Abstract
Vibrio natriegens is a fast-growing, non-pathogenic bacterium that is being considered as the next-generation workhorse for the biotechnology industry. However, little is known about the metabolism of this organism which is limiting our ability to apply rational metabolic engineering strategies. To address this critical gap in current knowledge, here we have performed a comprehensive analysis of V. natriegens metabolism. We constructed a detailed model of V. natriegens core metabolism, measured the biomass composition, and performed high-resolution 13C metabolic flux analysis (13C-MFA) to estimate intracellular fluxes using parallel labeling experiments with the optimal tracers [1,2-13C]glucose and [1,6-13C]glucose. During exponential growth in glucose minimal medium, V. natriegens had a growth rate of 1.70 1/h (doubling time of 24 min) and a glucose uptake rate of 3.90 g/g/h, which is more than two 2-fold faster than E. coli, although slower than the fast-growing thermophile Geobacillus LC300. 13C-MFA revealed that the core metabolism of V. natriegens is similar to that of E. coli, with the main difference being a 33% lower normalized flux through the oxidative pentose phosphate pathway. Quantitative analysis of co-factor balances provided additional insights into the energy and redox metabolism of V. natriegens. Taken together, the results presented in this study provide valuable new information about the physiology of V. natriegens and establish a solid foundation for future metabolic engineering efforts with this promising microorganism.
Keywords: Fast growth, glucose uptake, Vibrio natriegens, metabolism, metabolic network model
1. INTRODUCTION
Escherichia coli is the most widely studied microorganism in academia (Janssen et al., 2005; Long and Antoniewicz, 2014a). A wealth of knowledge has been generated over the past century for this model microbe, and many molecular tools have been developed for genetic engineering (Datsenko and Wanner, 2000; Gibson et al., 2009; Li et al., 2015; Wang et al., 2009). As a result, E. coli is often the go-to organism for metabolic engineering efforts. However, researchers are increasingly interested in selecting alternative hosts for various applications in biotechnology. One of the key physiological characteristics that impacts industrial performance is growth rate, or perhaps more importantly, biomass specific substrate uptake rate. This rate determines the maximum productivity that can be achieved for a given size bioreactor. As such, faster-growing organisms offer a clear advantage over slower-growing organisms. In this respect, wild-type E. coli has a relatively high growth rate of about 0.7 h−1 (60 min doubling time) when grown in minimal medium with glucose as the carbon source, and ~30 min doubling time when grown in rich medium. Faster-growing E. coli strains have also been generated through adaptive laboratory evolution with a maximum growth rate of about 1.0 h−1 (40 min doubling time) in glucose minimal medium (LaCroix et al., 2015; Sandberg et al., 2016); however, for yet unknown reasons, this appears to be the upper limit for E. coli growth rate.
To achieve even faster conversion rates, scientists are now interested in identifying alternative fast-growing organisms to replace E. coli as the workhorse host (Lee et al., 2016; Weinstock et al., 2016). One such organism is the Gram-negative, non-pathogenic marine bacterium Vibrio natriegens, which is commonly found in marine and coastal waters and sediments, and has a reported doubling time of 10 minutes or less when cultured under ideal conditions in rich medium (Eagon, 1962). V. natriegens (initially called Pseudomonas natriegens) was described for the first time by Payne et al. in the early 1960s (Payne et al., 1961). It is a moderate halophile, requiring about 1.5% NaCl for optimal growth, and grows well under laboratory conditions with glucose as the carbon source, with an optimal growth temperature of 37 °C (Eagon, 1962; Lee et al., 2016). Two annotated genomes are available for V. natriegens, for strains ATCC 12048 (Wang et al., 2013) and DSMZ 759 (Maida et al., 2013). Recently, a wide range of genetic tools were developed and described to engineer V. natriegens (Weinstock et al., 2016), and the fast growth of this species was shown to reduce the time needed to execute common cloning pipelines, which has clear advantages in highly iterative strain building efforts (Weinstock et al., 2016).
In this work, we have investigated the metabolism of V. natriegens using state-of-the-art tools for 13C metabolic flux analysis (13C-MFA) (Gonzalez and Antoniewicz, 2017). Currently, no information is available about intracellular metabolism of V. natriegens and this lack of knowledge significantly impacts our ability to apply rational strategies to engineer this organism. For example, in the absence of an experimentally validated flux map, constraint-based modeling and analysis (COBRA) approaches cannot be applied to guide new metabolic engineering designs (Becker et al., 2007; Schellenberger et al., 2011). To address this critical gap in knowledge, here we have characterized the growth physiology, constructed a detailed model of core metabolism, measured the biomass composition, and performed high-resolution 13C-MFA to estimate intracellular fluxes in V. natriegens. Analysis of co-factor balances provided additional insights into its metabolism. Taken together, the results presented in this study provide an important quantitative description of the physiology and metabolism of V. natriegens that can serve as the basis for informed host selection, future model building, and strain design efforts.
2. MATERIALS AND METHODS
2.1. Materials
Media and chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Tracers were purchased from Cambridge Isotope Laboratories, [1,2-13C]glucose (99.5 atom% 13C) and [1,6-13C]glucose (99.5%). The isotopic purity and enrichment of glucose tracers was validated by GC-MS analysis (Cordova et al., 2016). Wolfe’s minerals (Cat. No. MD-TMS) and Wolfe’s vitamins (Cat. No. MD-VS) were purchased from ATCC (Manassas, VA). The growth medium for V. natriegens was M9 minimal medium supplemented with (per liter of medium): 15 g of NaCl, 10 mL of Wolfe’s minerals, 10 mL of Wolfe’s vitamins, and 0.05 g of yeast extract. Glucose was added as indicated in the text. All media and stock solutions were sterilized by filtration.
