice at either 5 mo of age or 12 mo of age were anesthetized with 3% isofluorane and the left TA muscle was injected with 50 μL of 1.2% BaCl2. Intraperitoneal (IP) injections of 10 mM EdU (Carbosynth), resuspended in water, were given 2 h prior to collection at a volume of 100 µl/25 g mouse weight.
Immunofluorescence staining of tissue section
The TA muscle was dissected, fixed for 2 h on ice cold 4% paraformaldehyde, and then transferred to PBS with 30% sucrose at 4 °C overnight. Muscle was mounted in O.C.T. (Tissue-Tek®) and cryo-sectioning was performed on a Leica cryostat to generate 8 μm sections. Tissues and sections were stored at −80 °C until staining. Tissue sections were post-fixed in 4% paraformaldehyde for 10 min at room temperature (RT) and washed three times for 5 min in PBS. For heat-induced epitope retrieval, required for anti-Pax7 antibody staining, post-fixed slides were placed in citrate buffer, pH 6.0, and subjected to 6 min of high pressure-cooking in a Cuisinart model CPC-600 pressure cooker set on high pressure. For immunostaining, tissue sections were permeabilized with 0.25% Triton-X100 (Sigma) in PBS containing 3% bovine serum albumin (Sigma) for 45 min at RT, incubated with primary antibody at 4 °C overnight followed by incubation with a secondary antibody at RT for 1 h. Primary antibodies included mouse anti-Pax7 (DSHB) at 1:1000 and rabbit anti-laminin (Sigma-Aldrich) at 1:200. Alexa-488, 555, and 647 secondary antibodies (Molecular Probes) were used at a 1:750 dilution. For analysis that included EdU detection, EdU staining was completed prior to antibody staining using the Click-iT EdU Alexa fluor 488 detection kit (Molecular Probes) following the manufacturer’s protocols. Sections were incubated with 1 μg/mL DAPI for 10 min at room temperature then mounted in Mowiol supplemented with DABCO (Sigma-Aldrich) as an anti-fade agent.
Analysis of tissue sections
For analysis of immunostained TA muscle sections in Figs 1, 3, 4 and 7 one N is a biological replicate comparing a TA muscle from a Ts65Dn mouse with an aged-matched and sex-matched wild type mouse (disomic littermates). For biological replicates within a figure, both male and female mice were used. The sex of the mice used for the TA analysis are as follows: Fig. 1 (2 females and 1 male), Fig. 3 (1 female and 2 males), Fig. 4 (2 females and 1 male), Fig. 7 (3 males). Uninjured TA muscles are from the contralateral hindlimbs of BaCl2-injured mice in Fig. 1. For Pax7 cell quantification of TA muscle sections a minimum of eight images were scored from a minimum of 5 sections for each N and normalized to area. Images were blinded for quantification. When injured tissue was analyzed, Pax7+ cells were quantified only in areas of injury as identified by myofibers containing centrally located nuclei, scored and normalized as described for uninjured muscle. A minimum of 5 images from 5 sections were scored for myofiber size where images were blinded. In injured muscle, only myofibers with central nuclei were scored for size. For each N a minimum of 400 myofibers were scored.
Myofiber isolation, immunostaining and culture
The extensor digitorum longus (EDL) muscles were dissected, placed into 400 U/mL collagenase at 37 °C for 1.5 h with shaking and then placed into Ham’s F-12C supplemented with 15% horse serum to inactivate the collagenase. Individual EDL myofibers were separated and isolated using a glass pipet. For immediate analysis, myofibers were fixed in 4% paraformaldehyde for 10 min and stored in PBS for immunostaining. Intact myofibers were cultured in Ham’s F-12C media supplemented with 15% horse serum and 0.5 nM FGF-2 at 6% O2. Where assayed, EdU was added directly to the media at a final concentration of 10 μm. Myofibers were permeabilized with 0.25% Triton-X100 in PBS containing 3% bovine serum albumin (Sigma) for 45 min at RT and incubated with primary antibody at 4 °C overnight followed by incubation with secondary antibodies at room temperature for 1 h. Primary antibodies used were mouse anti-Pax7 (DSHB at 1:1000, rabbit anti-MyoD (Santa Cruz) at 1:1000, chicken anti-Syndecan4 at 1:1000, and mouse anti-phospho-H2A.X (EMD-Millipore 05–636) at 1:250. Alexa-488, 555 and 647 secondary antibodies (Molecular Probes) were used at a 1:750 dilution. For analysis that included EdU detection, EdU staining was completed prior to antibody staining using the Click-iT EdU Alexa fluor 488 detection kit (Molecular Probes) following the manufacturer’s protocol. Following immunostaining, myofibers were incubated with 1 μg/mL DAPI for 10 min at RT then mounted in Mowiol supplemented with DABCO (Sigma-Aldrich) as an anti-fade agent.
