Abstract
Adaptive laboratory evolution (ALE) is a widely-used method for improving the fitness of microorganisms in selected environmental conditions. It has been applied previously to Escherichia coli K-12 MG1655 during aerobic exponential growth on glucose minimal media, a frequently used model organism and growth condition, to probe the limits of E. coli growth rate and gain insights into fast growth phenotypes. Previous studies have described up to 1.6-fold increases in growth rate following ALE, and have identified key causal genetic mutations and changes in transcriptional patterns. Here, we report for the first time intracellular metabolic fluxes for six such adaptively evolved strains, as determined by high-resolution 13C-metabolic flux analysis. Interestingly, we found that intracellular metabolic pathway usage changed very little following adaptive evolution. Instead, at the level of central carbon metabolism the faster growth was facilitated by proportional increases in glucose uptake and all intracellular rates. Of the six evolved strains studied here, only one strain showed a small degree of flux rewiring, and this was also the strain with unique genetic mutations. A comparison of fluxes with two other wild-type (unevolved) E. coli strains, BW25113 and BL21, showed that inter-strain differences are greater than differences between the parental and evolved strains. Principal component analysis highlighted that nearly all flux differences (95%) between the nine strains were captured by only two principal components. The distance between measured and flux balance analysis predicted fluxes was also investigated. It suggested a relatively wide range of similar stoichiometric optima, which opens new questions about the path-dependency of adaptive evolution.
Keywords: Adaptive evolution, Escherichia coli, metabolic flux analysis, flux balance analysis, fast growth
1. INTRODUCTION
Adaptive laboratory evolution (ALE) is a method in which microorganisms are continuously cultured in a controlled environment over many generations, allowing for fitness improvement through the accumulation of beneficial mutations. ALE has been applied to increasing chemical tolerance (Atsumi et al., 2010; Horinouchi et al., 2010; Mundhada et al., 2017; Reyes et al., 2012), rates of growth on diverse substrates (Cordova et al., 2016; Herring et al., 2006; Lee and Palsson, 2010; Sandberg et al., 2017), and gaining general insight into microbial responses to environmental or genetic perturbations (Charusanti et al., 2010; Fong and Palsson, 2004; Tenaillon et al., 2012). Following an ALE experiment, the resulting strains are sequenced to identify genetic mutations (Herring et al., 2006). The difficulty inherent in identifying causal mutations has led to the practice of performing multiple independent ALE experiments and using the frequency of mutations to guide analysis (LaCroix et al., 2015). Phenotypic characterization is then necessary to quantitatively describe the extent of the fitness improvement (e.g. increase in growth rate) and the associated physiology. Detailed cellular characterizations involving omics techniques such as transcriptomics, proteomics, metabolomics, and fluxomics can then enable systems and pathway-level analysis of the phenotype and its enabling mechanisms. Ideally, such approaches would result in genotype-phenotype insights that improve our general scientific understanding of the cell system and inform future rational engineering efforts (Long and Antoniewicz, 2014a).
Exponential aerobic growth of E. coli K-12 MG1655 on glucose minimal media is arguably the most widely used combination of organism and condition in basic science and biotechnology (Janssen et al., 2005). ALE applied in this context, particularly serial passaging of batch cultures such that the exponential phase is maintained, selects for increased growth rate. Such efforts probe the limits of E. coli growth performance and allow for the study of fast growing phenotypes that might be useful in biotechnology. Previously, LaCroix et al. reported the phenotypes and transcriptional analysis of ten independent ALE experiments of E. coli MG1655 (LaCroix et al., 2015). They reported up to 1.6-fold increases in growth rate, and identified frequent causal mutations in rpoB and intergenic regions of hns/tdk and pyrE/rph. The pyrE/rph mutation ameliorates a well-characterized strain-specific defect in pyrimidine biosynthesis (Jensen, 1993), and the other two likely result in broad transcriptional changes as rpoB and hns are global regulators. Transcriptomic analyses revealed increases in expression of genes associated with protein production (amino acid metabolism, transcription, translation, folding), glucose transport, and glycolysis, and reductions in enzymes involved in the TCA cycle and glyoxylate shunt. Similarly, Sandberg et al. reported the results of six independent ALE experiments, also using E. coli MG1655 and aerobic exponential growth on glucose minimal media (Sandberg et al., 2016). Here, ALE was carried out over 40 days, or approximately 1000 generations, through serial passaging such that stationary phase was avoided. The glucose was 13C labeled to test the hypothesis that a subtle kinetic isotope effect may influence metabolism and the trajectory of ALE, but this was disproven in isotopic preference studies described previously (Sandberg et al., 2016). Furthermore, the final growth rates and most frequent mutations were very similar to those in (LaCroix et al., 2015), and the same key mutations (rpoB, pyrE/rph, and hsns/tdk) occurred with high frequency in the six ALE experiments.
The genetic and transcription level changes in these studies suggest the possibility of broad metabolic shifts in the adaptively evolved strains. However, to our knowledge, intracellular metabolic fluxes of such strains have not yet been reported. In this study, we applied high-resolution 13C-metabolic flux analysis (13C-MFA) (Antoniewicz, 2015) to the six ALE strains previously described in (Sandberg et al., 2016) to determine whether their high growth rate is enabled by or associated with rewiring of central carbon metabolism. For additional context, the fluxes of the ALE strains were compared to the parent MG1655 strain, a related K-12 strain (BW25113), and a more distantly related E. coli strain BL21. Finally, flux balance analysis (FBA) was performed to compare the calculated optimal stoichiometric solution to the measured in vivo fluxes.
2. MATERIALS AND METHODS
2.1. Materials
Chemicals and M9 minimal medium were purchased from Sigma-Aldrich (St. Louis, MO). Isotopic tracers were purchased from Cambridge Isotope Laboratories (Tewksbury, MA): [1,2-13C]glucose (99.7 %) and [1,6-13C]glucose (99.2 % 13C). The isotopic purity and enrichment of the tracers were validated by GC-MS analysis as described in (Sandberg et al., 2016) and (Cordova and Antoniewicz, 2016). All solutions were sterilized by filtration.