2.2. Strains and growth conditions
V. natriegens (ATCC Cat. No. 14048, Manassas, VA) was used in this study. For tracer experiments, cells from a frozen stock were first pre-cultured overnight at 37 °C in a shaker flask with 20 mM initial glucose. Next, 1 mL of this culture was washed with fresh medium and used to inoculate a new shaker flask with 10 mL of medium (20 mM initial glucose). After 1 hr, 100 uL of this culture was used to inoculate two mini-bioreactors, one containing 10 mM of [1,2-13C]glucose and one containing 10 mM of [1,6-13C]glucose. The optical density (OD600) of the inoculated cultures was about 0.015. We estimated that about 0.2 mM of unlabeled glucose was carried over from the inoculum to the labeling cultures. Cells were then grown at 37°C in mini-bioreactors as described before (Gonzalez et al., 2017). Air was sparged into the liquid at a rate of 12 mL/min to provide oxygen and to ensure sufficient mixing of the culture by the rising gas bubbles. Cell pellets were collected for GC-MS analysis during the mid-exponential growth phase when biomass concentration (OD600) was between 0.7 and 1.0.
2.3. Analytical methods
Samples were collected at multiple times during the growth phase to monitor cell growth, glucose uptake, and acetate accumulation. Cell growth was monitored by measuring the optical density at 600nm (OD600) using a spectrophotometer (Eppendorf BioPhotometer). The OD600 values were converted to cell dry weight concentrations using the following relationship for V. natriegens: 1.0 OD600 = 0.27 gDW/L, which was experimentally determined as described in (Long et al., 2016b). Acetate concentrations were determined using an Agilent 1200 Series HPLC (Whitaker et al., 2017), and glucose and lactate concentrations were determined using a YSI 2700 biochemistry analyzer (YSI, Yellow Springs, OH).
2.4. Biomass composition analysis
The methods used for quantifying biomass composition were described in (Long and Antoniewicz, 2014b). Briefly, samples were prepared by three respective methods: hydrolysis of protein and subsequent TBDMS derivatization of amino acids; hydrolysis of RNA and glycogen and subsequent aldonitrile propionate derivatization of sugars (ribose and glucose, respectively); and fatty acid methyl ester derivatization for analysis of fatty acids. In total, 17 amino acids were quantified. Glutamine and asparagine are deaminated during hydrolysis to glutamate and aspartate, respectively; thus, we report the combined pools of each. The amino acids arginine, cysteine and tryptophan are degraded during hydrolysis and thus were not quantified. Quantification of all measured components was achieved by isotope ratio analysis in duplicate using an isotopically labeled standard and a naturally labeled biomass sample. In this study, the standard was generated by growing wild-type E. coli on [U-13C]glucose and aliquoting identical (1 mL of an OD600 = 1.0) samples of this “fully labeled” biomass. These were centrifuged and washed twice with M9 medium. The composition of the fully labeled biomass was determined using unlabeled chemical standards as described in (Long and Antoniewicz, 2014b).
2.5. Gas chromatography-mass spectrometry
GC-MS analysis was performed on an Agilent 7890B GC system equipped with a DB-5MS capillary column (30 m, 0.25 mm i.d., 0.25 μm-phase thickness; Agilent J&W Scientific), connected to an Agilent 5977A Mass Spectrometer operating under ionization by electron impact (EI) at 70 eV. Helium flow was maintained at 1 mL/min. The source temperature was maintained at 230°C, the MS quad temperature at 150°C, the interface temperature at 280°C, and the inlet temperature at 250°C. GC-MS analysis of tert-butyldimethylsilyl (TBDMS) derivatized proteinogenic amino acids was performed as described in (Antoniewicz et al., 2007a). Labeling of glucose in the medium was determined after aldonitrile propionate derivatization as described in (Antoniewicz et al., 2011; Sandberg et al., 2016). Labeling of fatty acids was determined after derivatization to fatty acid methyl esters (FAME) (Crown et al., 2015b). Labeling of glucose (derived from glycogen) and ribose (derived from RNA) were determined as described in (Long et al., 2016a; McConnell and Antoniewicz, 2016). In all cases, mass isotopomer distributions were obtained by integration (Antoniewicz et al., 2007a) and corrected for natural isotope abundances (Fernandez et al., 1996).
2.6. Metabolic network model and 13C-metabolic flux analysis
A metabolic network model of V. natriegens core metabolism was constructed for 13C-MFA based on the reactions annotated in KEGG and BioCyc databases (Caspi et al., 2012; Kanehisa et al., 2012; Kanehisa and Goto, 2000). The complete 13C-MFA model is provided in Supplemental Materials. The model includes all major metabolic pathways of central carbon metabolism, including glycolysis, pentose phosphate pathway, Entner–Doudoroff pathway, TCA cycle, glyoxylate shunt, and various anaplerotic and cataplerotic reactions, lumped amino acid biosynthesis reactions, and a lumped biomass formation reaction, which was derived using the measured biomass composition for V. natriegens. Since it is not possible to distinguish between the fluxes of NAD-dependent malic enzyme (EC 1.1.1.38), NADP-dependent malic enzyme (EC 1.1.1.40), and oxaloacetate decarboxylase (EC 4.1.1.3) using 13C-MFA (Kappelmann et al., 2015), we included only one reaction in the model (the NADP-dependent malic enzyme) to describe the combined flux of all three reactions. The model also accounts for the exchange of intracellular and atmospheric unlabeled CO2 (Leighty and Antoniewicz, 2012), and G-value parameters to describe fractional labeling of amino acids. As described previously (Antoniewicz et al., 2007c), the G-value represents the fraction of a metabolite pool that is produced during the labeling experiment from glucose, while 1-G represents the fraction that is naturally labeled, i.e. from the incorporation of unlabeled amino acids from the yeast extract and from the inoculum. By default, one G-value parameter was included for each measured amino acid in each data set. Reversible reactions were modeled as separate forward and backward fluxes. Net and exchange fluxes were calculated as follows: vnet = vf−vb; vexch = min(vf, vb).
