ABSTRACT
Zika virus (ZIKV) is implicated in fetal stillbirth, microcephaly, intracranial calcifications, and ocular anomalies following vertical transmission from infected mothers. In adults, infection may trigger autoimmune inflammatory polyneuropathy. Transmission most commonly follows the bite of infected Aedes mosquitoes but may also occur through sexual intercourse or receipt of blood products. Definitive diagnosis through detection of viral RNA is possible in serum or plasma within 10 days of disease onset, in whole blood within 3 weeks of onset, and in semen for up to 3 months. Serological diagnosis is nonetheless critical because few patients have access to molecular diagnostics during the acute phase of infection and infection may be associated with only mild or inapparent disease that does not prompt molecular testing. Serological diagnosis is confounded by cross-reactivity of immune sera with other flaviviruses endemic in the areas where ZIKV has recently emerged. Accordingly, we built a high-density microarray comprising nonredundant 12-mer peptides that tile, with one-residue overlap, the proteomes of Zika, dengue, yellow fever, West Nile, Ilheus, Oropouche, and chikungunya viruses. Serological analysis enabled discovery of a ZIKV NS2B 20-residue peptide that had high sensitivity (96.0%) and specificity (95.9%) versus natural infection with or vaccination against dengue, chikungunya, yellow fever, West Nile, tick-borne encephalitis, or Japanese encephalitis virus in a microarray assay and an enzyme-linked immunosorbent assay (ELISA) of early-convalescent-phase sera (2 to 3 weeks after onset of symptomatic infection).
KEYWORDS: arbovirus, emerging infectious diseases, flavivirus, serology, Zika
IMPORTANCE
The emergence of Zika virus (ZIKV) as a teratogen is a profound challenge to global public health. Molecular diagnosis of infection is straightforward during the 3-week period when patients are viremic. However, serological diagnosis thereafter of historical exposure has been confounded by cross-reactivity. Using high-density peptide arrays that tile the proteomes of a selection of flaviviruses to identify a ZIKV-specific peptide, we established two assays that enable sensitive and specific diagnosis of exposure to ZIKV. These assays may be useful in guiding clinical management of mothers at risk for potential exposure to ZIKV and enable insights into the epidemiology of ZIKV infections.
INTRODUCTION
Zika virus (ZIKV) is an emerging mosquito-borne flavivirus that has become endemic in Africa, Asia, and the Americas and has been deemed a global threat by the World Health Organization (1, 2). ZIKV infection during pregnancy is causally linked to severe neurodevelopmental damage, including fetal microcephaly, intracranial calcifications, neurodevelopmental damage, and ocular anomalies (3). In adults, ZIKV may also trigger Guillain-Barré syndrome (4–6). In urban and suburban environments, ZIKV is transmitted in a human-mosquito-human transmission cycle. Transmission is also reported in sexual partners (7, 8) and via blood transfusion (9). Molecular assays for detection of ZIKV gene products in body fluids are the gold standard for definitive diagnosis of active infections (1, 10). However, the median duration of viral RNA is 22 days in whole blood and 10 days in plasma (11). Furthermore, many infections are asymptomatic, associated with only mild acute disease, or occur in areas where access to medical care is limited and blood collection is unlikely to be achieved during the acute phase of infection. Thus, teratological risk assessment during pregnancy and infection incidence and prevalence surveys also require serology for detection of gestational infections after resolution of viremia (12).
Cross-reactivity between ZIKV and other flaviviruses may complicate differential serodiagnosis and efforts to investigate the epidemiology of infection and linkage to disease. This is particularly challenging in areas where dengue virus (DENV) is endemic or Japanese encephalitis virus (JEV) or yellow fever virus (YFV) vaccination is common. Current recommendations from the U.S. Centers for Disease Control and Prevention (CDC) are that all samples found seropositive in a Zika virus IgM antibody capture enzyme-linked immunosorbent assay (MAC-ELISA) should be validated by plaque reduction neutralization testing (PRNT). PRNT is expensive and labor-intensive and requires live virus. Furthermore, antibodies to conserved flavivirus domains may confound assay specificity, particularly in the acute and early convalescent phases. Here, we report a sensitive, highly multiplexed, microarray-based assay that enables discrimination of antibody responses to linear epitopes specific to ZIKV, dengue virus (DENV) 1 to 4, chikungunya virus (CHIKV), West Nile virus (WNV), yellow fever virus (YFV), Ilheus virus (ILHV), and Oropouche virus (OROV). We also report the identification of a highly specific epitope for ZIKV located within the NS2B protein. We adapted this NS2B peptide to a synthetic biotinylated peptide IgG ELISA system with the goal of establishing an inexpensive, rapid, sensitive, specific, and point-of-care method for ZIKV IgG detection.
RESULTS
Arboviral peptide array design.
Our platform for epitope discovery is a programmable peptide microarray that can accommodate up to 3 million distinct linear peptides on a 75-mm by 26-mm slide (Roche). The array can also be divided into 12 subarrays, each containing approximately ~170,000 “12-mer peptides.” The 12-mer format is based on the observation that serum antibodies bind linear peptide sequences ranging from 5 to 9 amino acids (aa) and bind most efficiently when targets are flanked by additional amino acids (13). To enable differential detection of antibodies specific for arboviral infections in geographic regions where ZIKV has become endemic (14, 15), we created a custom NCBI GenBank database for ZIKV, DENV, CHIKV, YFV, WNV, OROV, and ILHV (Table 1). For each virus selected, we downloaded all available protein sequences available before October 2016 from the NCBI protein database. We then created a database comprising overlapping 12-mer peptides that tiled the whole proteome of each of these agents with 11-aa overlap in a sliding window pattern. The selected peptide sequences were passed through a redundancy filter to yield 88,212 unique peptide sequences and 83,748 nonunique peptide sequences (present in more than one virus) for a total of 171,960 peptides. Redundant peptides were excluded prior to synthesis. The individual peptides in the library were printed in random positions on the peptide array to minimize the impact of locational bias.
TABLE 1 .
Proteins and peptides represented on the arboviral peptide array
| Virus | Genus | No. of available protein sequences |
No. of nonredundant peptides printed |
|---|---|---|---|
| Zika virus | Flavivirus | 205 | 7,573 |
| Dengue virus | Flavivirus | 1,044 | 74,105 |
| Ilheus virus | Flavivirus | 33 | 7,697 |
| West Nile virus | Flavivirus | 3,650 | 34,842 |
| Yellow fever virus | Flavivirus | 3,650 | 34,842 |
| Chikungunya virus | Alphavirus | 2,534 | 18,262 |
| Oropouche virus | Orthobunyavirus | 390 | 16,041 |
| Total no. of peptides on the arbovirus array |
171,960 |
Arboviral peptide array data analysis.
The array data, in the form of fluorescence signal intensities (arbitrary units [AU]), were background and spatially corrected and quantile normalized using the R Package “preprocessCore” (16). Spatial correction was based on a two-dimensional local polynomial regression (LOESS) that adjusts for signal variation due to spatial effects. A cutoff threshold for peptide recognition was defined as mean + 2 times the standard deviation (SD) of the mean intensity value of all negative controls (17). The statistical comparison of peptide microarray intensities between groups was performed using the limma R package (18). Peptide intensities for all samples were loaded and normalized using the “normexp” method for cross-array normalization (19).
Identification and selection of reactive peptides.
To test the utility of the array, we used 308 sera immunoreactive to ZIKV, DENV, CHIKV, YFV, tick-borne encephalitis virus (TBEV), WNV, or JEV and 21 sera from controls from the greater New York City metropolitan area with no known history of flavivirus infection or flavivirus vaccination who had been tested for exposure to Borrelia burgdorferi, the causative agent of Lyme disease. Table 2 lists sources of sera and reference methods used to define immunoreactivity. We employed dilutions of 1:50 or 1:100 based on pilot experiments wherein intensity analysis demonstrated that 1:50, 1:100, and 1:200 dilutions with known positive sera produced signal above the threshold (mean + 2 SDs above the mean obtained with negative-control samples), whereas dilutions of >1:200 produced signals similar to those found with negative-control samples (mean + 2 SDs).
TABLE 2 .
