Abstract
Mycoplasma bovis is a major bovine pathogen that causes considerable economic losses in the cattle industry worldwide. Moreover, M. bovis biofilm can persist in the environment and its host. To date, M. bovis biofilm antigens recognized by bovine convalescent sera and their comparison with planktonic cells have not yet been explored. This study utilized an immunoproteomic approach using two-dimensional electrophoresis, immunoblotting using convalescent bovine serum, and subsequent matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometry (MALDI-TOF/TOF MS) to identify the immunoreactive proteins expressed in biofilm- and planktonic-grown M. bovis strain 08M. Results showed that M. bovis biofilms and planktonic cells demonstrate differential immunoreactivity to bovine convalescent serum for the first time. A total of 10 and 8 immunoreactive proteins were identified for biofilms and planktonic cells, respectively. To our knowledge, a total of 12 out of 15 had not been reported as immunoreactive proteins in M. bovis, and six were specific to M. bovis biofilms. Three proteins, namely, endoglucanase, thiol peroxidase, and one putative membrane protein, that is, mycoplasma immunogenic lipase A, were identified in planktonic cells and biofilms. Most of the identified proteins were cytoplasmic proteins that were mainly involved in transport and metabolism. Moreover, ATP binding, oxidoreductase activity, and GTP binding were their most representative molecular functions. DnaK and Tuf appeared to be the most interactive immunoreactive agent among the identified proteins. Furthermore, six proteins had potential as serodiagnostic antigens. These data will be helpful to improve our current understanding on the host response to M. bovis biofilms and planktonic cells, which may facilitate the development of novel molecular candidates of improved diagnostics and vaccines to prevent M. bovis infections.
Keywords: biofilms, Mycoplasma bovis, immunoprotein, immunoproteomics, planktonic cells
Introduction
Mycoplasma bovis is an important pathogen that can cause pneumonia, mastitis, arthritis, otitis, meningitis, and keratoconjunctivitis in cattle and result in considerable economic losses worldwide (Rosengarten and Citti, 1999; Nicholas et al., 2000; Nicholas and Ayling, 2003; Xin et al., 2008). This livestock pathogen is a member of the Mollicutes class. Many M. bovis strains can form biofilms, and the ability for biofilm formation varies among different strains (Mcauliffe et al., 2006). The biofilm growth provides M. bovis several advantages over its planktonic counterparts; these advantages include remarkable resistance to antimicrobials and persistence in the environment and host, which may result in the chronicity of a disease (Mcauliffe et al., 2006) and considerable difficulty of treatment. Biofilms are important in the pathogenesis of several chronic bacterial infections, and lead to persistent infections (Parsek and Singh, 2003). The growth of bacterium under biofilm conditions with increased drug resistance and virulence may be due to the altered metabolism and different protein expression profiles compared with those of planktonic cells (Sauer, 2003; Seneviratne et al., 2012).
Immunoproteins play a key role in host–bacteria interaction by eliciting host immunization and bacterial pathogenesis. Immunoproteomics has been used extensively to identify immunoproteins and virulence factors in several important livestock pathogenic mycoplasma species, such as Mycoplasma mycoides subsp. mycoides small-colony type (MmmSC) (Jores et al., 2009), M. capricolum subsp. capripneumoniae (Mccp) (Zhao et al., 2012), and Mycoplasma mycoides subsp. capri (Mmc) (Corona et al., 2013). Numerous studies investigated immunoproteins in different components of M. bovis in planktonic growth using immunoproteomic assays, in which many related proteins were identified. Variable surface lipoproteins, which vary in phase and size, are M. bovis immunogenic proteins that play a key role in evading the immune system of the host (Sachse et al., 1996, 2000). The conserved P26 (Sachse et al., 1993) and P48 lipoproteins (Robino et al., 2005), mycoplasma immunogenic lipase A (MilA) (Wawegama et al., 2014), and pMB67 (Behrens et al., 1996) are all surface proteins and are immunogenic. Several cytoplasmic proteins, including heat-shock protein 60 and glyceraldehyde-3-phosphate dehydrogenase, are M. bovis-immunogenic proteins. Sun et al. (2014) identified 19 immunoproteins from M. bovis strain PD via immunoproteomics with four naturally infected positive sera of M. bovis. Khan et al. (2016) recently identified 39 immunogens from M. bovis strain HB0801 by using whole-cell protein and membrane fraction analysis; among these identified proteins, 32 have not been reported. To date, M. bovis biofilm antigens that are recognized by bovine convalescent sera have not been explored and compared with planktonic cells.
The present study utilized an immunoproteomics approach using 2-DE, immunoblotting, and matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometry (MALDI-TOF/TOF MS) to identify the antigen profiles of M. bovis biofilms and planktonic cells that react with bovine convalescent sera. Bioinformatic analysis and immunogenicity confirming analysis were further performed. These data are expected to improve our current understanding on the host immune response to M. bovis biofilms and develop novel serological diagnostic markers and vaccine candidates for M. bovis infection.
Materials and Methods
Ethics Statement
All animals were handled in strict accordance with the requirements of the Animal Ethics Procedures and Guidelines of the People’s Republic of China. All efforts were exerted to minimize the suffering of animals. The animal experimental protocol was reviewed and approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Permit No. LVRIAEC2010-007, LVRIAEC2015-009).
Mycoplasma Isolate and Growth Conditions
Mycoplasma bovis strain 08M, which can form biofilms, was isolated from the pneumonia-infected lung of a calf in Ningxia province, China in 2008 (Chen et al., 2017). Biofilms were grown in 150 mm plates that contained 30 mL of PPLO broth (21 g/L PPLO broth, 1 g/L glucose, 2 g/L sodium pyruvate, 100 mL/L 25% yeast extract, 200 mL/L horse serum, pH 7.4–7.6), inoculated with a 1:10 dilution of a 20 h planktonic culture (about 7.40 × 108 CFU/mL), and left at 37°C for 72 h without shaking. The supernatants were removed, and the plates were washed gently twice with PBS (0.01 M) at pH 7.2. Biofilms were detached by scraping, collected by centrifugation at 12,000 × g for 30 min, and resuspended in 1 mL of Tris-HCl (25 mM) at pH 7.2. Planktonic cells were grown in a 1 L bottle that contained 500 mL of PPLO broth at 37°C for 72 h without shaking. The cells were pelleted and washed, as described previously. The planktonic and biofilm cells examined in this study were in the stationary phase.
Scanning Electron Microscopy Analysis
Scanning electron microscopy was conducted to observe the microstructure of the biofilms formed by M. bovis strain 08M on glass coverslips and M. bovis planktonic cells. Briefly, the biofilms on the glass coverslips were washed gently three times with PBS (0.01 M) at pH 7.2. Planktonic cells were collected by centrifugation at 12,000 × g for 30 min and washed three times with PBS (0.01 M) at pH 7.2. M. bovis biofilms and planktonic cells were fixed with 2.5% SEM-grade glutaraldehyde (Sigma-Aldrich, St. Louis, MO, United States), briefly rinsed with PBS twice for 5 min, washed with 4% sucrose, and dehydrated through a graded series of ethanol concentrations (30%, 50%, 70%, 80%, 90%, 95%, and 100%), with each series of ethanol concentrations lasting for 10 min. Finally, the fixed samples were dried by a Quorum K850 critical-point drying apparatus and sputter coated with gold palladium. Observations were performed at 20 kV with a FEI Inspect F SEM.