All 13C-MFA calculations were performed using the Metran software (Yoo et al., 2008) which is based on the elementary metabolite units (EMU) framework (Antoniewicz et al., 2007b). Fluxes were estimated by minimizing the variance-weighted sum of squared residuals (SSR) between the experimentally measured and model predicted mass isotopomer distributions of biomass amino acids, glucose derived from glycogen, ribose derived from RNA, and the measured acetate yield, using non-linear least-squares regression (Antoniewicz et al., 2006). All measured mass isotopomers are provided in Supplemental Materials. For integrated analysis of parallel labeling experiments, the data sets were fitted simultaneously to a single flux model (Antoniewicz, 2015). Flux estimation was repeated at least 10 times starting with random initial values for all fluxes to find a global solution. At convergence, accurate 95% confidence intervals were computed for all estimated fluxes by evaluating the sensitivity of the minimized SSR to flux variations. Precision of estimated fluxes was determined as follows (Antoniewicz et al., 2006):
| (7) |
2.7. Goodness-of-fit analysis
To determine the goodness-of-fit, 13C-MFA fitting results were subjected to a χ2-statistical test. In short, assuming that the model is correct and data are without gross measurement errors, the minimized SSR is a stochastic variable with a χ2-distribution (Antoniewicz et al., 2006). The number of degrees of freedom is equal to the number of fitted measurements n minus the number of estimated independent parameters p. The acceptable range of SSR values is between χ2α/2(n-p) and χ21-α/2(n-p), where α is a certain chosen threshold value, for example 0.05 for 95% confidence interval.
3. RESULTS AND DISCUSSION
3.1. Growth physiology
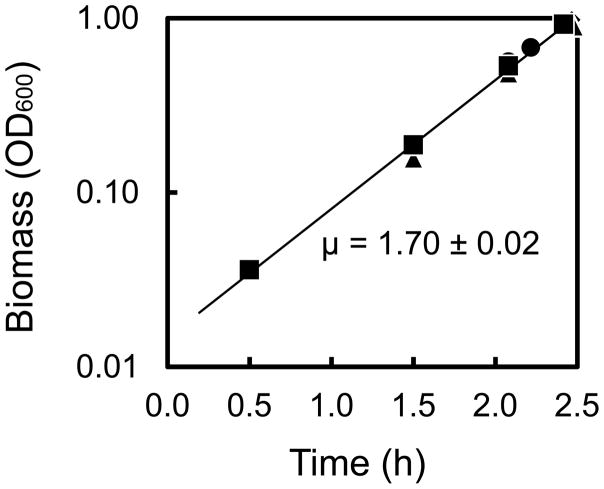
Growth characteristics of V. natriegens were determined in aerobic batch culture at 37 °C in medium containing glucose as the main carbon source (Fig. 1 and Supplemental Figures S1 and S2). The growth medium was M9 minimal medium supplemented with 1.5% NaCl, vitamins, minerals, and 0.05 g/L of yeast extract. The addition of small amount of yeast extract eliminated a short lag phase that was observed when cells were subcultured without it; however, the yeast extract did not impact glucose uptake rate or the specific growth rate (Fig. S1). During the exponential growth phase, the specific growth rate of V. natriegens was 1.70 ± 0.02 h−1, which corresponds to a doubling time of about 24 min (Fig 1). The biomass yield was 0.44 gDW/g and the biomass-specific glucose uptake rate was 21.4 ± 1.3 mmol/gDW/h. V. natriegens produced acetate as a byproduct during exponential growth, with about 0.8 ± 0.1 mol of acetate produced per mol of glucose consumed (Table 1). No other byproducts were detected by HPLC analysis.
Figure 1.
Characterization of V. natriegens growth rate in four replicate cultures. V. natriegens was grown aerobically in M9 medium supplemented with 1.5% NaCl, 1.8 g/L glucose and 0.05 g/L yeast extract.
Table 1.
Physiological characteristics of V. natriegens grown in aerobic batch culture in glucose minimal medium at 37°C (mean ± sem, n = 4, biological replicates).
| Growth rate | 1.70 ± 0.02 | 1/h |
| Biomass yield | 0.44 ± 0.03 | gDW/g |
| Acetate yield | 0.8 ± 0.1 | mol/mol |
| Glucose uptake rate | 21.4 ± 1.3 | mmol/gDW/h |
3.2. Metabolic model construction and biomass composition analysis
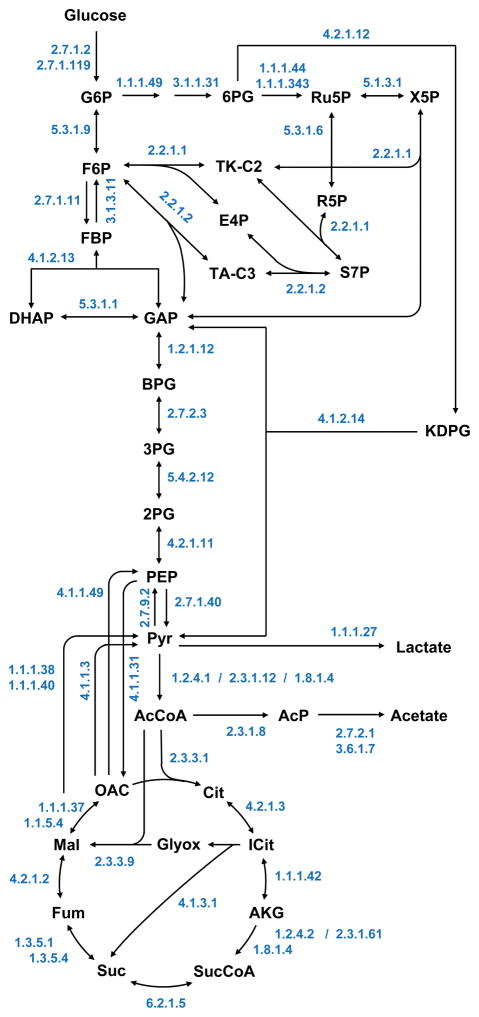
To facilitate quantitative studies of V. natriegens, a detailed network model of its core metabolism was constructed for 13C-MFA based on the reactions annotated in the KEGG and BioCyc databases. As illustrated in Fig. 2, central carbon metabolism of V. natriegens is similar to that of E. coli and includes the following core metabolic pathways: glycolysis (EMP pathway), pentose phosphate pathway, Entner-Doudoroff pathway, TCA cycle, glyoxylate shunt, and various anaplerotic and cataplerotic reactions. Compared to E. coli, V. natriegens has one additional cataplerotic reaction, oxaloacetate decarboxylase (EC 4.1.1.3), which is not present in E. coli. Further analysis of annotated genes revealed that V. natriegens engages the same canonical amino acid biosynthesis pathways as E. coli.
