Abstract
Background
Bamboo is an important member of the family Poaceae and has many inflorescence and flowering features rarely observed in other plant groups. It retains an unusual form of perennialism by having a long vegetative phase that can extend up to 120 years, followed by flowering and death of the plants. In contrast to a large number of studies conducted on the annual, reference plants Arabidopsis thaliana and rice, molecular studies to characterize flowering pathways in perennial bamboo are lacking. Since photoperiod plays a crucial role in flower induction in most plants, important genes involved in this pathway have been studied in the field grown Bambusa tulda, which flowers after 40-50 years.
Results
We identified several genes from B. tulda, including four related to the circadian clock [LATE ELONGATED HYPOCOTYL (LHY), TIMING OF CAB EXPRESSION1 (TOC1), ZEITLUPE (ZTL) and GIGANTEA (GI)], two circadian clock response integrators [CONSTANS A (COA), CONSTANS B (COB)] and four floral pathway integrators [FLOWERING LOCUS T1, 2, 3, 4 (FT1, 2, 3, 4)]. These genes were amplified from either gDNA and/or cDNA using degenerate as well as gene specific primers based on homologous sequences obtained from related monocot species. The sequence identity and phylogenetic comparisons revealed their close relationships to homologs identified in the temperate bamboo Phyllostachys edulis. While the four BtFT homologs were highly similar to each other, BtCOA possessed a full-length B-box domain that was truncated in BtCOB. Analysis of the spatial expression of these genes in selected flowering and non-flowering tissue stages indicated their possible involvement in flowering. The diurnal expression patterns of the clock genes were comparable to their homologs in rice, except for BtZTL. Among multiple BtCO and BtFT homologs, the diurnal pattern of only BtCOA and BtFT3, 4 were synchronized in the flower inductive tissue, but not in the non-flowering tissues.
Conclusion
This study elucidates the photoperiodic regulation of bamboo homologs of important flowering genes. The finding also identifies copy number expansion and gene expression divergence of CO and FT in bamboo. Further studies are required to understand their functional role in bamboo flowering.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4571-7) contains supplementary material, which is available to authorized users.
Keywords: Bamboo, Flowering genes, Circadian clock, Photoperiodism, Gene expression
Background
Controlling flowering time is one of the most important adaptations linked to the survival of angiosperms. Annual plants like A. thaliana or rice (Oryza sativa) undergo a short vegetative phase of a few weeks before the onset of flowering and then die. On the other hand, woody perennials such as Populus undergo years of vegetative growth before the onset of flowering and the flowering cycle then repeats for successive years. One extreme example of delayed flowering is bamboo, which has a vegetative phase of up to 120 years, followed by flowering and death of the plants [1]. This is a unique biological phenomenon known as semelparity/monocarpy. The flowering incidence may be restricted to few culms of a population (sporadic flowering) [2] or may happen across populations over a large geographical area (gregarious flowering) [3]. An important consequence of gregarious flowering is enormous seed setting, which results in a rapid increase in rat populations and thereby enormous crop loss in the vicinity that might culminate in famine [4]. The sudden induction of flowering also results in disappearance of large areas of vegetation that creates a major ecological imbalance in the surrounding plant community [5, 6]. Therefore, development of molecular markers for detecting possible induction of flowering will be of great help for proper forest management and ensuring food safety.
Flowering is a natural outcome of plant’s interaction with its surrounding environment. Depending on the nature of the external factors various flowering pathways such as photoperiodic (light as external cue) [7], vernalization (cold) [8], autonomous (endogenous factor/s) [9] and hormonal (GA3) [10] pathways have been characterized. Light is one of the most studied external cues and can control diverse physiological processes including flowering [11]. In photoperiodic regulation, the duration of day and night governs the timing of flowering, and plants can be categorized as long-day (LDP), short-day (SDP) or day neutral (DNP) [12]. The regulation of flowering as a consequence of day length is governed by the circadian oscillation of the expression of a group of genes known as circadian clock regulated genes [7]. The oscillation of the circadian clock regulated genes in response to light is synchronized by another set of genes called circadian clock genes [13]. In rice TIMING OF CAB EXPRESSION1 (TOC1), LATE ELONGATED HYPOCOTYL (LHY), ZEITLUPE (ZTL) and GIGANTEA (GI) are the major circadian clock genes that have been characterized so far [7–13]. CONSTANS (CO) is the gene that integrates the clock responses and subsequently passes the signal to the floral pathway integrator gene FLOWERING LOCUS T (FT) to induce flowering [14, 15]. CO is a B-box family gene, having a conserved CCT domain, while FT is a member of the phosphatidyl ethanolamine binding protein (PEBP) family.
All these studies have been conducted on the reference dicotyledonous plant A. thaliana and monocotyledonous plant rice [11, 16]. These plants have been preferred since they can be easily grown in the laboratory, their growth stages are defined, life cycles are short, germplasms easily accessible, genomes have been sequenced, and several gene mutants are available. However, it is an open question how much of the information generated from these reference plants can be translated to the non-reference plants such as bamboo that possesses striking differences in terms of growth and development. In spite of severe practical limitations such as infrequent tissue availability, low RNA yield, insufficient knowledge regarding floral histology, presence of multiple closely related paralogous flowering genes, woody bamboos offer a very interesting system to study the evolution and functional diversities of flowering genes [17].
Bamboo is a large plant group representing 1441 species within 116 genera and can grow in diverse tropical and temperate habitats [18]. Phyllostachys heterocycla, a temperate plant, is the only bamboo that has had its genome sequenced to date [19]. In addition to this small amount of genomic information, de novo transcriptome sequencing has been carried out to generate floral specific expressed sequence tags (ESTs) from different bamboo species such as, Bambusa oldhamii, Dendrocalamus latiflorus, P. heterocycla, P. edulis, P. aurea, B. edulis, Guadua inermis, Otatea acuminata and Lithachne pauciflora [20–27], and limited bamboo flowering genes were functionally characterized using transgenic approaches [28–32]. The transcriptome studies have identified millions of short ESTs of 75-250 bp long. However, in absence of the full-length gene sequences and their detailed functional characterization, understanding of their roles in flowering pathways remains incomplete.
The main objective of this study is to identify, characterize sequences, and analyse expression of important circadian clock and photoperiodic genes in bamboo. Taken together this study presents a comprehensive analysis of a set of flowering pathway genes in B. tulda, which flowers after 40-50 years [2].
Results
Study of B. tulda inflorescence to select appropriate flowering and associated leaf tissues
Photoperiodic genes are usually regulated by light and hence are expressed primarily in leaves or shoot apex regions [7]. Flowering B. tulda plants were observed closely to identify diverse types of leaves that could be studied to understand the photoperiodic regulation of the targeted genes. Like other Poaceae members, the bamboo inflorescence is primarily composed of spikelets, although pseudospikelets are often observed (Fig. 1). Although the bamboo inflorescence is broadly similar to the other two well characterized monocots, rice and maize, yet there exist differences with respect to the position and organization of the inflorescences. For example, in rice the typical flag leaf (FL) is located just beneath the single, terminal inflorescence, while in bamboo a single branch may bear multiple inflorescences, each of which is subtended by an individual FL (Fig. 1). At the advanced flowering stage, several inflorescences develop in a basipetal manner. Young bamboo inflorescences remain covered by the leaf sheath of the FL. As the young inflorescences remain invisible in the early developmental stage, these leaves were defined as possible flag leaves (PFL). Other than FL and PFL, young leaves located in the non-flowering branch of the flowering culm (YLF) may play a role in floral induction and therefore were included in the present study. In addition, a few more vegetative tissues were included such as culm sheath (CS), internodal region (IN), young leaf from non-flowering culm (YLN), root (R), rhizome (RH) and also reproductive tissues such as early staged inflorescence bud (E), middle staged inflorescence bud (M), late staged inflorescence bud (L, Fig. 1).
Fig. 1.
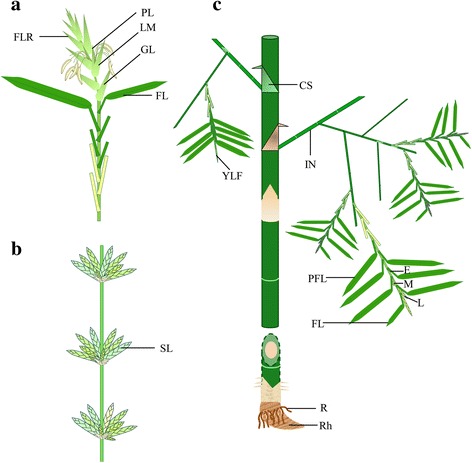
Study of B. tulda inflorescence and selection of appropriate flowering and vegetative tissue stages. a Morphology of a spikelet in B. tulda depicting multiple florets arranged on a single rachis. b Morphology of a pseudospikelet in B. tulda depicting multiple spikelets arranged in whorls on a rachis. Within each spikelet the florets are arranged on a single rachilla. c Different flowering and non-flowering tissue stages selected for studying expression pattern of important flowering genes in B. tulda. The Figures were prepared in Microsoft Power point 2016 based on the observations of the plant parts in their natural habitat. The abbreviations used: CS- culm sheath, IN- inter node, YLF- young leaf from flowering culm, YLN- young leaf from non-flowering culm, PFL- possible flag leaf, FL- flag leaf, E- early staged inflorescence bud, M- middle staged inflorescence bud, L- late staged inflorescence bud, R- root, RH- rhizome, PSL- pseudo spikelet, GL- glume, LM- lemma, PL- palea, FLR- floret
Molecular identification and sequence characterization of circadian clock genes
Single copies of the important circadian clock genes LHY, TOC1, ZTL and GI were identified in B. tulda (MF983713, KY249524, MF983715, MF983716). In order to obtain these genes and/or coding sequences, degenerate as well as gene specific primers were used for PCR amplification and subsequent sequencing (Additional file 1: Table S1). These sequences were used for BLAST analysis to identify their homologs in other monocot genomes. The best BLASTP hits obtained for BtLHY, BtTOC1, BtZTL and BtGI query sequences were Oryza brachyantha XP_006659145.1, O. sativa BAD38854.1, Thyridolepis multiculmis AML79118.1 and Setaria italica XP_004968438.1 having 78%, 85%, 93% and 94% sequence identities, respectively (Table 1). The translated B. tulda amino acid sequences were studied to identify the domains characteristics for these proteins. Indeed, the BtTOC1 sequence revealed the 127 amino acid receiver domain in the N-terminal end and 47 amino acid CCT domain in the C-terminal end (Fig. 2a). Like other ZTL proteins, BtZTL possessed N-terminal photo sensory light oxygen voltage (LOV) domain, F-box domain at the middle, and 4 kelch repeats at the C-terminal end (Fig. 2b). The other identified clock gene BtGI contained a transmembrane domain in the N-terminal region (Fig. 2c).