Preparation of Bovine Antisera Against M. bovis
Convalescent sera against M. bovis 08M were prepared as followed: Three 6-month-old, clinically healthy calves free of M. bovis were infected with M. bovis 08M strain (1.0 × 109 CCU/mL, 10 mL/calf, intratracheally). Bloods were collected from all calves on 0, 8, 17, 30, and 46 days after infection. Titers of the sera sample were evaluated by using a commercial M. bovis antibody ELISA Kit (Bio-X BIO K 302, Bio-X Diagnostics, Rochefort, Belgium). The pooled serum recovered from clinical illness with high titer (46 dpi, OD450nm = 1.068) was used in the immunoblotting experiments. The pooled pre-infected serum (0 dpi, OD450nm = 0.071) was used as a negative control.
Bovine-immune sera against M. bovis were prepared as described below: M. bovis 08M cells were grown in PPLO broth, collected, washed, and concentrated by PBS (0.15 M) at pH 7.2. The protein content was determined using the Bradford method to reach 0.46 and 1.48 mg/ml. Afterward, mycoplasma cells were inactivated with 4 mM binary ethyleneimine at 37°C for 24 h, following the addition of a final concentration of 4 mM Na2S2O3 at 37°C for 24 h, then emulsified with the same volumes of ISA 201 VG adjuvant (SEPPIC, France). Six 8-month-old, clinically healthy calves free of M. bovis were subcutaneously injected with 2 mL inactivated vaccine. A booster injection was administered 28 days after the initial immunization using the same method and doses. Sera samples were collected weekly from all calves from the time of initial immunization until day 28 after the booster injection. Titers of the sera sample were evaluated using a commercial M. bovis antibody ELISA Kit. Bovine antisera with over 2.0 ELISA titers were pooled and used for the Western blot analysis of the selected protein.
Preparation of Whole Proteins From M. bovis Cells
Protein was extracted from three independent cultures of biofilms and planktonic cells as previously described (Carvalhais et al., 2015), with several modifications. Briefly, cell pellets of M. bovis biofilms and planktonic cultures were resuspended in SDT lysis buffer (4% SDS, 100 mM DTT, and 100 mM Tris-HCL). The cell suspension was vortexed, placed in a boiling water bath for 5 min, and mechanically broken using MP Biomedicals FastPrep-24TM Instrument (6 m/s, 30 s per time, three times). Cell suspensions were sonicated on ice at 80 W for 10 cycles (10 s on, 15 s off), and placed in a boiling water bath for 5 min. Cell debris and unbroken cells were removed by centrifugation at 12,000 × g for 30 min at 25°C. The proteins in the supernatant were precipitated using 10 volumes of 10% chilled acetone at -20°C for 12 h. Precipitated protein was collected by centrifugation at 12,000 × g for 10 min at 4°C and washed two times with chilled acetone. The final pellet was air dried. The dried pellet was dissolved in 2D lysis buffer (8 M urea, 4% CHAPS, 40 mM Tris, 65 mM DTT, cocktail), incubated for 30 min at 25°C (vortexed every 10 min), and centrifuged at 12,000 × g for 20 min at 25°C. Prior to rehydration, the supernatant was treated with 2D Clean-up Kit (GE Healthcare) to remove contaminants that can interfere with electrophoresis. The protein content was determined by using the Bradford protein assay kit (Beijing Leagene Biotech, Beijing, China) according to the manufacturer’s instructions.
2-DE and Image Analysis
For the 2-DE, 200 μg of proteins were loaded onto analytical and preparative gels. Ettan IPGphor Isoelectric Focusing System (GE Amersham) and pH 3–10 IPG strips (13 cm, non-linear; GE Healthcare) were used for IEF. The IPG strips were rehydrated in 250 μL of rehydration buffer (8 M Urea, 2% CHAPS, 18 mM DTT, 0.5% IPG buffer, bromophenol blue trace), which contained the protein samples, for 30 min at room temperature. IEF was performed in the following steps: 30 V for 12 h, 500 V for 1 h, 1000 V for 1 h, 8000 V for 8 h, and 500 V for 4 h. After IEF, the IPG strips were equilibrated for 15 min in an equilibration buffer I (50 mM Tris-HCl pH 8.8, 6 M urea, 2% SDS, 30% glycerol, 1% DTT) and for another 15 min in an equilibration buffer II (50 mM Tris-HCl pH 8.8, 6 M urea, 2% SDS, 30% glycerol, 4% iodoacetamide). SDS-PAGE was conducted vertically in a Hofer SE 600 (Amersham Biosciences) using 12.5% polyacrylamide gels. The gels were then stained using modified silver staining methods that are compatible with subsequent mass spectrometric analysis (Yan et al., 2000). 2-DE was performed in triplicate for each growth condition.
The stained 2-DE gels were scanned with UMax Powerlook 2110XL (GE Amersham) and analyzed using ImageMasterTM 2D Platinum 5.0 software (GE Healthcare, United States) according to the manufacturer’s instruction. The gels of M. bovis biofilms and planktonic cells were compared using ImageMasterTM 2D Platinum 5.0 software (GE Healthcare, United States), as previously described (Wang et al., 2014). Briefly, the individual spots were quantified by calculating the relative spot volumes, which is the ratio of individual spot volume and total volume of all spots, and were expressed in volume percentage (vol.%). Changes in protein expression were calculated based on the overlapping measure ratios of corresponding spots by using ImageMasterTM 2D Platinum 5.0 software according to the manufacturer’s instruction. Spots that showed volume alteration greater than 50% (fold change ≥ 1.5) at 95% confidence interval (Student’s t-test; p < 0.05) were considered to be statistically significant, and five selected differentially expressed protein spots were subjected to subsequent identification by mass spectrometry.
Immunoblotting 2-DE Analysis
Immunoblotting analysis was conducted for each sample simultaneously using 2-DE analysis. The separated protein spots from 2-DE gels were transferred onto PVDF membrane (GE Healthcare, United States) using TE62 Tank Transfer Unit system (GE Healthcare, United States) at 100 V for 2 h. After blocking at room temperature for 2 h with 10% skimmed milk (Becton, Dickinson and Company, Franklin Lakes, NJ, United States) in TBST (0.5% v/v Tween) with gentle swinging, the membrane was incubated with the pooled convalescent sera against M. bovis at 4°C overnight. The membrane was washed three times with TBST buffer at 5 min each wash. Subsequently, the membrane was incubated with 1:8000 diluted HRP-conjugated goat anti-bovine IgG (Santa Cruz Biotech, United States) at room temperature for 1 h and washed five times with TBST buffer at 8 min each wash. Finally, the membranes were treated with Super Enhanced Chemiluminescent Substrate (ECL) Plus kit (Thermo Scientific, United States) to visualize the immunoreactive protein spots, according to the manufacturer’s instructions. Blots were scanned using Typhoon TMFLA 9500 (GE Amersham, United States). ImageMasterTM 2D Platinum 5.0 software (GE Healthcare, United States) was used to match the spots on the membranes with their homologs in 2-DE gels stained with a modified silver staining method that was compatible with the subsequent mass spectrometric analyses. The pooled pre-infected serum of healthy calves served as negative controls in the 2-DE immunoblotting analysis. The process was performed in triplicate.