2.2. Strains and growth conditions
E. coli BL21(DE3) was obtained from Invitrogen (Cat. No. C600003). E. coli BW25113 was obtained from the Keio collection (GE Healthcare Dharmacon, Cat. No. OEC5042). The MG1655 wild-type and adaptively evolved (ALE) strains were previously described in (Sandberg et al., 2016). The wild-type was K-12 MG1655 (ATCC 700926). Six independent cultures were adaptively evolved in M9 minimal glucose medium for 40 days, corresponding to an average of 963 generations, or 2.82 x 1012 cumulative cell divisions (CCD). Passaging was frequent enough to avoid glucose depletion and initiation of stationary phase. In this study, all strains were cultured aerobically in glucose M9 minimal medium at 37°C in mini-bioreactors with 10 mL working volume as previously described (Long et al., 2016b). Pre-cultures were grown overnight and then used to inoculate the experimental culture at an OD600 of 0.01. For 13C-MFA, glucose tracers were added at the beginning of the culture. Cells were harvested (1 mL samples) for GC-MS analysis at mid-exponential growth when OD600 was approximately 0.7. In all cases, parallel tracer experiments were performed using [1,2-13C]glucose and [1,6-13C]glucose. These tracers were previously determined to be optimal for high-resolution 13C-MFA of E. coli (Crown et al., 2016).
2.3. Analytical methods
Cell growth was monitored by measuring the optical density at 600nm (OD600) using a spectrophotometer (Eppendorf BioPhotometer). The OD600 values were converted to cell dry weight concentrations using previously determined OD600-dry cell weight relationship for E. coli (1.0 OD600 = 0.32 gDW/L; molecular weight of dry biomass = 24.6 gDW/C-mol (Long et al., 2016b). After centrifugation of the samples, the supernatant was separated from the biomass pellet. Acetate concentrations in the supernatant were determined using an Agilent 1200 Series HPLC (Gonzalez et al., 2017). Glucose concentrations were determined using a YSI 2700 biochemistry analyzer (YSI, Yellow Springs, OH). Growth rate was calculated using linear regression of the natural logarithm of the OD600 and time, and biomass yield via regression of biomass dry weight and glucose concentration in the medium.
2.4. Gas chromatography-mass spectrometry
GC-MS analysis was performed on an Agilent 7890B GC system equipped with a DB-5MS capillary column (30 m, 0.25 mm i.d., 0.25 μm-phase thickness; Agilent J&W Scientific), connected to an Agilent 5977A Mass Spectrometer operating under ionization by electron impact (EI) at 70 eV. Helium flow was maintained at 1 mL/min. The source temperature was maintained at 230°C, the MS quad temperature at 150°C, the interface temperature at 280°C, and the inlet temperature at 250°C. GC-MS analysis of tert-butyldimethylsilyl (TBDMS) derivatized proteinogenic amino acids was performed as described in (Long and Antoniewicz, 2014b). Labeling of glucose (derived from glycogen) and ribose (derived from RNA) were determined as described in (Long et al., 2016a; McConnell and Antoniewicz, 2016). In all cases, mass isotopomer distributions were obtained by integration (Antoniewicz et al., 2007a) and corrected for natural isotope abundances (Fernandez et al., 1996). Measurement errors of 0.3% were assumed for all measured mass isotopomers (Antoniewicz et al., 2007a).
2.5. Metabolic network model and 13C-metabolic flux analysis
The metabolic network model used for 13C-MFA is provided in Supplemental Materials. The model is based on the E. coli model described previously (Crown et al., 2015; Gonzalez et al., 2016), which includes all major metabolic pathways of central carbon metabolism, lumped amino acid biosynthesis reactions, and a lumped biomass formation reaction. Updates to the model include: i) making the reactions between PEP and pyruvate (Long et al., 2017), and between α-ketoglutarate and succinyl-CoA reversible; ii) allowing for deamination of serine to pyruvate; and iii) modeling atmospheric CO2 dilution of each labeling experiment independently (Leighty and Antoniewicz, 2012).
13C-MFA calculations were performed using the Metran software (Yoo et al., 2008), which is based on the elementary metabolite units (EMU) framework (Antoniewicz et al., 2007b). Fluxes were estimated by minimizing the variance-weighted sum of squared residuals (SSR) between the measured and model predicted mass isotopomer distributions and acetate yield using non-linear least-squares regression. All measured mass isotopomers are provided in Supplemental Materials. For integrated analysis of parallel labeling experiments, the data sets were fitted simultaneously to a single flux model as described previously (Leighty and Antoniewicz, 2013). Flux estimation was repeated 10 times starting with random initial values for all fluxes to find a global solution. At convergence, accurate 95% confidence intervals were computed for all estimated fluxes by evaluating the sensitivity of the minimized SSR to flux variations. Precision of estimated fluxes was determined as follows :
To describe fractional labeling of metabolites, G-value parameters were included in 13C-MFA. As described previously (Antoniewicz et al., 2007c), the G-value represents the fraction of a metabolite pool that is produced during the labeling experiment, while 1-G represents the fraction that is naturally labeled, i.e. from the inoculum. By default, one G-value parameter was included for each measured metabolite in each data set. Reversible reactions were modeled as separate forward and backward fluxes. Net and exchange fluxes were determined as follows: vnet = vf-vb; vexch = min(vf, vb).
2.6. Goodness-of-fit analysis
To determine the goodness-of-fit, 13C-MFA fitting results were subjected to a χ2-statistical test. In short, assuming that the model is correct and data are without gross measurement errors, the minimized SSR is a stochastic variable with a χ2-distribution (Antoniewicz et al., 2006). The number of degrees of freedom is equal to the number of fitted measurements n minus the number of estimated independent parameters p. The acceptable range of SSR values is between χ2α/2(n-p) and χ21-α/2(n-p), where α is a certain chosen threshold value, for example 0.05 for 95% confidence interval.
2.7. Flux balance analysis (FBA) and flux variability analysis (FVA)
For FBA and FVA calculations, the COBRA Toolbox 2.0 implemented in Matlab was used (Schellenberger et al., 2011). Gurobi was used for the linear solver (http://www.gurobi.com/). The E. coli iAF1260 genome scale model was used (Feist et al., 2007) for all calculations. The measured glucose and oxygen uptake rate were used as constraints as described in the text. All additional import and export fluxes, as well as internal constraints, were identical to those contained in the iAF1260 model file “Ec_iAF1260_flux1.xml”.