Table 1.
Identification of B. tulda homologous sequences of circadian clock, clock integrator and pathway integrator genes
| Genes | B. tulda query sequences | Best O. sativa hits | Identity (%) | Query cover (%) | E value | Best B. distachyon hits | Identity (%) | Query cover (%) | E value | Best P. heterocycla hits | Identity (%) | Query cover (%) | E value | Best hits in NCBI non-redundant database | Identity (%) | Query cover (%) | E value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LATE ELONGATED HYPOCOTYL (LHY) | MF983713 | Os08g06110 | 78.31 | 100 | 0 | Bradi3g16515 | 73 | 100 | 0 | PH01001283G0510 | 77 | 100 | 0 | Oryza brachyantha (XP_006659145.1) | 78 | 100 | 0 |
| TIMING OF CAB EXPRESSION 1 (TOC1) | KY249524 | Os02g40510 | 84 | 100 | 0 | Bradi3g48880 | 80 | 95.21 | 0 | PH01003618G0130 | 93 | 100 | 0 | O. sativa (BAD38854.1) | 85 | 100 | 0 |
| ZEITLUPE (ZTL) | MF983715 | Os02g05700 | 90.28 | 100 | 0 | Bradi3g04040 | 89 | 100 | 0 | PH01000114G1110 | 93 | 100 | 0 | Thyridolepis multiculmis (AML79118.1) | 93 | 100 | 0 |
| GIGANTEA (GI) | MF983716 | Os01g08700 | 91 | 100 | 0 | Bradi2g05226 | 88 | 100 | 0 | PH01002142G0290 | 70 | 100 | E− 129 | Setaria italica (XP_004968438.1) | 94 | 100 | 0 |
| CONSTANS A (COA) | KY249523 | Os06g16370 | 59 | 100 | 1.00E− 148 | Bradi1g43670 | 74.08 | 100 | 4.10E−120 | PH01005551G0030 | 51 | 97.64 | 3.00E−94 | Oryza. rufipogon (AFK31610.1) | 78 | 100 | 0 |
| CONSTANS B (COB) | MF983714 | Os09g06464 | 71.06 | 100 | 1.00E− 141 | Bradi3g56260 | 34.04 | 92.66 | 4.80E−17 | PH01000048G0270 | 68 | 100 | E−117 | Hordeum vulgare (AAM74066.1) | 70 | 100 | 2.00E−138 |
| FLOWERING LOCUS T (FT) | KT003820 | Os06g06320 | 89.71 | 98.31 | 1.00E−118 | Bradi1g48830 | 93 | 87.64 | 1.60E−106 | PH01002288G0050 | 62 | 85.95 | 7.00E−52 | Phyllostachys meyeri (BAI49899.1) | 94 | 100 | 5.00E−122 |
| KT003821 | Os06g06320 | 89.71 | 98.31 | 1.00E−118 | Bradi1g48830 | 93 | 87.64 | 1.60E−106 | PH01002288G0050 | 62 | 85.95 | 7.00E−52 | P. meyeri (BAI49899.1) | 94 | 100 | 5.00E−122 | |
| KU726232 | Os06g06320 | 87.43 | 98.31 | 1.00E−115 | Bradi1g48830 | 89 | 99.43 | 2.00E−116 | PH01002288G0050 | 62 | 94.94 | 5.00E−58 | P. meyeri (BAI49900.1) | 92 | 100 | 4.00E−118 | |
| KX290774 | Os06g06320 | 88 | 98.31 | 1.00E−116 | Bradi1g48830 | 90 | 99.43 | 2.30E−117 | PH01002288G0050 | 62 | 94.94 | 8.00E−59 | P. meyeri (BAI49900.1) | 92 | 100 | 7.00E− 119 |
The BLAST-P analyses was done against the reference monocot genome Oryza sativa, phylogenetically close Brachypodium distachyon, temperate bamboo Phyllostachys heterocycla and NCBI non-redundant database using B. tulda amino acid sequences as quarries. Only the top BLAST hit sequences are reported along with the respective identities (%), E values and coverage of the query sequences against the obtained hit sequences (%)
Fig. 2.
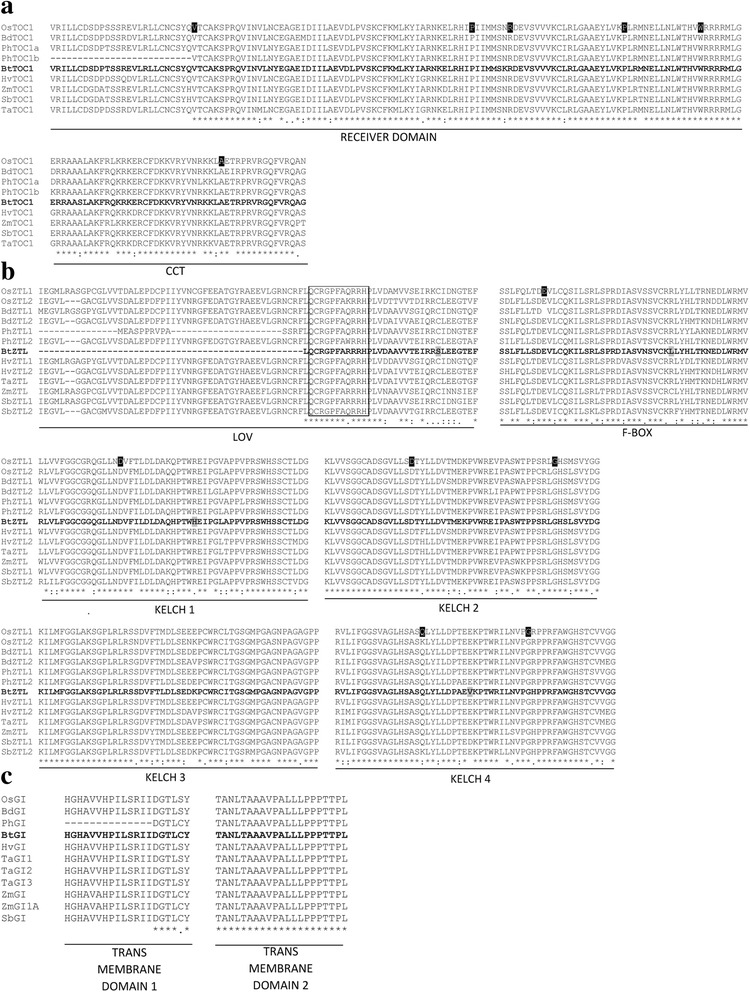
Multiple sequence alignment of BtTOC1, BtZTL and BtGI genes with homologous sequences from related monocots. Black highlighted amino acids are functionally important as evidenced by mutational analyses. a Detailed sequence characterization of BtTOC1 and identification of receiver and CCT domains. Sequences used are: OsTOC1: Os02g40510.1, BdTOC1: Bradi3g48880, PhTOC1a: PH01003618G0130, PhTOC1b: PH01000345G0790, BtTOC1: KY249524, HvTOC1: AEW48242.1, ZmTOC1: ADX60159.1, SbTOC1: SORBI_004G216700, TaTOC1: AMK48975.1 b Detailed sequence characterization of BtZTL and identification of Light Oxygen Voltage (LOV), F-box domain and four Kelch repeats. Sequences used are: OsZTL1: Os06g47890.2, OsZTL2: Os02g05700.2, BdZTL1: Bradi1g33610.2, BdZTL2: Bradi3g04040.2, PhZTL1: PH01007024G0030, PhZTL2: PH01000836G0340, BtZTL: MF983715, HvZTL1: HV273830G00010, HvZTL2: HV158755G00020, TaZTL: ABR14627.1, ZmZTL: GRMZM2G113244, SbZTL1: Sobic.010G243900.1, SbZTL2: Sobic.004G042200.2. c Detailed sequence characterization of BtGI and identification of two characteristic trans membrane domains. Sequences used are: OsGI: Os01g08700.2, BdGI: Bradi2g05226.1, PhGI: PH01002142G0290, BtGI: MF983716, HvGI: AAW66945.1, TaGI1: AAQ11738.1, TaGI2: AAT79486.1, TaGI3: AAT79487.1, ZmGI: ABZ81992.1, ZmGI1A: DAA06172.1, SbGI: Sobic.003G040900.3
Molecular identification, sequence characterization and phylogenetic analyses of BtCOA and BtCOB genes
CONSTANS (CO) is the circadian clock response integrator gene, which is a member of the B-box family [33]. Single copy BtCOA and BtCOB genes were amplified from gDNA and cDNA libraries, sequenced and analysed (KY249523, MF983714). The BtCOA protein sequence was most identical to Oryza rufipogon sequence (AFK31610.1) having 78% identity, while the highest identity (70%) of BtCOB was detected against barley (AAM74066.1, Table 1). Phylogenetic analyses based on the amino acid sequences revealed a clear split of BtCOA and BtCOB genes into two different clades (Fig. 3a). While BtCOA was more closely related to rice OsCOA than the temperate bamboo Phyllostachys PhCOA, BtCOB clustered with PhCOB. This indicated that with respect to gene sequences the two BtCOs were quite divergent. Prediction of gene models indicated that like other characterized CO sequences, BtCOA and BtCOB contained two exons and one intron each. The intron lengths of COB varied across species, while in COA it was more conserved (Fig. 3b). The translated BtCOA and BtCOB proteins were of 382 and 327 amino acids in length. Protein sequence analyses revealed that both BtCOA and BtCOB contained two B-boxes at their N-terminal ends (B-box 1, B-box 2) having conserved C and H residues (Fig. 3c). An intact 43 amino acid long B-boxes 1 and 2 was obtained for BtCOA. In contrast, 25 amino acids of the C-terminal end of B-box 1 and 18 amino acids in the N-terminal part of the B-box 2 were truncated in BtCOB (Fig. 3b, c). In addition to the N-terminal B-box domain, BtCOA and BtCOB possessed a 43 amino acid DNA binding CCT domain in their C-terminal ends (Fig. 3d). In plants the CCT domain interacts with other DNA binding proteins such as HAP3 and HAP5 with the help of nine conserved amino acids [34]. While all these amino acids were conserved in BtCOA, Arg33 was changed to Gln33 in BtCOB (Fig. 3d).