Sources of sera and methods used to define immunoreactivity, sensitivity, and specificity with Center for Infection and Immunity arboviral peptide array and ZIKV-NS2B-concat ELISAd
| Virus name and serotype | Phase (time point of sample collection after onset of illness)e | Immune status | No. of samples from source: |
No. of samples positive for ZIKV NS2B peptide/total no. (%) by assay: |
Reference assays used | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nicaragua | Thailand | Erasmus MC | WC/PHL | Banked sera, CII | CII-arboviral peptide array | ZIKV-NS2B-concat ELISA | ||||
| ZIKV | Acute | DENV immune | 9 | 5/9 (55) | 5/9 (55) | CDC Trioplex assay, ZCD triplex assay, CDC ZIKV monoplex assay, DENV-CHIKV multiplex assay, BOB-ELISAg | ||||
| DENV naive | 21 | 9/21 (43) | 7/21 (33) | |||||||
| Total | 30 | 14/30 (47)f | 12/30 (40) | |||||||
| Early convalescence | DENV immune | 25 | 24/25 (96) | 24/25 (96) | ||||||
| DENV naive | 65 | 64/65 (98) | 62/65 (96) | |||||||
| Total | 90 | 88/90 (98) | 86/90 (96) | |||||||
| Late convalescence | DENV immune | 8 | 6/8 (75) | 5/8 (62) | ||||||
| DENV naive | 10 | 6/10 (60) | 5/10 (50) | |||||||
| Total | 18 | 12/18 (67) | 10/18 (55) | |||||||
| Total ZIKV | 114/138 (82) | 108/138 (78) | ||||||||
| DENVa | CII-ArboViroPlex qPCR NAT assay, RVPs, DENV PRNTh | |||||||||
| NA | Acute | NA | 12 | 0/12 | 1/12 (8) | |||||
| NA | Early convalescence | Primary | 18 | 1/18 (5) | 1/18 (5) | |||||
| Secondary | 11 | 1/11 (9) | 1/11 (9) | |||||||
| Total | 29 | 2/29 (7) | 2/29 (7) | |||||||
| DENV1 | Late convalescence | Primary | 18 | 3 | 0/21 | 1/21 (5) | ||||
| Secondary | ||||||||||
| Total | 21 | 0/21 | 1/21 (5) | |||||||
| DENV2 | Late convalescence | Primary | 17 | 3 | 1/20 (5) | 2/20 (10) | ||||
| Secondary | 6 | 0/6 | 1/6 (17) | |||||||
| Total | 26 | 1/26 (4) | 3/26 (12) | |||||||
| DENV3 | Late convalescence | Primary | 9 | 3 | 0/12 | 0/12 | ||||
| Secondary | 9 | 0/9 | 1/9 (11) | |||||||
| Total | 21 | 0/21 | 1/21 (5) | |||||||
| DENV4 | Late convalescence | Primary | 4 | 3 | 0/7 | 0/7 | ||||
| Secondary | ||||||||||
| Total | 7 | 0/7 | 0/7 | |||||||
| NA | Late convalescence | Primary | 8 | 1/8 (12.5) | 1/8 (12.5) | |||||
| Secondary | ||||||||||
| Total | 8 | 1/8 (12.5) | 1/8 (12.5) | |||||||
| Total DENV | 4/124 (2) | 9/124 (7) | ||||||||
| CHIKVb | Acute | Natural infection | 6 | 6 | 1/12 (8) | 1/12 (8) | CDC Trioplex assay, method in reference 34, Euroimmun Arbo mosaic CHIKV IFAi | |||
| Convalescence | Natural infection | 6 | 6 | 0/12 | 0/12 | |||||
| Total CHIKV | 1/24 (4) | 1/24 (4) | ||||||||
| WNVc | Convalescence | Natural infection | 5 | 0/5 | 0/5 | Euroimmun anti-WNV ELISA (IgG/IgM) | ||||
| YFVc | NA | Vaccinated | 10 | 1/10 (10) | 1/10 (10) | Euroimmun flavivirus mosaic IFA | ||||
| TBEVc | NA | Vaccinated | 3 | 0/3 | 0/3 | Serion ELISA classic TBEV IgG and IgM tests | ||||
| JEVc | Convalescence | Natural infection | 1 | 0/1 | 0/1 | Euroimmun flavivirus mosaic IFA | ||||
| NA | Vaccinated | 3 | 1/3 (33) | 1/3 (33) | ||||||
| Total JEV | 1/4 (25) | 1/4 (25) | ||||||||
| Controlc | NA | Negative for above viruses | 21 | 1/21 (5) | 1/21 (5) | CII-Arbo ViroPlex qPCR NAT assay | ||||
DENV late-convalescent-phase (DENV-immune and ZIKV-naive) samples were collected before declaration of the ZIKV outbreak in Nicaragua. DENV acute-phase samples were collected from returning travelers during the 2016 ZIKV outbreak in Puerto Rico. DENV early-convalescent-phase samples were collected in 2016 during the ZIKV outbreak in Nicaragua.
CHIKV samples from Nicaragua were collected in 2014 to 2015 just before the ZIKV outbreak. CHIKV samples from WC/PHL were collected in 2016 from returning travelers during the ZIKV outbreak in Puerto Rico. CHIKV samples from Erasmus Medical Centre were collected in 2014 before declaration of the ZIKV outbreak in the Americas.
Other control group samples were collected before recognition of the ZIKV outbreak in the Americas.
Abbreviations: NA, no information available; CII, Center for Infection and Immunity; MC, Medical Centre; qPCR, quantitative PCR; NAT, nucleic acid test.
Acute phase, 1 to 6 days; early convalescent phase, ~14 to 21 days; late convalescent phase, ~6 months.
Total values by virus are highlighted in bold.
ZIKV cases were confirmed by real-time RT-PCR of serum, using the CDC Trioplex assay, the ZCD triplex assay (36), or in some cases the CDC ZIKV monoplex assay (37) in parallel with a DENV-CHIKV multiplex assay (38). Anti-ZIKV antibodies were detected by BOB-ELISA (23).
DENV acute-phase samples were tested by CII-ArboViroPlex qPCR NAT assay (Center for Infection and Immunity, 2017). In Nicaraguan samples, anti-DENV antibodies at the time of collection were detected by reporter virus particle neutralization assay (RVPs) (35). DENV convalescent-phase sera from Thailand were tested for positivity with DENV PRNT (40).
CHIKV acute-phase samples were tested by CDC Trioplex assay. Anti-CHIKV antibodies were detected by the method described in reference 34 or Euroimmun Arbo mosaic CHIKV IFA.
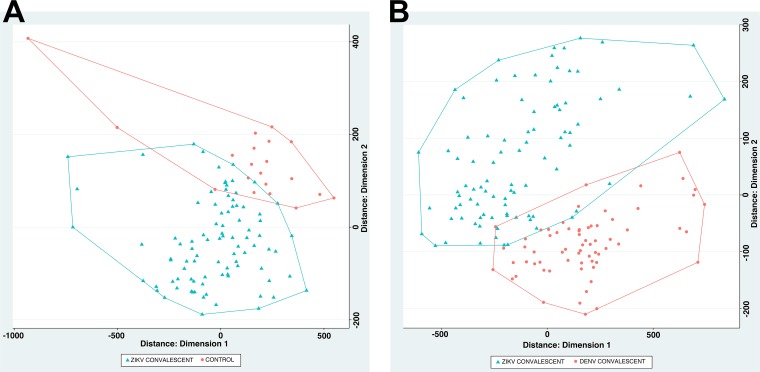
Signal data points were next filtered such that only peptides that showed signal in at least one sample (1/12 subarrays) with an intensity greater than the previously defined threshold (mean + 2 SDs, intensity of ~9,000 AU) were retained. This step reduced the initial number of peptides from 171,891 to 84,018 for data analysis. Fold changes and standard deviations were estimated by fitting a linear model for signal intensities generated by each peptide, applying empirical Bayes smoothing to the standard deviations, and then determining those peptides that yielded statistically significant signal by contrasting linear models for each peptide between conditions (20). The false-discovery rate was controlled at the 0.05 level using the Benjamini-Hochberg procedure (21). Each analysis was performed twice: once to differentiate peptides that were immunoreactive with ZIKV early-convalescent-phase versus control sera and once to differentiate peptides that were immunoreactive with ZIKV early-convalescent-phase versus DENV convalescent-phase sera. Six hundred twenty-four peptides yielded statistically significant differences (adjusted P < 0.05) in signal intensity between ZIKV convalescent-phase and control sera. Two thousand one hundred fifty-six peptides yielded statistically significant differences (adjusted P < 0.05) in signal intensity between ZIKV convalescent-phase and DENV convalescent-phase sera. Multidimensional scaling (MDS) demonstrated the capacity of these peptides to separate ZIKV from DENV sera and control group sera (Fig. 1).
FIG 1 .
Multidimensional scaling (MDS) of differential peptide signals in assays of sera from subjects with a history of infection with ZIKV, DENV, or neither (controls). Based on MDS analysis, ZIKV convalescent-phase samples (blue) versus healthy controls (red) clustered in two separate groups (A) and ZIKV convalescent-phase samples (blue) versus DENV convalescent-phase samples (red) clustered in two separate groups (B).