Figure 2.
Central carbon metabolism of V. natriegens reconstructed from genome annotations.
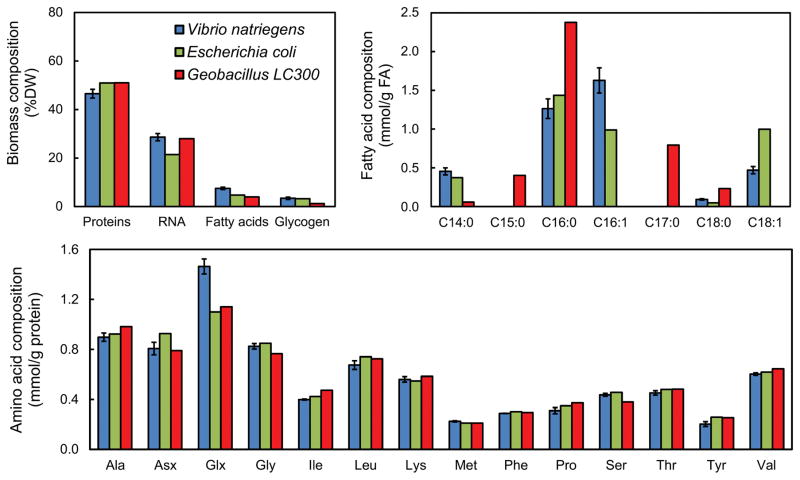
An important reaction the model for 13C-MFA is the lumped biomass reaction that captures the drain of precursor metabolites and cofactors needed for cell growth. To determine the coefficients in this biomass formation reaction, the biomass composition of V. natriegens was determined experimentally in this study. The results of this analysis are provided in Supplemental Materials and are shown in Fig. 3, where the biomass composition of V. natriegens is compared to the reported composition of E. coli (Long and Antoniewicz, 2014b) and the fast-growing thermophile Geobacillus LC300 (Cordova et al., 2015; Cordova and Antoniewicz, 2016). Proteins were the most abundant component of V. natriegens biomass (47% of dry weight), followed by RNA (29%), lipids (7%) and glycogen (3%). The RNA content was higher for V. natriegens (29%) compared to E. coli (21%), but similar to that of Geobacillus LC300 (28%). It has been observed previously that RNA content is often higher for fast growing strains (Long et al., 2016b; Pramanik and Keasling, 1997), which is thought to reflect the need for more ribosomes to support the higher growth rates. The ratio of RNA to protein of 0.6 g/g for V. natriegens is consistent with a previous report at a similar growth rate (Aiyar et al., 2002). The relative distribution of fatty acids in V. natriegens and E. coli were similar, with the most abundant fatty acids being C16:1 and C16:0 followed by C18:1 and C14:0. No odd-chain fatty acids (e.g., C15 or C17) were detected in V. natriegens. The relative distribution of amino acids in biomass was similar for the three microbes, with the notable exception of glutamate/glutamine (Glx), which was significantly elevated in V. natriegens compared to E. coli and Geobacillus LC300 (Fig. 3). The relative abundance of aspartate/asparagine (Asx) was lower for both V. natriegens and Geobacillus LC300 compared to E. coli.
Figure 3.
Biomass composition of V. natriegens compared to E. coli and the fast-growing thermophile Geobacillus LC300. The fractional amounts of major biomass components are shown, as well as the distributions of fatty acids and amino acids.
3.3. 13C Metabolic flux analysis
Next, we quantified intracellular metabolic fluxes for V. natriegens during exponential growth on glucose using high-resolution 13C-MFA. The analysis consisted of first performing two parallel labeling experiments with [1,2-13C]glucose and [1,6-13C]glucose (an experimental design previously identified as providing optimal flux precision (Crown et al., 2016)). Fluxes were estimated by simultaneously fitting the measured acetate yield and the labeling data from proteinogenic amino acids, the ribose moiety of RNA, and the glucose moiety of glycogen to the model described in the previous section. A statistically acceptable fit was obtained. The minimized SSR value of 188 was lower than the maximum acceptable SSR value of 224 at 95% confidence level, assuming a constant measurement error of 0.3 mol% for all GC-MS measurements (Antoniewicz et al., 2007a). The estimated metabolic fluxes and 95% flux confidence intervals are provided in Supplemental Materials.