Protein Identification by Mass Spectrometry
All of the immunoreactive proteins were selected and excised from 2D gels, which corresponded to the spots on the PVDF membranes and further in a gel digested with trypsin. MALDI-TOF-MS analysis was performed by using a MALDI-TOF/TOF instrument (5800 proteomics analyzer; Applied Biosystems) to identify proteins. Combined MS and MS/MS queries were conducted by using the Mascot search engine (Version 2.2; Matrix Science, Ltd.) on the database of UniProt M. bovis (downloaded on May 20, 2016; 1695 sequences) with the following parameter settings: Trypsin digestion, variable modification of oxidation (M), fixed modifications of carbamidomethyl (C), peptide mass tolerance for monoisotopic data of 100 ppm, one max missed cleavages, and MS/MS fragment tolerance of 0.4 Da. GPS Explorer protein with confidence index ≥ 95% (protein score C. I. %) was used for further manual validation.
Bioinformatics Analysis
Gene Ontology1, Clusters of Orthologous Groups of proteins (COGs)2, and KEGG3 databases were used for functional annotation of the identified proteins. The online software PSORTb v.3.04 was applied to predict the subcellular localization of identified proteins. Protein interaction analysis was conducted with STRING database5.
SDS-PAGE and Western Blot Analysis of Recombinant Selected Proteins
Six identified proteins, namely, endoglucanase, GTPase Era (Era), hydrolases of the HAD superfamily protein, phosphotransacetylase (Pta_1), chaperone protein DnaK (DnaK), and energy-coupling factor transporter ATP-binding protein EcfA (EcfA), were selected to confirm their immunogenicity by using SDS-PAGE and subsequent Western blot analysis. The complete genes of these selected proteins with single nucleotide mutation in the UGA codon (UGA to UGG) were synthesized by Genecreate (Wuhan, China) and cloned into pET-30a(+) vectors (Novagen, Darmstadt, Germany). The constructs were confirmed by sequencing on ABI 3730XL. The recombinant proteins were expressed in E. coli BL21(DE3) cells (Novagen, Darmstadt, Germany), purified by NTA-NI (Genscript, Nanjing, China), and examined by SDS-PAGE. The immunogenicity of the proteins was confirmed by Western blot analysis. Briefly, purified proteins were transferred onto 0.45 μm PVDF membrane (Millipore, United States) using a fast semi-dry eBlotTM protein transfer system (Genscript, Nanjing, China) for 7 min and blocked with 10% skimmed milk (Becton, Dickinson, and Company, Franklin Lakes, NJ, United States) in TBST (0.5% v/v Tween) with gentle swinging at 4°C overnight. After washing with TBST, the membrane was incubated with 1:100 dilution in the bovine-immune sera or convalescent sera against M. bovis for 1 h at room temperature. Afterward, the membrane was washed and incubated with 1:8000 diluted HRP-conjugated goat anti-bovine IgG (Santa Cruz Biotech, United States) for 1 h at room temperature. Finally, the membranes were washed and treated with Super ECL Plus kit (Thermo Scientific, United States) to visualize the immunoreaction according to the manufacturer’s instructions.
Statistical Analysis
Statistical analysis was performed by using SPSS 15.0 (SPSS Inc., Chicago, IL, United States). Student’s t-test was used for statistical analysis. Mean and standard deviations (SD) were determined for the independent experiments and the results were expressed as mean ± SD. p < 0.05 was considered statistically significant.
Results
Scanning Electron Microscopy Analysis of Biofilm Formation of M. bovis 08M
Scanning electron microscopy analysis showed that M. bovis 08M formed prolific biofilms after cultivation on the coverslips for 72 h. A large number of cell clusters formed a dense net enclosed by numerous mycoplasma cells and extracellular matrix for the biofilms. On the contrary, the planktonic cells were dispersed. The morphological features of M. bovis biofilms exhibited significant difference from those of their planktonic counterparts (Figure 1).
FIGURE 1.
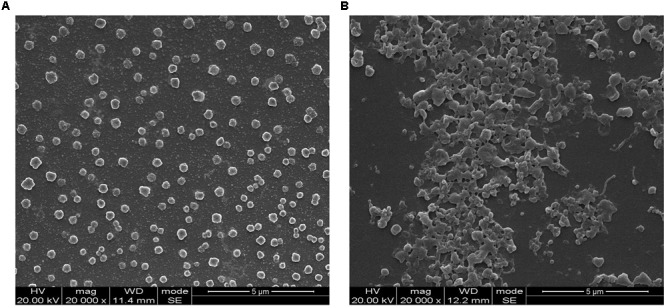
Scanning electron microscopy analysis on the Mycoplasma bovis 08M planktonic cells and biofilms formed on glass coverslips after 72 h of cultivation. (A) Planktonic cells and (B) biofilms.
2-DE Analysis of Whole-Cell Proteins From M. bovis Biofilms and Planktonic Cells
Mycoplasma bovis planktonic cells (2.67 ± 0.38 × 108 CFU/mL) and biofilms were harvested at 72 h, and then the whole-cell proteins were extracted. 2-DE analysis was performed to separate the whole-cell proteins of M. bovis planktonic cells and biofilms. This analysis was reproducible based on triplicate 2-DE gels of two group protein samples (M. bovis planktonic cells and biofilms) (Supplementary Figure S1). Approximately 1000–1300 spots were detected in 2D-E gels of M. bovis biofilms and planktonic cells. Approximately 596 matching protein spots were detected on the representative gels for M. bovis biofilms and planktonic cells with pH 3–10 IPG strips (Figures 2A,D), corresponding to 67.42% of the total number of coding genes of M. bovis 08M genome. The majority of these proteins exhibited molecular weights that ranged from 10 to 75 kDa.
FIGURE 2.
