Abstract
Vaccines are public health interventions aimed at preventing infections-related mortality, morbidity, and disability. While vaccines have been successfully designed for those infectious diseases preventable by preexisting neutralizing specific antibodies, for other communicable diseases, additional immunological mechanisms should be elicited to achieve a full protection. “New vaccines” are particularly urgent in the nowadays society, in which economic growth, globalization, and immigration are leading to the emergence/reemergence of old and new infectious agents at the animal–human interface. Conventional vaccinology (the so-called “vaccinology 1.0”) was officially born in 1796 thanks to the contribution of Edward Jenner. Entering the twenty-first century, vaccinology has shifted from a classical discipline in which serendipity and the Pasteurian principle of the three Is (isolate, inactivate, and inject) played a major role to a science, characterized by a rational design and plan (“vaccinology 3.0”). This shift has been possible thanks to Big Data, characterized by different dimensions, such as high volume, velocity, and variety of data. Big Data sources include new cutting-edge, high-throughput technologies, electronic registries, social media, and social networks, among others. The current mini-review aims at exploring the potential roles as well as pitfalls and challenges of Big Data in shaping the future vaccinology, moving toward a tailored and personalized vaccine design and administration.
Keywords: vaccine, Big Data, omics disciplines, Web 2.0, eHealth, history of vaccinology
Introduction: From the Classical 3Is “Isolate–Inactivate–Inject” Vaccinology 1.0 to Vaccinology 3.0, Vaccinomics, and Beyond
Vaccines are public health interventions aimed at preventing infections-related mortality, morbidity, and disability. As such, they represent a milestone of hygiene and preventive medicine (1). Since their implementation, they have managed to bring several health and economic benefits, both in developed and developing countries, significantly reducing the burden generated by infectious diseases (2). They have contributed to the eradication of smallpox and to the control of others infectious agents, such as polio. According to the estimates of the Global Alliance for Vaccines and Immunization (Gavi), they have contributed to avert up to 23.3 million projected deaths from 2011 to 2020, especially in Africa, Southeast Asia, and in the Eastern Mediterranean (3). Furthermore, they positively impact on perceived quality of life (3) and reduce inequity worldwide (1, 4).
While vaccines have been successfully designed for those infectious diseases preventable by preexisting neutralizing specific antibodies, for other communicable diseases, additional immunological mechanisms should be elicited to achieve a full protection. These additional mechanisms include the stimulation of effector and memory T lymphocytes, besides the release of antibodies by helper T cells-induced B cells (5). A better understanding of immune networks, their sophisticated tuning, and interactions is, as such, fundamental, in those vaccines against HIV/AIDS, malaria or tuberculosis, eluding classical vaccine development, which require new strategies and approaches (6).
“New vaccines” are particularly urgent in the nowadays society, in which economic growth, globalization, and immigration are leading to the emergence/reemergence of old and new infectious agents at the animal–human interface (7, 8).
Conventional vaccinology (the so-called “vaccinology 1.0”) was officially born in 1796 thanks to the contribution of Edward Jenner (1749–1823) and the pioneering discoveries of the New England Puritan minister Cotton Mather (1663–1728), and Lady Mary Wortley Montague (1689–1762), partially anticipated by Chinese and Indians different centuries before. The vaccine typical of vaccinology 1.0 is given by the rabies vaccine, the first human vaccine manufactured in 1885 in the laboratory (9). Other “first generation” vaccines are bacillus Calmette–Guérin (BCG), plague, pertussis, polio, and smallpox vaccines (9).
Entering the twenty-first century, vaccinology has shifted from a discipline in which serendipity and the Pasteurian principle of the three Is (isolate, inactivate, and inject) played a major role to a science, characterized by a rational design and plan (10).
If vaccinology 1.0 mainly consisted in isolating infectious agents, cultivating and inactivating them (as a whole or partially), and injecting the obtained product, vaccinology 2.0 utilizes purified microbial cell components. Example of “second generation” vaccines includes vaccines against tetanus, diphtheria, anthrax, pneumonia, influenza, hepatitis B, and Lyme disease (9). The transition from vaccinology 1.0 to vaccinology 2.0 has been made possible by several technological advancements, including genetic and protein engineering, recombinant DNA (11), polysaccharide and carbohydrate chemistry, combinatorial chemistry (12), among others.
Vaccinology 3.0 starts from the microbial genomic sequences (reverse vaccinology 1.0) or from the repertoire of protective human antibodies (reverse vaccinology 2.0) (13, 14). This shift has been possible thanks to omics data, which represent one type of Big Data, characterized by different aspects, such as enormous volume, velocity, and high variety of data (15).
High-throughput technologies-enabled omics disciplines [such as genomics and post-genomics specialties (16, 17), including transcriptomics, proteomics, metabolomics, cytomics, immunomics, secretomics, surfomics, or interactomics], briefly overviewed in Table 1, are able to produce a wealth of data and information, at a large-scale. Recently, these approaches have converged in what is termed vaccinomics, that is, to say the performance of large-scale, hypothesis-free, data-driven and holistic investigations. Poland and collaborators have defined vaccinomics as the “integration of immunogenetics and immunogenomics with systems biology and immune profiling” (18).
Table 1.
The different genomic/post-genomic specialties and their potential role in the field of vaccinology.
| Genomic/post-genomic specialty potentially relevant in vaccinology | Definition |
|---|---|
| Genomics | Systematic, genome-wide investigation of genes |
| Proteomics | Systematic, proteome-wide investigation of proteins |
| Transcriptomics | Systematic, transcriptome-wide investigation of gene transcription |
| Metabolomics | Systematic, metabolome-wide investigation of metabolites |
| Cytomics | Systematic, cytome-wide investigation of biochemical/biophysical events at a single cell level |
| Immunogenomics | Systematic, immunogenome-wide investigation of immunologically relevant genes |
| Immunoproteomics | Systematic, immunoproteome-wide investigation of immunologically relevant proteins |
| Immunometabolomics | Systematic, immunometabolome-wide investigation of immunologically relevant metabolites |
| Interactomics | Systematic, interactome-wide investigation of interactions among proteins and/or other cellular molecules/components |
| Secretomics | Systematic, secretome-wide investigation of all secreted proteins of a given cell/tissue/organism |
| Exoproteomics | Systematic, exoproteome-wide investigation of proteins in the extra-cellular proximity of a biological system |
| Surfomics | Systematic, surfome-wide investigation of surface proteins and other components, such as surface-exposed moieties |
| Immunomics | Systematic, immunome-wide investigation of immune system dynamics, regulation and response to a given pathogen |
| Protectomics | Systematic, protectome-wide investigation of the structural/functional protein motifs that confer immunological protection |
| Adversomics | Systematic, adversome-wide investigation of potential vaccine-related adverse events |
| Vaccinomics | Systematic, comprehensive integration of previously described omics disciplines for advancing vaccine discovery and development, as well as personalized vaccinology |
New cutting-edge technologies include next-generation sequencing (NGS) techniques [RNASeq (19) and large-scale B- and T-cell receptor sequencing (20, 21)], mass cytometry (CyTOF) (22), and peptide/protein arrays (23). Data produced by molecular biology and NGS as well as by bioinformatics (24) can be used to perform mechanistic reductionist studies but can be also exploited to comprehensively capture immune dynamics and interactions (25), carrying out, for instance, network analysis or systems biology (the so-called “systems vaccinology”). Novel bioinformatics tools and new approaches are needed to better integrate the enormous wealth of data originated from omics experiments, making the shift from single-omics to multi-omics possible.