Analysis of myofibers and myofiber cultures
Satellite cells for each N of Figs 1, 2, 5, 6 were scored from EDL myofibers isolated from separate Ts65DN and aged-matched and sex-matched wild type mice (each N are EDL myofibers from different mice). For biological replicates within a figure male and female mice were used. The sex of the mice used are: Fig. 1 (2 females and 1 male), Fig. 2 (2 females and 2 males), Fig. 5 (2 males and 2 females), Fig. 6 (2 females and 1 male). A minimum of 25 immunostained myofibers mounted on slides were scored blinded for Pax7 immunoreactivity. A minimum of 40 immunostained satellite cell images were scored blinded for ɣH2AX foci.
Dispersion cultures
Hindlimb muscles were dissected, minced, and digested in 400 U/mL collagenase type I in Ham’s F-12C at 37 °C for 1 h with gentle vortexing every 10 min. Digested muscle was placed into Ham’s F-12C supplemented with 15% horse serum to inactivate the collagenase and passed successively through three cell strainers: 100 μm, 70 μm, and 40 μm (BD Falcon). The flow through was centrifuged at 1500xg for 5 min and the cell pellets re-suspended in Ham’s F-12C supplemented with 15% horse serum and 0.5 nM FGF-2. The resuspended cells were pre-plated for 2 hours on plastic tissue culture plates to remove adherent fibroblasts and then the remaining suspended cells plated onto coverslips coated with 0.66% gelatin or onto 10 cm gelatin-coated tissue culture dishes (colonies were ≥90% satellite cells). Clonal analysis was performed with cells plated at ~100 cells/single well of a 6 well tissue culture plate containing a coverslip to permit identification of individual colonies. When required, EdU was added directly to the media for a final concentration of 10 μm. Satellite cell explant cultures were fixed and immunostained as described for myofibers.
Analysis of dispersion cultures
Each N is a biological replicate from an individual mouse where muscle was isolated from either a Ts65DN mouse or an aged-matched and sex matched-wild type mouse (disomic littermate). For biological replicates within a figure, both male and female mice were used. The sex of the mice used were: Fig. 2 (3 females and 2 males), Fig. 6 (1 female and 2 males). Scoring of coverslips was performed blinded where a minimum of 100 satellite cells or a minimum of 20 colonies for clonal analysis per N were scored, respectively.
Semi quantitative RT-PCR and real time qPCR
RNA was isolated from dispersion culture cells after 72 h of growth using the RNeasy kit (Qiagen) following manufacturers protocol. The Superscript III first strand synthesis system (ThermoFisher) was used for cDNA synthesis from 250 ng isolated RNA. Transcript expression analysis using standard semi-quantitative PCR analysis (FastStart Taq DNA polymerase, Sigma-Aldrich) at 25 cycles was done using a C1000 Touch Thermocycler (Bio-Rad). For real time quantitative PCR, iQ SYBR green supermix (BioRad) was used with a CFX384 Touch Real-Time Thermocycler (Bio-Rad).
Primer sequences: Usp16 For1-ctacacgacaccgagatccg, Usp16 Rev1-tgaagttcttacgccagcttg, Usp16 For2-gctggcgtaagaacttcaaaac, Usp16 Rev2-agtcctttcacagttatctggga, GAPDH For aggtcggtgtgaacggatttg, GAPDH Rev ggggtcgttgatggcaaca.
Microscopy and image analysis
All images were captured on a Nikon inverted spinning disk confocal microscope or Olympus IX81 Inverted wide-field microscope. Objectives used on the Nikon were: 10x/o.45NA Plan Apo, 20x/0.75NA Plan Apo and 40x/0.95 Plan Apo. Objectives used on the Olympus were 10x/0.40NA and 20x/0.70NA. Images were processed using Fiji ImageJ. Confocal stacks were projected as maximum intensity images for each channel and merged into a single image. Brightness and contrast were adjusted for the entire image as necessary. Both satellite cell numbers and average myofiber size were scored manually using Fiji ImageJ on blinded images.
Statistics
All statistical analyses were performed in Prism (GraphPad). To assess statistical significance, two-tailed, unpaired Student’s t test were performed and p < 0.05 was considered significant. At least three different animals per genotype and 3 animals per age group were used in all experiments.
