Abstract
Non-O157 Shiga toxin-producing Escherichia coli (STEC) is increasingly recognized as an important enteric foodborne pathogen. The hallmark of the disease is the production of Shiga toxins; however, there are other virulence factors that contribute to the pathogenesis of STEC. This study aimed to investigate the prevalence and genetic diversity of the enterohaemolysin gene, ehxA, among non-O157 STEC strains from human, animal, and food sources. The ehxA gene was amplified from 138 (31.8%) of 434 non-O157 STEC strains, among which 36 unique ehxA sequences were identified. Based on ehxA sequence analysis, three phylogenetic ehxA groups (I II, and III) were determined. Correlations between ehxA groups and sources, serotypes, and virulent gene profiles were observed. The ehxA group II strains were mostly diarrhoeal patient-derived and may demonstrate higher pathogenic potential compared with the ehxA group I and group III strains. Five types of replicons (I1-Ig, FIB, K, F, and B/O) were identified in the 138 ehxA-positive strains, and 3.6%, 5.8%, and 52.2% of the strains harboured toxB, katP and espP genes, respectively, implying marked genetic diversity of ehxA containing plasmids in non-O157 STEC strains. Sequence-based ehxA genotyping might be important in modern strain typing and in epidemiological surveillance of non-O157 STEC infections.
Introduction
Shiga toxin-producing Escherichia coli (STEC) is an important enteric foodborne pathogen causing mild human diarrhoea, haemorrhagic colitis (HC), and fatal haemolytic uremic syndrome (HUS) worldwide1. More than 400 serotypes have been detected in STEC, and O157:H7 is regarded as the most predominant and virulent serotype associated with severe human illness2. Nevertheless, recent studies revealed that non-O157 STEC serotypes, with O26, O45, O103, O111, O121, and O145 being the top six serogroups, are responsible for increasing numbers of outbreaks or sporadic cases worldwide3. Domestic or wild animals are the main natural reservoirs of STECs. Humans are the accidental host of STEC through contact with animals or ingestion of contaminated meat, milk, vegetables, fruit, and water4,5.
Shiga toxin (Stx) is regarded as the most critical virulence factor, which can be divided into two types: Stx1 and Stx26. Several subtypes and variants for each type have been described7. Another virulence factor, intimin (eae), is located on the locus of enterocyte effacement (LEE), and induces characteristic histopathological lesions, referred to as attaching and effacing lesions8. In addition, haemolysin plays an important role in STEC pathogenicity. To date, four different types of haemolysin (hlyA, ehxA, sheA, and e-hlyA) have been identified in E. coli, among which the plasmid-carried enterohaemolysin (ehxA) is widespread in STEC strains and is frequently associated with diarrhoeal disease and HUS9–11. The presence of enterohaemolysin correlates with that of Shiga toxin; therefore, it has been suggested as an epidemiological marker for the rapid and simple detection of STEC strains12. Six genetically distinct ehxA subtypes (A to F) were described in E. coli using PCR and restriction fragment length polymorphism (RFLP) analysis10.
The complete ehxA gene is about 3000 base pairs and resides in the ehx cluster, which contains four genes in the order of ehxC, ehxA, ehxB, and ehxD13. ehxC is associated with haemolysin activation, and ehxB and ehxD are related to the secretion of haemolysin13,14. In O157 STECs, the ehx cluster is located on a plasmid called pO157, a non-conjugative F-like plasmid with ranging in size from 92 to 104 kb. pO157 is a dynamic structure that contains other putative virulence-related factors, such as a catalase-peroxidase (katP) that increases the ability to colonize the host intestine in the absence of oxygen; a serine protease (espP) that influences the colonization and adhesion to intestinal epithelial cells; and a putative adhesin (toxB) that enhances adhesion by increasing the secretion of type three secretion system (TTSS)15–17. These genes may have important functions in the pathogenicity of STECs; however, their roles are not fully understood18.
In our previous studies, we systematically investigated the prevalence of STEC from various sources and geographical areas in China, and collected a broad variety of non-O157 STEC strains from cattle, goats, pigs, yaks, marmots, pika, antelopes, food, diarrhoeal patients, and healthy carriers19. The objective of the present study was to investigate the prevalence and genetic diversity of ehxA genes in correlation with the sources, serotypes, virulence profiles, haemolytic activities, and potential pathogenicity among various non-O157 STEC strains. Furthermore, using sequence-based analysis instead of the traditional PCR-RFLP method, which might produce possible misinterpretation, we intended to genotype the ehxA genes with high stability and accuracy, thus gaining a better understanding of their genetic diversity and relatedness.