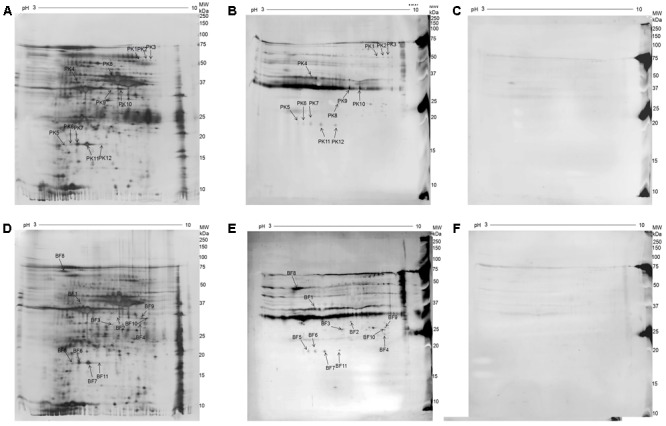
2D electrophoresis (2-DE) gel and Western blot analysis of whole-cell proteins of M. bovis 08M grown as planktonic cells and biofilms. (A) Silver-stained 2-DE gel of whole-cell proteins from M. bovis grown as planktonic cells (pH 3–10, 13 cm). (B) Western blot analysis of whole-cell proteins from M. bovis grown as planktonic cells using convalescent sera against M. bovis 08M. (C) Western blot analysis of whole-cell proteins from M. bovis grown as planktonic cells using pre-infected sera. (D) Silver-stained 2-DE gel analysis of whole-cell proteins from M. bovis grown as biofilms (pH 3–10, 13 cm). (E) Western blot analysis of whole-cell proteins from M. bovis grown as biofilm using convalescent sera against M. bovis 08M. (F) Western blot analysis of whole-cell proteins from M. bovis grown as biofilm using pre-infected sera.
Supplementary Figure S2 shows the representative silver-stained 2D-E gel images of proteins in M. bovis planktonic cells and biofilms. A total of 49 significantly differentially expressed protein spots with at least a 1.5-fold abundance change were detected between M. bovis planktonic cells and biofilms. Compared with M. bovis planktonic cells, 21 (42.86%) protein spots of the differential protein spots were increased in M. bovis biofilms, whereas 28 (57.14%) were decreased. Among which, five spots were selected and excised from representative 2-DE gels for identification by using MALDI-TOF/TOF MS analysis, corresponding to seven proteins (Supplementary Table S1). Four proteins, namely, thiol peroxidase (Tpx), segregation and condensation protein B, pyruvate dehydrogenase E1 component subunit beta (PdhB), and one putative lipoprotein, were increased, and three proteins, namely, enolase, endoglucanase, and elongation factor Ts (EF-Ts), were decreased.
Immunoblotting Analysis and Proteins Identified Using MALDI-TOF/TOF MS
A total of 12 and 11 immunoreactive protein spots that matched the protein spots detected in the 2-DE gels were observed on the immunoblot analysis of M. bovis planktonic cells and biofilms, respectively (Figures 2B,E). No specific immunoreactive protein spots reacted with the pre-infected sera for planktonic cells and biofilms (Figures 2C,F). MALDI-TOF/TOF MS analysis was performed to identify the protein spots. There were 12 immunoreactive protein spots for the planktonic cells and finally 8 different proteins were identified in the 11 protein spots of them. For the biofilms, 10 different proteins were identified in the 10 protein spots. For planktonic cells, six proteins corresponded to single spots and two proteins were represented by three and four isoforms, indicating the post-translational modification of these proteins. For M. bovis biofilms, eight proteins corresponded to single spots and two proteins were recognized by three and four isoforms. The detailed information of the identified immunoproteins for M. bovis planktonic cells and biofilms in this study are shown in Tables 1, 2. To the best of our knowledge, among these proteins, 12 proteins, including carbohydrate uptake ABC transporter-2 (CUT2) family ATP-binding protein (CUT2), endoglucanase, thiol peroxidase (Tpx), UvrABC system protein A, Pta_1, chromosome partition protein Smc, EcfA, DnaK, Era, lipoprotein, hydrolases of the HAD superfamily protein, and one uncharacterized protein, have not been reported as immunogen in M. bovis. Six proteins, namely, EcfA, DnaK, Era, lipoprotein, hydrolases of the HAD superfamily protein, and one uncharacterized protein, were specific to M. bovis biofilms. Moreover, endoglucanase, Tpx, and MilA were identified as common immunoreactive proteins in planktonic cells and biofilms.
Table 1.
Immunoreactive proteins of M. bovis planktonic cells identified by MALDI-TOF/TOF MS.
| Spot no. | Protein | Homology gene/ | Uniprot | Theoretical | Theoretical | No. of | Protein | Protein | PSORTb | PSORTb | COGb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| locus tag | accession no. | protein MW | protein PI | peptides | scores | score | localization | probability | |||
| matched | C.I. %a | ||||||||||
| PK1–PK2–PK3 | Carbohydrate uptake ABC transporter-2 (CUT2) family, ATP-binding protein | MBOVPG45_0018 | tr|E4PYW6| | 59.27 | 7.79 | 21 | 324 | 100 | Cytoplasmic Membrane |
9.99 | R |
| PK4 | Endoglucanase | MBOVPG45_0256 tpx/ |
tr|A0A059Y901| | 39.39 | 5.42 | 18 | 621 | 100 | Cytoplasmic | 7.50 | EG |
| PK6–PK7–PK11–PK12 | Thiol peroxidase (Tpx) | MBOVPG45_0640 tuf/ |
tr|A0A059Y3P0| | 18.47 | 5.44 | 15 | 774 | 100 | Cytoplasmic | 7.50 | O |
| PK8 | Elongation factor Tu (Tuf) | MBOVPG45_0411 uvrA/ |
tr|A0A059Y8L3| | 43.64 | 5.89 | 26 | 860 | 100 | Cytoplasmic | 9.97 | J |
| PK9 | UvrABC system protein A (UvrA) | MBOVPG45_0470 pta-1/ | tr|E4Q030| | 106.14 | 6.94 | 20 | 53 | 99.23 | Cytoplasmic | 7.50 | L |
| PK10 | Phosphotransacetylase (Pta_1) |
MBOVPG45_0153 milA |
tr|A0A059XYN4| | 34.37 | 6.10 | 23 | 1,090 | 100 | Cytoplasmic | 7.50 | C |
| PK10 | Putative membrane protein (MilA) | MBOVPG45_0710 smc/ |
tr|E4Q0M5| | 302.89 | 8.71 | 36 | 48 | 97.55 | Unknown | – | E |
| PK10 | Chromosome partition protein Smc (Smc) |
MBOVPG45_0520 | tr|A0A059XZ49| | 111.20 | 5.74 | 13 | 47 | 96.92 | Cytoplasmic | 7.50 | D |
aC.I. %, confidence interval.
b COGs database functional categories: (C) Energy production and conversion, (D) cell cycle control, cell division, chromosome partitioning, (E) amino acid transport and metabolism, (G) carbohydrate transport and metabolism, (J) translation, ribosomal structure and biogenesis, (L) replication, recombination and repair, (O) post-translational modification, protein turnover, chaperones, (P) inorganic ion transport and metabolism, (R) general function prediction only.
Table 2.