Identification of an immunoreactive, sensitive, and specific peptide sequence in the ZIKV NS2B protein.
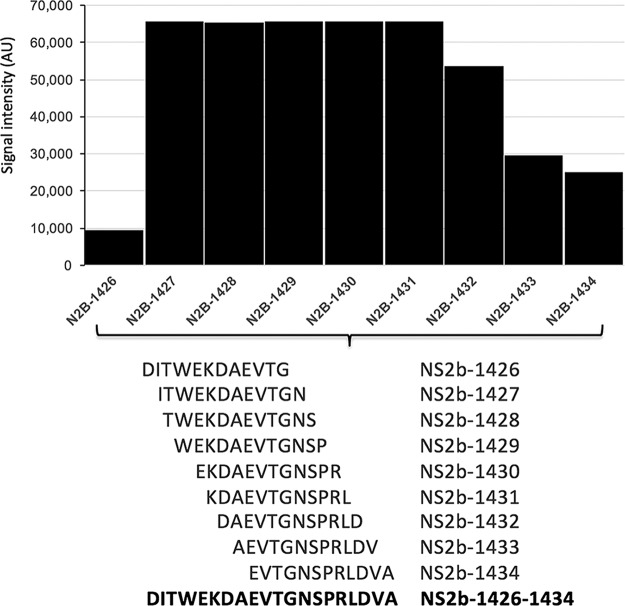
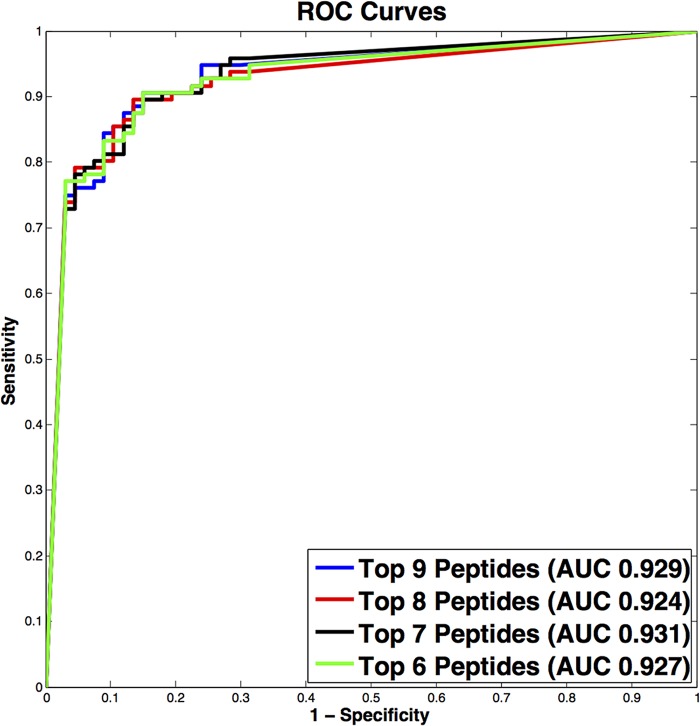
For each agent-positive sample examined, we cataloged all immunoreactive IgG linear epitopes. Immunoreactivity was highly reproducible, with minimal interchip variation (see Fig. S1 in the supplemental material). Peptides were considered agent specific if they were reactive only with their cognate sera, signal intensities were 2 SDs above the threshold established with negative-control sera, and a minimum of 3 continuous overlapping peptides were reactive. Based on these criteria, 9 overlapping peptides comprising a total of 20 aa were identified as agent specific in NS2B of ZIKV (Fig. 2). Nine peptides comprising a 20-aa region in NS2B of ZIKV were observed to be highly significant in differential analysis, displayed high signal intensities, and formed an epitope of 9 continuous overlapping peptides. To examine the potential predictive power of these 9 peptides when differentiating ZIKV convalescent-phase samples (early and late) from DENV (early and late) convalescent-phase samples, classification models were created using random forest (22) with signal intensities as predictors. Signal intensity data were obtained with 108 sera previously defined as containing antibodies to ZIKV (early and late convalescent phase) by ZIKV nonstructural protein 1 blockade-of-binding (BOB-ELISA) (23) and 114 DENV (early- and late-convalescent-phase) samples. The nine peptides from the 20-aa epitope in NS2B were first ranked with respect to their measurements of mean decrease in accuracy. The nested classifiers using the top-ranked peptides were evaluated by random resampling cross-validation with 1,000 iterations. Data were randomly split into a training set (80%) and a test set (20%) within each iteration. The area under the curve (AUC) values, together with their 95% confidence intervals (CIs), were calculated, and the corresponding receiver operating characteristic (ROC) curves were plotted. The best-performing classifier consisted of the top 6/9 ranked peptides and yielded a cross-validated AUC of 0.931 (95% CI, 0.818 to 0.975) (Fig. 3).
FIG 2 .
Identification of an immunoreactive 20-amino-acid ZIKV NS2B peptide. The overlapping and continuous amino acid sequences of the immunoreactive epitope are shown below the clustered column (peptides 1426 to 1434, NS2B-DITWEKDAEVTGNSPRLDVA; amino acid position based on sequence with accession no. ZIKV AY632535 and AAV34151).
FIG 3 .
Average receiver operating characteristic (ROC) curves over 1,000 runs using the 9 overlapping peptides identified (comprising 20-aa ZIKV NS2B peptide) for training and prediction. The top-performing model shows strong predictive power, with an average area under the curve (AUC) of 0.931.
Demonstration of minimal interchip variation and high signal reproducibility between two separate experiments (A and B). One DENV convalescent-phase, 1 ZIKV acute-phase, 1 ZIKV early-convalescent-phase, and 1 control serum were tested at a 1:100 dilution. Control sera were collected from individuals in the greater New York City metropolitan area who tested negative for Lyme disease and who had no known history of flavivirus or alphavirus infection or vaccination. Download FIG S1, TIF file, 7.8 MB (8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Testing and validation of 20-aa ZIKV NS2B peptide.
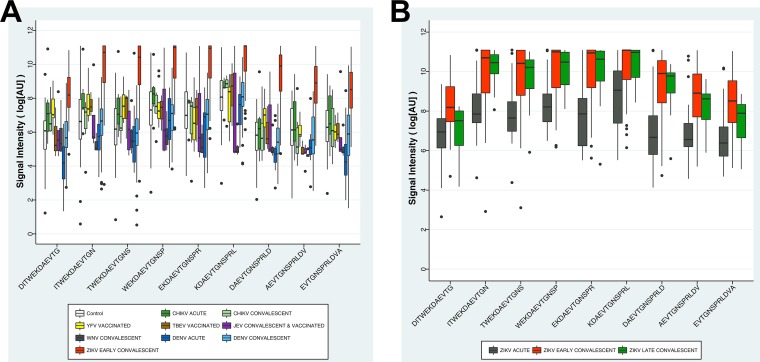
A total of 308 serum samples were tested with the arboviral peptide array: 138 for ZIKV, 124 for DENV, 24 for CHIKV, 5 for WNV, 10 for YFV, 3 for TBEV, and 4 for JEV and 21 healthy controls with no known history of flavivirus or alphavirus infection (Table 2). The ZIKV sera included samples collected from 15 patients at three time points: 1 to 6 days after onset of disease (acute phase), 2 to 3 weeks after onset of disease (early convalescence), and ~6 months after onset of disease (late convalescence). Other ZIKV sera included samples collected from individual subjects at a single time point: 15 acute-phase, 75 early-convalescent-phase, and 3 late-convalescent-phase sera. Based on previous exposure to DENV, we also divided ZIKV-positive samples into two groups, DENV immune and DENV naive. Thus, we included 30 ZIKV acute-phase (9 DENV-immune and 21 DENV-naive), 90 ZIKV early-convalescent-phase (25 DENV-immune and 65 DENV-naive), and 18 ZIKV late-convalescent-phase (8 DENV-immune and 10 DENV-naive) samples. Eighty-eight of 90 early-convalescent-phase ZIKV sera (98%) were strongly positive for the NS2B peptide with signal intensities exceeding 60,000 AU. Fourteen of 30 (47%) acute-phase ZIKV sera, 12/18 (67%) late-convalescent-phase ZIKV sera, and 1/3 (33%) JEV vaccinee sera were also strongly reactive. Samples with prior exposure to DENV had higher ZIKV reactivity with the 20-aa NS2B peptide: DENV-immune ZIKV-positive acute-phase sera had higher reactivity than DENV-naive ZIKV-positive acute-phase sera (55% versus 43%), and DENV-immune ZIKV-positive late-convalescent-phase sera had higher reactivity than DENV-naive ZIKV-positive late-convalescent-phase sera (75% versus 60%). Four of 124 (3%) DENV, 1/24 (4.1%) CHIKV, and 1/10 (10%) YFV sera were moderately reactive, with signal intensities below 20,000 AU. No cross-reactivity was observed with 12 DENV acute-phase sera, 1 JEV convalescent-phase serum, 5 WNV convalescent-phase sera, or 3 TBEV vaccinee sera. One positive acute-phase CHIKV sample from Nicaragua which was found moderately positive for ZIKV NS2B peptide was collected in November 2016 just before the first clinical ZIKV case was reported in Nicaragua (24). Detailed data for immunoreactivity, sensitivity, and specificity are given in Table 2. One of the 21 control sera from a patient residing in the greater New York City metropolitan area with an unknown travel history was also moderately reactive. ZIKV early-convalescent-phase samples showed signal intensity up to 60,000 AU for 20-aa NS2B peptides and were significantly higher (P < 0.005) than control groups except for one JEV vaccinee sample (Fig. 4A). ZIKV acute-phase and ZIKV late-convalescent-phase samples showed comparatively lower signal intensity (up to 40,000 AU) than ZIKV early-convalescent-phase samples (Fig. 4B).