Results
Proportion of ehxA in the non-O157 STEC collection
A total of 434 non-O157 STEC strains were screened for the presence of the ehxA gene. ehxA tested positive in 138 (31.8%) strains isolated from different sources including humans, animals, and foods: diarrhoeal patients (9), healthy carrier (1), yaks (66), pika (15), antelopes (4), marmots (4), goats (12), beef cattle/cow (6), pig (1), raw mutton (10), raw beef (8), raw chicken meat (1), and raw duck meat (1) (Tables 1 and S1).
Table 1.
The origin and location of 434 non-O157 STEC isolates used in this study.
| Source | Location | Sampling year | No. of samples | No. of stx-positive | No of ehxA-positive (%)* |
|---|---|---|---|---|---|
| Goat | Henan | 2009 | N/A | 28 | 12 (42.9%) |
| Yak | Qinghai | 2012 | 728 | 128 | 66 (51.6%) |
| Plateau Pika | Qinghai | 2012, 2012, 2015 | 1116 | 22 | 15 (68.2%) |
| Marmots | Qinghai | 2012, 2013 | 335 | 8 | 4 (50.0%) |
| Tibetan antelope | Qinghai | 2014 | 505 | 5 | 4 (80.0%) |
| Pig | Chongqing, Beijing, Guizhou, Shandong, Heilongjiang | 2011, 2012, 2013, 2015 | 1181 | 135 | 1 (0.7%) |
| Cattle/Cow | Shandong, Heilongjiang | 2009, 2013, 2015 | 605 | 14 | 6 (42.9%) |
| Raw meat | Beijing, Sichuan | 2013, 2014 | 853 | 60 | 20 (33.3%) |
| Diarrhoeal patient | Henan, Shenzhen, Shanghai, Sichuan | 2010, 2012, 2013, 2014, 2016 | N/A | 30 | 9 (33.3%) |
| Healthy carrier | Qinghai, Shenzhen | 2013, 2014 | 2058 | 4 | 1 (25.0%) |
| Total | 434 | 138 (31.8%) |
N/A: The number of samples was not applicable. *The percentage of ehxA gene was detected in the stx-positive strains.
Diversity of ehxA and ehxCABD
A total of 138 complete ehxA sequences were obtained and the sizes were all 2,997-bp. Among the 138 ehxA sequences, 36 unique ehxA sequences were identified, which were nominated as ehxA genotype 1 to genotype 36 (Table S2). The nucleotide identities among the 36 ehxA genotypes ranged from 96 to 99%.
Forty-two ehxCABD cluster sequences were obtained from the whole genome sequences. Similar to ehxA, the 42 complete ehxCABD clusters showed high similarity. The identities ranged from 93 to 100% for ehxC, 97 to 100% for ehxB, and 96 to 100% for ehxD.
ehxA groups based on the phylogenetic trees
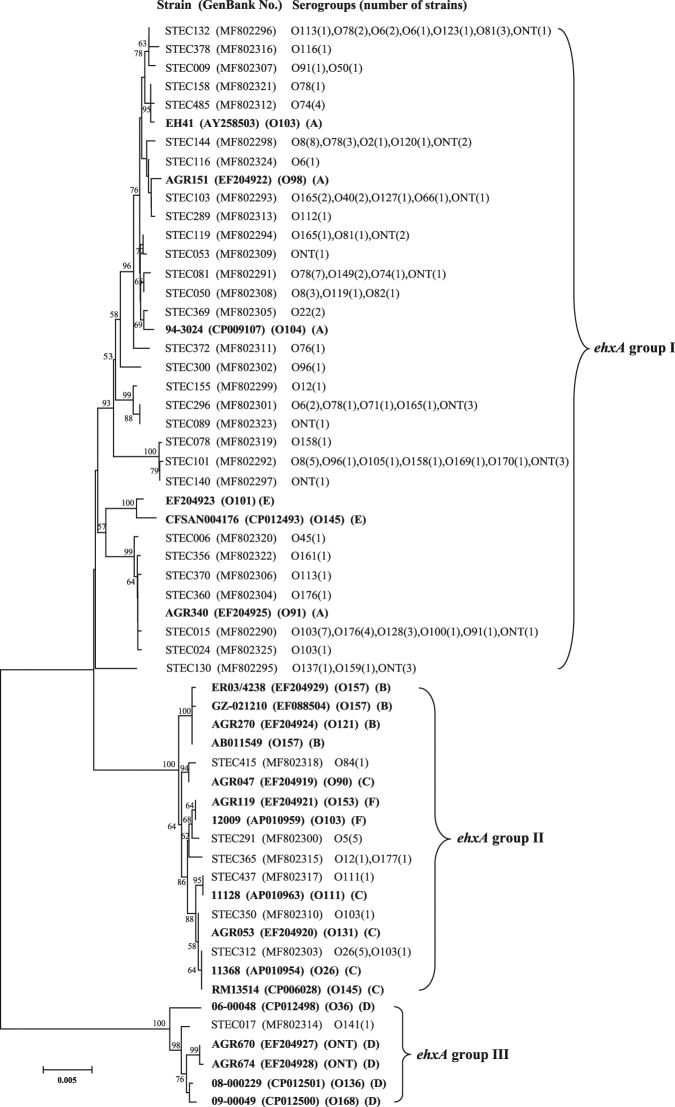
A neighbour-joining tree was constructed using the 36 unique ehxA sequences obtained in this study and 22 reference ehxA sequences (Fig. 1). The phylogenetic trees reconstructed using the neighbour-joining algorithm and the maximum-likelihood algorithms were quite similar (Fig. S1), which demonstrated that all ehxA sequences fell into three phylogenetic groups, named group I, group II, and group III. The sequence similarities within each group were higher than 98%. The majority (87.7%, 121 out of 138) of our ehxA-positive STEC strains clustered into group I; 16 (11.6%) strains were placed into group II; and only one strain belonged to group III (Fig. 1).