Electronic supplementary material
Acknowledgements
This work was supported by the Linda Crnic Institute for Down syndrome and the Anna and John J Sie Foundation, the National Intistutes of Health and the Glenn Foundation for Medical Research. The authors declare no competing interests. Anna and John J. Sie Foundation: Linda Crnic Institute Grand Challenge Grant; National Institutes of Health: AR049946, and Glenn Foundation for Medical Research.
Author Contributions
B.P., N.D., T.A., D.W. performed the experiments, analyzed the data and help draft the manuscript. B.B.O. supervised the research and helped to draft the manuscript. All authors read and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22342-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Antonarakis SE, Epstein CJ. The challenge of Down syndrome. Trends Mol Med. 2006;12:473–479. doi: 10.1016/j.molmed.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Parker SE, et al. Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004-2006. Birth Defects Res A Clin Mol Teratol. 2010;88:1008–1016. doi: 10.1002/bdra.20735. [DOI] [PubMed] [Google Scholar]
- 3.Rachidi M, Lopes C. Mental retardation in Down syndrome: from gene dosage imbalance to molecular and cellular mechanisms. Neurosci Res. 2007;59:349–369. doi: 10.1016/j.neures.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- 5.Korenberg JR, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci USA. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halevy T, Biancotti JC, Yanuka O, Golan-Lev T, Benvenisty N. Molecular Characterization of Down Syndrome Embryonic Stem Cells Reveals a Role for RUNX1 in Neural Differentiation. Stem Cell Reports. 2016;7:777–786. doi: 10.1016/j.stemcr.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soppa U, et al. The Down syndrome-related protein kinase DYRK1A phosphorylatesp27(Kip1) and Cyclin D1 and induces cell cycle exit and neuronal differentiation. Cell Cycle. 2014;13:2084–2100. doi: 10.4161/cc.29104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurabayashi N, Sanada K. Increased dosage of DYRK1A and DSCR1 delays neuronal differentiation in neocortical progenitor cells. Genes Dev. 2013;27:2708–2721. doi: 10.1101/gad.226381.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, et al. Hematopoietic Stem Cells from Ts65Dn Mice Are Deficient in the Repair of DNA Double-Strand Breaks. Radiat Res. 2016;185:630–637. doi: 10.1667/RR14407.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adorno M, et al. Usp16 contributes to somatic stem-cell defects in Down’s syndrome. Nature. 2013;501:380–384. doi: 10.1038/nature12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Filippi S, Roy A, Roberts I. Stem and progenitor cell dysfunction in human trisomies. EMBO Rep. 2015;16:44–62. doi: 10.15252/embr.201439583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cairney CJ, et al. A systems biology approach to Down syndrome: identification of Notch/Wnt dysregulation in a model of stem cells aging. Biochim Biophys Acta. 2009;1792:353–363. doi: 10.1016/j.bbadis.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Bianchi P, et al. Age-related impairment of olfactory bulb neurogenesis in the Ts65Dn mouse model of Down syndrome. Exp Neurol. 2014;251:1–11. doi: 10.1016/j.expneurol.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 14.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seale P, et al. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/S0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 16.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumont NA, Bentzinger CF, Sincennes MC, Rudnicki MA. Satellite Cells and Skeletal Muscle Regeneration. Compr Physiol. 2015;5:1027–1059. doi: 10.1002/cphy.c140068. [DOI] [PubMed] [Google Scholar]
- 19.Pawlikowski B, Pulliam C, Betta ND, Kardon G, Olwin BB. Pervasive satellite cell contribution to uninjured adult muscle fibers. Skeletal Muscle. 2015;5:1–13. doi: 10.1186/s13395-015-0067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernet JD, et al. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med. 2014;20:265–271. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cosgrove BD, et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nature Medicine. 2014;20:255–264. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sacco A, et al. Short Telomeres and Stem Cell Exhaustion Model Duchenne Muscular Dystrophy in mdx/mTR Mice. Cell. 2010;143:1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumont, N. A. et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med (2015). [DOI] [PMC free article] [PubMed]