Immunoreactive proteins of M. bovis biofilms identified by MALDI-TOF/TOF MS.
| Spot no. | Protein | Homology gene/locus tag | Uniprot accession no. | Theoretical protein MW | Theoretical protein PI | No. of peptides matched | Protein scores | Protein score CI %a | PSORTb localization | PSORTb probability | COGb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BF1 | Endoglucanase | MBOVPG45_0256 | tr|A0A059Y901| | 39.39 | 5.42 | 18 | 621 | 100 | Cytoplasmic | 7.50 | EG |
| BF3 | Energy-coupling factor transporter ATP-binding protein EcfA |
ecfA/ MBOVPG45_0295 |
tr|A0A059XZR6| | 29.41 | 6.02 | 7 | 95 | 100 | Cytoplasmic | 7.50 | PR |
| BF3 | Putative membrane protein | MBOVPG45_0710 | tr|E4Q0M5| | 302.89 | 8.71 | 43 | 64 | 99.94 | Unknown | – | E |
| BF4–BF9–BF10 | Pyruvate dehydrogenase E1 component subunit alpha (PdhA) |
pdhA/ MBOVPG45_0104 |
tr|A0A059XYH6| | 41.33 | 6.13 | 10 | 298 | 100 | Cytoplasmic | 9.97 | C |
| BF5–BF6–BF7–BF11 | Thiol peroxidase (Tpx) |
tpx/ MBOVPG45_0640 |
tr|A0A059Y3P0| | 18.47 | 5.44 | 13 | 906 | 100 | Cytoplasmic | 7.50 | O |
| BF8 | Chaperone protein DnaK (DnaK) |
dnaK/ MBOVPG45_0160 |
tr|A0A059Y7X3| | 65.30 | 4.95 | 28 | 692 | 100 | Cytoplasmic | 9.97 | O |
| BF8 | GTPase Era (Era) |
era/ MBOVPG45_0126 |
tr|A0A059Y7U9| | 32.94 | 8.54 | 13 | 54 | 99.31 | Cytoplasmic membrane | 8.78 | J |
| BF8 | Lipoprotein | MBOVPG45_0215 | tr|A0A059XZZ3| | 48.62 | 9.16 | 13 | 47 | 96.92 | Unknown | – | R |
| BF9 | Hydrolases of the HAD superfamily protein | MBOVPG45_0665 | tr|A0A059Y395| | 34.76 | 6.62 | 18 | 660 | 100 | Cytoplasmic | 7.50 | HR |
| BF10 | Uncharacterized protein | MBOVPG45_0718 | tr|A0A059Y955| | 34.91 | 8.13 | 14 | 61 | 99.87 | Cytoplasmic | 7.50 | X |
a C.I. %, confidence interval.
b COGs database functional categories: (C) Energy production and conversion, (D) cell cycle control, cell division, chromosome partitioning, (E) amino acid transport and metabolism, (G) carbohydrate transport and metabolism, (H) co-enzyme transport and metabolism, (J) translation, ribosomal structure and biogenesis, (L) replication, recombination and repair, (O) post-translational modification, protein turnover, chaperones, (P) inorganic ion transport and metabolism, (R) general function prediction only, (X) mobilome: prophages, transposons.
Bioinformatics Analysis
Gene Ontology and KEGG pathway analyses of the identified proteins are shown in Figure 3. For the biological process of GO, various biological processes, which mainly included metabolic process (GO:0008152), transport (GO:0006810), cell redox homeostasis (GO:0045454), and chromosome organization (GO:0051276), were observed. The most representative molecular functions of these proteins mainly included ATP binding (GO:0005524), oxidoreductase activity (GO:0016491), and GTP binding (GO:0005525). These proteins were largely involved in metabolic pathways, such as metabolism, carbohydrate metabolism, and amino acid metabolism. For the COG analysis of the identified immunoproteins, various categories, including amino acid transport and metabolism, general function predicted only, post-translational modification, protein turnover, and chaperone, energy production and conversion, translation, ribosomal structure and biogenesis, were also involved (Tables 1, 2). PSORTb v.3.0 database was used to predict the subcellular localization of the identified proteins. Results showed that CUT2 and Era were associated to the membrane. The remaining 11 immunoproteins were predicted to display cytoplasmic localizations (Tables 1, 2). STRING tool was used to predict the potential protein–protein interaction of the identified immunoproteins in Tables 1, 2 using M. bovis-type strain PG45 database as the default reference. The STRING network generated with the immunoproteins is shown in Figure 4. Results showed that the most significant proteins that are likely to interact are DnaK and elongation factor Tu (Tuf), suggesting that these proteins may function together.
FIGURE 3.
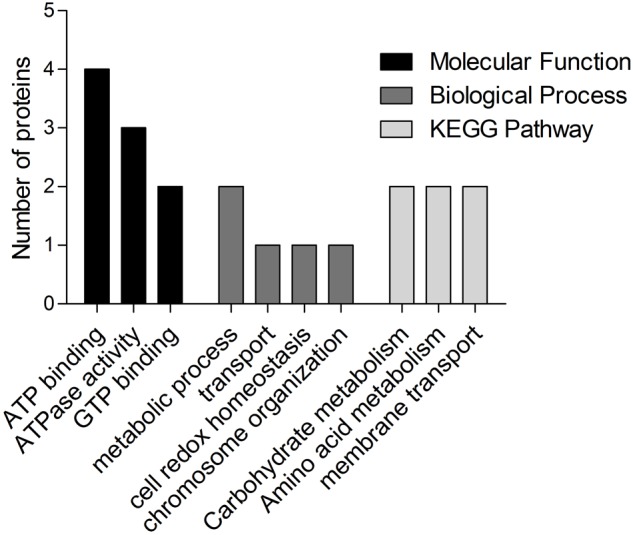
Most enriched categories of GO in terms of biological process, molecular function, and KEGG pathways of the identified immunoproteins.
FIGURE 4.
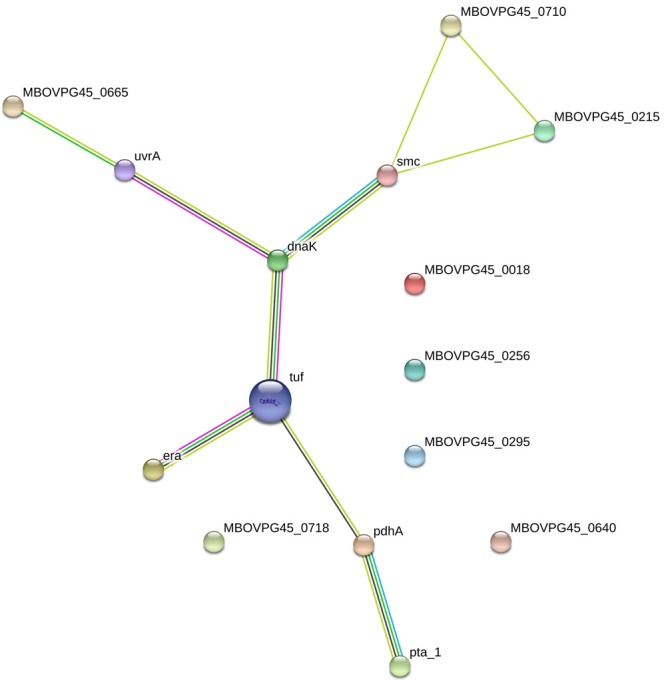
STRING network generated with the available immunoproteins from Tables 1, 2.