3. RESULTS AND DISCUSSION
3.1. Growth and physiology
In this study, three unevolved E. coli strains, i.e. BL21(DE3), BW25113, and MG1655, and six adaptively evolved MG1655 strains were investigated. The complete list of specific mutations observed in the six evolved strains, labeled ALE-1 through ALE-6, have been reported in (Sandberg et al., 2016). Briefly, the same key mutations previously identified as causal (LaCroix et al., 2015) were recapitulated in the evolved strains, particularly various insertion sequence mutations in the hns/tdk intergenic region (ALE-1, ALE-3, ALE-5, ALE-6), deletions in the pyrE/rph intergenic region of either 1 bp (ALE-2, ALE-5) or 82 bp (ALE-1, ALE-3, ALE-4) and SNP’s in rpoB (ALE-1, ALE-2, ALE-3, ALE-4, ALE-5). ALE-6 was noticeably distinct from the other five strains, as it was lacking pyrE/rph and rpoB mutations, but instead had a unique rpoC mutation.
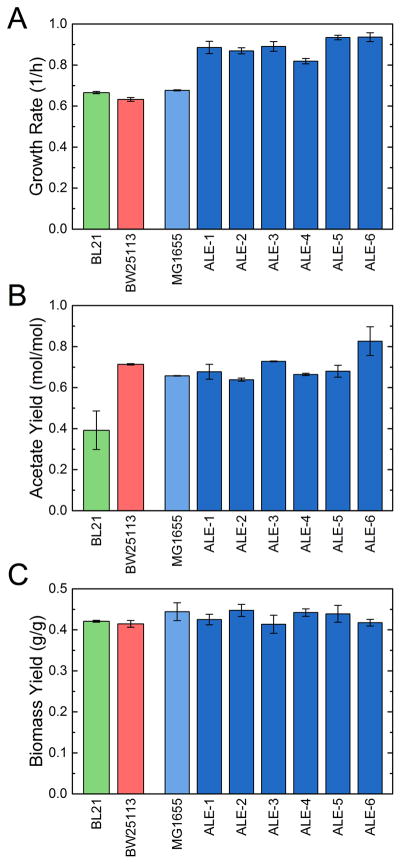
The growth rates, biomass and acetate yields, and glucose uptake rates for all nine strains investigated here (i.e. three unevolved and six evolved strains) are summarized in Figs. 1 and 2. The growth physiology of the BW25113 strain was previously described (Long et al., 2016b). Like MG1655, this strain is a K-12 derivative and thus is closely related, whereas BL21 is a more distantly related E. coli strain. All wild-type strains had similar growth rates (0.63 to 0.68 h−1) (Fig. 1A), and the ALE strains grew significantly faster (approximately 0.9 h−1) as expected and previously reported (Sandberg et al., 2016). This represents a 28–38% increase in fitness (i.e. growth rate) under our experimental conditions. Most of the strains had similar acetate production phenotypes (Fig. 1B). The K-12 strains produced approximately 0.7 mol acetate per mol glucose, in good agreement with previous reports (Chen et al., 2011; Leighty and Antoniewicz, 2013; Rahman and Shimizu, 2008). The BL21 strain produced significantly less acetate (0.39 mol/mol), a phenotype which has also been well characterized previously (Monk et al., 2016; Waegeman et al., 2012, 2011). In fact, previous studies have reported even lower acetate yields of 0.2 mol/mol and less, which may indicate a relatively larger variability in BL21 strains compared to K-12 strains. Interestingly, the acetate phenotypes following adaptive evolution were mostly unchanged, with the most significant change being an increase in acetate yield in ALE-6 from 0.66 to 0.83 mol/mol.
Figure 1.
Physiology of all strains during exponential growth on glucose minimal medium. The three E. coli wild-type strains, BL21 (green), BW25113 (red), and MG1655 (blue) are shown along with the adaptively evolved MG1655 strains (ALE, dark blue). Growth rates (A) were measured in triplicate cultures, and acetate yields (B) on media HPLC measurements of duplicate cultures. Error bars indicate standard errors of the mean. Biomass yield (C) was based on regression of substrate and biomass measurements in a culture, with error bars reflecting the uncertainty in the parameter fitting.
Figure 2.
Substrate uptake rates of all strains during exponential growth on glucose minimal medium. The three E. coli wild-type strains, BL21 (green), BW25113 (red), and MG1655 (blue) are shown along with the adaptively evolved MG1655 strains (ALE, dark blue). Glucose uptake rates (A) were calculated from the growth rate and biomass yield (Fig. 1), and oxygen uptake rate (B) was estimated by 13C-MFA. Error bars reflect standard errors.
The biomass yields were relatively consistent across all strains (0.41 to 0.44 gDW/gglc) (Fig. 1C). The directly measured yields and those estimated by 13C-MFA are shown in Fig. S2. While there was strong overall consistency, a larger difference was suggested between BL21 (0.45 g/g) and ALE-6 (0.38 g/g), as would be expected given the divergent acetate yield phenotypes. This suggests that increases in growth rate that were attained during adaptive evolution came not from increased carbon efficiency, but rather from increased overall metabolic rate. This was also reflected in the calculated glucose uptake rates (Fig. 2A), which increased from 8.5 mmol/gDW/h in the wild-type to up to 12.5 mmol/gDW/h in ALE-6. The oxygen uptake rates (Fig. 2B) also increased significantly in the ALE strains relative to the wild-type, but reached levels similar or only slightly higher (17 mmol/gDW/h) than the oxygen uptake rate of BL21 (15 mmol/gDW/h). Notably, the strain with the unique genetic mutations, i.e. ALE-6, had the highest rates of glucose uptake and acetate excretion.
3.2. 13C metabolic flux analysis
To quantify intracellular metabolic fluxes supporting the observed increases in growth and glucose uptake rates, high-resolution 13C-MFA was performed. For each strain, two parallel labeling experiments were performed with [1,2-13C]glucose and [1,6-13C]glucose, as this was previously identified to provide optimal precision in flux estimates throughout E. coli central carbon metabolism (Crown et al., 2016). Labeling of proteinogenic amino acids, labeling of ribose moiety of RNA, and glucose moiety of glycogen (Long et al., 2016a) from each parallel experiment were fitted simultaneously, along with the measured acetate yield, to estimate fluxes. The measured mass isotopomer distributions (MID’s) and the estimated metabolic fluxes are provided in Supplemental Materials. Statistically acceptable fits were achieved in all cases, assuming GC-MS measurement errors of 0.3% (Antoniewicz et al., 2007a).