- 24.He WA, et al. NF-κB-mediated Pax7 dysregulation in the muscle microenvironment promotes cancer cachexia. J Clin Invest. 2013;123:4821–4835. doi: 10.1172/JCI68523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowley PM, et al. Functional and biochemical characterization of soleus muscle in Down syndrome mice: insight into the muscle dysfunction seen in the human condition. Am J Physiol Regul Integr Comp Physiol. 2012;303:R1251–R1260. doi: 10.1152/ajpregu.00312.2012. [DOI] [PubMed] [Google Scholar]
- 26.Dey A, et al. Down Syndrome Related Muscle Hypotonia: Association with COL6A3 Functional SNP rs2270669. Front Genet. 2013;4:57. doi: 10.3389/fgene.2013.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shields N, et al. A community-based strength training programme increases muscle strength and physical activity in young people with Down syndrome: a randomised controlled trial. Res Dev Disabil. 2013;34:4385–4394. doi: 10.1016/j.ridd.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 28.Cisterna B, Costanzo M, Scherini E, Zancanaro C, Malatesta M. Ultrastructural features of skeletal muscle in adult and aging Ts65Dn mice, a murine model of Down syndrome. Muscles Ligaments Tendons J. 2013;3:287–294. [PMC free article] [PubMed] [Google Scholar]
- 29.Lin HC, Wuang YP. Strength and agility training in adolescents with Down syndrome: a randomized controlled trial. Res Dev Disabil. 2012;33:2236–2244. doi: 10.1016/j.ridd.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 30.Reeves RH, et al. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 31.Gupta M, Dhanasekaran AR, Gardiner KJ. Mouse models of Down syndrome: gene content and consequences. Mamm Genome. 2016;27:538–555. doi: 10.1007/s00335-016-9661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornelison DD, et al. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 2004;18:2231–2236. doi: 10.1101/gad.1214204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogler TO, Gadek KE, Cadwallader AB, Elston TL, Olwin BB. Isolation, Culture, Functional Assays, and Immunofluorescence of Myofiber-Associated Satellite Cells. Methods Mol Biol. 2016;1460:141–162. doi: 10.1007/978-1-4939-3810-0_11. [DOI] [PubMed] [Google Scholar]
- 34.Troy A, et al. Coordination of Satellite Cell Activation and Self-Renewal by Par-Complex-Dependent Asymmetric Activation of p38alpha/beta MAPK. Cell Stem Cell. 2012;11:541–553. doi: 10.1016/j.stem.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardy D, et al. Comparative Study of Injury Models for Studying Muscle Regeneration in Mice. PLoS One. 2016;11:e0147198. doi: 10.1371/journal.pone.0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webster MT, Manor U, Lippincott-Schwartz J, Fan CM. Intravital Imaging Reveals Ghost Fibers as Architectural Units Guiding Myogenic Progenitors during Regeneration. Cell Stem Cell. 2016;18:243–252. doi: 10.1016/j.stem.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zismanov, V., et al. Phosphorylation of eIF2alpha Is a Translational Control Mechanism Regulating Muscle Stem Cell Quiescence and Self-Renewal. Cell Stem Cell (2015). [DOI] [PubMed]
- 38.Hausburg MA, et al. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. eLife. 2015;4:e03390. doi: 10.7554/eLife.03390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shefer G, Yablonka-Reuveni Z. Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods Mol Biol. 2005;290:281–304. doi: 10.1385/1-59259-838-2:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bischoff R. Proliferation of muscle satellite cells on intact myofibers in culture. Dev. Biol. 1986;115:129–139. doi: 10.1016/0012-1606(86)90234-4. [DOI] [PubMed] [Google Scholar]
- 41.Bischoff R. Analysis of muscle regeneration using single myofibers in culture. Med Sci Sports Exerc. 1989;21:S164–S172. doi: 10.1249/00005768-198910001-00008. [DOI] [PubMed] [Google Scholar]
- 42.Joo HY, et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 43.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26. doi: 10.3389/fonc.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Citterio E. Fine-tuning the ubiquitin code at DNA double-strand breaks: deubiquitinating enzymes at work. Front Genet. 2015;6:282. doi: 10.3389/fgene.2015.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 47.Sousa-Victor P, et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- 48.Cioni M, et al. Strength deficit of knee extensor muscles of individuals with Down syndrome from childhood to adolescence. Am J Ment Retard. 1994;99:166–174. [PubMed] [Google Scholar]
- 49.Horvat M, Pitetti KH, Croce R. Isokinetic torque, average power, and flexion/extension ratios in nondisabled adults and adults with mental retardation. J Orthop Sports Phys Ther. 1997;25:395–399. doi: 10.2519/jospt.1997.25.6.395. [DOI] [PubMed] [Google Scholar]
- 50.Mercer VS, Lewis CL. Hip Abductor and Knee Extensor Muscle Strength of Children with and without Down Syndrome. Pediatr Phys Ther. 2001;13:18–26. [PubMed] [Google Scholar]