Figure 1.
Phylogenetic relationships of ehxA sequences based on the neighbour-joining method. Thirty-six unique ehxA sequences were obtained in this study. The serogroups for each ehxA genotype (representative strain) are given. Twenty-two sequences of six ehxA PCR-RFLP (polymerase chain reaction-restriction fragment length polymorphism) subtypes A to F downloaded from GenBank are indicated in bold. Bootstrap values > 50% are shown at the branch points.
STEC origins correlated with the ehxA groups
In this study, most animal-derived strains belong to group I, with exception of two marmot strains and one cattle strain that belonged to group II, and one goat strain that belonged to group III. Notably, all nine diarrhoeal patient-derived strains belonged to group II and one healthy carrier strain belonged to group I. Food-derived strains belonged to group I or II. Certain ehxA groups were associated with non-O157 STEC origins (P < 0.05) (Table 2).
Table 2.
The origins and virulence factors associated with ehxA groups in 138 non-O157 STEC strains.
| ehxA groups | No. of strains | No. of strains with virulence factor (s) from different sources | |||||
|---|---|---|---|---|---|---|---|
| Animal | Human | Food | |||||
| stx | stx + eae | stx | stx + eae | stx | stx + eae | ||
| I | 121 | 102 | 2 | 1 | 0 | 16 | 0 |
| II | 16 | 2 | 1 | 1 | 8 | 0 | 4 |
| III | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Total | 138 | 105 | 3 | 2 | 8 | 16 | 4 |
STEC serotypes in different ehxA groups
In total, 118 ehxA-positive non-O157 STEC strains were typed into 43 different O serogroups and 20 strains were O-untypable (ONT). ehxA groups I, II, and III contained 37, seven and one serogroup(s), respectively. Only strains of serogroup O103 and O12 were observed in both group I and II (Table 3); however, they could be further distinguished by their H types. Strains of O103:H8 and O12:H8 were in ehxA group I, while strains O103:H25 and O12:H-untypable were in group II (Table S2).
Table 3.
Serogroups, virulence factors, origin, and plasmid Inc (incompatibility) associated with ehxA groups.
| Serogroup | Number | stx | eae | Origin | Inc (number) | ehxA group | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stx 1 | stx 2 | stx1 + stx2 | H | A | F | I | II | III | ||||
| O2 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FIB(1);F(1) | 1 | 0 | 0 |
| O5 | 5 | 5 | 0 | 0 | 3 | 2 | 2 | 1 | FIB(5);F(3) | 0 | 5 | 0 |
| O6 | 5 | 3 | 2 | 0 | 0 | 0 | 5 | 0 | FIB(5);F(4);K(1) | 5 | 0 | 0 |
| O8 | 16 | 3 | 5 | 8 | 0 | 0 | 16 | 0 | I1-Ig(1);FIB(15);F(16);B/O(1) | 16 | 0 | 0 |
| O12 | 2 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | FIB(1);F(2);B/O(1) | 1 | 1 | 0 |
| O22 | 2 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | FIB(2);F(2) | 2 | 0 | 0 |
| O26 | 5 | 5 | 0 | 0 | 5 | 5 | 0 | 0 | FIB(5); B/O(2) | 0 | 5 | 0 |
| O40 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | FIB(2);F(2);B/O(1) | 2 | 0 | 0 |
| O45 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | FIB(1);K(1) | 1 | 0 | 0 |
| O50 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | FIB(1);F(1) | 1 | 0 | 0 |
| O66 | 3 | 1 | 2 | 0 | 0 | 0 | 3 | 0 | FIB(2);F(3);K(2) | 3 | 0 | 0 |
| O71 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FIB(1);F(1) | 1 | 0 | 0 |
| O74 | 5 | 0 | 4 | 1 | 0 | 0 | 5 | 0 | FIB(3);F(4);K(4) | 5 | 0 | 0 |
| O76 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | K(1) | 1 | 0 | 0 |
| O78 | 13 | 0 | 11 | 2 | 2 | 0 | 13 | 0 | FIB(11);F(5);K(3) | 13 | 0 | 0 |
| O81 | 4 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | FIB(4);F(3);K(1) | 4 | 0 | 0 |
| O82 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | FIB(1);F(1);B/O(1) | 1 | 0 | 0 |
| O84 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | FIB(1);F(1) | 0 | 1 | 0 |