FIG 4 .
(A) Immunoreactivity plots for the ZIKV NS2B 20-amino-acid peptide with ZIKV early-convalescent-phase, DENV acute-phase, DENV convalescent-phase, CHIKV, YFV, TBEV, WNV, JEV, and normal control sera. x axis, 9 peptides from ZIKV NS2B; y axis, logn values of signal intensities. Control sera were collected from individuals in the greater New York City metropolitan area who tested negative for Lyme disease and who had no known history of flavivirus or alphavirus infection or vaccination. (B) Immunoreactivity plots for the ZIKV NS2B 20-aa peptide with ZIKV acute-phase, ZIKV early-convalescent-phase, and ZIKV late-convalescent-phase sera. x axis, 9 peptides from ZIKV NS2B; y axis, logn values of signal intensities.
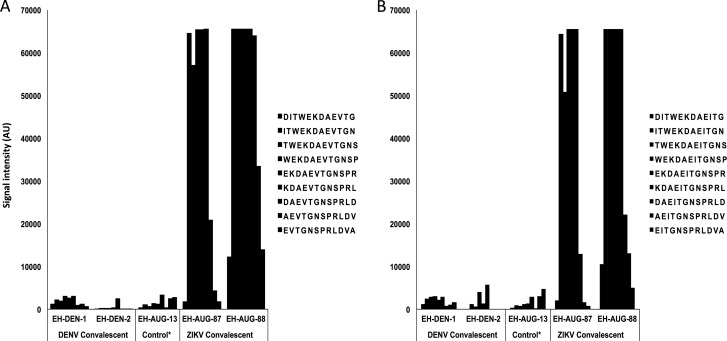
African ZIKV isolates differ from ZIKV isolates from Asia and the Americas in the presence of a valine rather than an isoleucine in the 20-aa ZIKV NS2B peptide region employed in our assays (10th amino acid in 20-aa ZIKV NS2B peptide, V>I). Both peptides were printed, allowing us to determine the impact of this mutation on immunoreactivity and assay performance. We found no differences in signals in sera from subjects infected in Asia or the Americas (Fig. 5). We have no African sera for reciprocal analyses.
FIG 5 .
Immunoreactivity of ZIKV acute-phase and ZIKV convalescent-phase sera from two Nicaraguan patients to the NS2B 20-aa peptide representing American isolates (A) (peptides 1426 to 1434, NS2B-DITWEKDAEVTGNSPRLDVA, accession no. ZIKV AAV34151) or African isolates (B) (peptides 1430 to 1438, NS2B-DITWEKDAEITGNSPRLDVA, accession no. ZIKV AMD61711). Controls include two DENV convalescent-phase sera and one control serum. Control sera were collected from individuals in the greater New York City metropolitan area who tested negative for Lyme disease and who had no known history of flavivirus or alphavirus infection or vaccination.
Development of a ZIKV NS2B peptide ELISA.
Due to costs for fabrication, instrument purchase, and protocol complexity, we view peptide arrays as platforms for discovery rather than as tools for routine serology. Accordingly, we used data acquired in array analyses to build a ZIKV NS2B peptide ELISA. Twenty-four-residue peptides were synthesized that included the immunoreactive 20-aa ZIKV NS2B peptide as well as two flanking amino acid residues (underlined) at both the amino and carboxyl termini to serve as spacer molecules for epitope presentation (AGDITWEKDAEVTGNSPRLDVALD). We also synthesized peptides ranging from 8 to 14 aa within the core of the region defined as most strongly immunoreactive in Fig. 2 and a concatemer of the 24-aa peptide joined by a central glutamic acid residue (AGDITWEKDAEVTGNSPRLDVALDEAGDITWEKDAEVTGNSPRLDVALD). All peptides were synthesized with biotin on either the amino or carboxyl terminus to facilitate binding to plates coated with antibodies to biotin. The performance of the carboxyl terminus biotinylated 49-aa peptide concatemer ELISA was superior to the other peptide ELISAs. It identified 96% of ZIKV sera positive in the peptide array with 94% specificity. The 8- to 14-aa peptide ELISA had less than 50% sensitivity and 80% specificity. The 24-aa peptide ELISA had ~80% sensitivity and specificity. The amino terminus biotinylated 49-aa peptide concatemer ELISA yielded 70% sensitivity and 60% specificity. Accordingly, we are here reporting only findings with the carboxyl terminus biotinylated 49-aa peptide concatemer ELISA (ZIKV-NS2B-concat ELISA).
Validation of the ZIKV-NS2B-concat ELISA.
All 329 serum samples (Table 2) previously tested with the arboviral peptide array were also tested with the ZIKV-NS2B-concat ELISA. Eighty-six of 90 early-convalescent-phase ZIKV sera (96%) were positive (optical density [OD] > 0.90) by ELISA. Eight of 30 (26.6%) acute-phase ZIKV sera, 8/18 (44%) late-convalescent-phase ZIKV sera, and 1/3 (33%) JEV vaccinee sera were also positive (>0.90 OD) with ELISA. Samples with prior exposure to DENV had higher ZIKV reactivity in ELISA also: DENV-immune ZIKV-positive acute-phase sera had higher reactivity than DENV-naive ZIKV-positive acute-phase sera (55% versus 33%), and DENV-immune ZIKV-positive late-convalescent-phase sera had higher reactivity than DENV-naive ZIKV-positive late-convalescent-phase sera (62% versus 50%). Two of 90 (2%) early-convalescent-phase ZIKV sera, 4/30 (13%) acute-phase ZIKV sera, 2/18 (11%) late-convalescent-phase ZIKV sera, 9/124 (7%) DENV sera, 1/24 CHIKV sera (4%), and 1/10 YFV sera (10%) were moderately reactive (0.65 to 0.90 OD). No cross-reactivity was observed with 5 WNV or 3 TBEV sera. One of 21 sera from the control group that was positive in the peptide array was also positive in the ELISA. Detailed data for immunoreactivity, sensitivity, and specificity are given in Table 2.