Furthermore, the actual era is characterized by the widespread diffusion of the new information and communication technologies (26): electronic health or eHealth refers to their exploitation as “a means to expand, to assist, or to enhance human activities, rather than as a substitute for them” (27). As omics experiments, eHealth generates as well an enormous wealth of data. Researchers have found that, usually, digital activities correlate with offline behaviors and other variables, such as vaccination knowledge and perception of own risk: for example, Betsch and Wicker (28), investigating a sample of 310 medical students found that explicitly surfing the Internet for vaccination risks-related websites led to fewer public health websites than generically searching for immunization practices.
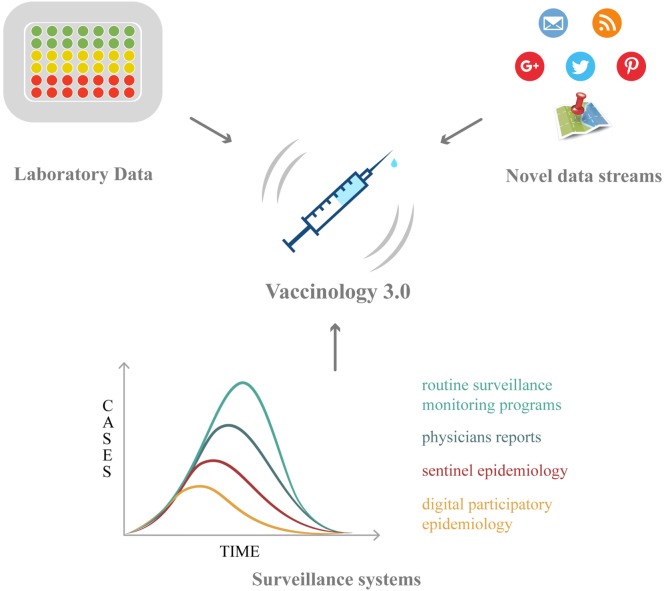
Vaccinology has now entered a new phase, characterized by new challenges: within this new framework, Big Data hold promises and opportunities, which will be overviewed in the following paragraphs (Table 2; Figure 1).
Table 2.
Potential applications of Big Data in the different subfields of vaccinology.
| Subfield of vaccinology | Examples of applications |
|---|---|
| Vaccine discovery and development | Structural/functional vaccinologySystems vaccinology Vaccine informatics/bioinformaticsIn silico/computational vaccinology Vaccine ontology Reverse vaccinology Vaccinomics/immunomics |
| Vaccine production and safety | Monitors and sensors |
| Vaccine campaigns | Evidence-based prevention and evidence-based vaccinology Immunization registry/information system Personalized vaccinology |
| Vaccine efficacy and effectiveness | Vaccine trials Vaccine ontology Digital epidemiology/infodemiology and infoveillance |
| Vaccine side effects | Vaccine adverse event reporting system (VAERS) Vaccine adverse event ontology Adversomics Digital epidemiology/infodemiology and infoveillance |
| Vaccine literacy/vaccine hesitancy | Digital epidemiology/infodemiology and infoveillance |
Figure 1.
The roles of Big Data in vaccination: from vaccine discovery and development (omics technologies and wet-lab approaches) to vaccination campaign and vaccine safety monitoring (electronic registries, social media/networks, and digital epidemiology).
Vaccine Discovery and Design: The Role of Big Data
Computational vaccinology (29, 30) and immunoinformatics (31), utilizing algorithms, enable experimental immunology to save time, focusing only on prescreened vaccine candidate antigens and, thus, avoiding cost, time-consuming, and labor intensive steps.
Different in silico tools exist, to aid and assist researchers in vaccine discovery and design (32, 33). Databases of vaccine candidates, such as MalVac (34), or Vaxar (35), software tools such as Vaxjo (36), VIOLIN (37–39), NERVE (40), Vaxign (41), Vacceed (42), Jenner-predict server (43), EpiToolKit (44), iVax (45), or VaxiJen (46), have been specifically implemented for vaccinology to enable prediction of vaccine antigens or adjuvants.
A successful example of rationally designed web-based vaccine is the vaccine against Neisseria meningitidis, commercially available with the trade name of Bexsero. For the selection of surface antigens, Masignani and collaborators (47) performed genome mining, using computational tools and algorithms, such as PSORT (48), PSI-BLAST (49), and FindPatterns to predict proteins with transmembrane domains, leader peptides, lipo-boxes and outer membrane anchoring motifs. At the end, 570 proteins were selected and GNA1870, a new surface-exposed lipoprotein inducing high levels of bactericidal antibodies, was discovered.
Reverse vaccinology technique is being applied also to other microorganisms, including Leptospira (50, 51), Streptococcus pneumoniae (52, 53), malaria (54), Schistosoma (55), Echinococcus granulosus (56), Rickettsia prowazekii (57), Mycobacterium (58), Acinetobacter baumannii (59), Escherichia coli (60), Staphylococcus aureus (61, 62), Corynebacterium pseudotuberculosis (63), Herpes simplex (64), Vibrio cholerae (65), and Cryptosporidium (65, 66), among others.
In the field of veterinary vaccinology, the reverse vaccinology approach is being applied, for instance, to organisms like cattle neosporosis (67, 68), Rhipicephalus microplus (69–71), Ehrlichia ruminantium (72, 73), and bovine herpesvirus 4 (74).