| O91 | 3 | 0 | 0 | 3 | 0 | 1 | 2 | 0 | FIB(3);F(3) | 3 | 0 | 0 |
| O96 | 2 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | FIB(1);F(1) | 2 | 0 | 0 |
| O100 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FIB(1);F(1);K(1) | 1 | 0 | 0 |
| O103 | 10 | 10 | 0 | 0 | 2 | 0 | 8 | 2 | FIB(10);F(8);K(3); B/O(5) | 8 | 2 | 0 |
| O105 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FIB(1);F(1);K(1) | 1 | 0 | 0 |
| O111 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | FIB(1);F(1);B/O(1) | 0 | 1 | 0 |
| O112 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | FIB(1);F(1) | 1 | 0 | 0 |
| O113 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | FIB(2);F(2) | 2 | 0 | 0 |
| O116 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | FIB(1);K(1) | 1 | 0 | 0 |
| O119 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | F(1);B/O(1) | 1 | 0 | 0 |
| O120 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | FIB(1);F(1) | 1 | 0 | 0 |
| O123 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | FIB(1);F(1) | 1 | 0 | 0 |
| O127 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | FIB(1);F(1) | 1 | 0 | 0 |
| O128 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | FIB(3);F(2);K(1); B/O(2) | 3 | 0 | 0 |
| O137 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | F(1);B/O(1) | 1 | 0 | 0 |
| O141 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FIB(1);F(1) | 0 | 0 | 1 |
| O149 | 2 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | FIB(2);F(1) | 2 | 0 | 0 |
| O158 | 2 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | FIB(2);F(2) | 2 | 0 | 0 |
| O159 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | Negative | 1 | 0 | 0 |
| O161 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | Negative | 1 | 0 | 0 |
| O165 | 4 | 2 | 1 | 1 | 0 | 0 | 4 | 0 | FIB(3);F(3);K(1) | 4 | 0 | 0 |
| O169 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FIB(1);F(1) | 1 | 0 | 0 |
| O170 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FIB(1);F(1) | 1 | 0 | 0 |
| O176 | 5 | 4 | 0 | 1 | 0 | 0 | 0 | 5 | FIB(4);F(4);K(1) | 5 | 0 | 0 |
| O177 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | FIB(1);F(1) | 0 | 1 | 0 |
| ONT | 20 | 8 | 6 | 6 | 0 | 0 | 20 | 0 | FIB(20);F(19);K(2); B/O(3) | 20 | 0 | 0 |
H: human; A: animal; F: food. Strains of serogroup O103 and O12 in both group I and group II are indicated in bold.
Co-occurrence of ehxA and other virulence genes
The intimin gene eae and three putative pO157-derived virulence genes (toxB, katP, and espP) were screened among the 138 ehxA-positive non-O157 STEC isolates. In total, only 15 (10.9%) isolates harboured eae, among which two (1.7%) were placed into ehxA group II and 13 (81.2%) into group II (Table 2). Among the 13 isolates that were both eae and ehxA-positive, eight were from diarrhoeal patients. These results showed that ehxA group II strains were mostly eae-positive and clinically related, while strains in ehxA group I and group III might be eae-negative (P < 0.05). In addition, espP was present in 72 (52.2%) isolates, but only eight (5.8%) and five (3.6%) isolates were katP and toxB positive.
Haemolytic activity on SHIBAM plates
Among the 138 ehxA-positive non-O157 STEC strains, 93 (67.4%) strains showed haemolytic activity against washed sheep red blood cells. The haemolysis ability varied among the strains. Fifty-five strains showed relatively clear transparent zones on the SHIBAM plates, while only narrow haemolytic zones were observed for 38 strains (Table S2). Haemolytic strains in ehxA groups I and II were 69.4% (84/121) and 56.3% (9/16), respectively. The ehxA group III strain did not show haemolytic activity. No statistical significance was observed for the correction between haemolytic activity and ehxA groups (P > 0.05).