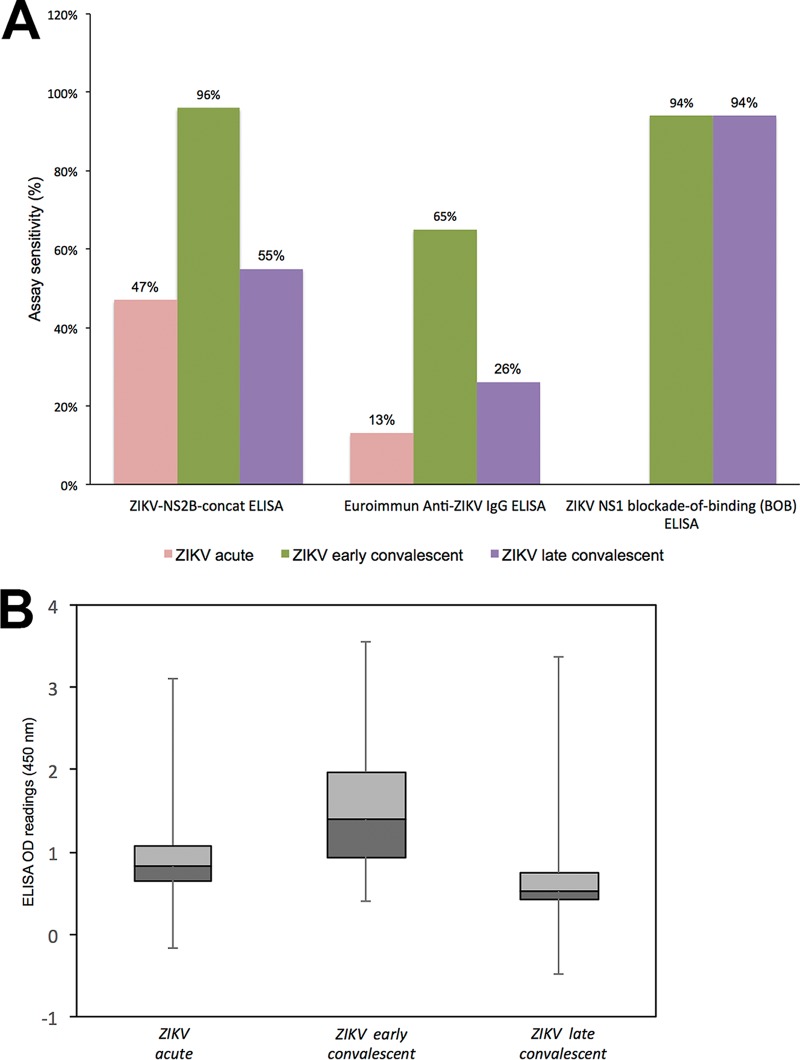
A total of 154 ZIKV sera (30 acute-phase, 70 early-convalescent-phase, and 54 late-convalescent-phase sera) were also tested side by side by the ZIKV-NS2B-concat ELISA, a commercial NS1 recombinant IgG ELISA (anti-ZIKV ELISA IgG assays; Euroimmun, Lübeck, Germany) (25), and a Zika nonstructural protein 1 (NS1) BOB-ELISA (23) (Fig. 6A). The sensitivity of the ZIKV-NS2B-concat ELISA was superior to the recombinant commercial NS1 IgG ELISA with acute-phase (47% versus 13%), early-convalescent-phase (96% versus 65%), and late-convalescent-phase (55% versus 26%) sera. The BOB-ELISA had similar sensitivity with early-convalescent-phase sera (94%) and superior sensitivity with late-convalescent-phase sera (94%) but detected no immunoreactivity in acute-phase sera (data not shown). The immunoreactivity of ZIKV-NS2B-concat ELISA with sera collected from 28 ZIKV patients over the time course of infection was higher in early convalescent phase than in the acute and late convalescent phases and consistent with findings obtained by using the peptide array (Fig. 4B and 6B).
FIG 6 .
(A) ZIKV-NS2B-concat ELISA sensitivity comparison with Euroimmun anti-ZIKV IgG ELISA and ZIKV-NS1 blockade-of-binding (BOB) ELISA. (B) ZIKV-NS2B-concat ELISA demonstrates higher anti-ZIKV antibody titer with ZIKV early-convalescent-phase group than with ZIKV acute-phase and ZIKV late-convalescent-phase groups.
DISCUSSION
Since the emergence of ZIKV in the Americas in 2015, 583,144 cases have been reported to the World Health Organization. On 6 April 2017, the United Nations Development Programme, in partnership with the International Federation of Red Cross, Red Crescent Societies and the Pan-American Health Organization, estimated the economic cost of Zika disease in Latin America and the Caribbean alone as up to $18 billion between 2015 and 2017 (26). Costs will ultimately be higher because sequelae of congenital infection require lifetime support and include more subtle neurodevelopmental disabilities than the severe abnormalities observed at birth, such as microcephaly (27, 28).
Accurate estimates of the public health burden of ZIKV and medical management will require easily accessible, sensitive, and specific assays for detection of infection, which has been challenging. The efficacy of molecular methods for detection of ZIKV in serum, plasma, and whole blood is limited to the first few weeks of infection. Serology has been confounded by cross-reactivity of antibody responses due to genetic similarity and structural homology of ZIKV with other flaviviruses, especially DENV. Across the entire genomic sequence, ZIKV shares 55.6% amino acid identity with DENV, 46.0% with YFV, 56.1% with JEV, and 57.0% with WNV (29). False-positive results in ZIKV ELISA and PRNT assays have been reported after natural infection with other flaviviruses as well as after YFV and JEV vaccination (30).
The advent of peptide microarrays provides new opportunities to finely map discrete regions within the proteomes of viruses and other microbes that can facilitate differential diagnosis of infectious diseases. We used highly multiplexed, programmable peptide arrays to identify discriminant epitopes for serodiagnosis of infection. The ZIKV-NS2B-concat ELISA, generated based on results obtained with the peptide array, has utility for diagnosis of ZIKV infection. In our protocol using a threshold of an 0.90 OD, the ELISA detected IgG antibodies in 47% of sera from subjects with acute disease (1 to 6 days after onset), more than 95% of sera from subjects in early convalescence (2 to 3 weeks after onset), and 55% of sera from subjects in late convalescence (more than 6 months after onset).
The specificity of the ZIKV-NS2B-concat ELISA was 94%. One of three JEV vaccinee sera was seropositive; however, a subject with natural JEV infection was not seropositive. One individual from Nicaragua with acute CHIKV infection was also seropositive. Chikungunya virus is an alphavirus that would not be anticipated to elicit antibodies cross-reactive with flaviviruses, and the samples analyzed were all collected before the introduction of ZIKV. We do not know whether this individual also had a history of exposure to ZIKV. The closest homologue in NCBI-BlastP to the 20-aa ZIKV NS2B peptide sequence is a sequence in Spondweni virus (SPONV) (17/20 aa, 85% identity) (see Fig. S2 in the supplemental material). Spondweni virus has been reported in sub-Saharan Africa; only 6 cases are documented (31). The SPONV serogroup includes both ZIKV and SPONV. We have not tested sera from individuals infected with SPONV and acknowledge that differential diagnosis may not be feasible with the ZIKV-NS2B-concat ELISA alone. JEV and St. Louis encephalitis virus (SLEV) have 80% amino acid homology (16/20 amino acid residues) with the ZIKV NS2B peptide sequence. This may account for false-positive results with sera from the subject recently vaccinated for JEV wherein the antibody titers for the 20-aa ZIKV NS2B peptide sequence were high and underscores the importance of obtaining an accurate vaccine history. We did not have access to sera from subjects exposed to SLEV to test for immunoreactivity to the ZIKV NS2B peptide. We observed lower-titer cross-reactivity that did not cross the 0.90-OD threshold for definitive serological ZIKV diagnosis with other flaviviruses, including DENV1, DENV2, DENV3, DENV4, WNV, and TBEV.
Amino acid alignment showing residues in the 20-aa NS2B ZIKV peptide homologous with other flaviviruses. Download FIG S2, TIF file, 7.8 MB (8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Preexisting dengue virus antibodies may modulate immune responses to ZIKV infection (32). Priyamvada et al. have proposed that this is due to reactivation of preexisting memory B cells that target conserved epitopes (33). We found that prior exposure to DENV was associated with higher ZIKV reactivity to NS2B peptide in both the peptide array and ZIKV-NS2B-concat ELISA. We cannot ascertain whether the converse is true as we have no sera from subjects infected with ZIKV who were subsequently infected with DENV. However, we found no reactivity to the DENV NS2B using DENV sera.
One of 21 control sera obtained from patients in the greater New York City metropolitan area tested for Borrelia burgdorferi, the causative agent of Lyme disease, also had reactivity in the ZIKV NS2B peptide ELISA and peptide array. We do not have travel history or vaccine records for these control patients and cannot discern whether reactivity was due to flavivirus infection or vaccination.
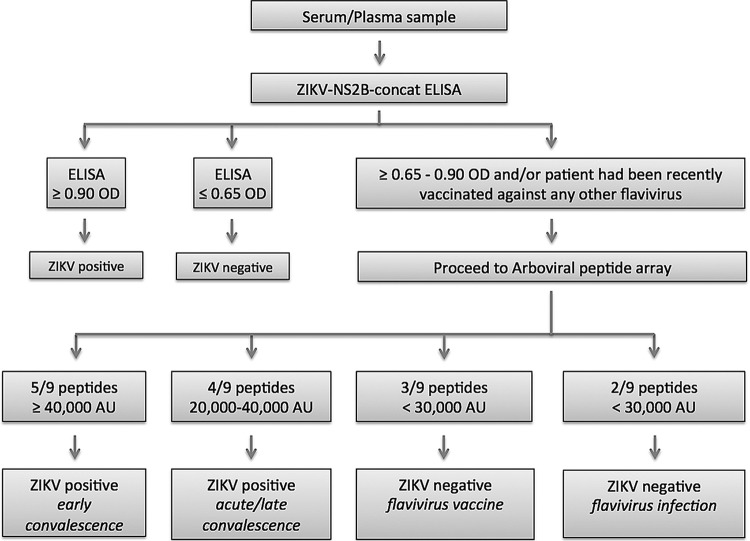
The peptide array provided a modest improvement over the ELISA in sensitivity with acute-phase (47% versus 40%), early-convalescent-phase (98% versus 96%), and late-convalescent-phase (67% versus 55%) sera. It was also more specific and allowed us to resolve the JEV and CHIKV immunoreactivity observed in ELISA. Whereas all early-convalescent-phase ZIKV sera detected 5 or more peptides, each with signal in excess of 40,000 AU, the JEV vaccine and CHIKV sera detected only 3 peptides; none yielded signal greater than 30,000 AU. Nine of 124 DENV sera had intermediate reactivity in ELISA (OD of 0.60 to 0.90). None reacted with 5 peptides or had signal in excess of 40,000 AU. The single serum from the 21-subject control group that was positive in ELISA remained positive on the array (detecting 5 peptides with more than 40,000 AU). An algorithm for serodiagnosis of ZIKV exposure based on the limited sample set employed in this work is shown in Fig. 7.