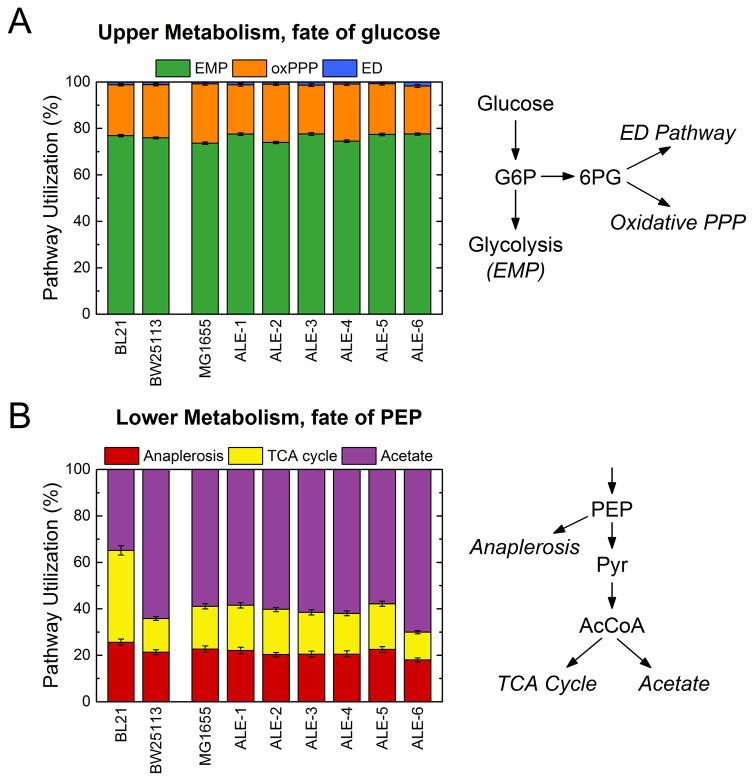
The results of 13C-MFA are summarized in Figs. 3 and 4. In Fig. 3, the distributions of fluxes through two key branch points in central carbon metabolism are shown for all strains. The first branch point (Fig. 3A) describes the split in upper central carbon metabolism between glycolysis (EMP pathway), the oxidative pentose phosphate pathway (oxPPP), and the Entner Doudoroff (ED) pathway. The relative usage of these three pathways was remarkably consistent among the nine strains studied here. The main route of glucose catabolism being the EMP pathway (74 to 78%), with almost all the rest going to the oxPPP. ED pathway usage was minimal, not exceeding 2% in any of these strains. There was a small increase in EMP usage in four of the six ALE strains relative to the parental strain. More variations between strains were observed in lower metabolism (Fig. 3B). The branch point here describes the fate of the lower glycolytic intermediate phosphoenolpyruvate (PEP), into anaplerosis (conversion to oxaloacetate via PPC), the TCA cycle via citrate synthase, or acetate production. The differences in the pathway usage here also reflect the differing acetate secretion phenotypes (Fig. 1B). Particularly, the BL21 strain has a much lower acetate flux (35% of PEP) and a correspondingly higher TCA cycle flux (40%) than all K-12 strains (15% for BW25113 and 18% for MG1655). In the adaptively evolved strains, the relative pathway usage is once again remarkably unchanged (compared to the parental strain), with the only significant difference being a higher acetate secretion and reduced TCA cycle flux in ALE-6. The constant relative intracellular pathway usage (i.e., fluxes normalized to these branch points or to glucose uptake rate) in the ALE strains corresponded to substantial and proportional increases in absolute flux throughout central carbon metabolism, as the glucose uptake rates were significantly elevated (Fig. 2A).
Figure 3.
Key flux branch points for all strains as measured by 13C-MFA. The upper flux branch point (A) reflects the fate of glucose into one of the EMP pathway (glycolysis), the oxidative pentose phosphate pathway (oxPPP) or the ED pathway. The lower branch point shown (B) reflects the fate of phosphoenolpyruvate (PEP) into anaplerosis, the TCA cycle via citrate synthase, or to acetate production. Error bars reflect the 95% confidence interval of the flux estimates.
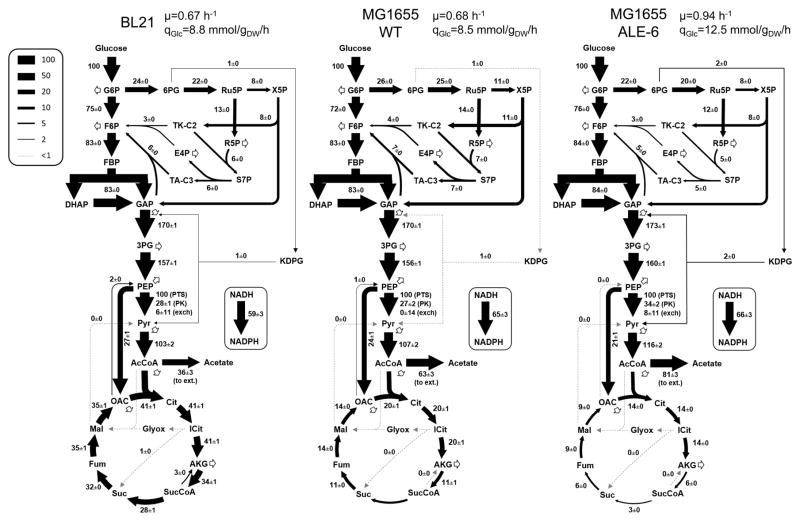
Figure 4.
Complete flux maps of selected strains. All fluxes in central carbon metabolism are shown for the BL21 strain, as well as the wild-type MG1655 and one of its evolved descendants (ALE-6). Growth and glucose uptake rates for each strain are listed above, and all intracellular fluxes shown are then normalized to 100 units of glucose uptake. The indicated uncertainties for fluxes represent standard errors of the estimates.
Detailed flux maps of central carbon metabolism are shown for three selected strains in Fig. 4. The wild-type BL21 and MG1655 are compared along with ALE-6, which was the fastest growing strain and exhibited the most unique phenotype of the evolved strains. The fluxes shown were normalized to 100 units of glucose uptake, with the growth and glucose uptake rates for each strain noted. As discussed above, there was a slight increase in normalized EMP flux from the wild-type to ALE-6, from 72% to 76% of glucose, at the expense of the oxPPP. The flux differences in lower metabolism, particularly the relative rate of TCA cycle and acetate secretion in these three strains, can also be seen here. No significant fluxes were observed in the ED pathway, glyoxylate shunt, malic enzyme, or PCK reactions in any of the studied strains. Given this, the two branch points described in Fig. 3 captured the main variations in the normalized intracellular fluxes of the strains. As noted above, ALE-6 was the most different from the wild-type, with elevated acetate yield and reduced TCA cycle usage. In absolute terms, however, the citrate synthase flux of ALE-6 (1.74 mmol/gDW/h) was quite similar to that of the wild-type (1.67 mmol/gDW/h). The normalized fluxes of the other ALE strains were highly conserved from the parental strain.