Confirmation of the Immunogenicity for Selected Proteins
To confirm the immunogenicity of endoglucanase, Era, hydrolases of the HAD superfamily protein, Pta_1, DnaK, and EcfA, the recombinant selected proteins were successfully cloned and expressed in E. coli BL21(DE3) cells (Figure 5A). Western blot analysis showed that the purified recombinant proteins can react with bovine-immune and convalescent sera against M. bovis (Figures 5B,C), but not with healthy bovine sera (data not shown). The results indicated the good immunogenicity of these identified proteins. A very big spot was observed for Pta_1 (Figure 5C) upon immunoblotting using the sera obtained from immunized animals, demonstrating that among the selected proteins, Pta_1 had the highest reactivity with bovine-immune sera against M. bovis.
FIGURE 5.
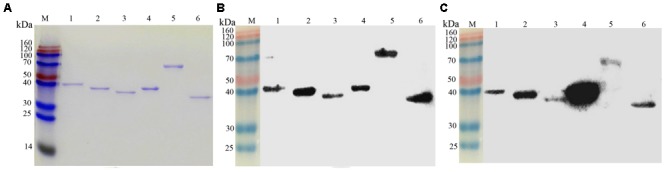
Analysis of six recombinant expressed proteins. (A) SDS-PAGE of six purified recombinant expressed proteins. (B) Western blot analysis of recombinant expressed proteins using convalescent sera against M. bovis 08M. (C) Western blot analysis of recombinant expressed proteins using bovine immune-sera against M. bovis. “M” represents the protein markers, and the numbers on the left indicate the molecular weight standards (kDa). Lines 1–6: Endoglucanase, Era, hydrolases of the HAD superfamily protein, Pta_1, DnaK, and EcfA.
Discussion
Mycoplasma bovis infection has become a major threat to the cattle industry in many countries. In the past few decades, considerable efforts focused on identifying the immunogenic proteins of M. bovis to develop diagnostic kits and vaccines to control its infection (Perez-Casal et al., 2017). Several immunogenic proteins were identified in different components of M. bovis planktonic cells (Sun et al., 2014; Khan et al., 2016). To the best of our knowledge, the immunogenic component of M. bovis biofilm remains unknown. In the present study, the immunogenic proteins of M. bovis biofilms were identified and compared with their planktonic counterparts.
Mycoplasma bovis can form biofilms in vitro (Mcauliffe et al., 2006); however, whether the biofilm formation can occur in vivo remains unknown. In the present study, whole-cell proteins of M. bovis biofilms and planktonic cells were prepared in vitro. 2-DE, immunoblotting, and MALDI-TOF/TOF MS were performed to identify the immunogens of M. bovis in these two growth modes. The early generation of a virulent Chinese strain 08M was used to avoid antigenic variation. Moreover, whole-cell proteins were prepared as protein samples for 2-DE instead of membrane proteins to prevent the loss of important cytoplasmic antigen proteins. Report showed that some additional promising antigens for diagnostics could be obtained in the membrane fraction of M. bovis planktonic cells (Khan et al., 2016). Thus, identifying the membrane immunoproteomic surface of M. bovis biofilms and planktonic cells will be useful and should be studied further.
Mycoplasma bovis biofilms and planktonic cells presented different antigen profiles. This phenomenon was also observed in Streptococcus pneumoniae, and it may be related to protein expression (Sanchez et al., 2011). Significantly differentially expressed proteins were found between M. bovis planktonic cells and biofilms in the 2D-E analysis. For example, Tpx, which is related to oxidative stress response, was increased in M. bovis biofilms. Kalmokoff et al. (2006) used 2D-E to reveal that Tpx was up-regulated in Campylobacter jejuni biofilms. Enolase and EF-Ts, which are involved in glycolysis and carbohydrate metabolism, and protein synthesis, respectively, were down-regulated in M. bovis biofilms; this finding was also described in Lactobacillus plantarum (De Angelis et al., 2015) biofilms compared to planktonic cells. In the present study, endoglucanase, Tpx, and MilA were identified in planktonic and biofilm-grown M. bovis. Common immunoreactive proteins may be promising candidates for the development of a vaccine that can prevent biofilm formation and M. bovis infection. A total of 12 novel immunogens may also be molecular candidates for the development of improved diagnostics and vaccines to control M. bovis infections. The remaining pyruvate dehydrogenase E1 component subunit alpha (PdhA) (Sun et al., 2014), Tuf (Khan et al., 2016), and MilA (Wawegama et al., 2014) were previously reported as antigens in M. bovis. Pta-1, which is involved in the synthesis of acetyl-CoA from acetate, was an immunogen in Mmc (Corona et al., 2013) but is not reported as an immunogen in M. bovis. DnaK was identified as an immunogenic protein in several mycoplasma species in planktonic mode, such as MmmSC (Jores et al., 2009), Mccp (Zhao et al., 2012), and Mmc (Churchward et al., 2015). However, DnaK is not reported as an immunogen in M. bovis planktonic cells, to the best of our knowledge. This discrepancy might due to the different species of Mycoplasma or the serum used. Notably, DnaK was identified as immunogenic in the M. bovis biofilms, same with other bacteria biofilms, such as S. pneumoniae (Sanchez et al., 2011), Streptococcus suis (Wang et al., 2012), and Staphylococcus epidermidis (Carvalhais et al., 2015). Tuf promotes the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein biosynthesis. It was shown to be an highly immunogenic protein in many Mycoplasma species (Jores et al., 2009; Churchward et al., 2015), and was also identified in M. bovis planktonic cells in this study. Moreover, Tuf was predicted by PSORTb database to be cytoplasmically localized in many Mycoplasma species (Churchward et al., 2015; Khan et al., 2016). In M. pneumoniae, Tuf was observed to be surface-localized protein that can mediate binding to fibronectin (Dallo et al., 2002). Furthermore, DnaK was reported to be a surface-localized protein that can mediate binding to host receptors (Boulanger et al., 1995). In this study, DnaK and Tuf were predicted to be cytoplasmically localized in M. bovis and appeared to be the most interactive immunoreactive and may be related to M. bovis biofilm formation. DnaK and Tuf were reported to be differently expressed in biofilms and planktonic cells for some bacteria, such as Streptococcus equi ssp. zooepidemicus (Yi et al., 2016) and L. plantarum (De Angelis et al., 2015), and may be related to biofilm formation. The subcellular localization and role of these immunoreactive proteins in the M. bovis biofilm formation should be investigated further.
Several antigens, such as pMB67, glyceraldehyde-3-phosphate dehydrogenase, the pyruvate dehydrogenase beta subunit, and other antigens identified in previous studies (Sachse et al., 1993; Robino et al., 2005; Sun et al., 2014; Khan et al., 2016), were not detected in the planktonic cells in the present study. This phenomenon was previously described (Khan et al., 2016) and might be due to the differences in methods and antiserum selection. Moreover, antigen profiles may exhibit several differences due to various strains, such as antigen variation, as reported previously in R. anatipestifer (Hu et al., 2012). The selection of antiserum is crucial in the screening of immunogenic proteins. Antisera from natural and experimental infections may present different antigen profiles for M. bovis because of different antibody titers, doses, and courses in natural and experimental infections. In the present study, immunoblotting with different sera obtained from cattle experimentally infected with M. bovis was performed in the preliminary study. Finally, pooled sera of 46 dpi were selected to obtain considerable amount of immunogenic proteins.