These computational approaches, using massive data mining techniques, rely on brute force (the so-called “test-all-to-lose-nothing” approach). Altindis and collaborators (75) have recently attempted to refine this framework, based on the idea that protective antigens share specific structural/functional features, termed as “protective signatures” or “immunosignatures,” differing from other pathogen components, in terms of immunological properties. Instead of focusing on protein localization, as in previous investigations, Altindis and coworkers concentrated their computational analyses on protein biological role and function. In this sense, their approach, termed as “protectome,” is protein localization unbiased, in that it leads to the identification of surface-exposed and secreted or cytoplasmic protective antigens.
Big Data and Vaccine Production and Delivery
After production, to properly preserve, store, handle, ship, and deliver vaccine supplies, it is fundamental to maintain cold chain from the manufacturer to the point of use, keeping temperatures within a precise range of values, and avoiding temperature excursions or fluctuations. Vaccines need, indeed, to be stored within a safe zone, namely, between 2 and 8°C (76–78), otherwise their quality is compromised, and their potency cannot be restored. According to the World Health Organization (WHO) and the United Nations Children’s Fund, approximately two percent of health-care facilities in low- and middle-income countries are equipped with proper functional technology for maintaining cold chain.
To solve these issues, Merck and Microsoft have, for example, established a collaboration, in which Merck exploits Microsoft R Server for Hadoop for analyzing, monitoring, and predicting variables that could affect the cold chain, including origin, destination, and delivery route as well as external weather and logistics providers, utilizing special thermal-protection containers equipped with temperature-recording sensors and temperature-sensitive vaccine vial monitors.
Nexleaf has produced ColdTrace (currently, ColdTrace version 5), which has already been implemented in more than 7,000 health-care facilities worldwide, and has recently established a new partnership with www.Google.org and Gavi.
The benefits provided by these technologies are the fact they are low-cost and particularly useful in developing countries, which often rely on stem thermometers or 30-day temperate loggers.
Big Data and Vaccine Campaigns
Other major sources of Big Data are immunization registries and surveillance systems such as SmiNet-2 (79), or SurvNet@RKI (80). These enormous databases are precious databanks, which can be mined to capture data concerning vaccination coverage rate and its determinants.
Non-conventional data sources or novel data streams, such as Internet search data and tools monitoring web queries, like Google Trends (GT) (81), social media (YouTube, Facebook, Google Plus, Twitter, Pinterest, Instagram, and so on), or news source scraping like HealthMap (82), provide researchers and public health workers with real-time information concerning public reaction to epidemic outbreaks. Novel data streams can track different vaccine-preventable infectious diseases, such as influenza (83–85), pertussis (86, 87), or measles (88), among others. As such, they can be exploited to predict epidemiological figures as well as monitor the effect of vaccine campaigns.
Big Data and Vaccine Efficacy/Effectiveness
Big Data enable also to individuate molecular signatures and predictors of the outcomes of vaccination, being correlates of vaccine efficacy/effectiveness in different populations (89). Haks and collaborators (89), for instance, utilized transcriptomics to quantitatively assess the immunogenetic signature of immunization response. Dunachie and coworkers (90) explored the differentially expressed genes induced by a malaria candidate vaccine and found that most genes conferring immunological protection belonged to the interferon-gamma and to the proteasome/antigen presentation pathways, differently from genes associated with hemopoietic stem cells, regulatory monocytes, and the myeloid lineage modules.
Novel data streams, such as mobile/smartphone applications, can be utilized in the monitoring and management of vaccine-related data (91).
Big Data and Vaccine Side Effects
Vaccine adverse events and reactions are very rare. As such, most studies are statistically underpowered to capture the rate of rare/very rare side effects. Meta-analytical approaches and data mining have emerged as useful strategies with this regard. As claimed by Chandler (92), the classical paradigm of the actual pharmacovigilance/vaccine vigilance system based on three stage-approach (namely, signal detection, development of a causality hypothesis, and testing of the causality hypothesis) is plagued by some limitations, in that “routine vaccine pharmacovigilance practice is not sufficient to understand suspected harms that are poorly defined and whose pathophysiology are not completely understood. Furthermore, estimations of risk at the population level fail to acknowledge that vaccines may cause harm in subgroups with individual-level risk factors” for adverse events following immunization. As such new approaches are needed to capture new side effects and, also in this case, Big Data could play a major role.
“Adversomics” is a term coined by Poland in 2009 and is an emerging discipline defined as “the study of vaccine adverse reactions using immunogenomics and systems biology approaches” (93, 94).
Berendsen and coworkers (95) exploited Big Data, to explore BCG-related “non-specific effects,” that is to say effects induced by the vaccination on health beyond its target disease. In particular, they evaluated the effect of timing of BCG on stunting in Sub-Saharan African children under 5 years, analyzing cross-sectional data for 368,450 subjects from 33 controls. Authors found that BCG vaccination did not affect stunting, with timing of BCG vaccination being statistically significant. Similar patterns could be detected for diphtheria–tetanus–pertussis and measles vaccinations.
Vaccine ontology (96, 97), a class of biomedical ontologies, that is to say a consensus-based computer and human interpretable set of terms and relations indicating specific biomedical entities, is another valuable approach. It enables support integrative adverse events-related data collection and analysis, utilizing a normalization strategy more effective than other controlled terminologies. These include the Medical Dictionary for Regulatory Activities, the Common Terminology Criteria for Adverse Events, and the WHO Adverse Reactions Terminology, among others. Using Ontology-Based Vaccine Adverse Event representation, Xie and He (96) explored the adverse events related to Flublok, a recombinant hemagglutinin influenza vaccine.
Novel data streams can be used to see how often people Google for vaccination and for vaccination-related adverse events. Bragazzi and collaborators (98) utilized GT for monitoring the interest toward preventable infections and related vaccines. Authors found that, generally speaking, vaccine was not a popular topic, with the valuable exception of the vaccine against Human Papillomavirus, with vaccines-related queries being approximately one third of the volumes regarding preventable infections. Users tended to search information about possible vaccine-related side effects.
Big Data and Vaccine Literacy/Vaccine Hesitancy
Big Data enable to track and monitor interest toward vaccination practices (99). The increasing phenomenon of vaccine hesitancy (an umbrella term that includes indecision, uncertainty, delay and reluctance) is multifactorial, and closely linked to social contexts, with different determinants, ranging from geographical area, to political situation, complacency, convenience and confidence in vaccines. Novel data streams, providing a snapshot of perceptions of vaccination in a given place and at a specific time, could be used to assess lay-people’s perceptions of vaccination, enabling health-care workers to actively engage citizens and to plan ad hoc communication strategies and plans to contain vaccine hesitancy and to promote vaccine literacy (100).