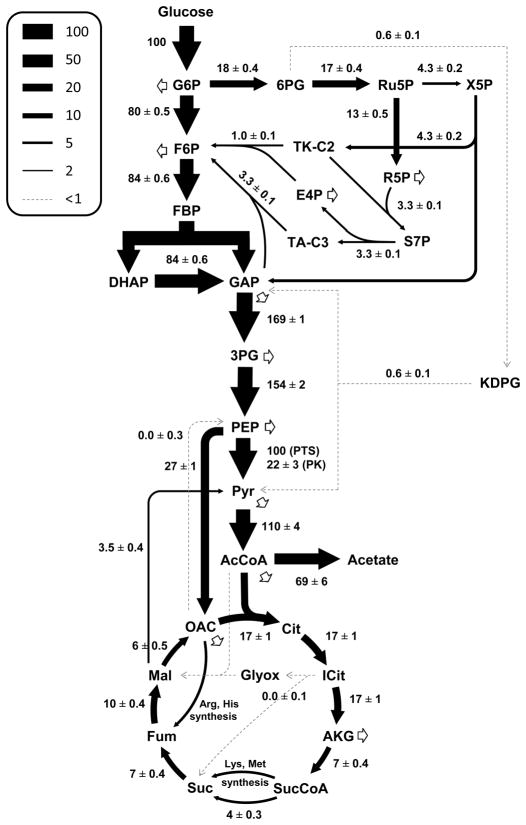
Fig. 4 shows the estimated fluxes in central carbon metabolism for V. natriegens during aerobic growth on glucose (fluxes normalized to glucose uptake rate of 100). The flux map of V. natriegens was characterized by a high glycolytic flux (80 ± 0.5 for phosphoglucose isomerase, and 169 ± 1 for glyceraldehyde 3-phosphate dehydrogenase), relatively high anaplerosis flux via phosphoenolpyruvate carboxylase (27 ± 1), and moderate oxidative PPP flux (18 ± 0.4) and TCA cycle fluxes (e.g. 17 ± 1 for citrate synthase; and 7 ± 0.4 for α-ketoglutarate dehydrogenase). Several pathways were found to be inactive (or nearly inactive) during aerobic growth on glucose, including the Entner-Doudoroff pathway (0.6 ± 0.1), the glyoxylate shunt (0.0 ± 0.1), and gluconeogenesis via phosphoenolpyruvate carboxykinase (0.0 ± 0.3). Interestingly, a previous study reported that a majority of marine bacteria prefer the ED pathway over glycolysis for glucose metabolism, with the notable exception of Vibrio (Klinger et al., 2015). In our model, the NAD-dependent malic enzyme (EC 1.1.1.38), the NADP-dependent malic enzyme (EC 1.1.1.40), and oxaloacetate decarboxylase (EC 4.1.1.3) were lumped into a single reaction since it was not possible to independently estimate these three fluxes. The combined flux of these three reactions was estimated to be very low (3.5 ± 0.4). Taken together, we determined that the V. natriegens flux map was similar to that of E. coli during aerobic growth on glucose (Crown et al., 2015a; Leighty and Antoniewicz, 2013; Long et al, 2017), with the main difference being a 33% lower flux of glucose through oxidative PPP, i.e. 27% of glucose for E. coli compared to 18% for V. natriegens. Given approximately 2.5-fold higher glucose uptake rate in V. natriegens compared to E. coli, however, the absolute carbon fluxes (i.e., mmol/gDW/h) through the central carbon metabolic pathways are much higher in this organism.
Figure 4.
Central carbon metabolic fluxes of V. natriegens, as determined by high-resolution 13C-MFA (estimated flux ± SD). Fluxes were normalized to glucose uptake. The two malic enzymes and oxaloacetate decarboxylase were lumped into a single reaction, shown here as the flux from malate to pyruvate.
3.4. Quantitative analysis of co-factor balances
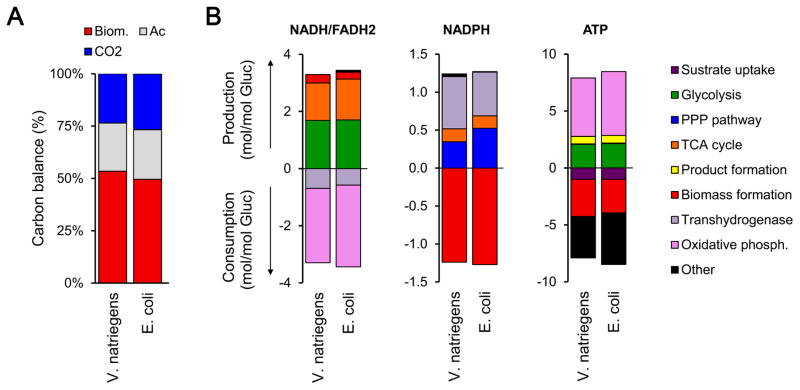
To provide additional insights into the physiology of V. natriegens, we analyzed the production and consumption rates of key co-factors in metabolism based on the 13C-MFA estimated fluxes, and calculated the overall carbon balance. In Fig. 5, the results are summarized (see Supplemental Materials for additional details) and compared to E. coli based on previous 13C-MFA results (Crown et al., 2015a), on a glucose-normalized basis. The overall carbon balance for V. natriegens was similar to that of E. coli, with about 50% of glucose being converted to biomass, 25% to acetate, and 25% to CO2. Fig. 5B shows the normalized production and consumption rates of NADH/FADH2, NADPH and ATP, with contributions by the various pathways (absolute contributions in units of mmol/gDW/h are shown in Supplemental Figure S3). The uncertainty introduced by the lumping of the NAD and NADP-dependent cataplerotic reaction (malic enzymes and oxaoloacetate decarboxylase) was minimal given the small measured flux. The amount of NADH/FADH2 produced (per mol of glucose) was similar for V. natriegens and E. coli, with glycolysis (~50% contribution) and the TCA cycle (~40% contribution) being the main contributing pathways. The vast majority of NADH/FADH2 was oxidized to generate ATP via oxidative phosphorylation (~80%), and 20% of NADH was converted to NADPH by the transhydrogenases.
Figure 5.
Overview of carbon and cofactor balances for V. natriegens and E. coli. (A) The overall carbon balance reflects the fate of glucose on a C-mol basis. (B) Metabolic pathways responsible for the production and consumption of the cofactors NADH/FADH2, NADPH, and ATP are summarized, on a normalized (per unit glucose) basis. For the ATP balance, the production rate from oxidative phosphorylation and the maintenance cost (‘Other’) are based on an assumed effective P/O ratio of 2. Since the P/O ratio has not been reliably measured in V. natriegens, this result should be interpreted with caution. The results for E. coli were taken from a previous reported 13C-flux study (Crown et al., 2015).
There were relatively larger differences in the production of NADPH between V. natriegens and E. coli. For V. natriegens, the majority of NADPH was produced from NADH via transhydrogenases (56%), with oxPPP contributing 25%, and TCA cycle 14% to NADPH production, and negligible contribution from malic enzyme (<3%). For comparison, in E. coli, transhydrogenases and oxPPP contribute about equally to NADPH production (45% and 41%, respectively), with the TCA cycle contributing the remaining 13%. Based on the genome annotations, both V. natriegens and E. coli are believed to contain two pyridine nucleotide transhydrogenases, the membrane-bound PntAB (EC 1.6.1.2) which primarily converts NADH to NADPH, and the soluble form SthA (EC 1.6.1.1, also referred to as UdhA) which primarily converts NADPH to NADH (Sauer et al., 2004). The absolute net transhydrogenase flux from NADH to NADPH was 3-fold higher for V. natriegens (14.7 mmol/gDW/h) compared to E. coli (5.0 mmol/gDW/h), thus suggesting a much more active PntAB enzyme.