FIG 7 .
An algorithm for serodiagnosis of ZIKV exposure based on the limited sample set employed in this work.
ZIKV infection in any trimester can result in stillbirth or congenital birth defects (12). This includes clinically inapparent infections. It is important, therefore, that the diagnostic armamentarium for risk assessment have the capacity to detect evidence of current as well as past infection over the entire course of gestation. The ZIKV-NS2B-concat ELISA is most sensitive for detection of infection during early convalescence. We do not have the samples required to assess performance in the interval between early and late convalescence (3 weeks to 6 months); nonetheless, based on data obtained during early convalescence, monthly testing during each of the three trimesters should detect more than 95% of ZIKV infections. Where results are equivocal, the peptide array can be used to increase the sensitivity for detection of ZIKV infection to 98% and delineate triggers for cross-reactivity, including other flavivirus infections or vaccination. The ZIKV-NS2B-concat ELISA can be readily implemented with equipment employed in routine clinical microbiology laboratories found in the developing world where the burden of infection is highest. As the peptide array requires more elaborate imaging equipment and bioinformatics support for analysis, it will be important to explore whether less resource-intensive serological tests can be established that quantitate responses to the multiple peptides that enhance sensitivity and specificity in the peptide array.
The positive and negative predictive values of a serological assay vary with the prevalence of infection in the population. Our data concerning sensitivity and specificity reflect findings in subjects considered to be at high risk for infection with ZIKV or other arboviruses. Additional studies will be required to establish the positive and negative predictive values of the assays in other populations.
The peptide array platform that we employed displays only linear amino acid sequences. It is possible that a system that displays conformational epitopes might have revealed epitopes with better diagnostic performance profiles than ZIKV NS2B. We nonetheless predict that due to flexibility and high-throughput screening capacity, the programmable peptide array platform will become a vital tool in the discovery and development of serodiagnostics.
MATERIALS AND METHODS
Serum or plasma samples for the study.
One hundred thirty-eight serum samples were collected from 90 children in the Nicaraguan Pediatric Dengue Cohort Study (PDCS) (23) who were reverse transcription-PCR (RT-PCR) positive for ZIKV in serum and had signs and symptoms of ZIKV infection between January and 20 September 2016. Acute-phase (1 to 6 days after onset of symptoms), early-convalescent-phase (14 to 21 days after onset of symptoms), and late-convalescent-phase (~6 months after onset of symptoms) blood samples were drawn for DENV, CHIKV, and ZIKV diagnostic testing and annual sample collection as part of the cohort study (Table 2) (34). We included confirmed ZIKV cases with known prior DENV infection history status (DENV naive or DENV immune) (35). Confirmed ZIKV-positive cases were classified as DENV naive if they entered the cohort study with no detectable anti-DENV antibodies (as measured by DENV inhibition ELISA) and had no documented DENV infections (symptomatic or inapparent) during their time in the cohort. Confirmed ZIKV-positive cases were classified as DENV immune if they either entered the cohort study with detectable anti-DENV inhibition ELISA antibodies or entered the cohort study with no detectable anti-DENV antibodies and had one or more documented DENV infections during their time in the cohort study. All suspected ZIKV cases were confirmed by real-time RT-PCR in serum, using the CDC Trioplex assay, the ZCD triplex assay (36), or in some cases the CDC ZIKV monoplex assay (37), in parallel with a DENV-CHIKV multiplex assay (38). We also included as a control group a total of 124 samples with DENV (12 acute-, 29 early-convalescent-, and 83 late-convalescent-phase) infection confirmed by RT-PCR between 2005 and 2013, from the New York State Department of Health Wadsworth Center Laboratory (WC), New York City Department of Health and Mental Hygiene Public Health Laboratory (PHL) (10), Nicaraguan PDCS (34), or the Nicaraguan Hospital-Based Dengue Study at the National Pediatric Reference Hospital in Managua (39). Twelve additional DENV early-convalescent-phase sera (3 for each of DENV1 to -4) that tested positive with PRNT were collected from Thailand (40). Six acute- and 12 early-convalescent-phase CHIKV sera were collected from real-time RT-PCR-confirmed chikungunya cases in the Nicaraguan hospital-based study (41); 6 acute-phase CHIKV sera were provided by WC and PHL (10). Convalescent-phase CHIKV sera were tested for anti-CHIKV antibodies with Euroimmun anti-chikungunya virus immunofluorescent assay (IFA) (IgG). Five banked WNV early-convalescent-phase sera after natural infection were also used (10). Ten YFV (vaccinated), 3 TBEV (vaccinated), and 4 JEV (1 natural infection convalescent-phase and 3 vaccinated) sera were collected by the Erasmus University Medical Centre (Rotterdam, Netherlands). Sera from 21 subjects in the greater New York City metropolitan area tested for Lyme disease who had no known history of flavivirus or alphavirus infection or vaccination were provided by the Lyme Disease Laboratory (School of Medicine, Stony Brook University) for use as controls.
Arboviral peptide array synthesis, sample binding, and processing.
Peptide synthesis was accomplished by light-directed array synthesis in a Roche maskless array synthesizer (MAS) using an amino functionalized substrate coupled with 6-aminohexanoic acid as a spacer and amino acid derivatives carrying a photosensitive 2-(2-nitrophenyl)propyloxycarbonyl (NPPOC) group. Coupling of amino acids was done using preactivated amino acid with activator (1-hydroxybenzotriazole [HOBT]/2-(1H-benzotriazol-1-yl)-1,1,3,3- tetramethyluronium [HBTU]) and ethyldiisopropylamine in dimethylformamide (DMF) for 5 to 7 min before flushing the substrate. Cycles of coupling were repeated until 12-mer peptides were synthesized. Intermediate washes on the arrays were done with N-methyl-2-pyrrolidone (NMP), and site-specific cleavage of the NPPOC group was accomplished by irradiation of an image created by a digital micromirror device (Texas Instruments; SXGA+ graphics format), projecting light with a 365-nm wavelength. Final deprotection to cleave off the side chain protecting groups of the amino acids was done with trifluoroacetic acid (TFA)-water-triisopropylsilane for 30 min.
Sera were diluted (1:50 or 1:100) with binding buffer (0.1 M Tris-Cl, 1% alkali-soluble casein, 0.05% Tween 20, and water). The peptide arrays were incubated overnight at 4°C on a flat surface with individual sample/subarray. Overnight sample incubation was followed by three 10-min washes with 1× Tris-buffered saline with Tween 20 (TBST) (0.05% Tween 20) on a Little Dipper automatic washer (catalog no. 1080-40-1; SciGene) at room temperature (RT). Secondary antibodies (catalog no. 109-605-098; Alexa Fluor 647-AffiniPure goat anti-human IgG, Fcγ fragment specific; Jackson ImmunoResearch Labs) were diluted in binding buffer at a concentration of 0.1 µg/ml, and arrays were incubated in a plastic Coplin jar (catalog no. S90130; Fisher Scientific) for 3 h at RT with gentle shaking on a rocker shaker. Secondary antibody incubation was also followed by three 10-min washes on a Little Dipper washer with 1× TBST at RT. After a final wash, the arrays were dried and read using a Roche MS 200 microarray scanner (Roche), and signals were extracted using Roche internally developed image extraction software. The array slides were scanned at 2-μm resolution, with an excitation wavelength of 635 nm. The images were analyzed using the PepArray analysis program. The fluorescent signals were converted into arbitrary unit (AU) intensity plots ranging in minimum to maximum intensity from 0 to 65,000 AU.
ZIKV-NS2B-concat ELISA.