3.3 Cofactor metabolism
The measured intracellular fluxes can be used to calculate the contributions of individual pathways to the production or consumption of key cofactors in metabolism, including NADH and FADH2 (Fig. 5A), NADPH (Fig. 5B), and ATP (Fig. 5C) (shown here in absolute flux units). This analysis highlights that NADH is roughly evenly produced in glycolysis and the TCA cycle (approximately 40% and 50%, respectively, for BL21, and the reverse for the K-12 strains), and mostly consumed by the electron transport chain as part of oxidative phosphorylation in all strains. 15–23% of NADH is converted by transhydrogenase to NADPH, accounting for 41–55% of NADPH produced and supplementing the oxPPP and TCA cycle. The large TCA cycle flux in BL21 makes it a particularly significant source of NADPH in that strain (28%). All NADPH is utilized for biomass synthesis. Fig. 5C illustrates that ATP is mainly produced by oxidative phosphorylation (64–70%), with a smaller contribution from glycolysis (23–27%), and is consumed for biosynthesis and maintenance costs (under the category of “Other” in Fig. 5C). A constant P/O ratio of 2 was assumed for all strains to calculate ATP production, and the maintenance costs were estimated as the difference between total production and consumption for substrate uptake and growth. As a more conservative P/O ratio of 1.5 (Noguchi et al., 2004; Taymaz-Nikerel et al., 2010) would reduce the estimated ATP production and maintenance consumption rates somewhat, these results should be interpreted with caution. The inter-strain differences remain relevant though, as no mutations were observed in oxidative phosphorylation genes that would indicate a changing P/O ratio in the ALE strains. Given the conservation of normalized fluxes in the MG1655 strains, much of the variation in absolute cofactor rates is due to differences in glucose uptake and overall metabolic rate. In Supplemental Fig. S1, the cofactor balances are shown normalized to glucose uptake, where it is apparent that there are only very subtle differences in relative cofactor metabolism across the K-12 wild-types and ALE strains. The elevated TCA cycle of BL21 does contribute significantly more to cofactor production, and the increased use of oxidative phosphorylation results in a higher overall ATP yield.
Figure 5.
Quantitative cofactor balances. For each strain, the contributions of metabolic pathways to the production and consumption of cofactors are calculated in absolute units. Positive values reflect production of cofactor, and negative values reflect consumption. Shown are balances for NADH/FADH2 (lumped) (A), NADPH (B), and ATP (C). “Other” in the ATP panel represents the estimated ATP maintenance cost (here, assuming P/O ratio=2.0).
3.4. Principal component analysis and flux balance analysis
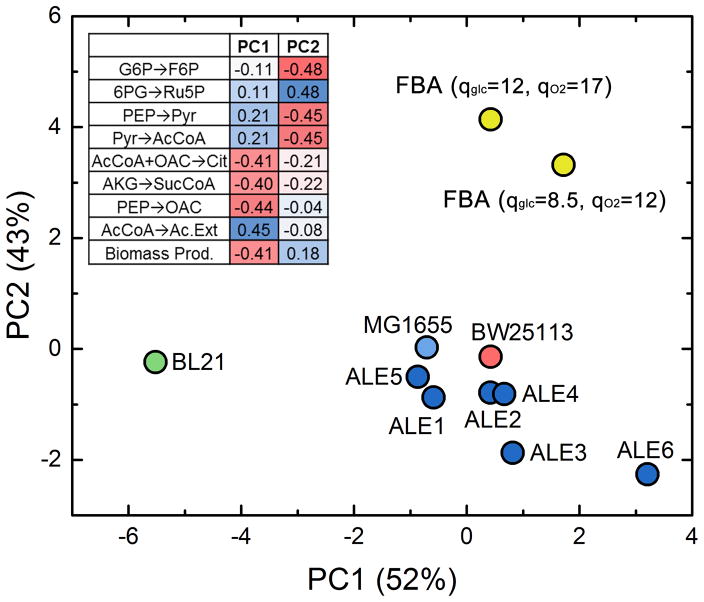
To further assess the degree of similarity between the metabolic flux profiles of the investigated strains, principal component analysis (PCA) was performed using nine key normalized fluxes in central carbon metabolism. PCA is a data reduction technique in which large multivariate data sets can be described in a new lower dimensional space in terms of principal components, which are linear combinations of the original variables. These principal components capture the maximum amount of original variation in the data. The results of PCA are shown in Fig. 6. The fluxes used for the analysis were selected from various representative intracellular and extracellular pathways, and excluded those with minimal flux (e.g. the ED pathway or glyoxylate shunt) where the inter-strain variance was not meaningful. The first two principal components capture nearly all (95%) of the flux variation, with PC1 (52%) reflecting the lower metabolism split between acetate production and the TCA cycle, and PC2 (43%) the upper split between glycolysis and oxPPP. This confirms our analysis above in Fig. 3, suggesting that these were the major areas of flux variance among the strains. All the K-12 strains, including BW25113, the MG1655 wild-type and ALE strains, clustered together in the PCA plot. ALE-6 was positioned at the extreme end of the group with a high PC1 value, reflecting its particularly low TCA cycle and high acetate flux, while BL21 was positioned at the opposite end with a low PC1 value.
Figure 6.
Principal component analysis of key normalized intracellular metabolic fluxes in measured strains and two flux balance analysis (FBA) simulations. The simulations were based on two different sets of substrate uptake constraints as noted. The coefficients of the top two principal components are shown in the table.