Fig. 3.
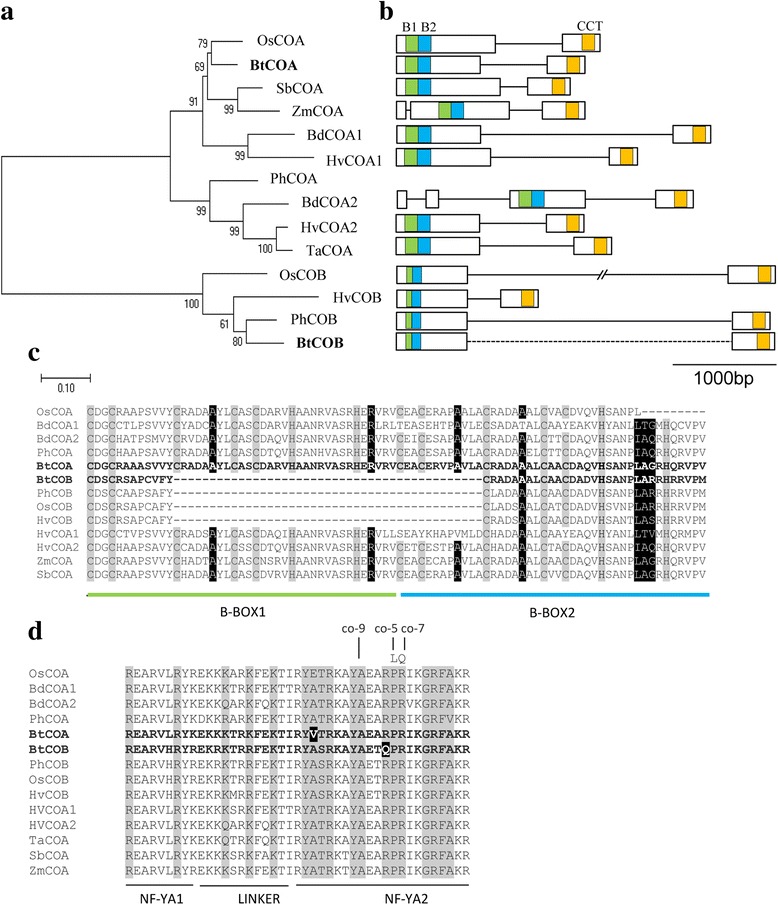
Phylogenetic and sequence characterization of BtCOA and BtCOB genes. a Phylogenetic comparison of BtCOA and BtCOB coding sequences with homologous sequences in related monocot species. The Neighbour Joining (NJ) tree was developed by Mega 7.0 using default parameters and bootstrap value 1000. b Predicted exon-intron structures of BtCOA, BtCOB genes and comparison with other monocot genes. Exons are marked as rectangles and introns as solid lines. c Multiple sequence alignment of the B-box domains of BtCOA and BtCOB protein sequences, which indicates presence of two full length B-boxes in BtCOA, while it is truncated in BtCOB. The characteristic C and H residues of B-box domains are highlighted in grey. Black highlighted amino acids are functionally important as evidenced by mutational analyses. d Sequence comparison of CCT domains between BtCOA, BtCOB and other related monocot members. Amino acids conserved for HAP3 and HAP5 binding are highlighted in grey. Amino acids not conserved in B. tulda are highlighted in black. NF-YA1 interacts with HAP3 and NF-YA2 interacts with CCAAT DNA sequences. Sequences used are: OsCOA: Os06g16370.1, BdCOA1: Bradi1g43670.1, BdCOA2: Bradi3g56260.1, PhCOA: PH01005551G0030, BtCOA: KY249523, HvCOA1: AF490467.1, HvCOA2: AF490469.1, ZmCOA: GRMZM2G405368, SbCOA: Sobic.010G115800.1, OsCOB: Os09g06464.1, PhCOB: PH01000048G0270, BtCOB: MF983714, HvCOB: AF490473.1
Molecular identification, sequence characterization, phylogenetic analyses of four BtFT genes
Flowering locus T (FT), a member of PEBP family, is one of the most important floral pathway integrator genes. In the present study, four alleles of BtFT genes were identified (Additional file 2: Figure S1, BtFT1: KT003820, BtFT2: KT003821, BtFT3: KU726232, BtFT4: KX290774). A homology search using translated coding sequences of the BtFT1, 2, 3 and 4 revealed very high identity (92-94%) with FT sequences of another bamboo P. meyeri (Table 1). The four BtFT sequences were phylogenetically separated into two different clades (Fig. 4a). While BtFT1 clustered with BtFT2, BtFT3 clustered with BtFT4, indicating that the two groups of genes are distinct based on their sequences. This finding was also supported by their predicted exon-intron organization (Fig. 4b). Each of the four BtFT genes contained four exons and three introns. Exon 4 was the longest (233 bp), while exon 3 was the shortest (41 bp). Although the exon lengths were highly conserved among the 4 BtFT homologs, the length of intron 1 was longer in BtFT1, 2 than that of BtFT3, 4 (Fig. 4b). Each predicted BtFT protein was 178 amino acid long, having a PEBP domain that retained seven conserved amino acid residues and two C-terminal amino acid stretches, which are important for maintaining the floral inducing function. Incidentally, another PEBP member is TERMINAL FLOWER1 (TFL1), which is a floral repressor and is highly similar in sequence to FT. Among the differences are two signature amino acids, Tyr85 and Gln140 present in FT (Fig. 4c), while His88 and Asp144 in TFL1 instead [35]. The present analysis confirmed that all the identified sequences are indeed FT, not TFL1 (Fig. 4c).
Fig. 4.

Phylogenetic and sequence characterization of four BtFT genes. a Phylogenetic comparison of BtFT1, BtFT2, BtFT3 and BtFT4 coding sequences with homologous sequences in related monocot species. The Neighbour Joining (NJ) tree was developed by Mega 7.0 using default parameters and bootstrap value 1000. b Predicted exon-intron structures of four BtFT genes and comparison with other monocot genes. Exons are marked as rectangles having PEBP domains marked in solid black boxes and introns as solid lines. c Sequence comparison of the PEBP domains of BtFT and other related monocot sequences. Two residues marked with arrow heads are characteristics for either FT or TFL1 identity. Residues having important biological functions are marked in asterisks. Sequences used are: OsHd3a: Os06g06320.1, OsRFT1: Os06g06300.1, BdFT: Bradi1g48830.1, PhFT: PH01002288G0050, PmFT1: AB498760.1, PmFT2: AB240578.1, PmFT3: AB498761.1, PmFT4: AB498762.1, BtFT1: KT003820, BtFT2: KT003821, BtFT3: KU726232, BtFT4: KX290774, PvFT1: Guo et al. (2015), PvFT2: Guo et al. (2015), HvFT1: DQ100327, HvFT2: DQ297407.1, HvFT3: DQ411319, HvFT4: DQ411320, TaFT: DQ890162.1, ZmFT: EU241924, SbFT1: XP_002436509.1, SbFT8: XP_002456354.1, SbFT10: Sb09g025760
In silico study on the molecular interactions between individual BtFT and Os14-3-3 proteins
The rice FT homologue Hd3a interacts with 14-3-3 proteins at the shoot apical meristem (SAM) to form the Hd3a-14-3-3 complex, which is translocated to the nucleus to interact with rice FD1, a bZIP transcription factor [36]. The resulting “florigen activation complex” (FAC), promotes the conversion of the SAM to an inflorescence meristem [36]. Out of seven conserved amino acids located within the PEBP domain of FT that contribute to the direct interaction between FT and 14-3-3 (Fig. 5a), two substitutions, from Phe101 to Ile101 in BtFT1, 2 and Phe64 to Leu64 in BtFT3, 4 were observed. In silico protein-protein interaction analyses were conducted to understand the overall interaction efficiency between individual BtFT and 14-3-3 sequences and to detect whether these changes affect the interaction. Since no crystal structures were available for BtFT proteins and no sequence or structure of Bt14-3-3, the interaction between BtFT and Os14-3-3 pairs were investigated. Homology models of BtFT1, 2, 3 and 4 were developed, and these were 86-88% identical to their rice homologue OsHd3a. Given the profound homologies among all BtFT alleles, their interaction with Os14-3-3 remained mostly conserved (Fig. 5b), with interaction interface remaining interdigitated (Fig. 5c). Similar to OsHd3a-Os14-3-3 interaction [36], BtFT1, 2, 3, 4 and Os14-3-3 interaction interface consisted of a hydrophobic cavity as well as an in-between acidic lobe (Asp208 and Glu212 of 14-3-3), interacting with Arg130 and Arg62 of BtFT1, 2, 3 and 4 (Fig. 5d) through conserved salt-bridge interactions. These interactions are essential not only for FT binding with 14-3-3 but also with FD. In contrast to OsHd3a sequence (Phe66 and Phe103), Leu64 was present in BtFT3, 4 and Ile101 was present in BtFT1, 2, respectively. In BtFT1, 2, Phe64 stabilized the hydrophobic interaction with Ile204 of Os14-3-3, similar to the OsHd3a interaction. In BtFT1, 2, Ile101 made hydrophobic contact with Phe200 of Os14-3-3, in BtFT3, 4, but there was a possibility of a stacking interaction between Phe101 and Phe200 of Os14-3-3, similar to Hd3a. Although the hydrophobic interactions (Fig. 5d) were subtly different in BtFT1, 2 and BtFT3, 4 compared to OsHd3a-Os14-3-3 interactions, such changes might influence the specificity of BtFT interactions with 14-3-3.