Plasmid incompatibility (Inc)/replicon (Rep) profiles in ehxA-positive non-O157 STEC strains
Eighteen different plasmid replicons were screened by PCR, including HI1, HI2, I1-Ig, X, L/M, N, FIA, FIB, W, Y, P, FIC, T, A/C, FIIA, K, F, and B/O. Five types of replicons (I1-Ig, FIB, K, F, and B/O) were detected (Table S2). Among them, 123 (89.1%) and 107 (77.5%) strains were positive for FIB and F replicons, respectively. K and B/O replicons were present in 24 (17.4%) and 21 (15.2%) strains, respectively. Only one strain contained the I1-Ig replicon. Most strains (82.6%) harboured two or more replicons; however, four strains did not harbour any replicons.
All five plasmid replicons were observed in ehxA group I strains and three plasmid replicons were detected in group II. The only strain in group III contained replicons FIB and F.
Discussion
STEC related virulence factors, such as Shiga toxin and intimin, have been well investigated worldwide, especially for O157:H7 and other predominant serotypes20,21. Nevertheless, enterohaemorrhagic E. coli haemolysin (Ehx) is an increasingly recognized putative virulence factor22. In the present study, we systematically investigated the prevalence of the ehxA gene in non-O157 STEC strains isolated from a variety of sources and different geographical locations in China. Our study showed that the ehxA gene was present in 31.8% (138/434) of the non-O157 STEC strains, which was lower than that of a previous report (69.2%)23, indicating that the prevalence of ehxA among non-O157 STEC might vary in different sources or geographical locations.
The ehxA gene is relatively conserved in STEC strains, and six ehxA subtypes have been reported based on PCR-RFLP subtyping methods23. ehxA subtypes may provide information on the epidemiology and evolution of E. coli; however, the traditional PCR-RFLP subtyping method is time-consuming and laborious. Furthermore, cross-reactions sometimes occurred and appeared as ghost bands on gel electrophoresis, especially for those strains with high sequence similarity, which call for additional stringency to differentiate them. Notably, PCR-RFLP protocols have generated contradictory results for the subtyping of some toxins, for example, Shiga toxin subtypes7. Here, a sequence-based phylogenetic approach was used for ehxA subtyping. By comparing the complete ehxA sequences of 138 ehxA-positive STEC isolates from in this study and those assigned to six existing subtypes23, three clear phylogenetic groups were formed. Additionally, the alignment of all known subtypes allowed us to evaluate the PCR-RFLP subtyping method and identify possible misinterpretations of PCR-RFLP results, as this method has never really been validated on ehxA against a representative number of strains. Our results showed that the phylogenic relatedness of the three ehxA groups assigned in this study were in agreement with that of the A-F subtypes reported previously23. ehxA group I comprised ehxA sequences that were assigned as the A and E subtypes; group II comprised sequences assigned as ehxA subtype B, C, and F, which were found to show a close relationship; while group III contained only the ehxA D subtype, which formed the most divergent subdivision and was well separated from the other groups/subtypes (Fig. 1). It was deduced that group III or subtype D strains might carry an ehxA-containing plasmid that is different from the other subtypes24.
The current study demonstrated a correlation between ehxA groups and their sources, which was in agreement with a previous study10. Healthy carrier strains, and most animal (96.2%) and food-derived strains (80.0%) fell into ehxA group I, where toxB and espP gene were detected in five (4.1%) and 60 (49.6%) isolates, and the serotypes were diverse. Remarkably, all strains from diarrhoeal patients belonged to group II, in which most strains harboured eae (81.3%), katP (50.0%), and espP (50.0%), and high pathogenic serotypes: i.e, O26:H11, O111:H8, and O103:H25 were observed. It can be inferred that ehxA group II strains showed higher pathogenic potential compared with those in group I, while strains in group III were less associated with human illness. It should be noted that 20.0% of food-derived strain and 2.7% of the animal strains were classified into group II, further enhancing our previous finding that some of the food- and animal-derived strains showed high pathogenic potential; thus, humans might be infected by the consumption of foods or contamination from animals19.
The ehxA positive strains have been demonstrated to have an enterohaemolytic phenotype in vitro on washed sheep blood agar10. In this study, 93 (67.4%) strains showed enterohaemolytic activity, which was less than that described in a previous study, where 276 (82.8%) of ehxA-positive STEC strains showed enterohaemolytic capacity23. It could be inferred that some non-O157 STECs showed lower tendency to cause haemolysis, especially among those less predominant serotypes. No obvious correlation between haemolytic capacity and ehxA groups was observed. By contrast, the absence of an enterohaemolytic phenotype indicated that the precise conditions for the regulation and optimum expression of enterohaemolysin among the non-O157 STEC strains remains to be further determined22.