Shah and colleagues (101) compared time series of rotavirus-related Internet searches as captured by GT with rotavirus laboratory reports from the United States and United Kingdom and with hospitalizations for acute gastroenteritis in the United States and Mexico, before and after national vaccine introductions. Authors found a strong positive correlation between web queries and laboratory reports in the United States (R2 = 0.79) and United Kingdom (R2 = 0.60) and between the Internet searches and acute gastroenteritis hospitalizations in the United States (R2 = 0.87) and Mexico (R2 = 0.69). Correlations were stronger in the prevaccine period and after vaccine introduction, the mean Internet queries decreased by 40–70% in the United States and Mexico, with a loss of seasonal variation in the United Kingdom.
Bakker and coworkers (102) exploited GT to monitor the interest toward chicken pox, over an 11-year period, from 36 countries. Authors found seasonal peaks with striking latitudinal variation in information seeking behavior. Authors concluded that novel data streams are able to track the global burden of childhood disease as well as to investigate effects of immunization at population level.
Goldlust and collaborators (103) investigated the use of large-scale medical claims data for local surveillance of under-immunization for childhood infections in the United States, developing a statistical framework for integrating disparate data sources on surveillance of vaccination behavior. In this way, authors were able to identify the determinants of vaccine hesitancy behavior. Within the “Vaccine Confidence Project,” Larson and colleagues (104) extensively analyzed data from 10,380 reports (from 144 countries) and found that 7,171 (69%) contained positive or neutral content whereas 3,209 (31%) contained negative content (related to vaccine programs and disease outbreaks, vaccine-related beliefs, awareness, and perceptions; vaccine safety; and vaccine delivery programs).
Within the ambitious “Project Tycho” (freely accessible at www.tycho.pitt.edu) launched by the University of Pittsburgh, United States (105, 106), authors have digitized all weekly surveillance reports of notifiable diseases for United States cities and states published in the period between 1888 and 2011. This data set consists of 87,950,807 reported individual cases and has been used to derive a quantitative history of disease dynamics and transmission in the United States. Pattern analysis has documented, in a statistically robust way, a significant reduction of infections-generated burden, underlining the positive effect of vaccination programs (105). This use of big data emphasizes the dimension of “veracity,” through which is possible to contrast vaccine-related “fake news” and “post-modern, post-factual truths,” disseminated by the anti-vaccination movements (107).
Conclusion: State-of-the-Art, Current Challenges, and Future Prospects
Big Data have contributed and are expected to continue contributing toward facilitating the discovery, development, production, and delivery of rationally designed vaccines. Further, enabling to identify predictive biomolecular signatures of response to vaccination, vaccination will shift from the classical “one-size-fits-all” paradigm to a personalized approach. Moreover, Big Data can be used to track the success of vaccination campaigns, in term of vaccination coverage rate, as well as the rare/very rate vaccine-related adverse events, for which “classical epidemiological studies” would be statistically underpowered.
However, a number of pitfalls and challenges should be properly recognized to be addressed by future research: Big Data and Big Data sources, as previously overviewed, are highly heterogeneous and should be effectively integrated and harmonized together. Moreover, some algorithms underlying novel data streams need to be refined in that, sometimes, do not exactly predict epidemic outbreaks (108), even though some scholars have shown that, in principle, is possible to correct them to achieve higher predictive power (109). Further, efforts should be done to preserve and protect privacy, confidentiality, and identity. The emerging field of “Big Data Ethics” is trying to address all these issues (110, 111). Currently, we are only witnessing the very beginning of the ongoing “Big Data revolution.”
Author Contributions
NB, MM, and MB conceived the study. NB, VG, MV, RR, AN, AH, MM, and MB drafted and revised the manuscript; read and approved the last version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Ms. Valeria Parisi, University of Milan, for help in preparing the figure.
References
- 1.Andre FE, Booy R, Bock HL, Clemens J, Datta SK, John TJ, et al. Vaccination greatly reduces disease, disability, death and inequity worldwide. Bull World Health Organ (2008) 86(2):140–6. 10.2471/BLT.07.040089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee LA, Franzel L, Atwell J, Datta SD, Friberg IK, Goldie SJ, et al. The estimated mortality impact of vaccinations forecast to be administered during 2011-2020 in 73 countries supported by the GAVI Alliance. Vaccine (2013) 31(Suppl 2):B61–72. 10.1016/j.vaccine.2012.11.035 [DOI] [PubMed] [Google Scholar]
- 3.Lee BY, Bartsch SM, Brown ST, Cooley P, Wheaton WD, Zimmerman RK. Quantifying the economic value and quality of life impact of earlier influenza vaccination. Med Care (2015) 53(3):218–29. 10.1097/MLR.0000000000000302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehreth J. The global value of vaccination. Vaccine (2003) 21:596–600. 10.1016/S0264-410X(02)00623-0 [DOI] [PubMed] [Google Scholar]
- 5.Hagan T, Nakaya HI, Subramaniam S, Pulendran B. Systems vaccinology: enabling rational vaccine design with systems biological approaches. Vaccine (2015) 33:5294–301. 10.1016/j.vaccine.2015.03.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rappuoli R, Aderem A. A 2020 vision for vaccines against HIV, tuberculosis and malaria. Nature (2011) 473(7348):463–9. 10.1038/nature10124 [DOI] [PubMed] [Google Scholar]
- 7.Gutiérrez AH, Spero DM, Gay C, Zimic M, De Groot AS. New vaccines needed for pathogens infecting animals and humans: One Health. Hum Vaccin Immunother (2012) 8(7):971–8. 10.4161/hv.20202 [DOI] [PubMed] [Google Scholar]