Fig. 5.

In silico study on the molecular interactions between individual BtFT and Os14-3-3 proteins. a Detailed sequence analysis of BtFT protein regions responsible for interacting with 14-3-3 protein. Seven amino acids conserved for 14-3-3 interaction are highlighted in grey. Amino acids not conserved in B. tulda are highlighted in black. Sequences used are: OsHd3a: Os06g06320.1, OsRFT1: Os06g06300.1, BdFT: Bradi1g48830.1, PhFT: PH01002288G0050, PmFT1: AB498760.1, PmFT2: AB240578.1, PmFT3: AB498761.1, PmFT4: AB498762.1, BtFT1: KT003820, BtFT2: KT003821, BtFT3: KU726232, BtFT4: KX290774, PvFT1: Guo et al. (2015), PvFT2: Guo et al. (2015), HvFT1: DQ100327, HvFT2: DQ297407.1, HvFT3: DQ411319, HvFT4: DQ411320, TaFT: DQ890162.1, ZmFT: EU241924, SbFT1: XP_002436509.1, SbFT8: XP_002456354.1, SbFT10: Sb09g025760. b Conserved interaction pattern between BtFT and 14-3-3. Given more than 86% homology with each other as well as rice counterpart Hd3a, all BtFT proteins (1-4) are almost perfectly superimposable to each other. Their interaction pattern with 14-3-3 also remains mostly conserved. c Surface analysis showing interdigitated interface between a pair of BtFT and 14-3-3. d Conserved salt bridge interactions between BtFT and 14-3-3. Asp208-Arg130 and Glu212-Arg62 salt bridges could be essential for BtFT’s interaction not only with 14-3-3 but also with FD. e Difference in hydrophobic cavity lining BtFT proteins. Subtle alterations e.g. Phe101 to Ile101in BtFT1, 2 and Phe64 to Leu64 in BtFT3, 4 might alter the specificity of BtFT1,2/3,4 interaction with 14-3-3
Tissue specific expression analyses of circadian clock, CO and FT genes
The transcriptional expression of the circadian clock (BtLHY, BtTOC1, BtZTL, BtGI), circadian clock integrator (BtCOA, BtCOB) and floral pathway integrator (BtFT1, BtFT2, BtFT3 and BtFT4) genes were investigated in ten selected flowering and non-flowering tissue stages to understand their possible role in flowering. Higher transcript abundance of all these genes was detected in young leaves isolated from the flowering culm (YLF) than that of the non-flowering culm (YLN). However, when the expression levels were compared among ten tissues, the highest expression of BtLHY and BtTOC1 was obtained in early stage inflorescence bud and internodal tissues (Fig. 6a, b), while it was YLF in case of BtZTL and BtGI (Fig. 6c, d). In the case of BtCOA and BtCOB, higher transcriptional expression was detected in YLF and culm sheath (CS) respectively, while the expression level was consistently low in all other eight tissues. The expression of BtCOA was much higher in YLF than CS, although such a clear distinction in expression levels was absent in BtCOB (Fig. 6e, f). This is an important indication of the possible involvement of BtCOA in floral induction, because YLF is biologically associated with the floral induction while CS is mostly vegetative in nature. Such distinctions in expression patterns between flowering and non-flowering tissue stages were not observed for the two groups of BtFT genes that were suggested by the phylogenetic analysis (Fig. 6g, h). The highest expression of all of the four homologs was observed in CS. However, in case of BtFT3, 4 the expression was also quite high in YLF, which was not the case for BtFT1, 2.
Fig. 6.
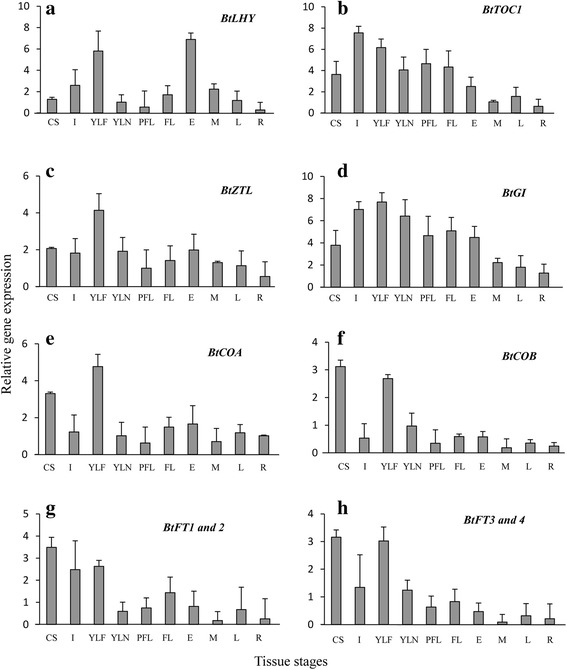
Study of relative gene expression levels in ten tissue stages of B. tulda. a-h spatial gene expression levels of BtLHY; BtTOC1; BtZTL; BtGI; BtCOA; BtCOB; BtFT1, 2 and BtFT3, 4. Transcript expression of eIF4α was used to normalize expression data of the targeted flowering genes. The relative fold change was calculated by 2-∆∆CT method using the expression level observed in rhizome as the calibrator. Each bar represents mean of three biological replicates ± SE
Study on the diurnal expression patterns of circadian clock genes
Although tissue specific expression patterns can provide important clues about gene functionality, the majority of genes studied here need to follow a circadian rhythm in order to perform their developmental role in the plant. Therefore, the diurnal expression patterns of the circadian clock genes (BtLHY, BtTOC1, BtZTL and BtGI) were studied at four different time points (morning: 8 am, noon: 12 pm, afternoon: 4 pm, night: 8 pm) under the short-day (11 h light) and long-day (14 h light) conditions of the natural habitat of the plants. Two sets of leaf tissues were selected for this study. The leaves collected from a flowering culm (YLF) were selected due to their anticipated involvement in floral induction, which is supported by obtaining higher level of expression of the clock genes compared to the other leaf tissues. On the contrary, the leaves from a non-flowering culm (YLN) were selected as the comparable tissue representing the non-inductive stage. In general, the transcript abundance of all these genes was detected at higher levels under SD than LD, both for YLF and YLN (Fig. 7a-h). The diurnal expression patterns of BtTOC1 and BtGI attained a peak in the afternoon while BtLHY and BtZTL transcripts were abundant in the morning, followed by a gradual decrease under SD (Fig. 7a-h).
Fig. 7.
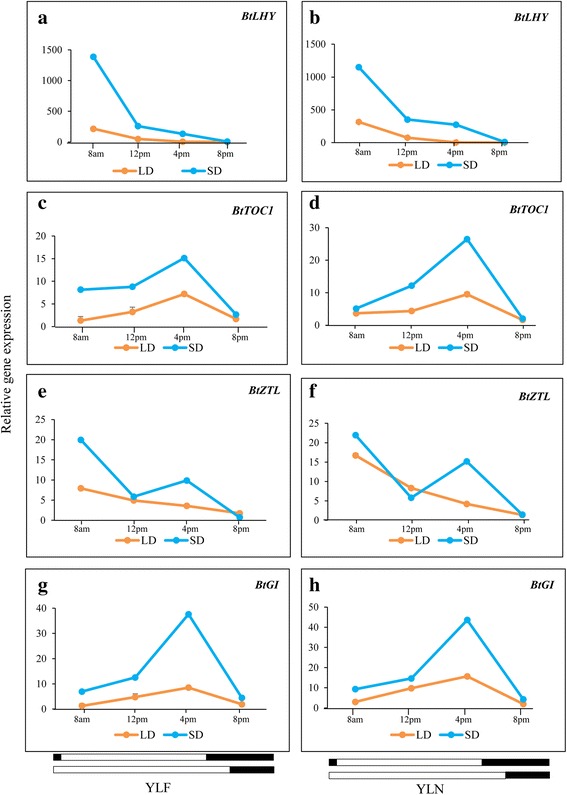
Comparison of diurnal expressions of circadian clock genes in YLF and YLN during SD and LD. a, b BtLHY, c, d BtTOC1, e, f BtZTL and g, h BtGI. Transcript expression of eIF4α was used to normalize expression data of targeted flowering genes in different tissues. The relative fold change was calculated by 2-∆∆CT method using the expression data in rhizome as calibrator and is plotted using two Y axis. Each data point in the line graph represents mean of three biological replicates ± SE in case of LD and one biological replicate in case of SD
Study on the diurnal expression patterns of BtCO and BtFT genes
The circadian oscillations acquired by the circadian clock genes are transmitted to CO, which eventually interacts with FT to induce flowering. Therefore, the circadian rhythm of CO should be followed by FT in order to perform their assigned biological functions. In bamboo, the situation was not straightforward since multiple CO and FT gene copies/alleles were present. Therefore, diurnal expression of two BtCO and four BtFT homologs were measured in YLF and YLN under SD and LD conditions. Similar to the clock genes, the expression of BtCOA was higher in both YLF and YLN under SD than LD (Fig. 8a, b). In contrast, the opposite trend was observed for BtCOB, the homolog of which acts as a floral repressor in rice. The diurnal expression pattern of BtCOA reached a peak in the afternoon followed by a sudden decrease. In contrast, the maximum expression level of BtCOB was observed in the morning and gradually decreased throughout the day (Fig. 8a, b). The diurnal expression patterns of BtCOA and BtCOB were compared to that of four BtFT alleles. Similar to BtCOA, the diurnal expression pattern of BtFT3, 4 revealed its highest expression in the afternoon in both SD and LD condition in YLF, but not in YLN. In contrast, the diurnal expression pattern of BtFT1, 2 did not follow that of BtCOA or BtCOB.