E. coli possess a variety of plasmids, and many plasmids are associated with pathogenicity. Plasmids are defined as incompatibility groups when they have the same replication mechanisms25,26. The ehxA gene is located on the F-like plasmid pO157 in E. coli O157:H718. We tested eighteen plasmid incompatibility groups, and five groups were detected in 138 ehxA-positive non-O157 STEC. FIB and F were the major plasmid incompatibility groups detected in this study. Furthermore, plasmid pO157 contains others genes, such as katP, espP, and toxB18. In a previous report, plasmids resembling pO157 were found in non-O157 STEC strains that carried ehxA, katP and espP genes in less than 50% of strains27. In this study, only 3.6% and 5.8% of strains harboured the toxB and katP genes, respectively, while espP was positive in 52.2% of isolates. These results implied a marked genetic diversity of ehxA containing plasmids in non-O157 STEC strains, which requires further analysis.
In conclusion, this study systematically investigated the prevalence of the enterohaemolysin gene ehxA in non-O157 STEC strains from a variety of sources collected from different geographical locations in China. A sequence-based phylogenetic approach was used for ehxA genotyping, based on a wide variety of strains. In total, 138 ehxA-positive non-O157 STEC strains were identified from among 434 strains, which were phylogenetically clustered into three groups (I, II, and III). Correlations between the ehxA groups and the sources, serotypes, and virulent gene profiles were observed. The ehxA group II non-O157 STEC strains were mostly diarrhoeal patient-derived and may demonstrate a higher pathogenic potential compared with group I and group III strains. These results are of great importance in modern strain typing and in epidemiological surveillance of non-O157 STEC infections.
Methods
Bacterial strains
A total of 434 non-O157 STEC strains representing 95 different O-serogroups were collected during April 2009 to August 2016 in ten geographical regions in China (Tables 1 and S1). Most strains were obtained and reported in our previous investigations19,28–31. Only a small number of strains were isolated from samples collected by local centres for disease control and prevention. These strains were isolated from various sources, including cattle/cow, goat, pig, yak, marmot, pika, antelope, food, patients with diarrhoea, and healthy carriers. All strains were confirmed to be STEC using previously described methods19.
Serotyping and detection of virulence genes
The O:H serotyping of each strain was determined using previously described methods29. The enterohaemolysin gene ehxA of all 434 non-O157 STEC strains, and eae, katP, espP and toxB genes of all ehxA-positive strains were determined using PCR with primer described previously30.
Haemolysis test
All ehxA-positive STEC strains were inoculated overnight on Luria-Bertani (LB) agar (Oxoid, UK). A single fresh colony was picked and inoculated onto SHIBAM plates and incubated aerobically at 37 °C. SHIBAM plates are brain-heart infusion medium supplemented with 10 mM CaCl2, 5% phosphate-buffered saline (PBS)-washed defibrinated sheep blood, and 0.5 μg/ml mitomycin C32. Hemolysis was observed after 6 and 24 h, as described previously10. Each strain was tested in duplicate.
Sequencing of the complete ehxA gene
The complete ehxA gene was obtained by PCR using previously described methods10,23. Two other pairs of primers designed in this study were used for sequencing: ehxA F-W1F (5′-TGGGCTGGATGTTGTCTC-3′) and ehxA R-W1R (5′-TTCCACTACCACCAAATAAC-3′); ehxA R-W1F-low (5′-GTTATAACAGATAAAGATGGTCG-3′) and ehxA F-W1R-low (5′-CTGGTTTGCAATCGCTGTATCAT-3′). The PCR product was purified using a QIAquick PCR Purification kit (Qiagen, Hilden, Germany) and sequenced using the BigDye™ Terminator V3.1 Cycle Sequencing kit (Applied Biosystems, USA).
Phylogenetic analysis
The sequenced ~2997-bp ehxA was assembled using SeqMan II (DNASTAR Inc., USA). The representatives of reference ehxA sequences of six ehxA subtypes A to F were downloaded from GenBank10,24. The ehxA sequences obtained in this study and the reference sequences were aligned using MEGA 7 software (www.megasoftware.net)33. Phylogenetic trees were constructed with two algorithms, neighbour-joining (NJ), and maximum-likelihood (ML), using MEGA 7. The stability was estimated by bootstrap analysis (1000 replications), and genetic distances were calculated by the maximum composite likelihood method. A sequence was designated to a specific ehxA group based on its phylogenetic placement.