Biological energy in the form of ATP is needed for three key cellular processes: 1) transport of substrates and nutrients into the cells; 2) anabolism (i.e. cell growth); and 3) maintenance. The overall ATP balances were similar for V. natriegens and E. coli (Fig 5), with the oxidative phosphorylation contributing the most towards ATP production, approximately 65% for both organisms, followed by substrate level phosphorylation in glycolysis (~25%), and acetate production (~10%). The contribution of the TCA cycle to ATP production was negligible for both organisms (<5%). It should be noted that the absolute ATP production rate for V. natriegens calculated here should be viewed with caution, since little information is available about the effective P/O ratio for V. natriegens. For simplicity, we assumed the same P/O ratio of 2.0 for both organisms. Once a reliable estimate of the P/O ratio for V. natriegens is available, the ATP analysis results reported here should be re-evaluated.
3.5. Comparison of V. natriegens physiology and other fast growing bacteria
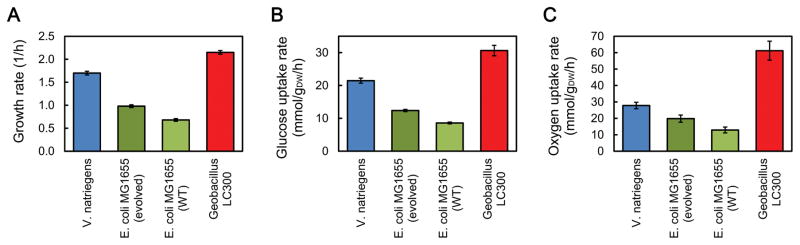
Finally, we compared key physiological characteristics (i.e. growth rate, glucose uptake rate, and oxygen uptake rate) of V. natriegens to three other strains (Fig 6): wild-type E. coli (Leighty and Antoniewicz, 2013), adaptively evolved E. coli (Sandberg et al., 2016; Long et al, 2017), and the fast-growing extreme thermophile Geobacillus LC300 (Cordova et al., 2015; Cordova and Antoniewicz, 2016). Like V. natriegens, the E. coli characteristics reflect growth at 37 °C, while those of Geobacillus LC300 are from growth at 72 °C. The growth rate of V. natriegens on minimal glucose medium (1.70 h−1, doubling time 24 min) was significantly higher than the growth rate of wild-type E. coli (~0.7 h−1, doubling time 60 min) and evolved E. coli (~1.0 h−1, doubling time 40 min), but significantly lower than the growth rate of the thermophile Geobacillus LC300, which has a growth rate on minimal glucose medium of 2.15 h−1 (doubling time of 19 min) (Cordova et al., 2016). Since biomass yields are similar for all four strains, the biomass-specific glucose uptake rates followed the same pattern as the growth rates (Fig 6B), with Geobacillus LC300 having the highest glucose uptake rate of 31 mmol/gDW/h, followed by V. natriegens (21 mmol/gDW/h), the evolved E. coli (12 mmol/gDW/h), and wild-type E. coli (8.5 mmol/gDW/h). Even more striking were the differences between the three mesophilic strains and the thermophilic Geobacillus LC300 when the biomass-specific oxygen uptake rates were compared. The oxygen uptake rate of Geobacillus LC300 (~60 mmol/gDW/h) was more than 2-fold higher than the oxygen uptake rate of V. natriegens (28 mmol/gDW/h), 3-fold higher compared to evolved E. coli (19 mmol/gDW/h), and 5-fold higher compared to wild-type E. coli (12 mmol/gDW/h). These values suggest that the gram-positive thermophile Geobacillus LC300 has a significantly more active electron transport chain compared to the gram-negative mesophiles V. natriegens and E. coli. Unfortunately, the glucose metabolism of Geobacillus LC300 has not been elucidated by 13C-MFA, so a more comprehensive comparison of metabolic fluxes between the four strains is not possible at this time.
Figure 6.
Overall physiological assessment of four potential host strains for biotechnology applications: V. natriegens, E. coli (evolved and wild-type), and the thermophilic Geobacillus LC300. The values reflect growth at 37 °C for the mesophilic strains, and at 72 °C for Geobacillus LC300. The growth rates (A), and biomass specific glucose (B) and oxygen (C) uptake rates are shown. The results for E. coli and Geobacillus LC300 were taken from previous reported studies (Crown et al., 2015; Cordova et al., 2016).
4. CONCLUSION
The physiological and fluxomic analyses presented here are important foundational components of a developing knowledge base from which the community will be able to study and engineer V. natriegens. The verified intracellular metabolic pathways and wild-type reference flux map will enable the use of several COBRA model-based analyses and designs. Additional systems measurements, particularly of the proteome and transcriptome, will complement these results and help begin to answer the fundamental question of how this organism achieves such fast growth. For example, these measurements may determine whether V. natriegens more efficiently allocates its proteome for growth. In adaptively evolved E. coli, mutations in global regulation are frequently observed which shift gene expression toward catabolic processes at the expense of stress response functions (LaCroix et al., 2015; Utrilla et al., 2016). There is some evidence of less robust stress responses already, with the observation of substantially lower intrinsic catalase activity in V. natriegens than in E. coli (Weinstock et al., 2016). Other molecular mechanisms may also be investigated, such as the DNA replication and protein synthesis machinery (the high RNA content measured is believed to be related to high rRNA levels and ribosome number (Aiyar et al., 2002)). As more biological knowledge is developed, advanced in silico approaches including ME models (Liu et al., 2014; O’Brien et al., 2013), kinetic models (Khodayari and Maranas, 2016; Tran et al., 2008), and even whole cell models (Karr et al., 2012) may be applied to develop a systems-level understanding.