Fig. 8.
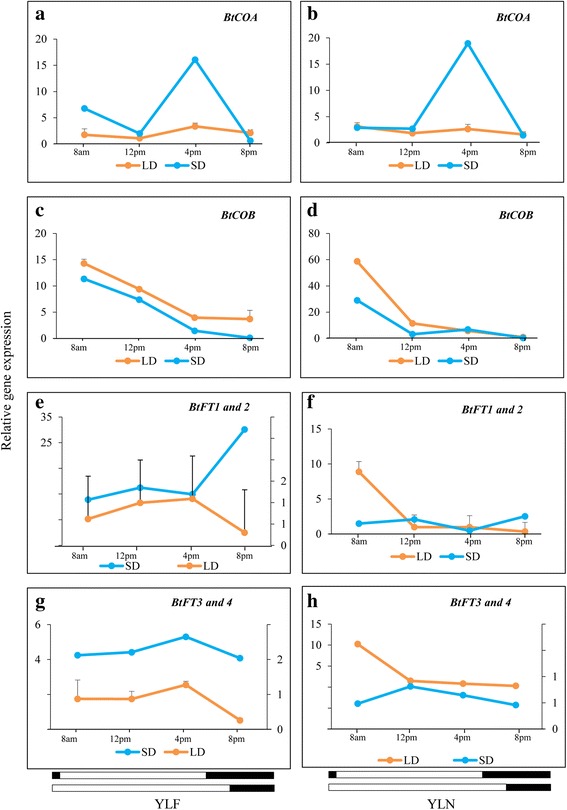
Comparison of diurnal expressions of BtCO and BtFT homologs in YLF and YLN during SD and LD. a, b BtCOA, c, d BtCOB, e, f BtFT1, 2 and g, h BtFT3, 4. Transcript expression of eIF4α was used to normalize expression data of the targeted flowering genes in different tissues. The relative fold change was calculated by 2-∆∆CT method using the expression data in rhizome as calibrator and is plotted using two Y axis. Each data point in the line graph represents mean of three biological replicates ± SE in case of LD and one biological replicate in case of SD
Discussion
Molecular studies on bamboo flowering are limited and the primary reasons are the unavailability of sufficient reproductive tissues and undefined developmental stages [17]. The possible alternatives, such as use of annual flowering bamboo (e.g., Indocalamus wightianus, Ochlandra sp.) or use of in vitro induced flowering tissues [37] are not credible since they either lack the extended vegetative phase or the plants are grown under artificial conditions and therefore regulation of the genes might be different. Consequently, apart from a few exceptions [20], the majority of studies have relied on field-grown flowering plants, undertook de novo transcriptome sequencing of floral tissues and annotated short ESTs based on BLAST based sequence homology [20–27]. All these studies yielded important but partial understanding of the genes and their regulation, as they do not provide full-length gene sequences or detailed expression profiles. In the absence of those data, collective characterization of genes involved in a particular flowering pathway remains elusive in bamboo.
Important diurnally regulated circadian clock genes are identified in B. tulda
Plant circadian rhythms in response to light, are regulated by a series of interconnected transcriptional and translational loops of clock related genes. The roles of these genes have been extensively studied in reference plants, A. thaliana and rice, which are mostly annual [7, 13]. In rice OsLHY, OsTOC1, OsZTL and OsGI are the key components of the core feedback loop of the circadian clock [38–41]. The OsLHY is up regulated in the morning via red light [42]. This elevated OsLHY transcript supressed the expression of OsTOC1 in the morning [39, 43], but by evening OsTOC1 regained transcriptional peak. Eventually OsTOC1 upregulated OsLHY and simultaneously suppressed OsGI. On the other hand, the upregulation of OsGI in the evening was caused by the blue light mediated degradation of OsTOC1 by OsZTL [38, 44]. In our study the identified B. tulda gene homologs were highly identical to sequences obtained from other monocots including Phyllostachys. The overall diurnal rhythms of BtLHY, BtTOC1 and BtGI, but not BtZTL, were comparable to that of rice [39, 44–46]. The OsZTL showed a unimodal expression peak in the morning under SD, but was bimodal (morning and afternoon) under LD [39, 45]. However, this trend was reversed in B. tulda, where the observed diurnal peak was unimodal (only morning) under LD and bimodal (morning and afternoon) under SD. This could be a significant clue for future studies as because it is established that the function of ZTL is primarily flower specific, while the other circadian clock genes such as LHY and TOC1 perform pleiotropic functions including leaf movement, maintenance of hypocotyl length, expression of antenna protein, cell elongation and UV-B protection [47–49].
Distinct sequence and expression divergence observed for the two types of CONSTANS genes identified
A large number of CONSTANS like genes (COLs) are present in plants. For example, there are 17 COLs in A. thaliana, 16 in rice, and 26 in soybean [33, 50]. Depending on the number of B-boxes present, all these COLs can be grouped into four different clusters, which are indicated as I, II, III and IV [33]. The members of group I COLs primarily act as floral regulators and may act either as floral activators or repressors [51–54]. In B. tulda two CO genes have been identified, BtCOA and BtCOB, which are the members of the group I cluster. The B-box domain organization (two intact B-boxes in BtCOA vs. truncated B-boxes in BtCOB) and tissue specific expression patterns (BtCOA expression is high in YLF, while BtCOB is highest in CS) indicate that BtCOA is possibly involved in photoperiodic regulation of flowering, while BtCOB is not. This was further supported by the diurnal circadian rhythm. BtCOA exhibited a transcript expression peak in the afternoon, which is similar to the flower inductive rice OsCOA homolog HEADING DATE1 [55, 56]. On the contrary BtCOB demonstrated an expression peak in the morning. The rice COB homolog OsCO3, which is a negative regulator of OsHd3a, also demonstrated a similar diurnal rhythm [57]. All this evidence suggests that BtCOA contains biologically important sequence elements and characteristic diurnal expression patterns, which were not observed in BtCOB. Further studies are required to verify whether additional CO like genes exist in bamboo and, if so, how they contribute to flower induction.
High sequence similarity, but differential diurnal regulation indicates possible functional divergence of four BtFT homologs
FT is a member of the PEBP family and is present in multiple copies in different plant species [58–68]. In B. tulda four FT alleles have been identified, which are more than 98% similar in terms of their amino acid sequences. The individual amino acid differences in the four BtFT sequences, particularly in context to 14-3-3 interactions, were carefully considered to predict their possible influence on FT functioning. Phylogenetic as well as in silico interaction analyses clearly indicated that BtFT1 was more homologous to BtFT2, while BtFT3 was closer to BtFT4. Though most essential salt bridge interactions between 14-3-3-BtFT pairs, Asp208-Arg130 and Glu212-Arg62 were conserved, there was little change in the composition of the hydrophobic cavity lining BtFT. Such a subtle change in hydrophobicity, though apparently not drastic, might influence the specificity of BtFT and 14-3-3 interactions.
There exists wide diversity with respect to the roles of FT gene copies in flowering. In poplar, expression divergence leading to distinct subfunctionalization has been noticed between the two FT genes [65]. While PtFT1 is primarily responsible for inducing reproductive development, PtFT2 is involved in the vegetative growth of the plant. Similarly, expression diversification of the two FT genes was also reported in the temperate bamboo P. violascens [68]. PvFT1 is expressed in leaves and induces flowering, while PvFT2 possibly plays an important role in floral organ differentiation. Since flowering is an environmentally regulated biological process, the regulation of genes related to flowering is likely to be different in temperate and tropical bamboos. Circadian oscillation of BtFT3 and 4 in YLF revealed highest expression in the afternoon, while no such pattern was observed in case of YLN under either SD or LD conditions. In A. thaliana, barley and soybean the diurnal expression rhythm of FT showed a transcriptional peak in the afternoon [69, 70], while for rice it was in the morning [44]. The diurnal expression pattern of BtFT1 and 2 in both YLF and YLN was quite divergent to that of BtFT3 and 4. The expression divergence of BtFT genes might have been caused by changes in the promoter regions since such observations have been made in rice and Brassica [71, 72]. Therefore, native bamboo gene promoters should be sequenced in order to understand the expression regulation of these genes.