Whole genome sequencing
Forty-two ehxA-positive strains representing diverse sources, stx subtypes, and virulence profiles were selected for whole genome sequencing (WGS). For each strain, a 500–2000 bp library was constructed and then sequenced on an Illumina HiSeq. 2500-PE125 (Illumina, San Diego, CA, USA) to produce 150 bp paired-end reads. These qualified reads were then assembled into scaffolds using the program SOAP de novo (http://soap.genomics.org.cn/soapdenovo.html). Open reading frames (ORFs) were identified and annotated using the Artemis program (www.sanger.ac.uk) and homology searches were performed against several databases, including GenBank (www.ncbi.nlm.nih.gov/GenBank)34. The ehxCABD gene cluster was extracted from the draft genome sequences using a perl script. Three genes, katP, toxB and espP, were searched against the 42 genome sequences using BLAST.
PCR-based replicon typing
Eighteen pairs of primers were used to determine the plasmid incompatibility (Inc) groups in all ehxA-positive strains, as described previously25. The assays were performed in five multiplex-PCR and three simplex-PCR reactions. PCR conditions are as follows: initial denaturation of 94 °C for 5 min; followed by 30 cycles of 94 °C for 1 min, 60 °C for 30 s and 72 °C for 1 min; and a final extension at 72 °C for 5 min, with the exception of the IncF-simplex PCR, where the annealing temperature was set at 52 °C.
Statistical analysis
The correlations between ehxA groups and STEC origins, serogroups, haemolytic ability, or virulence genes were analysed using Fisher’s exact test. SAS® 9.3 (SAS Institute Inc., USA) was used to perform the calculations. A P value < 0.05 was considered significant statistically.
Nucleotide sequence accession numbers
Thirty-six complete ehxA sequences obtained in this study were submitted to GenBank under the accession numbers MF802290–MF802325.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81371762, 81701977, 81772152), the National Basic Research Priorities Program of China (2015CB554201), and the State Key Laboratory of Infectious Disease Prevention and Control (2015SKLID504). We would like to thank the native English speaking scientists of Elixigen Company (Huntington Beach, California) for editing our manuscript.
Author Contributions
X.B. and Y.X. designed the research. R.F., H.S., and Y. Xu. prepared the samples. S.F. conducted the experiments and analysed the data. S.F., X.B., and Y.X. wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22699-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Karmali MA. Emerging public health challenges of Shiga toxin-producing Escherichia coli related to changes in the pathogen, the population, and the environment. Clin Infect Dis. 2017;64:371–376. doi: 10.1093/cid/ciw708. [DOI] [PubMed] [Google Scholar]
- 2.Miko A, et al. Emerging types of Shiga toxin-producing E. coli (STEC) O178 present in cattle, deer, and humans from Argentina and Germany. Front Cell Infect Microbiol. 2014;4:78. doi: 10.3389/fcimb.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad CC, Stanford K, McAllister TA, Thomas J, Reuter T. Further Development of sample preparation and detection methods for O157 and the top 6 non-O157 STEC serogroups in cattle feces. J Microbiol Methods. 2014;105:22–30. doi: 10.1016/j.mimet.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 4.Johnson KE, Thorpe CM, Sears CL. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin Infect Dis. 2006;43:1587–1595. doi: 10.1086/509573. [DOI] [PubMed] [Google Scholar]
- 5.Kappeli U, Hachler H, Giezendanner N, Beutin L, Stephan R. Human infections with non-O157 Shiga toxin-producing Escherichia coli, Switzerland, 2000-2009. Emerg Infect Dis. 2011;17:180–185. doi: 10.3201/eid1702.100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergan J, Dyve Lingelem AB, Simm R, Skotland T, Sandvig K. Shiga toxins. Toxicon. 2012;60:1085–1107. doi: 10.1016/j.toxicon.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 7.Scheutz F, et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol. 2012;50:2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith JL, Fratamico PM, Gunther NWT. Shiga toxin-producing Escherichia coli. Adv Appl Microbiol. 2014;86:145–197. doi: 10.1016/B978-0-12-800262-9.00003-2. [DOI] [PubMed] [Google Scholar]
- 9.Lehmacher A, Meier H, Aleksic S, Bockemuhl J. Detection of hemolysin variants of Shiga toxin-producing Escherichia coli by PCR and culture on vancomycin-cefixime-cefsulodin blood agar. Appl Environ Microbiol. 1998;64:2449–2453. doi: 10.1128/aem.64.7.2449-2453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorenz SC, et al. Prevalence of hemolysin genes and comparison of ehxA subtype patterns in Shiga toxin-producing Escherichia coli (STEC) and non-STEC strains from clinical, food, and animal sources. Appl Environ Microbiol. 2013;79:6301–6311. doi: 10.1128/AEM.02200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandhu KS, Clarke RC, Gyles CL. Hemolysin phenotypes and genotypes of eaeA-positive and eaeA-negative bovine verotoxigenic Escherichia coli. Adv Exp Med Biol. 1997;412:295–302. doi: 10.1007/978-1-4899-1828-4_49. [DOI] [PubMed] [Google Scholar]