We were also interested to compare the flux phenotypes of the adaptively evolved strains to an ‘optimal’ flux distribution as predicted by flux balance analysis (FBA), a widely-used tool in metabolic engineering. FBA finds a set of fluxes that optimizes the maximal growth rate given substrate uptake constraints, the genome scale network stoichiometry, and a biomass growth equation. This is commonly justified as reflecting the selection pressure and result of evolution in laboratory strains (Edwards and Palsson, 2000; García Sánchez and Torres Sáez, 2014; Segre et al., 2002). FBA calculations were performed using the E. coli iAF1260 genome scale model (Feist et al., 2007) with two sets of glucose and oxygen uptake constraints (all others were set to simulate the glucose minimal media environment): one corresponding to the MG1655 wild-type (qglc=8.5 mmol/gDW/h, qO2=12 mmol/gDW/h), and one corresponding to the ALE strains (qglc=12, qO2=17). The predicted growth rates from FBA agreed well with the measured growth rates (i.e., unevolved predicted 0.63 h−1, and evolved predicted 0.92 h−1) and acetate yields (unevolved 0.7 mol/mol, evolved 0.63 mol/mol). The corresponding normalized flux predictions from FBA were included in the PCA plot (Fig. 6). There were some notable disagreements between the predicted and measured metabolic fluxes, especially in upper metabolism, where FBA predicted in both cases (i.e. unevolved and evolved scenarios) that approximately half of glucose flux was catabolized through oxPPP (54–55%). As discussed previously and shown in Figs. 3 and 4, this flux was measured by 13C-MFA as 21–25% of glucose flux in all strains studied here. Since the measured and FBA-predicted growth rates agreed well, it appears that the optimal growth rate is not strongly affected by upper pathway usage and alternate optimal solutions may enable the observed growth rates.
The hypothesis of alternate optimal solutions was confirmed with flux variability analysis (FVA) (Mahadevan and Schilling, 2003), which calculates a range of possible flux values that can support a given rate of growth. FVA was applied to both sets of glucose and oxygen uptake rate constraints, yielding very similar normalized flux variabilities in each case. The results described below are from the higher uptake rate case, corresponding to the ALE phenotype. Focusing again on the oxPPP flux, stepping down the optimal growth constraint slightly to 99% of the optimal growth rate, the oxPPP flux varied from 34%–67% of glucose uptake. Stepping down further, this range increased to 14–74% of glucose uptake for 98% of optimal growth rate. Interestingly, in looking at another major central metabolic pathway, the branch point between the TCA cycle and acetate production, was more constrained: the citrate synthase (TCA) fluxes varied only from 8–19% at 99% optimal growth, and 8–27% at 98% optimal growth. Thus, these results demonstrate that alternate optimal flux distributions can support rapid of E. coli and some pathways have more flexibility than others.
4. CONCLUSIONS
In this work, intracellular fluxes of E. coli subject to adaptive laboratory evolution were analyzed for the first time using 13C-MFA. Given the numerous genetic mutations (Sandberg et al., 2016) and previously reported transcriptional changes in evolved strains (LaCroix et al., 2015), it was expected that significant intracellular metabolic rewiring would be occurring in these strains. Instead, we show here that normalized intracellular metabolic fluxes change very little in six independently evolved MG1655 strains. In absolute terms, intracellular fluxes increased proportionally and substantially, along with the glucose uptake rate, to support faster growth. The one significant change, a 26% increase in acetate yield in ALE-6, corresponded to a unique set of mutations. Interestingly, it was previously reported that in similarly evolved strains, enzymes involved in the TCA cycle were broadly transcriptionally repressed (LaCroix et al., 2015), but this did not correspond to reductions in normalized (5 out of 6 strains) or absolute (6 out of 6) TCA fluxes in the strains analyzed here. Future studies may explore whether some TCA cycle enzymes (or others in central metabolism) have excess flux capacity in the wild-type, allowing for increases in absolute flux under certain conditions without commensurate increases in expression. Overall, it was found that the magnitude of the differences between wild-type E. coli strains, particularly between BL21 and the K-12 strains (MG1655 and BW25113), exceeded the variation in unevolved and adaptively evolved MG1655 strains. The broad similarities, but notable differences between E. coli strains, should further inform analyses of cell metabolism rigidity across species (Tang et al., 2009; Wu et al., 2016).
Principal component analysis of the differences in normalized intracellular fluxes highlighted the similarity between all K-12 strains and the uniqueness of BL21 strain. It also showed that the particular solution found using FBA optimization of the growth rate function predicted a high oxPPP flux, differing significantly from that measured here using 13C-MFA. This apparently reflects alternate optima or near-optima, which was further supported through FVA. While growth rate optimization is a commonly used objective function, and reflects the selective pressure in the ALE experiment, alternative objective functions could also be explored (García Sánchez and Torres Sáez, 2014; Schuetz et al., 2007). Overall, in the case of K-12 MG1655, 13C-MFA demonstrated there was no strong selective pressure to change fluxes from the starting flux distribution. As the research community accumulates more results for adaptively evolved strains with different initial metabolic phenotypes (e.g. different wild-type or gene knockout strains (Fong et al., 2006)), it may become possible to elucidate the path dependency of the evolved ‘optima’ achieved through ALE. For example, it would be interesting to determine whether the high TCA flux of BL21 strain is reduced upon evolution or is maintained during fast growth.
The knowledge that faster growth of adaptively evolved strains was not enabled by any particular change in metabolic pathway usage adds some clarity to the picture previously presented via genetic and transcriptomic analysis (LaCroix et al., 2015). Along with increases in protein producing machinery, i.e. transcription and translation, changes to expression levels of central carbon metabolic enzymes were also reported. These included increases in PTS glucose transporters, enzymes in glycolysis and acetate production, and decreases in TCA cycle and glyoxylate shunt enzymes. The fact that the overall state of the metabolic network remained the same, despite adjustment in expression of metabolic enzymes, may indicate that the adaptive evolution responses are a matter of proteomic allocation rather than optimization of cellular processes. For example, the rpoB mutation has been shown to affect the balance between growth and stress functions (Utrilla et al., 2016). Moving forward, combined multi-omics analysis of ALE strains will be useful in advancing cellular modeling of kinetics (Khodayari et al., 2014) and physical and macromolecular constraints on phenotype (O’Brien et al., 2013). Identifying a growth-optimal proteome and regulatory mechanisms by which it can be achieved will be useful in engineering efficient strains. Understanding the hard constraints of E. coli performance, which in addition to the proteome can also include membrane space limitations (Liu et al., 2014), may motivate the development of alternative high-performance organisms for future applications (Cordova et al., 2015; Lee et al., 2016).