Existence of CO-FT regulon in bamboo
For the induction of flowering, the specific diurnal rhythm of CO has to be followed by FT. It has been observed in many plants such as soybean and rice that out of multiple copies of CO and FT genes, only a few candidates follow the diurnal expression pattern necessary for flower induction [70, 73]. In Glycine max, among the 28 CO and 11 FT like genes, the diurnal expression pattern of GmCOL5 and GmCOL13 synchronized with 6 GmFT genes [70]. In poplar the co-expression of PtCO2 and PtFT1 gene pairs controls the timing of flowering and is known as the CO-FT regulon [74]. In bamboo four FT genes have been identified in P. meyeri and their tissue specific expression patterns have been studied [67]. However, no information could be obtained on any bamboo CO homologs and their expression patterns in different tissues and diurnal conditions. In the absence of such data the possible existence of CO-FT regulon in bamboo flowering could not be tested. Therefore, the synchronization of the diurnal expression patterns of BtCO and BtFT gene copies were investigated. Indeed, the diurnal oscillation of BtCOA was followed by BtFT3 and 4 in YLF, but not in YLN suggesting the possible existence of CO-FT regulon in bamboo. Further studies are required to confirm the functional significance of this finding in terms of regulation of flowering in bamboo.
Conclusion
Sequence comparison, phylogeny, and expression analyses of the studied genes indicate existence of an active photoperiodic pathway in bamboo. The findings also indicate that an increase in gene copy numbers and expression divergences of CO and FT play an important role in photoperiodic regulation of flowering in bamboo. Involvement of many more additional factors such as physiological maturity [75], micro RNAs [76] or RNA splicing [77] may ultimately determine the timing of flowering. Further studies are required to characterize many of the genes identified here by loss-of-function or overexpression analyses to understand their functional role in bamboo flowering. Taken together, the present findings would not only be useful for future research on bamboo but also for the non-reference plants that remain neglected.
Methods
Identification and collection of appropriate flowering and vegetative tissues in B. tulda
Floral tissue samples of B. tulda were collected from sporadic flowering events that happened at Shyamnagar (22.83° N, 88.40° E) and Bandel (22.93° N, 88.38° E), West Bengal, India during April, 2013 to July, 2017. Voucher specimen were submitted to the Botanical Survey of India (B.S.I), Shibpur (deposition nos.-56A, 56B, 57A, 57B, 58A. 58B, 59A, 59B, 59C dated 05.06.2015). Tissues from diverse vegetative and floral developmental stages were snap frozen in liquid nitrogen in the field, transported to the laboratory and stored in the -80 °C freezer. Three biological replicates were collected for each tissue stage. Vegetative tissues selected for tissue specific expression analyses were leaf from both flowering and non-flowering branches of a flowering culm, leaf from non-flowering culm, flag leaf, culm sheath, rhizome, root and internodal region (Fig. 1). Three defined floral tissues stages such as early, middle and late developmental stages were selected based on the histological observations of the developing floral primordia [17]. For diurnal analyses, leaf tissues were selected from non-flowering culm and non-flowering branches of flowering culm. Tissues were collected from naturally grown plants at four different time points of a day- morning (8 am), noon (12 pm), afternoon (4 pm) and night (8 pm) for both long-day (LD, 14 h light exposure, sunrise at 4:30 am and sunset at 6:30 pm) and short-day (SD, 11 h light exposure, sunrise at 6 am and sunset at 5 pm). LD experiments were conducted using three biological replicates, while only one replicate was available for SD analyses.
Isolation of nucleic acids and preparation of cDNA libraries
Genomic DNA was isolated from the young, healthy leaves using DNeasy Plant Mini Kit (Qiagen, Germany). Total RNA was extracted from the selected tissues using a combination of Trizol (Invitrogen, USA) and RNAeasy Plant Mini Kit (Qiagen, Germany) [78, 79]. DNase I (Thermo Scientific, USA) was added to avoid any genomic DNA contamination. Quality and quantity of the isolated samples were determined in a BioSpectrometer (Eppendorf, Germany) and agarose-formamide gel elctrophoresis. Around 1 μg total RNA was used for cDNA synthesis using Verso cDNA Synthesis Kit (Thermo Scientific) following manufacturer’s protocol. 2 μl of 1/20th diluted cDNA sample was used for real time RT-qPCR analyses.
Primer designing, PCR amplification, cloning and sequencing of homologous genes
Gene specific degenerate primers were designed by aligning multiple sequences retrieved from related close monocot genomes (Additional file 1: Table S1). Coding sequences were multiple aligned in MUSCLE and gene specific primers were designed by using Primer3 program. PCR amplification was done using high fidelity Phusion Taq DNA polymerase (Thermo Scientific). Amplified bands of desired molecular weight were eluted from agarose gel by using GeneJET gel elution kit (Thermo Scientific) and cloned into TA vector (pGEM®-T Easy Vector Systems, Promega, USA) or blunt end vector (pJET PCR cloning kit, Thermo Scientific) following the instructions of the manufacturers. Positively transformed colonies were selected on blue-white selection and/or ampicillin medium and plasmids were purified using plasmid isolation kit (GeneJET Plasmid Miniprep Kit, Thermo Scientific). Sequencing was done by Sanger’s method, trimmed to remove vector sequences, assembled by CAP3 [80] and used for all further bioinformatics analyses. Comparisons with other known sequences revealed identification of full length BtTOC1; BtCOA; BtFT1, 2, 3, 4 genes. Although, the other four genes (BtLHY, BtZTL, BtGI and BtCOB) could be partially sequenced, biologically important domain regions were mostly present in the sequenced regions. All sequence data were deposited at NCBI (http://www.ncbi.nlm.nih.gov/) BtFT1 (KT003820), BtFT2 (KT003821), BtFT3 (KU726232), BtFT4 (KX290774), BtCOA (KY249523), BtCOB (MF983714), BtTOC1 (KY249524), BtLHY (MF983713), BtZTL (MF983715), BtGI (MF983716).
Sequence data and phylogenetic analyses
The amino acid sequences of the identified B. tulda genes were aligned with other related sequences using the Clustal W program. The sequences were compared to that of available sequences from related monocots genomes such as Oryza sativa, Phyllostachys meyeri, P. heterocycla, P. violascens, Brachypodium distachyon, Sorghum bicolor, Hordeum vulgare, Zea mays and Triticum aestivum. The phylogenetic tree was constructed by the NJ method with Mega 7 software [81]. Bootstrap analysis with values for 1000 replicates was conducted to estimate nodal support. All available literatures were consulted to identify specific amino acid residues within the target genes that are involved in significant biological functions.
In silico study on the molecular interactions between individual BtFT and Os14-3-3 proteins
Due to unavailability of crystal structures of BtFT1-4 and sequence/and structure of Bt-14-3-3, interaction between the BtFT-14-3-3 pairs was investigated, keeping 14-3-3 structural coordinates [36] constant from rice Os14-3-3. Homology models of BtFT1-4, which were 86-88% identical to their rice homologue OsHd3a, were built using the web version of MODELLER [82]. Interaction analyses were carried out using PyMOL.
Gene expression analyses by real time RT-qPCR method
Gene specific primers were designed from the coding sequences of the targeted genes to measure their transcriptional expression level by real time RT-qPCR analyses (Additional file 1: Table S1). Sequences of four BtFT gene alleles were so similar that it was rather impossible to design individual primers for each. Therefore, one pair of primers was designed for BtFT1 and 2, while another was designed for BtFT3 and 4 and that too were designed only based on one nucleotide sequence divergence at the 3′ end. The identity of the amplified gene products was confirmed by sequencing the amplified PCR products. SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad, USA) was used to measure the expression level of the targeted genes in CFX connect real-time PCR detection system (Bio Rad). The amplification conditions were 30 s at 95 °C, 40 cycles of 10 s at 94 °C and 40 s at 55 or 64 °C. A standard dissociation curve analyses was conducted to confirm the absence of any primer dimers in the amplified products. Data were normalized using eIF4α as the reference gene and relative fold change in gene expression was estimated following the 2-ΔΔCt method [83]. In a comprehensive study we have shown that elF4α is one of the most stable reference genes in B. tulda (data unpublished), therefore was used for data normalization in the current study.
Additional files
Table S1. A summary of the oligonucleotides used in this study for different purposes. (DOC 90 kb)
Figure S1. Two PCR amplified bands using degenerate primers specific for FLOWERING LOCUS T (FT). (TIFF 472 kb)
Acknowledgements
The research results reported in this paper are funded by Council of Scientific and Industrial Research, India [38(1386)/14/EMR-II], Department of Biotechnology, India (BT/PR10778/PBD/16/1070/2014) and FRPDF grant of Presidency University. PB acknowledges a JRF fellowship from University Grant Commission, India. We also thank Prof. Anuradha Lohia for her constant encouragement and Prof. James Westwood, Virginia Tech for carefully editing the manuscript.
Funding
This work was supported by Council of Scientific and Industrial Research, India [38(1386)/14/EMR-II], Department of Biotechnology (BT/PR10778/PBD/16/1070/2014), India and FRPDF grant, Presidency University. Funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All sequence data that support the findings of this study have been deposited in NCBI (http://www.ncbi.nlm.nih.gov/) with the accession numbers KT003820 (BtFT1), KT003821 (BtFT2), KU726232 (BtFT3), KX290774 (BtFT4), KY249523 (BtCOA), MF983714 (BtCOB), KY249524 (BtTOC1), MF983713 (BtLHY), MF983715 (BtZTL), MF983716 (BtGI).