- 51.Casabona A, Valle MS, Pisasale M, Pantò MR, Cioni M. Functional assessments of the knee joint biomechanics by using pendulum test in adults with Down syndrome. J Appl Physiol. 2012;113:1747–1755. doi: 10.1152/japplphysiol.00960.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keefe AC, et al. Muscle stem cells contribute to myofibres in sedentary adult mice. Nat Commun. 2015;6:7087. doi: 10.1038/ncomms8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsimaras VK, Fotiadou EG. Effect of training on the muscle strength and dynamic balance ability of adults with down syndrome. J Strength Cond Res. 2004;18:343–347. doi: 10.1519/R-12832.1. [DOI] [PubMed] [Google Scholar]
- 54.Abreu P, Mendes SV, Ceccatto VM, Hirabara SM. Satellite cell activation induced by aerobic muscle adaptation in response to endurance exercise in humans and rodents. Life Sci. 2017;170:33–40. doi: 10.1016/j.lfs.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 55.Smith HK, Merry TL. Voluntary resistance wheel exercise during post-natal growth in rats enhances skeletal muscle satellite cell and myonuclear content at adulthood. Acta Physiol (Oxf) 2012;204:393–402. doi: 10.1111/j.1748-1716.2011.02350.x. [DOI] [PubMed] [Google Scholar]
- 56.Bellamy LM, et al. The acute satellite cell response and skeletal muscle hypertrophy following resistance training. PLoS One. 2014;9:e109739. doi: 10.1371/journal.pone.0109739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Souroullas GP, Sharpless NE. Stem cells: Down’s syndrome link to ageing. Nature. 2013;501:325–326. doi: 10.1038/nature12558. [DOI] [PubMed] [Google Scholar]
- 58.Roper RJ, VanHorn JF, Cain CC, Reeves RH. A neural crest deficit in Down syndrome mice is associated with deficient mitotic response to Sonic hedgehog. Mech Dev. 2009;126:212–219. doi: 10.1016/j.mod.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zigman WB. Atypical aging in Down syndrome. Dev Disabil Res Rev. 2013;18:51–67. doi: 10.1002/ddrr.1128. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Z, Yang H, Wang H. The histone H2A deubiquitinase USP16 interacts with HERC2 and fine-tunes cellular response to DNA damage. J Biol Chem. 2014;289:32883–32894. doi: 10.1074/jbc.M114.599605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 62.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Micali N, et al. Down syndrome fibroblasts and mouse Prep1-overexpressing cells display increased sensitivity to genotoxic stress. Nucleic Acids Res. 2010;38:3595–3604. doi: 10.1093/nar/gkq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zana M, et al. Age-dependent oxidative stress-induced DNA damage in Down’s lymphocytes. Biochem Biophys Res Commun. 2006;345:726–733. doi: 10.1016/j.bbrc.2006.04.167. [DOI] [PubMed] [Google Scholar]
- 65.Morawiec Z, et al. DNA damage and repair in children with Down’s syndrome. Mutat Res. 2008;637:118–123. doi: 10.1016/j.mrfmmm.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 66.Lorenzo LP, et al. Defective hematopoietic stem cell and lymphoid progenitor development in the Ts65Dn mouse model of Down syndrome: potential role of oxidative stress. Antioxid Redox Signal. 2011;15:2083–2094. doi: 10.1089/ars.2010.3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanij E, Hatzistavrou T, Hertzog P, Kola I, Wolvetang EJ. Ets-2 is induced by oxidative stress and sensitizes cells to H(2)O(2)-induced apoptosis: implications for Down’s syndrome. Biochem Biophys Res Commun. 2001;287:1003–1008. doi: 10.1006/bbrc.2001.5680. [DOI] [PubMed] [Google Scholar]
- 68.Kusters MA, Verstegen RH, Gemen EF, de Vries E. Intrinsic defect of the immune system in children with Down syndrome: a review. Clin Exp Immunol. 2009;156:189–193. doi: 10.1111/j.1365-2249.2009.03890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ram G, Chinen J. Infections and immunodeficiency in Down syndrome. Clin Exp Immunol. 2011;164:9–16. doi: 10.1111/j.1365-2249.2011.04335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aurora AB, Olson EN. Immune modulation of stem cells and regeneration. Cell Stem Cell. 2014;15:14–25. doi: 10.1016/j.stem.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saclier M, Cuvellier S, Magnan M, Mounier R, Chazaud B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 2013;280:4118–4130. doi: 10.1111/febs.12166. [DOI] [PubMed] [Google Scholar]
- 72.Chazaud B. Macrophages: supportive cells for tissue repair and regeneration. Immunobiology. 2014;219:172–178. doi: 10.1016/j.imbio.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 73.Chazaud B. Inflammation during skeletal muscle regeneration and tissue remodeling: application to exercise-induced muscle damage management. Immunol Cell Biol. 2016;94:140–145. doi: 10.1038/icb.2015.97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.