- 8.Neiderud CJ. How urbanization affects the epidemiology of emerging infectious diseases. Infect Ecol Epidemiol (2015) 5:27060. 10.3402/iee.v5.27060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhee JH. Towards vaccine 3.0: new era opened in vaccine research and industry. Clin Exp Vaccine Res (2014) 3(1):1–4. 10.7774/cevr.2014.3.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy RB, Poland GA. The top five “game changers” in vaccinology: toward rational and directed vaccine development. OMICS (2011) 15(9):533–7. 10.1089/omi.2011.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lepenies B, Yin J, Seeberger PH. Applications of synthetic carbohydrates to chemical biology. Curr Opin Chem Biol (2010) 14(3):404–11. 10.1016/j.cbpa.2010.02.016 [DOI] [PubMed] [Google Scholar]
- 12.Pardee K, Slomovic S, Nguyen PQ, Lee JW, Donghia N, Burrill D, et al. Portable, on-demand biomolecular manufacturing. Cell (2016) 167(1):248–59.e12. 10.1016/j.cell.2016.09.013 [DOI] [PubMed] [Google Scholar]
- 13.Burton DR. What are the most powerful immunogen design vaccine strategies? Reverse vaccinology 2.0 shows great promise. Cold Spring Harb Perspect Biol (2017) 9(11). 10.1101/cshperspect.a030262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rappuoli R, Bottomley MJ, D’Oro U, Finco O, De Gregorio E. Reverse vaccinology 2.0: human immunology instructs vaccine antigen design. J Exp Med (2016) 213(4):469–81. 10.1084/jem.20151960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blohmke CJ, O’Connor D, Pollard AJ. The use of systems biology and immunological big data to guide vaccine development. Genome Med (2015) 7:114. 10.1186/s13073-015-0236-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horvatić A, Kuleš J, Guillemin N, Galan A, Mrljak V, Bhide M. High-throughput proteomics and the fight against pathogens. Mol Biosyst (2016) 12(8):2373–84. 10.1039/c6mb00223d [DOI] [PubMed] [Google Scholar]
- 17.Dwivedi P, Alam SI, Tomar RS. Secretome, surfome and immunome: emerging approaches for the discovery of new vaccine candidates against bacterial infections. World J Microbiol Biotechnol (2016) 32(9):155. 10.1007/s11274-016-2107-3 [DOI] [PubMed] [Google Scholar]
- 18.Poland GA, Ovsyannikova IG, Kennedy RB, Haralambieva IH, Jacobson RM. Vaccinomics and a new paradigm for the development of preventive vaccines against viral infections. OMICS (2011) 15(9):625–36. 10.1089/omi.2011.0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Gerstein M, Snyder MRNA- Seq: a revolutionary tool for transcriptomics. Nat Rev Genet (2009) 10(1):57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yaari G, Kleinstein SH. Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Med (2015) 7:121. 10.1186/s13073-015-0243-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodsworth DJ, Castellarin M, Holt RA. Sequence analysis of T-cell repertoires in health and disease. Genome Med (2013) 5(10):98. 10.1186/gm502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeves PM, Sluder AE, Paul SR, Scholzen A, Kashiwagi S, Poznansky MC. Application and utility of mass cytometry in vaccine development. FASEB J (2018) 32(1):5–15. 10.1096/fj.201700325R [DOI] [PubMed] [Google Scholar]
- 23.Chase BA, Johnston SA, Legutki JB. Evaluation of biological sample preparation for immunosignature-based diagnostics. Clin Vaccine Immunol (2012) 19(3):352–8. 10.1128/CVI.05667-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y, Rappuoli R, De Groot AS, Chen RT. Emerging vaccine informatics. J Biomed Biotechnol (2010) 2010:218590. 10.1155/2010/218590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chattopadhyay PK, Roederer M. A mine is a terrible thing to waste: high content, single cell technologies for comprehensive immune analysis. Am J Transplant (2015) 15(5):1155–61. 10.1111/ajt.13193 [DOI] [PubMed] [Google Scholar]
- 26.Bragazzi NL. From P0 to P6 medicine, a model of highly participatory, narrative, interactive, and “augmented” medicine: some considerations on Salvatore Iaconesi’s clinical story. Patient Prefer Adherence (2013) 7:353–9. 10.2147/PPA.S38578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh H, Rizo C, Enkin M, Jadad A. What is eHealth (3): a systematic review of published definitions. J Med Internet Res (2005) 7(1):e1. 10.2196/jmir.7.1.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betsch C, Wicker S. E-health use, vaccination knowledge and perception of own risk: drivers of vaccination uptake in medical students. Vaccine (2012) 30(6):1143–8. 10.1016/j.vaccine.2011.12.021 [DOI] [PubMed] [Google Scholar]
- 29.Khalili S, Jahangiri A, Borna H, Ahmadi Zanoos K, Amani J. Computational vaccinology and epitope vaccine design by immunoinformatics. Acta Microbiol Immunol Hung (2014) 61(3):285–307. 10.1556/AMicr.61.2014.3.4 [DOI] [PubMed] [Google Scholar]
- 30.Söllner J, Heinzel A, Summer G, Fechete R, Stipkovits L, Szathmary S, et al. Concept and application of a computational vaccinology workflow. Immunome Res (2010) 6(Suppl 2):S7. 10.1186/1745-7580-6-S2-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegde NR, Gauthami S, Sampath Kumar HM, Bayry J. The use of databases, data mining and immunoinformatics in vaccinology: where are we? Expert Opin Drug Discov (2018) 13(2):117–30. 10.1080/17460441.2018.1413088 [DOI] [PubMed] [Google Scholar]
- 32.He Y, Xiang Z. Databases and in silico tools for vaccine design. Methods Mol Biol (2013) 993:115–27. 10.1007/978-1-62703-342-8_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y. Vaccine adjuvant informatics: from data integration and analysis to rational vaccine adjuvant design. Front Immunol (2014) 5:32 10.3389/fimmu.2014.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaudhuri R, Ahmed S, Ansari FA, Singh HV, Ramachandran S. MalVac: database of malarial vaccine candidates. Malar J (2008) 7:184. 10.1186/1475-2875-7-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todd T, Dunn N, Xiang Z, He Y. Vaxar: a web-based database of laboratory animal responses to vaccinations and its application in the meta-analysis of different animal responses to tuberculosis vaccinations. Comp Med (2016) 66(2):119–28. [PMC free article] [PubMed] [Google Scholar]