Abbreviations
- BLAST
Basic Local Alignment Search Tool
- CCT
CONSTANS, CONSTANS -like, TIMING OF CAB EXPRESSION 1
- COA
CONSTANS A
- COB
CONSTANS B
- COLs
CONSTANS like genes
- CS
Culm sheath
- DNP
Day neutral plant
- E
Early staged inflorescence bud
- eIF4α
Eukaryotic initiation factor 4α
- ESTs
Expressed sequence tags
- FL
Flag leaf
- FLR
Floret
- FT
FLOWERING LOCUS T
- GA3
Gibberellic acid 3
- GI
GIGANTEA
- GL
Glume
- HAP
Heme activator protein
- IN
Inter node
- L
Late staged inflorescence bud
- LDP
Long-day plant
- LHY
LATE ELONGATED HYPOCOTYL
- LM
Lemma
- LOV
Light oxygen voltage
- M
Middle staged inflorescence bud
- PEBP
Phosphatidyl ethanolamine binding protein
- PFL
Possible flag leaf
- PL
Palea
- PSL
Pseudo spikelet
- R
Root
- RH
Rhizome
- SAM
Shoot apical meristem
- SDP
Short-day plant
- SE
Standard error
- TFL1
TERMINAL FLOWER1
- TOC1
TIMING OF CAB EXPRESSION1
- YLF
Young leaf from flowering culm
- YLN
Young leaf from non-flowering culm
- ZTL
ZEITLUPE
Authors’ contributions
MD and AP conceptualized the research and designed the experiments. SD performed gene cloning, sequencing and diurnal experiments. PB performed tissue collection, nucleic acids extraction and all sequence data analyses. SC carried out the real time RT-qPCR gene expression analyses. DM performed in silico molecular interaction studies. MD wrote the article with help from all the co-authors. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The plant materials used in this study have been collected from their natural habitat located at Shyamnagar (22.83° N, 88.40° E) and Bandel (22.93° N, 88.38° E), West Bengal, India during April, 2013 to July, 2017. As per the Convention on the International Trade in Endangered Species of Wild Fauna and Flora (CITES) guidelines, Bambusa does not fall under the category of endangered species. The studied plant Bambusa tulda is a tropical bamboo wildly grown and profusely available in West Bengal.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4571-7) contains supplementary material, which is available to authorized users.
Contributor Information
Smritikana Dutta, Email: smritikana.rs@presiuniv.ac.in.
Prasun Biswas, Email: prasun.rs@presiuniv.ac.in.
Sukanya Chakraborty, Email: sukanya.rs@presiuniv.ac.in.
Devrani Mitra, Email: devrani.dbs@presiuniv.ac.in.
Amita Pal, Email: amita@jcbose.ac.in.
Malay Das, Email: malay.dbs@presiuniv.ac.in.
References
- 1.Janzen DH. Why bamboos wait so long to flower. Annu Rev Ecol Syst. 1976;7:347–391. doi: 10.1146/annurev.es.07.110176.002023. [DOI] [Google Scholar]
- 2.Bhattacharya S, Das M, Bar R, Pal A. Morphological and molecular characterization of Bambusa tulda with a note on flowering. Ann Bot. 2006;98:529–535. doi: 10.1093/aob/mcl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharya S, Ghosh JS, Das M, Pal A. Morphological and molecular characterization of Thamnocalamus spathiflorus subsp. spathiflorus at population level. Plant Syst Evol. 2009;282:13–20. doi: 10.1007/s00606-008-0092-1. [DOI] [Google Scholar]
- 4.John CK, Nadgauda RS. Bamboo flowering and famine. Curr Sci. 2002;82:261–262. [Google Scholar]
- 5.Giordano CV, Sánchez RA, Austin AT. Gregarious bamboo flowering opens a window of opportunity for regeneration in a temperate forest of Patagonia. New Phytol. 2009;181:880–889. doi: 10.1111/j.1469-8137.2008.02708.x. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Franklin SB, Lu Z, Rude BJ. Delayed flowering in bamboo: evidence from Fargesia qinlingensis in the Qinling mountains of China. Front Plant Sci. 2016;7:151. doi: 10.3389/fpls.2016.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol. 2015;66:441–464. doi: 10.1146/annurev-arplant-043014-115555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouché F, Woods DP, Amasino RM. Winter memory throughout the plant kingdom: different paths to flowering. Plant Physiol. 2017;173:27–35. doi: 10.1104/pp.16.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng JZ, Zhou YP, Lv TX, Xie CP, Tian CE. Research progress on the autonomous flowering time pathway in Arabidopsis. Physiol Mol Biol Plants. 2017;23:477. doi: 10.1007/s12298-017-0458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung CY, Qiu J, Sun YH, Chen J, Kittur FS, Henny RJ, Jin G, Fan L, Xie J. Gibberellin deficiency is responsible for shy-flowering nature of Epipremnum aureum. Sci Rep. 2016;6:28598. doi: 10.1038/srep28598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Putterill J, Laurie R, Macknight R. It’s time to flower: the genetic control of flowering time. Bioessays. 2004;26:363–373. doi: 10.1002/bies.20021. [DOI] [PubMed] [Google Scholar]
- 12.Garner WW, Allard HA. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J Agr Res. 1920;18:553–606. [Google Scholar]
- 13.McClung CR. Circadian rhythms in plants. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:139–162. doi: 10.1146/annurev.arplant.52.1.139. [DOI] [PubMed] [Google Scholar]
- 14.Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- 15.Valverde F. CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J Exp Bot. 2011;62:2453–2463. doi: 10.1093/jxb/erq449. [DOI] [PubMed] [Google Scholar]
- 16.Colasanti J, Coneva V. Mechanisms of floral induction in grasses: something borrowed something new. Plant Physiol. 2009;149:56–62. doi: 10.1104/pp.108.130500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biswas P, Chakraborty S, Dutta S, Pal A, Das M. Bamboo flowering from the perspective of comparative genomics and transcriptomics. Front Plant Sci. 2016;7:1900. doi: 10.3389/fpls.2016.01900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kellogg EA. Flowering plants. Monocots: Poaceae. NewYork, NY: Springer; 2015. [Google Scholar]
- 19.Peng Z, Lu Y, Li L, Zhao Q, Feng Q, Gao Z, et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla) Nat Genet. 2013;45:456–461. doi: 10.1038/ng.2569. [DOI] [PubMed] [Google Scholar]
- 20.Lin XC, Chow TY, Chen HH, Liu CC, Chou SJ, Huang BL, et al. Understanding bamboo flowering based on large-scale analysis of expressed sequence tags. Genet Mol Res. 2010;9:1085–1093. doi: 10.4238/vol9-2gmr804. [DOI] [PubMed] [Google Scholar]
- 21.Liu M, Qiao G, Jiang J, Yang H, Xie L, Xie J, et al. Transcriptome sequencing and de novo analysis for ma bamboo (Dendrocalamus latiflorus Munro) using the illumina platform. PLoS One. 2012;7:e46766. doi: 10.1371/journal.pone.0046766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang XM, Zhao L, Larson-Rabin Z, Li DZ, Guo ZH. De novo sequencing and characterization of the floral transcriptome of Dendrocalamus latiflorus (Poaceae: Bambusoideae) PLoS One. 2012;7:e42082. doi: 10.1371/journal.pone.0042082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Zhang Y, Zhang C, Qi F, Li X, Mu S, et al. Characterization of the floral transcriptome of moso bamboo (Phyllostachys edulis) at different flowering developmental stages by transcriptome sequencing and RNA-seq analysis. PLoS One. 2014;9:e98910. doi: 10.1371/journal.pone.0098910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shih MC, Chou ML, Yue JJ, Hsu CT, Chang WJ, Ko SS, et al. BeMADS1 is a key to delivery MADSs into nucleus in reproductive tissues- De novo characterization of Bambusa edulis transcriptome and study of MADS genes in bamboo floral development. BMC Plant Biol. 2014;14:179. doi: 10.1186/1471-2229-14-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge W, Zhang Y, Cheng Z, Hou D, Li X, Gao J. Main regulatory pathways, key genes, and microRNAs involved in flower formation and development of moso bamboo (Phyllostachys edulis) Plant Biotechnol J. 2016;15:82–96. doi: 10.1111/pbi.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wysocki WP, Ruiz-Sanchez E, Yin Y, Duvall MR. The floral transcriptomes of four bamboo species (Bambusoideae; Poaceae): support for common ancestry among woody bamboos. BMC Genomics. 2016;17:384. doi: 10.1186/s12864-016-2707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao H, Dong L, Sun H, Li L, Lou Y, et al. Comprehensive analysis of multitissue transcriptome data and the genome-wide investigation of GRAS family in Phyllostachys edulis. Sci Rep. 2016;6:27640. doi: 10.1038/srep27640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin E, Peng H, Jin Q, Deng M, Li T, Xiao XC, et al. Identification and characterization of two bamboo (Phyllostachys praecox) AP1/SQUA-like MADS-box genes during floral transition. Planta. 2009;231:109–120. doi: 10.1007/s00425-009-1033-0. [DOI] [PubMed] [Google Scholar]
- 29.Liu S, Ma T, Ma L, Lin X. Ectopic expression of PvSOC1, a homolog of SOC1 from Phyllostachys violascens, promotes flowering in Arabidopsis and rice. Acta Physiol Plantarum. 2016;38:166. doi: 10.1007/s11738-016-2186-7. [DOI] [Google Scholar]
- 30.Liu S, Qi TT, Ma JJ, Ma T, Ma L, Lin X. Ectopic expression of a SOC1 homolog from Phyllostachys violascens alters flowering time and identity of floral organs in Arabidopsis thaliana. Trees. 2016;30:1–13. doi: 10.1007/s00468-015-1346-5. [DOI] [Google Scholar]
- 31.Liu SN, Zhu LF, Lin XC, Ma LY. Overexpression of the repressor gene PvFRI-L from Phyllostachys violascens delays flowering time in transgenic Arabidopsis thaliana. Biol Plantarum. 2016;60:401–409. doi: 10.1007/s10535-016-0614-6. [DOI] [Google Scholar]
- 32.Zhu L, Shi Y, Zang Q, Shi Q, Liu S, Xu Y, et al. Functional analysis of PI-like gene in relation to flower development from bamboo (Bambusa oldhamii) J Genet. 2016;95:71–78. doi: 10.1007/s12041-015-0605-y. [DOI] [PubMed] [Google Scholar]