- 12.Beutin L, et al. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J Clin Microbiol. 1989;27:2559–2564. doi: 10.1128/jcm.27.11.2559-2564.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt H, Maier E, Karch H, Benz R. Pore-forming properties of the plasmid-encoded hemolysin of enterohemorrhagic Escherichia coli O157:H7. Eur J Biochem. 1996;241:594–601. doi: 10.1111/j.1432-1033.1996.00594.x. [DOI] [PubMed] [Google Scholar]
- 14.Welch RA, et al. Battling against host phagocytes: the wherefore of the RTX family of toxins? Infect Agents Dis. 1995;4:254–272. [PubMed] [Google Scholar]
- 15.Brunder W, Schmidt H, Karch H. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142(Pt 11):3305–3315. doi: 10.1099/13500872-142-11-3305. [DOI] [PubMed] [Google Scholar]
- 16.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 17.Tatsuno I, et al. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect Immun. 2001;69:6660–6669. doi: 10.1128/IAI.69.11.6660-6669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim JY, Yoon J, Hovde CJ. A brief overview of Escherichia coli O157:H7 and its plasmid O157. J Microbiol Biotechnol. 2010;20:5–14. [PMC free article] [PubMed] [Google Scholar]
- 19.Bai X, et al. Molecular and phylogenetic characterization of non-O157 Shiga toxin-producing Escherichia coli strains in China. Front Cell Infect Microbiol. 2016;6:143. doi: 10.3389/fcimb.2016.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruger A, Lucchesi PM. Shiga toxins and stx phages: highly diverse entities. Microbiology. 2015;161:451–462. doi: 10.1099/mic.0.000003. [DOI] [PubMed] [Google Scholar]
- 21.Franzin FM, Sircili MP. Locus of enterocyte effacement: a pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation. Biomed Res Int. 2015;2015:534738. doi: 10.1155/2015/534738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bielaszewska M, Aldick T, Bauwens A, Karch H. Hemolysin of enterohemorrhagic Escherichia coli: structure, transport, biological activity and putative role in virulence. Int J Med Microbiol. 2014;304:521–529. doi: 10.1016/j.ijmm.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Cookson AL, Bennett J, Thomson-Carter F, Attwood GT. Molecular subtyping and genetic analysis of the enterohemolysin gene (ehxA) from Shiga toxin-producing Escherichia coli and atypical enteropathogenic E. coli. Appl Environ Microbiol. 2007;73:6360–6369. doi: 10.1128/AEM.00316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz SC, Monday SR, Hoffmann M, Fischer M, Kase JA. Plasmids from Shiga toxin-producing Escherichia coli strains with rare enterohemolysin gene (ehxA) subtypes reveal pathogenicity potential and display a novel evolutionary path. Appl Environ Microbiol. 2016;82:6367–6377. doi: 10.1128/AEM.01839-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carattoli A, et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Francia MV, et al. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 2004;28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Caprioli A, Morabito S, Brugere H, Oswald E. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet Res. 2005;36:289–311. doi: 10.1051/vetres:2005002. [DOI] [PubMed] [Google Scholar]
- 28.Bai X, et al. Prevalence and characteristics of Shiga toxin-producing Escherichia coli isolated from retail raw meats in China. Int J Food Microbiol. 2015;200:31–38. doi: 10.1016/j.ijfoodmicro.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 29.Bai X, et al. Shiga toxin-producing Escherichia coli in plateau pika (Ochotona curzoniae) on the Qinghai-Tibetan plateau, China. Front Microbiol. 2016;7:375. doi: 10.3389/fmicb.2016.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai X, et al. Shiga toxin-producing Escherichia coli in yaks (Bos grunniens) from the Qinghai-Tibetan plateau, China. PloS one. 2013;8:e65537. doi: 10.1371/journal.pone.0065537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng Q, et al. Characterization of Shiga toxin-producing Escherichia coli isolated from healthy pigs in China. BMC Microbiol. 2014;14:5. doi: 10.1186/1471-2180-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin A, et al. Isolation of Shiga toxin-producing Escherichia coli from fresh produce using STEC heart infusion washed blood agar with mitomycin-C. J Food Prot. 2012;75:2028–2030. doi: 10.4315/0362-028X.JFP-12-157. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, et al. Defining the genetic features of O-antigen biosynthesis gene cluster and performance of an O-antigen serotyping scheme for Escherichia albertii. Front Microbiol. 2017;8:1857. doi: 10.3389/fmicb.2017.01857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.