Supplementary Material
Acknowledgments
This work was supported by NSF MCB-1616332 grant. C.P.L. and J.E.G. were also supported by the University of Delaware Graduate Fellows Award. A.M.F and B.O.P. were supported under Novo Nordisk Foundation Center for Biosustainability grant no. NNF10CC1016517.
References
- Antoniewicz MR. Parallel labeling experiments for pathway elucidation and 13C metabolic flux analysis. Curr Opin Biotechnol. 2015;36:91–97. doi: 10.1016/j.copbio.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Accurate Assessment of Amino Acid Mass Isotopomer Distributions for Metabolic Flux Analysis. Anal Chem. 2007a;79:7554–7559. doi: 10.1021/ac0708893. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab Eng. 2007b;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–337. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kraynie DF, Laffend La, González-Lergier J, Kelleher JK, Stephanopoulos G. Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol. Metab Eng. 2007c;9:277–92. doi: 10.1016/j.ymben.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Wu TY, Machado IMP, Huang WC, Chen PY, Pellegrini M, Liao JC. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol. 2010;6:1–11. doi: 10.1038/msb.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charusanti P, Conrad TM, Knight EM, Venkataraman K, Fong NL, Xie B, Gao Y, Palsson BØ. Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene. PLoS Genet. 2010;6:e1001186. doi: 10.1371/journal.pgen.1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Alonso AP, Allen DK, Reed JL, Shachar-Hill Y. Synergy between 13C-metabolic flux analysis and flux balance analysis for understanding metabolic adaptation to anaerobiosis in E. coli. Metab Eng. 2011;13:38–48. doi: 10.1016/j.ymben.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Cordova LT, Antoniewicz MR. 13C metabolic flux analysis of the extremely thermophilic, fast growing, xylose-utilizing Geobacillus strain LC300. Metab Eng. 2016;33:148–157. doi: 10.1016/j.ymben.2015.06.004. [DOI] [PubMed] [Google Scholar]
- Cordova LT, Long CP, Venkataramanan P, Antoniewicz MR. Complete genome sequence, metabolic model construction and phenotypic characterization of Geobacillus LC300, an extremely thermophilic, fast growing, xylose-utilizing bacterium. Metab Eng. 2015;32:74–81. doi: 10.1016/j.ymben.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova LT, Lu J, Cipolla RM, Sandoval NR, Long CP, Antoniewicz MR. Co-utilization of glucose and xylose by evolved Thermus thermophilus LC113 strain elucidated by 13C metabolic flux analysis and whole genome sequencing. Metab Eng. 2016;37:63–71. doi: 10.1016/j.ymben.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Long CP, Antoniewicz MR. Optimal tracers for parallel labeling experiments and 13C metabolic flux analysis: A new precision and synergy scoring system. Metab Eng. 2016;38:10–18. doi: 10.1016/j.ymben.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Long CP, Antoniewicz MR. Integrated 13C-metabolic flux analysis of 14 parallel labeling experiments in Escherichia coli. Metab Eng. 2015;28:151–158. doi: 10.1016/j.ymben.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JS, Palsson BO. The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc Natl Acad Sci U S A. 2000;97:5528–5533. doi: 10.1073/pnas.97.10.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BØ. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Ca, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–62. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Fong SS, Nanchen A, Palsson BO, Sauer U. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J Biol Chem. 2006;281:8024–8033. doi: 10.1074/jbc.M510016200. [DOI] [PubMed] [Google Scholar]
- Fong SS, Palsson BO. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat Genet. 2004;36:1056–1058. doi: 10.1038/ng1432. [DOI] [PubMed] [Google Scholar]
- García Sánchez CE, Torres Sáez RG. Comparison and analysis of objective functions in flux balance analysis. Biotechnol Prog. 2014;30:985–991. doi: 10.1002/btpr.1949. [DOI] [PubMed] [Google Scholar]
- Gonzalez JE, Long CP, Antoniewicz MR. Comprehensive analysis of glucose and xylose metabolism in Escherichia coli under aerobic and anaerobic conditions by 13C metabolic flux analysis. Metab Eng. 2017;39:9–18. doi: 10.1016/j.ymben.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, Palsson BO. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- Horinouchi T, Tamaoka K, Furusawa C, Ono N, Suzuki S, Hirasawa T, Yomo T, Shimizu H. Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genomics. 2010;11:579. doi: 10.1186/1471-2164-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen P, Goldovsky L, Kunin V, Darzentas N, Ouzounis Ca. Genome coverage, literally speaking. EMBO Rep. 2005;6:397–399. doi: 10.1038/sj.embor.7400412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KF. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodayari A, Zomorrodi AR, Liao JC, Maranas CD. A kinetic model of Escherichia coli core metabolism satisfying multiple sets of mutant flux data. Metab Eng. 2014;25:50–62. doi: 10.1016/j.ymben.2014.05.014. [DOI] [PubMed] [Google Scholar]
- LaCroix RA, Sandberg TE, O’Brien EJ, Utrilla J, Ebrahim A, Guzman GI, Szubin R, Palsson BO, Feist AM. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol. 2015;81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Palsson BO. Adaptive evolution of escherichia coli K-12 MG1655 during growth on a nonnative carbon source, L-l,2-propanediol. Appl Environ Microbiol. 2010;76:4158–4168. doi: 10.1128/AEM.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Ostrov N, Wong BG, Gold MA, Khalil AS, Church GM. Vibrio natriegens, a new genomic powerhouse. bioRxiv 2016 [Google Scholar]