- 33.Griffiths S, Dunford RP, Coupland G, Laurie DA. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003;131:1855–1867. doi: 10.1104/pp.102.016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, et al. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006;46:462–476. doi: 10.1111/j.1365-313X.2006.02706.x. [DOI] [PubMed] [Google Scholar]
- 35.Hanzawa Y, Money T, Bradley D. A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci U S A. 2005;102:7748–7753. doi: 10.1073/pnas.0500932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- 37.Yuan JL, Yue JJ, Gu XP, Lin CS. Flowering of woody bamboo in tissue culture systems. Front Plant Sci. 2017;8:1589. doi: 10.3389/fpls.2017.01589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 39.Murakami M, Tago Y, Yamashino T, Mizuno T. Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48:110–121. doi: 10.1093/pcp/pcl043. [DOI] [PubMed] [Google Scholar]
- 40.Tsuji H, Taoka K, Shimamoto K. Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol. 2010;14:45–52. doi: 10.1016/j.pbi.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 41.Zhao J, Huang X, Ouyang X, Chen W, Du A, Zhu L, et al. OsELF3-1, an ortholog of Arabidopsis EARLY FLOWERING 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS One. 2012;7:e43705. doi: 10.1371/journal.pone.0043705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim JY, Song HR, Taylor BL, Carre IA. Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J. 2003;22:935–944. doi: 10.1093/emboj/cdg075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 44.Sheng P, Wu F, Tan J, Zhang H, Ma W, Chen L, et al. A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice. Plant Mol Biol. 2016;92:209–222. doi: 10.1007/s11103-016-0506-3. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Peng Q, Chen GX, Li XH, Wu CY. OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Mol Plant. 2013;6:202–215. doi: 10.1093/mp/sss062. [DOI] [PubMed] [Google Scholar]
- 46.Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T. The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003;44:1229–1236. doi: 10.1093/pcp/pcg135. [DOI] [PubMed] [Google Scholar]
- 47.Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA. A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell. 2001;13:2659–2670. doi: 10.1105/tpc.13.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yanovsky MJ, Kay SA. Signaling networks in the plant circadian system. Curr Opin Plant Biol. 2001;4:429–435. doi: 10.1016/S1369-5266(00)00196-5. [DOI] [PubMed] [Google Scholar]
- 49.Kim J, Geng R, Gallenstein RA, Somers DE. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development. 2013;140:4060–4069. doi: 10.1242/dev.096651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu F, Price BW, Haider W, Seufferheld G, Nelson R, Hanzawa Y. Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. PLoS One. 2014;9:e85754. doi: 10.1371/journal.pone.0085754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong AC, Hecht VF, Picard K, Diwadkar P, Laurie RE, Wen J, et al. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front Plant Sci. 2014;5:486. doi: 10.3389/fpls.2014.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao D, Li Y, Lu S, Wang J, Nan H, Li X, et al. GmCOL1a and GmCOL1b function as flowering repressors in soybean under long-day conditions. Plant Cell Physiol. 2015;56:2409–2422. doi: 10.1093/pcp/pcv152. [DOI] [PubMed] [Google Scholar]
- 53.Mulki MA, von Korff M. CONSTANS controls floral repression by up-regulating VERNALIZATION2 (VRN-H2) in barley. Plant Physiol. 2016;170:325–337. doi: 10.1104/pp.15.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan J, Jin M, Wang J, Wu F, Sheng P, Cheng Z, et al. OsCOL10, a CONSTANS-Like gene, functions as a flowering time repressor downstream of Ghd7 in rice. Plant Cell Physiol. 2016;57:798–812. doi: 10.1093/pcp/pcw025. [DOI] [PubMed] [Google Scholar]
- 55.Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 2002;16:2006–2020. doi: 10.1101/gad.999202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- 57.Kim SK, Yun CH, Lee JH, Jang YH, Park HY, Kim JK. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta. 2008;228:355–365. doi: 10.1007/s00425-008-0742-0. [DOI] [PubMed] [Google Scholar]
- 58.Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- 59.Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci U S A. 2006;103:6398–6403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carmona MJ, Calonje M, Martínez-Zapater JM. The FT/TFL1 gene family in grapevine. Plant Mol Biol. 2007;63:637–650. doi: 10.1007/s11103-006-9113-z. [DOI] [PubMed] [Google Scholar]
- 61.Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- 62.Kong F, Liu B, Xia Z, Sato S, Kim BM, Watanabe S, et al. Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 2010;154:1220–1231. doi: 10.1104/pp.110.160796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kotoda N, Hayashi H, Suzuki M, Igarashi M, Hatsuyama Y, Kidou S, et al. Molecular characterization of FLOWERING LOCUS T-like genes of apple (Malus x domestica Borkh.) Plant Cell Physiol. 2010;51:561–575. doi: 10.1093/pcp/pcq021. [DOI] [PubMed] [Google Scholar]
- 64.Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJ, et al. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science. 2010;330:1397–1400. doi: 10.1126/science.1197004. [DOI] [PubMed] [Google Scholar]
- 65.Hsu CY, Adams JP, Kim H, No K, Ma C, et al. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc Natl Acad Sci U S A. 2011;108:10756–10761. doi: 10.1073/pnas.1104713108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harig L, Beinecke FA, Oltmanns J, Muth J, Müller O, Rüping B, et al. Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J. 2012;72:908–921. doi: 10.1111/j.1365-313X.2012.05125.x. [DOI] [PubMed] [Google Scholar]
- 67.Hisamoto Y, Kobayashi M. Flowering habit of two bamboo species, Phyllostachys meyeri and Shibataea chinensis, analyzed with flowering gene expression. Plant Species Biol. 2013;28:109–117. doi: 10.1111/j.1442-1984.2012.00369.x. [DOI] [Google Scholar]
- 68.Guo X, Wang Y, Wang Q, Xu Z, Lin X. Molecular characterization of FLOWERING LOCUS T (FT) genes from bamboo (Phyllostachys violascens) J Plant Biochem Biotechnol. 2015;25:168–178. doi: 10.1007/s13562-015-0322-x. [DOI] [Google Scholar]
- 69.Lu KJ, Huang NC, Liu YS, Lu CA, Yu TS. Long-distance movement of Arabidopsis FLOWERING LOCUS T RNA participates in systemic floral regulation. RNA Biol. 2012;9:653–662. doi: 10.4161/rna.19965. [DOI] [PubMed] [Google Scholar]
- 70.Fan C, Hu R, Zhang X, Wang X, Zhang W, Zhang Q, et al. Conserved CO-FT regulons contribute to the photoperiod flowering control in soybean. BMC Plant Biol. 2014;14:9. doi: 10.1186/1471-2229-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takahashi Y, Teshima KM, Yokoi S, Innan H, Shimamoto K. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc Natl Acad Sci U S A. 2009;106:4555–4560. doi: 10.1073/pnas.0812092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Hopkins CJ, Hou J, Zou X, Wang C, Long Y, Kurup S, King GJ, Meng J. Promoter variation and transcript divergence in Brassicaceae lineages of FLOWERING LOCUS T. PLoS One. 2012;7:e47127. doi: 10.1371/journal.pone.0047127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ballerini ES, Kramer EM. In the light of evolution: a reevaluation of conservation in the CO-FT regulon andiIts role in photoperiodic regulation of flowering time. Front Plant Sci. 2011;2:81. doi: 10.3389/fpls.2011.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 75.Wang JW. Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot. 2014;65:4723–4730. doi: 10.1093/jxb/eru246. [DOI] [PubMed] [Google Scholar]
- 76.Wu L, Liu D, Wu J, Zhang R, Qin Z, Liu D, et al. Regulation of FLOWERING LOCUS T by a microRNA in Brachypodium distachyon. Plant Cell. 2013;25:4363–4377. doi: 10.1105/tpc.113.118620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qin Z, Wu J, Geng S, Feng N, Chen F, Kong X, et al. Regulation of FT splicing by an endogenous cue in temperate grasses. Nat Commun. 2017;8:14320. doi: 10.1038/ncomms14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Das M, Reichman JR, Haberer G, Welzl G, Aceituno FF, Mader MT, et al. A composite transcriptional signature differentiates responses towards closely related herbicides in Arabidopsis thaliana and Brassica napus. Plant Mol Biol. 2010;72:545–556. doi: 10.1007/s11103-009-9590-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Das M, Haberer G, Panda A, Das Laha S, Ghosh TC, Schäffner AR. Expression pattern similarities support the prediction of orthologs retaining common functions after gene duplication events. Plant Physiol. 2016;171:2343–2357. doi: 10.1104/pp.15.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanchez R, Sali A. Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc Natl Acad Sci U S A. 1998;95:13597–13602. doi: 10.1073/pnas.95.23.13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. A summary of the oligonucleotides used in this study for different purposes. (DOC 90 kb)
Figure S1. Two PCR amplified bands using degenerate primers specific for FLOWERING LOCUS T (FT). (TIFF 472 kb)
Data Availability Statement
All sequence data that support the findings of this study have been deposited in NCBI (http://www.ncbi.nlm.nih.gov/) with the accession numbers KT003820 (BtFT1), KT003821 (BtFT2), KU726232 (BtFT3), KX290774 (BtFT4), KY249523 (BtCOA), MF983714 (BtCOB), KY249524 (BtTOC1), MF983713 (BtLHY), MF983715 (BtZTL), MF983716 (BtGI).