- 36.Sayers S, Ulysse G, Xiang Z, He Y. Vaxjo: a web-based vaccine adjuvant database and its application for analysis of vaccine adjuvants and their uses in vaccine development. J Biomed Biotechnol (2012) 2012:831486. 10.1155/2012/831486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang Z, Todd T, Ku KP, Kovacic BL, Larson CB, Chen F, et al. VIOLIN: vaccine investigation and online information network. Nucleic Acids Res (2008) 36(Database issue):D923–8. 10.1093/nar/gkm1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y, Xiang Z. Bioinformatics analysis of Brucella vaccines and vaccine targets using VIOLIN. Immunome Res (2010) 6(Suppl 1):S5. 10.1186/1745-7580-6-S1-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He Y, Racz R, Sayers S, Lin Y, Todd T, Hur J, et al. Updates on the web-based VIOLIN vaccine database and analysis system. Nucleic Acids Res (2014) 42(Database issue):D1124–32. 10.1093/nar/gkt1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vivona S, Bernante F, Filippini F. NERVE: new enhanced reverse vaccinology environment. BMC Biotechnol (2006) 6:35 10.1186/1472-6750-6-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y, Xiang Z, Mobley HL. Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J Biomed Biotechnol (2010) 2010:297505. 10.1155/2010/297505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodswen SJ, Kennedy PJ, Ellis JT. Vacceed: a high-throughput in silico vaccine candidate discovery pipeline for eukaryotic pathogens based on reverse vaccinology. Bioinformatics (2014) 30(16):2381–3. 10.1093/bioinformatics/btu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaiswal V, Chanumolu SK, Gupta A, Chauhan RS, Rout C. Jenner-predict server: prediction of protein vaccine candidates (PVCs) in bacteria based on host-pathogen interactions. BMC Bioinformatics (2013) 14:211. 10.1186/1471-2105-14-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert B, Brachvogel HP, Jürges C, Kohlbacher O. EpiToolKit – a web-based workbench for vaccine design. Bioinformatics (2015) 31(13):2211–3. 10.1093/bioinformatics/btv116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moise L, Gutierrez A, Kibria F, Martin R, Tassone R, Liu R, et al. iVAX: an integrated toolkit for the selection and optimization of antigens and the design of epitope-driven vaccines. Hum Vaccin Immunother (2015) 11(9):2312–21. 10.1080/21645515.2015.1061159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics (2007) 8:4. 10.1186/1471-2105-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masignani V, Comanducci M, Giuliani MM, Bambini S, Adu-Bobie J, Arico B, et al. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med (2003) 197(6):789–99. 10.1084/jem.20021911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins (1991) 11(2):95–110. 10.1002/prot.340110203 [DOI] [PubMed] [Google Scholar]
- 49.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res (1997) 25(17):3389–402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang HL, Zhu YZ, Qin JH, He P, Jiang XC, Zhao GP, et al. In silico and microarray-based genomic approaches to identifying potential vaccine candidates against Leptospira interrogans. BMC Genomics (2006) 7:293. 10.1186/1471-2164-7-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grassmann AA, Kremer FS, Dos Santos JC, Souza JD, Pinto LDS, McBride AJA. Discovery of novel leptospirosis vaccine candidates using reverse and structural vaccinology. Front Immunol (2017) 8:463. 10.3389/fimmu.2017.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barocchi MA, Censini S, Rappuoli R. Vaccines in the era of genomics: the pneumococcal challenge. Vaccine (2007) 25(16):2963–73. 10.1016/j.vaccine.2007.01.065 [DOI] [PubMed] [Google Scholar]
- 53.Talukdar S, Zutshi S, Prashanth KS, Saikia KK, Kumar P. Identification of potential vaccine candidates against Streptococcus pneumoniae by reverse vaccinology approach. Appl Biochem Biotechnol (2014) 172(6):3026–41. 10.1007/s12010-014-0749-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuju J, Kamuyu G, Murungi LM, Osier FHA. Vaccine candidate discovery for the next generation of malaria vaccines. Immunology (2017) 152(2):195–206. 10.1111/imm.12780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merrifield M, Hotez PJ, Beaumier CM, Gillespie P, Strych U, Hayward T, et al. Advancing a vaccine to prevent human schistosomiasis. Vaccine (2016) 34(26):2988–91. 10.1016/j.vaccine.2016.03.079 [DOI] [PubMed] [Google Scholar]
- 56.Gan W, Zhao G, Xu H, Wu W, Du W, Huang J, et al. Reverse vaccinology approach identify an Echinococcus granulosus tegumental membrane protein enolase as vaccine candidate. Parasitol Res (2010) 106(4):873–82. 10.1007/s00436-010-1729-x [DOI] [PubMed] [Google Scholar]
- 57.Caro-Gomez E, Gazi M, Goez Y, Valbuena G. Discovery of novel cross-protective Rickettsia prowazekii T-cell antigens using a combined reverse vaccinology and in vivo screening approach. Vaccine (2014) 32(39):4968–76. 10.1016/j.vaccine.2014.06.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pandey K, Sharma M, Saarav I, Singh S, Dutta P, Bhardwaj A, et al. Analysis of the DosR regulon genes to select cytotoxic T lymphocyte epitope specific vaccine candidates using a reverse vaccinology approach. Int J Mycobacteriol (2016) 5(1):34–43. 10.1016/j.ijmyco.2015.10.005 [DOI] [PubMed] [Google Scholar]
- 59.Hassan A, Naz A, Obaid A, Paracha RZ, Naz K, Awan FM, et al. Pangenome and immuno-proteomics analysis of Acinetobacter baumannii strains revealed the core peptide vaccine targets. BMC Genomics (2016) 17(1):732. 10.1186/s12864-016-2951-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moriel DG, Tan L, Goh KG, Phan MD, Ipe DS, Lo AW, et al. Novel protective vaccine antigen from the core Escherichia coli genome. mSphere (2016) 1(6):1–13. 10.1128/mSphere.00326-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holtfreter S, Kolata J, Stentzel S, Bauerfeind S, Schmidt F, Sundaramoorthy N, et al. Omics approaches for the study of adaptive immunity to Staphylococcus aureus and the selection of vaccine candidates. Proteomes (2016) 4(1):1–25. 10.3390/proteomes4010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oprea M, Antohe F. Reverse-vaccinology strategy for designing T-cell epitope candidates for Staphylococcus aureus endocarditis vaccine. Biologicals (2013) 41(3):148–53. 10.1016/j.biologicals.2013.03.001 [DOI] [PubMed] [Google Scholar]