- Leighty RW, Antoniewicz MR. COMPLETE-MFA: Complementary parallel labeling experiments technique for metabolic flux analysis. Metab Eng. 2013;20:49–55. doi: 10.1016/j.ymben.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Leighty RW, Antoniewicz MR. Parallel labeling experiments with [U-13C]glucose validate E. coli metabolic network model for 13C metabolic flux analysis. Metab Eng. 2012;14:533–541. doi: 10.1016/j.ymben.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Liu JK, O’Brien EJ, Lerman Ja, Zengler K, Palsson BO, Feist AM. Reconstruction and modeling protein translocation and compartmentalization in Escherichia coli at the genome-scale. BMC Syst Biol. 2014;8:110. doi: 10.1186/s12918-014-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR. Metabolic flux analysis of Escherichia coli knockouts: lessons from the Keio collection and future outlook. Curr Opin Biotechnol. 2014a;28:127–133. doi: 10.1016/j.copbio.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR. Quantifying Biomass Composition by Gas Chromatography/Mass Spectrometry. Anal Chem. 2014b;86:9423–7. doi: 10.1021/ac502734e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Au J, Gonzalez JE, Antoniewicz MR. 13C metabolic flux analysis of microbial and mammalian systems is enhanced with GC–MS measurements of glycogen and RNA labeling. Metab Eng. 2016a;38:65–72. doi: 10.1016/j.ymben.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Au J, Sandoval NR, Gebreselassie NA, Antoniewicz MR. Enzyme I facilitates reverse flux from pyruvate to phosphoenolpyruvate in Escherichia coli. Nat Commun. 2017;8:14316. doi: 10.1038/ncomms14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Gonzalez JE, Sandoval NR, Antoniewicz MR. Characterization of physiological responses to 22 gene knockouts in Escherichia coli central carbon metabolism. Metab Eng. 2016b;37:102–113. doi: 10.1016/j.ymben.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R, Schilling CH. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng. 2003;5:264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- McConnell BO, Antoniewicz MR. Measuring the Composition and Stable-Isotope Labeling of Algal Biomass Carbohydrates via Gas Chromatography/Mass Spectrometry. Anal Chem. 2016 doi: 10.1021/acs.analchem.6b00779. [DOI] [PubMed] [Google Scholar]
- Monk JM, Koza A, Campodonico MA, Machado D, Seoane JM, Palsson BO, Herrgård MJ, Feist AM. Multi-omics Quantification of Species Variation of Escherichia coli Links Molecular Features with Strain Phenotypes. Cell Syst. 2016:238–251. doi: 10.1016/j.cels.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundhada H, Seoane JM, Schneider K, Koza A, Christensen HB, Klein T, Phaneuf PV, Herrgard M, Feist AM, Nielsen AT. Increased production of L-serine in Escherichia coli through Adaptive Laboratory Evolution. Metab Eng. 2017;39:141–150. doi: 10.1016/j.ymben.2016.11.008. [DOI] [PubMed] [Google Scholar]
- Noguchi Y, Nakai Y, Shimba N, Toyosaki H, Kawahara Y, Sugimoto S, Suzuki EI. The energetic conversion competence of Escherichia coli during aerobic respiration studied by31P NMR using a circulating fermentation system. J Biochem. 2004;136:509–515. doi: 10.1093/jb/mvh147. [DOI] [PubMed] [Google Scholar]
- O’Brien EJ, Lerman Ja, Chang RL, Hyduke DR, Palsson BØ. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol Syst Biol. 2013;9:693. doi: 10.1038/msb.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Shimizu K. Altered acetate metabolism and biomass production in several Escherichia coli mutants lacking rpoS-dependent metabolic pathway genes. Mol Biosyst. 2008;4:160–169. doi: 10.1039/b712023k. [DOI] [PubMed] [Google Scholar]
- Reyes LH, Almario MP, Winkler J, Orozco MM, Kao KC. Visualizing evolution in real time to determine the molecular mechanisms of n-butanol tolerance in Escherichia coli. Metab Eng. 2012;14:579–590. doi: 10.1016/j.ymben.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Sandberg TE, Lloyd CJ, Palsson BO, Feist AM. Laboratory Evolution to Alternating Substrate Environments Yields Distinct Phenotypic and Genetic Adaptive Strategies. Appl Environ Microbiol. 2017 doi: 10.1128/AEM.00410-17. AEM.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg TE, Long CP, Gonzalez JE, Feist AM, Antoniewicz MR, Palsson BO. Evolution of E. coli on [U-13C]Glucose Reveals a Negligible Isotopic Influence on Metabolism and Physiology. PLoS One. 2016;11:e0151130. doi: 10.1371/journal.pone.0151130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger J, Que R, Fleming RMT, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, Kang J, Hyduke DR, Palsson BØ. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc. 2011;6:1290–307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz R, Kuepfer L, Sauer U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol. 2007;3:119. doi: 10.1038/msb4100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YJ, Martin HG, Dehal PS, Deutschbauer A, Llora X, Meadows A, Arkin A, Keasling JD. Metabolic flux analysis of shewanella spp. Reveals evolutionary robustness in central carbon metabolism. Biotechnol Bioeng. 2009;102:1161–1169. doi: 10.1002/bit.22129. [DOI] [PubMed] [Google Scholar]
- Taymaz-Nikerel H, Borujeni AE, Verheijen PJT, Heijnen JJ, van Gulik WM. Genome-derived minimal metabolic models for Escherichia coli MG1655 with estimated in vivo respiratory ATP stoichiometry. Biotechnol Bioeng. 2010;107:369–381. doi: 10.1002/bit.22802. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. The Molecular Diversity of Adaptive Convergence. Science (80- ) 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- Utrilla J, O’Brien EJ, Chen K, McCloskey D, Cheung J, Wang H, Armenta-Medina D, Feist AM, Palsson BO. Global Rebalancing of Cellular Resources by Pleiotropic Point Mutations Illustrates a Multi-scale Mechanism of Adaptive Evolution. Cell Syst. 2016;2:260–271. doi: 10.1016/j.cels.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegeman H, Beauprez J, Moens H, Maertens J, De Mey M, Foulquié-Moreno MR, Heijnen JJ, Charlier D, Soetaert W. Effect of iclR and arcA knockouts on biomass formation and metabolic fluxes in Escherichia coli K12 and its implications on understanding the metabolism of Escherichia coli BL21 (DE3) BMC Microbiol. 2011:11. doi: 10.1186/1471-2180-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegeman H, Maertens J, Beauprez J, De Mey M, Soetaert W. Effect of iclR and arcA deletions on physiology and metabolic fluxes in Escherichia coli BL21 (DE3) Biotechnol Lett. 2012;34:329–337. doi: 10.1007/s10529-011-0774-6. [DOI] [PubMed] [Google Scholar]
- Wu SG, Wang Y, Jiang W, Oyetunde T, Yao R, Zhang X, Shimizu K, Tang YJ, Bao FS. Rapid Prediction of Bacterial Heterotrophic Fluxomics Using Machine Learning and Constraint Programming. PLoS Comput Biol. 2016:12. doi: 10.1371/journal.pcbi.1004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Antoniewicz MR, Kelleher JK, Stephanopoulos G. J Biol Chem. 2008;283(30):20621–7. doi: 10.1074/jbc.M706494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.