Insights into fast growing strains like V. natriegens (as well as Geobacillus LC300, adaptively evolved E. coli, and others) may enable rational engineering approaches of conventional organisms for improved performance. Given its natural advantages, however, it has been proposed that V. natriegens may be a superior next-generation host for biotechnological applications. Its similar flux distribution to E. coli in glucose minimal medium during aerobic growth is encouraging in that raises the prospect that some metabolic engineering strategies previously developed in E. coli may also be successful in V. natriegens. Further study of metabolic kinetics and regulation in V. natriegens, and the nature of responses to genetic and environmental perturbations (e.g. for microaerobic or anaerobic conditions), will be critical. Finally, we have shown that although V. natriegens has large advantages in rate over its mesophilic counterpart E. coli, it lags behind the thermophilic Geobacillus LC300. Similar development would be needed in systems knowledge and genetic tools to develop this (and any other non-model organism) into a viable host, after which an informed choice based on desired process temperature and other characteristics would be possible.
Supplementary Material
Acknowledgments
This work was supported by NSF MCB-1616332 grant.
References
- Aiyar SE, Gaal T, Gourse RL. rRNA Promoter Activity in the Fast-Growing Bacterium Vibrio natriegens. J Bacteriol. 2002;184:1349–1358. doi: 10.1128/JB.184.5.1349-1358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR. Parallel labeling experiments for pathway elucidation and 13C metabolic flux analysis. Curr Opin Biotechnol. 2015;36:91–97. doi: 10.1016/j.copbio.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Measuring deuterium enrichment of glucose hydrogen atoms by gas chromatography/mass spectrometry. Anal Chem. 2011;83:3211–6. doi: 10.1021/ac200012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Accurate Assessment of Amino Acid Mass Isotopomer Distributions for Metabolic Flux Analysis. Anal Chem. 2007a;79:7554–7559. doi: 10.1021/ac0708893. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab Eng. 2007b;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–337. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kraynie DF, Laffend La, González-Lergier J, Kelleher JK, Stephanopoulos G. Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol. Metab Eng. 2007c;9:277–92. doi: 10.1016/j.ymben.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker SA, Feist AM, Mo ML, Hannum G, Palsson BØ, Herrgard MJ. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dreher K, Fulcher CA, Pallavi S, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Pujar A, Shearer AG, Travers M, Weerasinghe D, Zhang P, Karp PD. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D472–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova LT, Antoniewicz MR. 13C metabolic flux analysis of the extremely thermophilic, fast growing, xylose-utilizing Geobacillus strain LC300. Metab Eng. 2016;33:148–157. doi: 10.1016/j.ymben.2015.06.004. [DOI] [PubMed] [Google Scholar]
- Cordova LT, Long CP, Venkataramanan P, Antoniewicz MR. Complete genome sequence, metabolic model construction and phenotypic characterization of Geobacillus LC300, an extremely thermophilic, fast growing, xylose-utilizing bacterium. Metab Eng. 2015;32:74–81. doi: 10.1016/j.ymben.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova LT, Lu J, Cipolla RM, Sandoval NR, Long CP, Antoniewicz MR. Co-utilization of glucose and xylose by evolved Thermus thermophilus LC113 strain elucidated by 13C metabolic flux analysis and whole genome sequencing. Metab Eng. 2016;37:63–71. doi: 10.1016/j.ymben.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Long CP, Antoniewicz MR. Optimal tracers for parallel labeling experiments and 13C metabolic flux analysis: A new precision and synergy scoring system. Metab Eng. 2016;38:10–18. doi: 10.1016/j.ymben.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Long CP, Antoniewicz MR. Integrated 13C-metabolic flux analysis of 14 parallel labeling experiments in Escherichia coli. Metab Eng. 2015a;28:151–158. doi: 10.1016/j.ymben.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Marze N, Antoniewicz MR. Catabolism of Branched Chain Amino Acids Contributes Significantly to Synthesis of Odd-Chain and Even-Chain Fatty Acids in 3T3-L1 Adipocytes. PLoS One. 2015b;10:e0145850. doi: 10.1371/journal.pone.0145850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko Ka, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagon RG. Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes. J Bacteriol. 1962;83:736–737. doi: 10.1128/jb.83.4.736-737.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Ca, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–62. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison Ca, Smith HO, CAH, America N. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gonzalez JE, Antoniewicz MR. Tracing metabolism from lignocellulosic biomass and gaseous substrates to products with stable-isotopes. Curr Opin Biotechnol. 2017;43:86–95. doi: 10.1016/j.copbio.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Gonzalez JE, Long CP, Antoniewicz MR. Comprehensive analysis of glucose and xylose metabolism in Escherichia coli under aerobic and anaerobic conditions by 13C metabolic flux analysis. Metab Eng. 2017;39:9–18. doi: 10.1016/j.ymben.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen P, Goldovsky L, Kunin V, Darzentas N, Ouzounis Ca. Genome coverage, literally speaking. EMBO Rep. 2005;6:397–399. doi: 10.1038/sj.embor.7400412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappelmann J, Wiechert W, Noack S. Cutting the Gordian Knot: Identifiability of anaplerotic reactions in Corynebacterium glutamicum by means of 13C-metabolic flux analysis. Biotechnol Bioeng. 2015;113:661–674. doi: 10.1002/bit.25833. [DOI] [PubMed] [Google Scholar]
- Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B, Assad-garcia N, Glass JI, Covert MW. A Whole-Cell Computational Model Predicts Phenotype from Genotype. 2012:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodayari A, Maranas CD. A genome-scale Escherichia coli kinetic metabolic model satisfying flux data for multiple mutant strains. Nat Commun. 2016;7:1–12. doi: 10.1038/ncomms13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingner A, Bartsch A, Dogs M, Wagner-Döbler I, Jahn D, Simon M, Brinkhoff T, Becker J, Wittmann C. Large-Scale 13C flux profiling reveals conservation of the Entner-Doudoroff pathway as a glycolytic strategy among marine bacteria that use glucose. Appl Environ Microbiol. 2015;81:2408–2422. doi: 10.1128/AEM.03157-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCroix RA, Sandberg TE, O’Brien EJ, Utrilla J, Ebrahim A, Guzman GI, Szubin R, Palsson BO, Feist AM. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol. 2015;81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Ostrov N, Wong BG, Gold MA, Khalil AS, Church GM. Vibrio natriegens, a new genomic powerhouse. bioRxiv 2016 [Google Scholar]
- Leighty RW, Antoniewicz MR. COMPLETE-MFA: Complementary parallel labeling experiments technique for metabolic flux analysis. Metab Eng. 2013;20:49–55. doi: 10.1016/j.ymben.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Leighty RW, Antoniewicz MR. Parallel labeling experiments with [U-13C]glucose validate E. coli metabolic network model for 13C metabolic flux analysis. Metab Eng. 2012;14:533–541. doi: 10.1016/j.ymben.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Li Y, Lin Z, Huang C, Zhang Y, Wang Z, Tang Yjie, Chen T, Zhao X. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Liu JK, OBrien EJ, Lerman Ja, Zengler K, Palsson BO, Feist AM. Reconstruction and modeling protein translocation and compartmentalization in Escherichia coli at the genome-scale. BMC Syst Biol. 2014;8:110. doi: 10.1186/s12918-014-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR. Metabolic flux analysis of Escherichia coli knockouts: lessons from the Keio collection and future outlook. Curr Opin Biotechnol. 2014a;28:127–133. doi: 10.1016/j.copbio.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR. Quantifying Biomass Composition by Gas Chromatography/Mass Spectrometry. Anal Chem. 2014b;86:9423–7. doi: 10.1021/ac502734e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Au J, Gonzalez JE, Antoniewicz MR. 13C metabolic flux analysis of microbial and mammalian systems is enhanced with GC–MS measurements of glycogen and RNA labeling. Metab Eng. 2016a;38:65–72. doi: 10.1016/j.ymben.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Gonzalez JE, Sandoval NR, Antoniewicz MR. Characterization of physiological responses to 22 gene knockouts in Escherichia coli central carbon metabolism. Metab Eng. 2016b;37:102–113. doi: 10.1016/j.ymben.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Gonzalez JE, Feist AM, Palsson BO, Antoniewicz MR. Fast growth phenotype of E. coli K-12 from adaptive laboratory evolution does not require intracellular flux rewiring. Metab Eng. 2017;44:100–107. doi: 10.1016/j.ymben.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maida I, Bosi E, Perrin E, Papaleo MC, Orlandini V, Fondi M, Fani R, Wiegel J, Bianconi G, Canganella F. Complete Genome Sequence of the Fast-Growing Bacterium Vibrio natriegens Strain DSMZ 759. Genome Announc. 2013;1:e00648–13. doi: 10.1186/1471-2164-9-75.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BO, Antoniewicz MR. Measuring the Composition and Stable-Isotope Labeling of Algal Biomass Carbohydrates via Gas Chromatography/Mass Spectrometry. Anal Chem. 2016;88:4624–4628. doi: 10.1021/acs.analchem.6b00779. [DOI] [PubMed] [Google Scholar]
- O’Brien EJ, Lerman Ja, Chang RL, Hyduke DR, Palsson BØ. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol Syst Biol. 2013;9:693. doi: 10.1038/msb.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne WJ, Eagon RG, Williams AK. Some observations on the physiology of Pseudomonas natriegens nov. spec. Antonie Van Leeuwenhoek. 1961;27:121–128. doi: 10.1007/BF02538432. [DOI] [PubMed] [Google Scholar]
- Pramanik J, Keasling JD. Stoichiometric model of Escherichia coli metabolism: incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol Bioeng. 1997;56:398–421. doi: 10.1002/(SICI)1097-0290(19971120)56:4<398::AID-BIT6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Sandberg TE, Long CP, Gonzalez JE, Feist AM, Antoniewicz MR, Palsson BO. Evolution of E. coli on [U-13C]Glucose Reveals a Negligible Isotopic Influence on Metabolism and Physiology. PLoS One. 2016;11:e0151130. doi: 10.1371/journal.pone.0151130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E. The Soluble and Membrane-bound Transhydrogenases UdhA and PntAB Have Divergent Functions in NADPH Metabolism of Escherichia coli. J Biol Chem. 2004;279:6613–6619. doi: 10.1074/jbc.M311657200. [DOI] [PubMed] [Google Scholar]
- Schellenberger J, Que R, Fleming RMT, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, Kang J, Hyduke DR, Palsson BØ. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc. 2011;6:1290–307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LM, Rizk ML, Liao JC. Ensemble modeling of metabolic networks. Biophys J. 2008;95:5606–5617. doi: 10.1529/biophysj.108.135442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utrilla J, O’Brien EJ, Chen K, McCloskey D, Cheung J, Wang H, Armenta-Medina D, Feist AM, Palsson BO. Global Rebalancing of Cellular Resources by Pleiotropic Point Mutations Illustrates a Multi-scale Mechanism of Adaptive Evolution. Cell Syst. 2016;2:260–271. doi: 10.1016/j.cels.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–8. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Lin B, Iv WJH, Vora GJ. Draft Genome Sequence of the Fast-Growing Marine Bacterium Vibrio natriegens Strain ATCC 14048. Genome Announc. 2013;1:e00589–13. doi: 10.1128/genomeA.00589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock MT, Hesek ED, Wilson CM, Gibson DG. Vibrio natriegens as a fast-growing host for molecular biology - supplementary data. Nat Methods. 2016;13:1–39. doi: 10.1038/nmeth.3970. [DOI] [PubMed] [Google Scholar]
- Whitaker WB, Jones JA, Bennett RK, Gonzalez JE, Vernacchio VR, Collins SM, Palmer MA, Schmidt S, Antoniewicz MR, Koffas MA, Papoutsakis ET. Engineering the biological conversion of methanol to specialty chemicals in Escherichia coli. Metab Eng. 2017;39:49–59. doi: 10.1016/j.ymben.2016.10.015. [DOI] [PubMed] [Google Scholar]
- Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem. 2008;283:20621–7. doi: 10.1074/jbc.M706494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.