- 63.Soares SC, Trost E, Ramos RT, Carneiro AR, Santos AR, Pinto AC, et al. Genome sequence of Corynebacterium pseudotuberculosis biovar equi strain 258 and prediction of antigenic targets to improve biotechnological vaccine production. J Biotechnol (2013) 167(2):135–41. 10.1016/j.jbiotec.2012.11.003 [DOI] [PubMed] [Google Scholar]
- 64.Xiang Z, He Y. Genome-wide prediction of vaccine targets for human herpes simplex viruses using Vaxign reverse vaccinology. BMC Bioinformatics (2013) 14(Suppl 4):S2. 10.1186/1471-2105-14-S4-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barh D, Barve N, Gupta K, Chandra S, Jain N, Tiwari S, et al. Exoproteome and secretome derived broad spectrum novel drug and vaccine candidates in Vibrio cholerae targeted by Piper betel derived compounds. PLoS One (2013) 8(1):e52773. 10.1371/journal.pone.0052773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ifeonu OO, Simon R, Tennant SM, Sheoran AS, Daly MC, Felix V, et al. Cryptosporidium hominis gene catalog: a resource for the selection of novel Cryptosporidium vaccine candidates. Database (Oxford) (2016) 2016:1–13. 10.1093/database/baw137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodswen SJ, Kennedy PJ, Ellis JT. Discovering a vaccine against neosporosis using computers: is it feasible? Trends Parasitol (2014) 30(8):401–11. 10.1016/j.pt.2014.06.004 [DOI] [PubMed] [Google Scholar]
- 68.Goodswen SJ, Kennedy PJ, Ellis JT. On the application of reverse vaccinology to parasitic diseases: a perspective on feature selection and ranking of vaccine candidates. Int J Parasitol (2017) 47(12):779–90. 10.1016/j.ijpara.2017.08.004 [DOI] [PubMed] [Google Scholar]
- 69.Andreotti R, Giachetto PF, Cunha RC. Advances in tick vaccinology in Brazil: from gene expression to immunoprotection. Front Biosci (Schol Ed) (2018) 10:127–42. 10.2741/s504 [DOI] [PubMed] [Google Scholar]
- 70.Aguirre Ade A, Lobo FP, Cunha RC, Garcia MV, Andreotti R. Design of the ATAQ peptide and its evaluation as an immunogen to develop a Rhipicephalus vaccine. Vet Parasitol (2016) 221:30–8. 10.1016/j.vetpar.2016.02.032 [DOI] [PubMed] [Google Scholar]
- 71.Maritz-Olivier C, van Zyl W, Stutzer C. A systematic, functional genomics, and reverse vaccinology approach to the identification of vaccine candidates in the cattle tick, Rhipicephalus microplus. Ticks Tick Borne Dis (2012) 3(3):179–87. 10.1016/j.ttbdis.2012.01.003 [DOI] [PubMed] [Google Scholar]
- 72.Liebenberg J, Pretorius A, Faber FE, Collins NE, Allsopp BA, van Kleef M. Identification of Ehrlichia ruminantium proteins that activate cellular immune responses using a reverse vaccinology strategy. Vet Immunol Immunopathol (2012) 145(1–2):340–9. 10.1016/j.vetimm.2011.12.003 [DOI] [PubMed] [Google Scholar]
- 73.Sebatjane SI, Pretorius A, Liebenberg J, Steyn H, Van Kleef M. In vitro and in vivo evaluation of five low molecular weight proteins of Ehrlichia ruminantium as potential vaccine components. Vet Immunol Immunopathol (2010) 137(3–4):217–25. 10.1016/j.vetimm.2010.05.011 [DOI] [PubMed] [Google Scholar]
- 74.Palmeira L, Machiels B, Lété C, Vanderplasschen A, Gillet L. Sequencing of bovine herpesvirus 4 v.test strain reveals important genome features. Virol J (2011) 8:406. 10.1186/1743-422X-8-406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Altindis E, Cozzi R, Di Palo B, Necchi F, Mishra RP, Fontana MR, et al. Protectome analysis: a new selective bioinformatics tool for bacterial vaccine candidate discovery. Mol Cell Proteomics (2015) 14(2):418–29. 10.1074/mcp.M114.039362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lloyd J, Cheyne J. The origins of the vaccine cold chain and a glimpse of the future. Vaccine (2017) 35(17):2115–20. 10.1016/j.vaccine.2016.11.097 [DOI] [PubMed] [Google Scholar]
- 77.Vangroenweghe F. Good vaccination practice: it all starts with a good vaccine storage temperature. Porcine Health Manag (2017) 3:24. 10.1186/s40813-017-0071-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hatchett R. The medicines refrigerator and the importance of the cold chain in the safe storage of medicines. Nurs Stand (2017) 32(6):53–63. 10.7748/ns.2017.e10960 [DOI] [PubMed] [Google Scholar]
- 79.Rolfhamre P, Jansson A, Arneborn M, Ekdahl K. SmiNet-2: description of an internet-based surveillance system for communicable diseases in Sweden. Euro Surveill (2006) 11(5):103–7. 10.2807/esm.11.05.00626-en [DOI] [PubMed] [Google Scholar]
- 80.Faensen D, Claus H, Benzler J, Ammon A, Pfoch T, Breuer T, et al. SurvNet@RKI – a multistate electronic reporting system for communicable diseases. Euro Surveill (2006) 11(4):100–3. 10.2807/esm.11.04.00614-en [DOI] [PubMed] [Google Scholar]
- 81.Nuti SV, Wayda B, Ranasinghe I, Wang S, Dreyer RP, Chen SI, et al. The use of Google Trends in health care research: a systematic review. PLoS One (2014) 9(10):e109583. 10.1371/journal.pone.0109583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Althouse BM, Scarpino SV, Meyers LA, Ayers JW, Bargsten M, Baumbach J, et al. Enhancing disease surveillance with novel data streams: challenges and opportunities. EPJ Data Sci (2015) 4:17. 10.1140/epjds/s13688-015-0054-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seo DW, Shin SY. Methods using social media and search queries to predict infectious disease outbreaks. Healthc Inform Res (2017) 23(4):343–8. 10.4258/hir.2017.23.4.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Samaras L, García-Barriocanal E, Sicilia MA. Syndromic surveillance models using web data: the case of influenza in Greece and Italy using Google Trends. JMIR Public Health Surveill (2017) 3(4):e90. 10.2196/publichealth.8015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang S, Santillana M, Kou SC. Accurate estimation of influenza epidemics using Google search data via ARGO. Proc Natl Acad Sci U S A (2015) 112(47):14473–8. 10.1073/pnas.1515373112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y, Milinovich G, Xu Z, Bambrick H, Mengersen K, Tong S, et al. monitoring pertussis infections using Internet search queries. Sci Rep (2017) 7(1):10437. 10.1038/s41598-017-11195-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pollett S, Wood N, Boscardin WJ, Bengtsson H, Schwarcz S, Harriman K, et al. Validating the use of Google Trends to enhance pertussis surveillance in California. PLoS Curr (2015) 7. 10.1371/currents.outbreaks.7119696b3e7523faa4543faac87c56c2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Warren KE, Wen LS. Measles, social media and surveillance in Baltimore City. J Public Health (Oxf) (2017) 39(3):e73–8. 10.1093/pubmed/fdw076 [DOI] [PubMed] [Google Scholar]