A 49-aa-long concatemer of the 24-aa peptide ZIKV NS2B peptide joined by a central glutamic acid residue and carboxyl-terminal biotin labeled with a junction lysine residue, AGDITWEKDAEVTGNSPRLDVALDEAGDITWEKDAEVTGNSPRLDVALD(Lys[biotin]), was synthesized with high-performance liquid chromatography (HPLC) purity of ≥75.0% (GenScript, Piscataway, NJ). The peptide was dissolved in 3% ammonia water and stored at −20°C in small aliquots. Peptide ELISAs were performed on Corning 96-well clear-bottom plates (Sigma-Aldrich). Preadsorbed rabbit polyclonal antibiotin antibody-IgG heavy plus light chains (H+L) (catalog no. ab53494; Abcam) were diluted with 1× ELISA coating buffer (catalog no. BUF030A; Bio-Rad), applied as a coating on 96-well ELISA plates (0.6 μg/100 μl per well), and incubated at 37°C overnight under a sealed condition. After incubation, the plate was washed three times with phosphate-buffered saline (PBS)–0.05% Tween 20 (PBS-T). Following three washes with PBS-T, plates were incubated for 1 h at RT with 200 µl/well of blocking buffer (catalog no. 37539; Thermo Fisher Scientific). Afterward, plates were washed again three times with PBS-T. ZIKV NS2B peptide was diluted in blocking buffer (0.4 μg/100 μl), and 100 μl of diluted ZIKV NS2B peptide was applied as a coating on plate-wells and incubated at 37°C for 90 min. Following 3 washes with PBS-T, serum/plasma samples were diluted 1:50 or 1:100 in blocking buffer, coated (200 µl/well), and incubated at 37°C for 90 min. After 3 washes with PBS-T, preadsorbed goat anti-human IgG H+L–horseradish peroxidase (catalog no. ab97175; Abcam) was diluted (1:5,000) and coated (100 µl/well) for 90 min at 37°C. Plates were washed again 3 times with PBS-T, 1-Step Ultra tetramethylbenzidine (TMB)-ELISA substrate solution (catalog no. 34028; Thermo Fisher Scientific) was added, and plates were incubated at RT and in the dark. The reaction was stopped with 50 μl Wash and Stop solution for ELISA without sulfuric acid (catalog no. MK021; TaKaRa Clontech). Plates were read on an automated plate reader at 450 nm. The OD readings represent the reactivity of peptide ELISA. The ELISA cutoff was established by running 48 healthy control human sera with ZIKV-NS2B-concat ELISA. The nonparametric method of two times the mean of ODs (0.65) of the sera from uninfected healthy controls was established as moderate positive, and three times the mean of ODs (0.90) of the sera from uninfected healthy controls was used as the definitive positive cutoff (42).
ACKNOWLEDGMENTS
We are grateful to the patients and clinicians who provided samples without which this work could not have proceeded. We are also grateful for technical support from Dominick Centurioni and Amy Dean (New York State Department of Health) and Dakai Liu, Taryn Burke, and Aaron Olsen (New York City Department of Health and Mental Hygiene). We thank past and present members of the study team based at the Hospital Infantil Manuel de Jesús Rivera, the National Virology Laboratory in the Centro Nacional de Diagnóstico y Referencia of the Nicaraguan Ministry of Health, and the Sustainable Sciences Institute in Managua, Nicaragua, for their dedication and high-quality work.
We are grateful for financial support from National Institutes of Health award U19AI109761 (CETR [Center for Research in Diagnostics and Discovery]). E.H. received funding support from National Institutes of Health grants P01AI106695, U19AI118610, and R01AI099631 (to A.B.). C.B.E.M.R. received funding support from ZIKAlliance H2020 (73548). N.M. was partially supported by 2016–2017 Calderone Junior Faculty Awards awarded by the Mailman School of Public Health, Columbia University.
The manuscript has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense.
Eric H. Sullivan and Jigar J. Patel are full-time employees of Roche, Madison, WI. The other authors declare no competing financial interests.
Footnotes
Citation Mishra N, Caciula A, Price A, Thakkar R, Ng J, Chauhan LV, Jain K, Che X, Espinosa DA, Montoya Cruz M, Balmaseda A, Sullivan EH, Patel JJ, Jarman RG, Rakeman JL, Egan CT, Reusken CBEM, Koopmans MPG, Harris E, Tokarz R, Briese T, Lipkin WI. 2018. Diagnosis of Zika virus infection by peptide array and enzyme-linked immunosorbent assay. mBio 9:e00095-18. https://doi.org/10.1128/mBio.00095-18.
REFERENCES
- 1.World Health Organization 2016. Laboratory testing for Zika virus infection: interim guidance. WHO/ZIKV/LAB/16.1. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/204671/1/WHO_ZIKV_LAB_16.1_eng.pdf. [Google Scholar]
- 2.Gulland A. 2016. Zika virus is a global public health emergency, declares WHO. BMJ 352:i657. doi: 10.1136/bmj.i657. [DOI] [PubMed] [Google Scholar]
- 3.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. 2016. Zika virus and birth defects—reviewing the evidence for causality. N Engl J Med 374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 4.de Vasconcelos ZFM, Azevedo RC, Thompson N, Gomes L, Guida L, Moreira MEL. 2018. Challenges for molecular and serological ZIKV infection confirmation. Childs Nerv Syst 34:79–84. doi: 10.1007/s00381-017-3641-5. [DOI] [PubMed] [Google Scholar]
- 5.Broutet N, Krauer F, Riesen M, Khalakdina A, Almiron M, Aldighieri S, Espinal M, Low N, Dye C. 2016. Zika virus as a cause of neurologic disorders. N Engl J Med 374:1506–1509. doi: 10.1056/NEJMp1602708. [DOI] [PubMed] [Google Scholar]
- 6.Krauer F, Riesen M, Reveiz L, Oladapo OT, Martínez-Vega R, Porgo TV, Haefliger A, Broutet NJ, Low N, WHO Zika Causality Working Group . 2017. Zika virus infection as a cause of congenital brain abnormalities and Guillain-Barré syndrome: systematic review. PLoS Med 14:e1002203. doi: 10.1371/journal.pmed.1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winkler CW, Woods TA, Rosenke R, Scott DP, Best SM, Peterson KE. 2017. Sexual and vertical transmission of Zika virus in anti-interferon receptor-treated Rag1-deficient mice. Sci Rep 7:7176. doi: 10.1038/s41598-017-07099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson A, Slavinski S, Komoto K, Rakeman J, Weiss D. 2016. Suspected female-to-male sexual transmission of Zika virus—New York City, 2016. MMWR Morb Mortal Wkly Rep 65:716–717. doi: 10.15585/mmwr.mm6528e2. [DOI] [PubMed] [Google Scholar]
- 9.Motta IJ, Spencer BR, Cordeiro da Silva SG, Arruda MB, Dobbin JA, Gonzaga YB, Arcuri IP, Tavares RC, Atta EH, Fernandes RF, Costa DA, Ribeiro LJ, Limonte F, Higa LM, Voloch CM, Brindeiro RM, Tanuri A, Ferreira OC Jr.. 2016. Evidence for transmission of Zika virus by platelet transfusion. N Engl J Med 375:1101–1103. doi: 10.1056/NEJMc1607262. [DOI] [PubMed] [Google Scholar]
- 10.US Food and Drug Administration 11 August 2017. CII-ArboViroPlex rRT-PCR assay: authorization of the emergency use of in vitro diagnostics for detection of Zika virus and/or diagnosis of Zika virus infection. US Food and Drug Administration, Washington, DC: https://www.fda.gov/downloads/MedicalDevices/Safety/EmergencySituations/UCM571320.pdf. [Google Scholar]