Endoglucanase, Era, hydrolases of the HAD superfamily protein, Pta_1, DnaK, and EcfA that were identified in the present study were selected to express the recombinant protein in E. coli, to be further evaluated as molecular candidates for diagnostics or vaccines. The results showed that these selected proteins can all react with bovine-immune and convalescent sera against M. bovis. Pta_1 had the highest reactivity with sera obtained from immunized animals among the selected proteins, possibly because Pta_1 was a dominant immunoreactive protein in M. bovis and was also described in Mmc (Churchward et al., 2015). The selected proteins may be as novel molecular candidates of diagnostics or vaccines but this finding needs further evaluation.
Conclusion
In the present study, immunoproteomic analysis was used to compare the antigen profiles of M. bovis biofilms and planktonic cells reacted with bovine convalescent sera. Differential immunoreactivity to bovine convalescent serum between M. bovis biofilms and planktonic cells were observed. A total of 12 novel immunoreactive proteins were identified. Most of the identified proteins were cytoplasmic proteins that were mainly involved in transport and metabolism. DnaK and Tuf appeared to be the most interactive immunoreactive proteins and may be related to M. bovis biofilm formation. Six proteins had potential as serodiagnostic antigens but still needs further evaluation. These results lay the foundation for the understanding of the host response to M. bovis in two growth modes and may facilitate the development of novel molecular candidates for the improved diagnostics and vaccines for controlling M. bovis infections.
Author Contributions
SC conceived and designed the experiments. SC, HH, PZ, WJ, and ML performed the experiments: SC and HH wrote the manuscript and analyzed the data. YC and YL contributed reagents/materials/analysis tools. SC, HH, and YC critically revised the manuscript. YC and YL supervised all work. All authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer SA and handling Editor declared their shared affiliation.
Acknowledgments
The authors thank Shanghai Applied Protein Technology Co., Ltd. for their technical help.
Abbreviations
- 2D
two dimensional
- 2-DE
two dimensional gel electrophoresis
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- DTT
dithiothreitol
- GO
Gene Ontology
- IEF
isoelectric focusing
- IPG
immobilized pH gradient
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MALDI-TOF/TOF MS
matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometry
- MS
mass spectrometry
- PBS
phosphate buffered saline
- PPLO broth
pleuropneumonia-like organism broth
- PVDF
polyvinylidene fluoride
- SDS
sodium dodecyl sulfate
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEM
scanning electron microscopy
- TBST
tris buffered saline with tween
Funding. This work was supported by the National Natural Science Foundation of China (Grant Nos. 31402223 and 31602088), National Key Technologies R&D Program (Grant No. 2015BAD12B02), the National Key Research and Development Plan (Grant Nos. 2016YFD0500907 and 2017YFD0500905), the Fundamental Research Funds for CAAS (Grant No. 1610312016027), and the International Scientific and Technological Cooperation Projects of Gansu Province (No. 17YF1WA170).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00379/full#supplementary-material
2-DE gel images of proteins in M. bovis planktonic cells and biofilms in triplicates.
Image of proteins of representative 2D-E gel in M. bovis planktonic cells and biofilms. (A) The representative silver-stained 2D-E gel image of M. bovis planktonic cells and biofilms. Differentially expressed protein spots were circled with numbers by using ImageMasterTM 2D Platinum 5.0 software. (B) Expanded region of differentially expressed proteins spot with circles.
Selected differentially expressed proteins spots identified by MALDI-TOF/TOF MS between M. bovis planktonic cells and biofilms.
References
- Behrens A., Poumarat F., Le G. D., Heller M., Rosengarten R. (1996). A newly identified immunodominant membrane protein (pMB67) involved in Mycoplasma bovis surface antigenic variation. Microbiology 142(Pt 9) 2463–2470. 10.1099/00221287-142-9-2463 [DOI] [PubMed] [Google Scholar]
- Boulanger J., Faulds D., Eddy E. M., Lingwood C. A. (1995). Members of the 70 kDa heat shock protein family specifically recognize sulfoglycolipids: role in gamete recognition and mycoplasma-related infertility. J. Cell Physiol. 165 7–17. 10.1002/jcp.1041650103 [DOI] [PubMed] [Google Scholar]
- Carvalhais V., Cerveira F., Vilanova M., Cerca N., Vitorino R. (2015). An immunoproteomic approach for characterization of dormancy within Staphylococcus epidermidis biofilms. Mol. Immunol. 65 429–435. 10.1016/j.molimm.2015.02.024 [DOI] [PubMed] [Google Scholar]
- Chen S., Hao H., Zhao P., Gao P., He Y., Ji W., et al. (2017). Complete genome sequence of Mycoplasma bovis strain 08M. Genome Announc. 5:e00324-17. 10.1128/genomeA.00324-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward C. P., Rosales R. S., Gielbert A., Dominguez M., Nicholas R. A., Ayling R. D. (2015). Immunoproteomic characterisation of Mycoplasma mycoides subspecies capri by mass spectrometry analysis of two-dimensional electrophoresis spots and western blot. J. Pharm. Pharmacol. 67 364–371. 10.1111/jphp.12344 [DOI] [PubMed] [Google Scholar]
- Corona L., Cillara G., Tola S. (2013). Proteomic approach for identification of immunogenic proteins of Mycoplasma mycoides subsp. capri. Vet. Microbiol. 167 434–439. 10.1016/j.vetmic.2013.08.024 [DOI] [PubMed] [Google Scholar]
- Dallo S. F., Kannan T. R., Blaylock M. W., Baseman J. B. (2002). Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46 1041–1051. 10.1046/j.1365-2958.2002.03207.x [DOI] [PubMed] [Google Scholar]
- De Angelis M., Siragusa S., Campanella D., Di Cagno R., Gobbetti M. (2015). Comparative proteomic analysis of biofilm and planktonic cells of Lactobacillus plantarum DB200. Proteomics 15 2244–2257. 10.1002/pmic.201400363 [DOI] [PubMed] [Google Scholar]
- Hu Q., Ding C., Tu J., Wang X., Han X., Duan Y., et al. (2012). Immunoproteomics analysis of whole cell bacterial proteins of Riemerella anatipestifer. Vet. Microbiol. 157 428–438. 10.1016/j.vetmic.2012.01.009 [DOI] [PubMed] [Google Scholar]
- Jores J., Meens J., Buettner F. F. R., Linz B., Naessens J., Gerlach G. F. (2009). Analysis of the immunoproteome of Mycoplasma mycoides subsp. mycoides small colony type reveals immunogenic homologues to other known virulence traits in related Mycoplasma species. Vet. Immunol. Immunopathol. 131 238–245. 10.1016/j.vetimm.2009.04.016 [DOI] [PubMed] [Google Scholar]