- 89.Haks MC, Bottazzi B, Cecchinato V, De Gregorio C, Del Giudice G, Kaufmann SHE, et al. Molecular signatures of immunity and immunogenicity in infection and vaccination. Front Immunol (2017) 8:1563. 10.3389/fimmu.2017.01563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dunachie S, Berthoud T, Hill AV, Fletcher HA. Transcriptional changes induced by candidate malaria vaccines and correlation with protection against malaria in a human challenge model. Vaccine (2015) 33(40):5321–31. 10.1016/j.vaccine.2015.07.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Atkinson KM, El-Khatib Z, Barnum G, Bell C, Turcotte MC, Murphy MSQ, et al. Using mobile apps to communicate vaccination records: a city-wide evaluation with a national immunization app, maternal child registry and public health authorities. Healthc Q (2017) 20(3):41–6. 10.12927/hcq.2017.25289 [DOI] [PubMed] [Google Scholar]
- 92.Chandler RE. Safety concerns with HPV vaccines continue to linger: are current vaccine pharmacovigilance practices sufficient? Drug Safety (2017) 40(12):1167–70. 10.1007/s40264-017-0610-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Poland GA, Ovsyannikova IG, Jacobson RM. Adversomics: the emerging field of vaccine adverse event immunogenetics. Pediatr Infect Dis J (2009) 28(5):431–2. 10.1097/INF.0b013e3181a6a511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Whitaker JA, Ovsyannikova IG, Poland GA. Adversomics: a new paradigm for vaccine safety and design. Expert Rev Vaccines (2015) 14(7):935–47. 10.1586/14760584.2015.1038249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Berendsen MLT, Smits J, Netea MG, van der Ven A. Non-specific effects of vaccines and stunting: timing may be essential. EBioMedicine (2016) 8:341–8. 10.1016/j.ebiom.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xie J, He Y. Ontology-based vaccine adverse event representation and analysis. Adv Exp Med Biol (2017) 1028:89–103. 10.1007/978-981-10-6041-0_6 [DOI] [PubMed] [Google Scholar]
- 97.He Y, Ong E, Xie J. Integrative representations and analyses of vaccine-induced intended protective immunity and unintended adverse events using ontology-based and theory-guided approaches. Glob Vaccines Immunol (2016) 1(2):37–9. 10.15761/GVI.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bragazzi NL, Barberis I, Rosselli R, Gianfredi V, Nucci D, Moretti M, et al. How often people Google for vaccination: qualitative and quantitative insights from a systematic search of the web-based activities using Google Trends. Hum Vaccin Immunother (2017) 13(2):464–9. 10.1080/21645515.2017.1264742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Amicizia D, Domnich A, Gasparini R, Bragazzi NL, Lai PL, Panatto D. An overview of current and potential use of information and communication technologies for immunization promotion among adolescents. Hum Vaccin Immunother (2013) 9(12):2634–42. 10.4161/hv.26010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rosselli R, Martini M, Bragazzi NL. The old and the new: vaccine hesitancy in the era of the Web 2.0. Challenges and opportunities. J Prev Med Hyg (2016) 57(1):E47–50. [PMC free article] [PubMed] [Google Scholar]
- 101.Shah MP, Lopman BA, Tate JE, Harris J, Esparza-Aguilar M, Sanchez-Uribe E, et al. Use of internet search data to monitor rotavirus vaccine impact in the United States, United Kingdom, and Mexico. J Pediatric Infect Dis Soc (2018) 7(1):56–63. 10.1093/jpids/pix004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bakker KM, Martinez-Bakker ME, Helm B, Stevenson TJ. Digital epidemiology reveals global childhood disease seasonality and the effects of immunization. Proc Natl Acad Sci U S A (2016) 113(24):6689–94. 10.1073/pnas.1523941113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goldlust S, Lee E, Bansal S. Assessing the distribution and drivers of vaccine hesitancy using medical claims data. Online J Public Health Inform (2017) 9(1):e012 10.5210/ojphi.v9i1.7590 [DOI] [Google Scholar]
- 104.Larson HJ, Smith DM, Paterson P, Cumming M, Eckersberger E, Freifeld CC, et al. Measuring vaccine confidence: analysis of data obtained by a media surveillance system used to analyse public concerns about vaccines. Lancet Infect Dis (2013) 13(7):606–13. 10.1016/S1473-3099(13)70108-7 [DOI] [PubMed] [Google Scholar]
- 105.van Panhuis WG, Grefenstette J, Jung SY, Chok NS, Cross A, Eng H, et al. Contagious diseases in the United States from 1888 to the present. N Engl J Med (2013) 369(22):2152–8. 10.1056/NEJMms1215400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shrestha S, Foxman B, Berus J, van Panhuis WG, Steiner C, Viboud C, et al. The role of influenza in the epidemiology of pneumonia. Sci Rep (2015) 5:15314. 10.1038/srep15314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kata A. A postmodern Pandora’s box: anti-vaccination misinformation on the Internet. Vaccine (2010) 28(7):1709–16. 10.1016/j.vaccine.2009.12.022 [DOI] [PubMed] [Google Scholar]
- 108.Butler D. When Google got flu wrong. Nature (2013) 494(7436):155–6. 10.1038/494155a [DOI] [PubMed] [Google Scholar]
- 109.Santillana M, Zhang DW, Althouse BM, Ayers JW. What can digital disease detection learn from (an external revision to) Google Flu Trends? Am J Prev Med (2014) 47(3):341–7. 10.1016/j.amepre.2014.05.020 [DOI] [PubMed] [Google Scholar]
- 110.Milton CL. The ethics of big data and nursing science. Nurs Sci Q (2017) 30(4):300–2. 10.1177/0894318417724474 [DOI] [PubMed] [Google Scholar]
- 111.Lipworth W, Mason PH, Kerridge I. Ethics and epistemology of big data. J Bioeth Inq (2017) 14(4):485–8. 10.1007/s11673-017-9815-8 [DOI] [PubMed] [Google Scholar]