- 11.Mansuy JM, Mengelle C, Pasquier C, Chapuy-Regaud S, Delobel P, Martin-Blondel G, Izopet J. 2017. Zika virus infection and prolonged viremia in whole-blood specimens. Emerg Infect Dis 23:863–865. doi: 10.3201/eid2305.161631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brasil P, Pereira JP Jr, Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, Zin AA, Horovitz D, Daltro P, Boechat M, Raja Gabaglia C, Carvalho de Sequeira P, Pilotto JH, Medialdea-Carrera R, Cotrim da Cunha D, Abreu de Carvalho LM, Pone M, Machado Siqueira A, Calvet GA, Rodrigues Baião AE, Neves ES, Nassar de Carvalho PR, Hasue RH, Marschik PB, Einspieler C, Janzen C, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. 2016. Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med 375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buus S, Rockberg J, Forsström B, Nilsson P, Uhlen M, Schafer-Nielsen C. 2012. High-resolution mapping of linear antibody epitopes using ultrahigh-density peptide microarrays. Mol Cell Proteomics 11:1790–1800. doi: 10.1074/mcp.M112.020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez JP, Souris M, Valdivia-Granda W. 2018. Global spread of hemorrhagic fever viruses: predicting pandemics. Methods Mol Biol 1604:3–31. doi: 10.1007/978-1-4939-6981-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Travassos da Rosa JF, de Souza WM, Pinheiro FP, Figueiredo ML, Cardoso JF, Acrani GO, Nunes MRT. 2017. Oropouche virus: clinical, epidemiological, and molecular aspects of a neglected Orthobunyavirus. Am J Trop Med Hyg 96:1019–1030. doi: 10.4269/ajtmh.16-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 17.Valentini D, Rao M, Rane L, Rahman S, Axelsson-Robertson R, Heuchel R, Löhr M, Hoft D, Brighenti S, Zumla A, Maeurer M. 2017. Peptide microarray-based characterization of antibody responses to host proteins after bacille Calmette-Guerin vaccination. Int J Infect Dis 56:140–154. doi: 10.1016/j.ijid.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oshlack A, Emslie D, Corcoran LM, Smyth GK. 2007. Normalization of boutique two-color microarrays with a high proportion of differentially expressed probes. Genome Biol 8:R2. doi: 10.1186/gb-2007-8-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. 2016. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat 10:946–963. doi: 10.1214/16-AOAS920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Stat Methodol 57:289–300. [Google Scholar]
- 22.Breiman L. 2001. Random forests. Mach Learn 45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 23.Balmaseda A, Stettler K, Medialdea-Carrera R, Collado D, Jin X, Zambrana JV, Jaconi S, Cameroni E, Saborio S, Rovida F, Percivalle E, Ijaz S, Dicks S, Ushiro-Lumb I, Barzon L, Siqueira P, Brown DWG, Baldanti F, Tedder R, Zambon M, de Filippis AMB, Harris E, Corti D. 2017. Antibody-based assay discriminates Zika virus infection from other flaviviruses. Proc Natl Acad Sci U S A 114:8384–8389. doi: 10.1073/pnas.1704984114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El Nuevo Diario 27 January 2016. Confirman dos casos positivos de Zika en Nicaragua. El Nuevo Diario, Managua, Nicaragua: https://www.elnuevodiario.com.ni/nacionales/383401-nicaragua-confirma-dos-casos-positivos-zika/. [Google Scholar]
- 25.L’Huillier AG, Hamid-Allie A, Kristjanson E, Papageorgiou L, Hung S, Wong CF, Stein DR, Olsha R, Goneau LW, Dimitrova K, Drebot M, Safronetz D, Gubbay JB. 2017. Evaluation of Euroimmun anti-Zika virus IgM and IgG enzyme-linked immunosorbent assays for Zika virus serologic testing. J Clin Microbiol 55:2462–2471. doi: 10.1128/JCM.00442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.United Nations Development Programme 6 April 2017. Social and economic costs of Zika can reach up to US$18 billion in Latin America and the Caribbean. United Nations Development Programme, New York, NY: http://www.undp.org/content/undp/en/home/presscenter/pressreleases/2017/04/06/social-and-economic-costs-of-zika-can-reach-up-to-us-18-billion-in-latin-america-and-the-caribbean.html. [Google Scholar]
- 27.Melo AS, Aguiar RS, Amorim MM, Arruda MB, Melo FO, Ribeiro ST, Batista AG, Ferreira T, Dos Santos MP, Sampaio VV, Moura SR, Rabello LP, Gonzaga CE, Malinger G, Ximenes R, de Oliveira-Szejnfeld PS, Tovar-Moll F, Chimelli L, Silveira PP, Delvechio R, Higa L, Campanati L, Nogueira RM, Filippis AM, Szejnfeld J, Voloch CM, Ferreira OC Jr, Brindeiro RM, Tanuri A. 2016. Congenital Zika virus infection: beyond neonatal microcephaly. JAMA Neurol 73:1407–1416. doi: 10.1001/jamaneurol.2016.3720. [DOI] [PubMed] [Google Scholar]
- 28.Lipkin WI. 6 September 2016. The coming trials of generation Zika: we may see an increase in the incidence of mental illness, Parkinson’s and dementia. The Wall Street Journal, New York, NY: https://www.wsj.com/articles/the-coming-trials-of-generation-zika-1473203849. [Google Scholar]
- 29.Chang HH, Huber RG, Bond PJ, Grad YH, Camerini D, Maurer-Stroh S, Lipsitch M. 2017. Systematic analysis of protein identity between Zika virus and other arthropod-borne viruses. Bull World Health Organ 95:517–525I. doi: 10.2471/BLT.16.182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haug CJ, Kieny MP, Murgue B. 2016. The Zika challenge. N Engl J Med 374:1801–1803. doi: 10.1056/NEJMp1603734. [DOI] [PubMed] [Google Scholar]
- 31.Haddow AD, Woodall JP. 2016. Distinguishing between Zika and Spondweni viruses. Bull World Health Organ 94:711–711A. doi: 10.2471/BLT.16.181503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan MJ, Wilson PC, Ahmed R, Suthar MS, Wrammert J. 2016. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A 113:7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priyamvada L, Suthar MS, Ahmed R, Wrammert J. 2017. Humoral immune responses against Zika virus infection and the importance of preexisting flavivirus immunity. J Infect Dis 216:S906–S911. doi: 10.1093/infdis/jix513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuan G, Gordon A, Aviles W, Ortega O, Hammond SN, Elizondo D, Nuñez A, Coloma J, Balmaseda A, Harris E. 2009. The Nicaraguan pediatric dengue cohort study: study design, methods, use of information technology, and extension to other infectious diseases. Am J Epidemiol 170:120–129. doi: 10.1093/aje/kwp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattia K, Puffer BA, Williams KL, Gonzalez R, Murray M, Sluzas E, Pagano D, Ajith S, Bower M, Berdougo E, Harris E, Doranz BJ. 2011. Dengue reporter virus particles for measuring neutralizing antibodies against each of the four dengue serotypes. PLoS One 6:e27252. doi: 10.1371/journal.pone.0027252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waggoner JJ, Gresh L, Mohamed-Hadley A, Ballesteros G, Davila MJ, Tellez Y, Sahoo MK, Balmaseda A, Harris E, Pinsky BA. 2016. Single-reaction multiplex reverse transcription PCR for detection of Zika, chikungunya, and dengue viruses. Emerg Infect Dis 22:1295–1297. doi: 10.3201/eid2207.160326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waggoner JJ, Ballesteros G, Gresh L, Mohamed-Hadley A, Tellez Y, Sahoo MK, Abeynayake J, Balmaseda A, Harris E, Pinsky BA. 2016. Clinical evaluation of a single-reaction real-time RT-PCR for pan-dengue and Chikungunya virus detection. J Clin Virol 78:57–61. doi: 10.1016/j.jcv.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narvaez F, Gutierrez G, Perez MA, Elizondo D, Nuñez A, Balmaseda A, Harris E. 2011. Evaluation of the traditional and revised WHO classifications of dengue disease severity. PLoS Negl Trop Dis 5:e1397. doi: 10.1371/journal.pntd.0001397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas SJ, Nisalak A, Anderson KB, Libraty DH, Kalayanarooj S, Vaughn DW, Putnak R, Gibbons RV, Jarman R, Endy TP. 2009. Dengue plaque reduction neutralization test (PRNT) in primary and secondary dengue virus infections: how alterations in assay conditions impact performance. Am J Trop Med Hyg 81:825–833. doi: 10.4269/ajtmh.2009.08-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balmaseda A, Gordon A, Gresh L, Ojeda S, Saborio S, Tellez Y, Sanchez N, Kuan G, Harris E. 2016. Clinical attack rate of chikungunya in a cohort of Nicaraguan children. Am J Trop Med Hyg 94:397–399. doi: 10.4269/ajtmh.15-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Classen DC, Morningstar JM, Shanley JD. 1987. Detection of antibody to murine cytomegalovirus by enzyme-linked immunosorbent and indirect immunofluorescence assays. J Clin Microbiol 25:600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demonstration of minimal interchip variation and high signal reproducibility between two separate experiments (A and B). One DENV convalescent-phase, 1 ZIKV acute-phase, 1 ZIKV early-convalescent-phase, and 1 control serum were tested at a 1:100 dilution. Control sera were collected from individuals in the greater New York City metropolitan area who tested negative for Lyme disease and who had no known history of flavivirus or alphavirus infection or vaccination. Download FIG S1, TIF file, 7.8 MB (8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Amino acid alignment showing residues in the 20-aa NS2B ZIKV peptide homologous with other flaviviruses. Download FIG S2, TIF file, 7.8 MB (8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.