- Kalmokoff M., Lanthier P., Tremblay T. L., Foss M., Lau P. C., Sanders G., et al. (2006). Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 188 4312–4320. 10.1128/JB.01975-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F. A., Faisal M., Chao J., Liu K., Chen X., Zhao G., et al. (2016). Immunoproteomic identification of MbovP 579, a promising diagnostic biomarker for serological detection of Mycoplasma bovis infection. Oncotarget 7 39376–39395. 10.18632/oncotarget.9799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcauliffe L., Ellis R. J., Miles K., Ayling R. D., Nicholas R. A. (2006). Biofilm formation by mycoplasma species and its role in environmental persistence and survival. Microbiology 152 913–922. 10.1099/mic.0.28604-0 [DOI] [PubMed] [Google Scholar]
- Nicholas R., Baker S., Ayling R., Stipkovits L. (2000). Mycoplasma infections in growing cattle. Cattle Pract. 8 115–118. 17582119 [Google Scholar]
- Nicholas R. A., Ayling R. D. (2003). Mycoplasma bovis: disease, diagnosis, and control. Res. Vet. Sci. 74 105–112. 10.1016/S0034-5288(02)00155-8 [DOI] [PubMed] [Google Scholar]
- Parsek M. R., Singh P. K. (2003). Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57 677–701. 10.1146/annurev.micro.57.030502.090720 [DOI] [PubMed] [Google Scholar]
- Perez-Casal J., Prysliak T., Maina T., Suleman M., Jimbo S. (2017). Status of the development of a vaccine against Mycoplasma bovis. Vaccine 35 2902–2907. 10.1016/j.vaccine.2017.03.095 [DOI] [PubMed] [Google Scholar]
- Robino P., Alberti A., Pittau M., Chessa B., Miciletta M., Nebbia P., et al. (2005). Genetic and antigenic characterization of the surface lipoprotein P48 of Mycoplasma bovis. Vet. Microbiol. 109 201–209. 10.1016/j.vetmic.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Rosengarten R., Citti C. (1999). “Mycoplasmas of ruminants:pathogenicity, diagnostics, epidemiology and molecular genetics,” in The Role of Ruminant Mycoplasmas in Systemic Infection eds Stipkovits L., Rosengarten R., Frey J. (Brussels: European Commission; ) 14–17. [Google Scholar]
- Sachse K., Grajetzki C., Rosengarten R., Hanel I., Heller M., Pfutzner H. (1996). Mechanisms and factors involved in Mycoplasma bovis adhesion to host cells. Zentralbl. Bakteriol. 284 80–92. 10.1016/S0934-8840(96)80157-5 [DOI] [PubMed] [Google Scholar]
- Sachse K., Helbig J. H., Lysnyansky I., Grajetzki C., Muller W., Jacobs E., et al. (2000). Epitope mapping of immunogenic and adhesive structures in repetitive domains of Mycoplasma bovis variable surface lipoproteins. Infect. Immun. 68 680–687. 10.1128/IAI.68.2.680-687.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse K., Pfutzner H., Heller M., Hanel I. (1993). Inhibition of Mycoplasma bovis cytadherence by a monoclonal antibody and various carbohydrate substances. Vet. Microbiol. 36 307–316. 10.1016/0378-1135(93)90097-Q [DOI] [PubMed] [Google Scholar]
- Sanchez C. J., Hurtgen B. J., Lizcano A., Shivshankar P., Cole G. T., Orihuela C. J. (2011). Biofilm and planktonic pneumococci demonstrate disparate immunoreactivity to human convalescent sera. BMC Microbiol. 11:245. 10.1186/1471-2180-11-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K. (2003). The genomics and proteomics of biofilm formation. Genome Biol. 4:219. 10.1186/gb-2003-4-6-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne C. J., Wang Y., Jin L., Wong S. S. W., Herath T. D. K., Samaranayake L. P. (2012). Unraveling the resistance of microbial biofilms: Has proteomics been helpful? Proteomics 12 651–665. 10.1002/pmic.201100356 [DOI] [PubMed] [Google Scholar]
- Sun Z., Fu P., Wei K., Zhang H., Zhang Y., Xu J., et al. (2014). Identification of novel immunogenic proteins from Mycoplasma bovis and establishment of an indirect ELISA based on recombinant E1 beta subunit of the pyruvate dehydrogenase complex. PLoS One 9:e88328. 10.1371/journal.pone.0088328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yuan C., Wang S., Song X., Xu L., Yan R., et al. (2014). Transcriptional and proteomic analysis reveal recombinant galectins of Haemonchus contortus down-regulated functions of goat PBMC and modulation of several signaling cascades in vitro. J. Proteomics 98 123–137. 10.1016/j.jprot.2013.12.017 [DOI] [PubMed] [Google Scholar]
- Wang Y., Yi L., Wu Z., Shao J., Liu G., Fan H., et al. (2012). Comparative proteomic analysis of Streptococcus suis biofilms and planktonic cells that identified biofilm infection-related immunogenic proteins. PLoS One 7:e33371. 10.1371/journal.pone.0033371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawegama N. K., Browning G. F., Kanci A., Marenda M. S., Markham P. F. (2014). Development of a recombinant protein-based enzyme-linked immunosorbent assay for diagnosis of Mycoplasma bovis infection in cattle. Clin. Vaccine Immunol. 21 196–202. 10.1128/CVI.00670-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin J. Q., Li Y., Guo D., Song N. H., Hu S. P., Chen C., et al. (2008). First isolation of Mycoplasma bovis from calf lung with pneumoniae in China. Chin. J. Prev. Vet. Med. 30 661–664. [Google Scholar]
- Yan J. X., Wait R., Berkelman T., Harry R. A., Westbrook J. A., Wheeler C. H., et al. (2000). A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis 21 3666–3672. [DOI] [PubMed] [Google Scholar]
- Yi L., Wang Y., Ma Z., Lin H., Xu B., Grenier D., et al. (2016). Identification and characterization of a Streptococcus equi ssp. zooepidemicus immunogenic GroEL protein involved in biofilm formation. Vet. Res 47:50. 10.1186/s13567-016-0334-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., He Y., Chu Y. F., Gao P. C., Zhang X., Zhang N. Z. (2012). Identification of novel immunogenic proteins in Mycoplasma capricolum subsp. Capripneumoniae strain M1601. J. Vet. Med. Sci. 74 1109–1115. 10.1292/jvms.12-0095 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2-DE gel images of proteins in M. bovis planktonic cells and biofilms in triplicates.
Image of proteins of representative 2D-E gel in M. bovis planktonic cells and biofilms. (A) The representative silver-stained 2D-E gel image of M. bovis planktonic cells and biofilms. Differentially expressed protein spots were circled with numbers by using ImageMasterTM 2D Platinum 5.0 software. (B) Expanded region of differentially expressed proteins spot with circles.
Selected differentially expressed proteins spots identified by MALDI-TOF/TOF MS between M. bovis planktonic cells and biofilms.


