Abstract
The use of parallel labeling experiments for 13C metabolic flux analysis (13C-MFA) has emerged in recent years as the new gold standard in fluxomics. The methodology has been termed COMPLETE-MFA, short for complementary parallel labeling experiments technique for metabolic flux analysis. In this contribution, we have tested the limits of COMPLETE-MFA by demonstrating integrated analysis of 14 parallel labeling experiments with Escherichia coli. An effort on such a massive scale has never been attempted before. In addition to several widely used isotopic tracers such as [1,2-13C]glucose and mixtures of [1-13C]glucose and [U-13C]glucose, four novel tracers were applied in this study: [2,3-13C]glucose, [4,5,6-13C]glucose, [2,3,4,5,6-13C]glucose and a mixture of [1-13C]glucose and [4,5,6-13C]glucose. This allowed us for the first time to compare the performance of a large number of isotopic tracers. Overall, there was no single best tracer for the entire E. coli metabolic network model. Tracers that produced well-resolved fluxes in the upper part of metabolism (glycolysis and pentose phosphate pathways) showed poor performance for fluxes in the lower part of metabolism (TCA cycle and anaplerotic reactions), and vice versa. The best tracer for upper metabolism was 75% [1-13C]glucose + 25% [U-13C]glucose, while [4,5,6-13C]glucose and [5-13C]glucose both produced optimal flux resolution in the lower part of metabolism. COMPLETE-MFA improved both flux precision and flux observability, i.e. more independent fluxes were resolved with smaller confidence intervals, especially exchange fluxes. Overall, this study demonstrates that COMPLETE-MFA is a powerful approach for improving flux measurements and that this methodology should be considered in future studies that require very high flux resolution.
Keywords: Isotopic labeling, 13C-tracers, metabolism, combined flux analysis, mass spectrometry
1. INTRODUCTION
Quantitative determination of metabolic fluxes using 13C metabolic flux analysis (13C-MFA) has become a key activity in many research fields, including in metabolic engineering, systems biology and biomedical investigations (Crown and Antoniewicz, 2013b; Hiller and Metallo, 2013; Long and Antoniewicz, 2014a; Toya and Shimizu, 2013; Young, 2013). The emergence of 13C-MFA as the preferred flux analysis method was spearheaded in 1990s by the development of advanced computational approaches to comprehensively simulate all isotopomers in complex network models (Wiechert et al., 2001). With the isotopomer framework in place (Schmidt et al., 1997), and later replaced by the EMU framework (Antoniewicz et al., 2007b), fluxes could be finally determined for large-scale systems that encompassed most of central metabolism, as well as amino acids metabolism and fatty acid metabolism (Suthers et al., 2010).
One of the key challenges in 13C-MFA remains the selection of optimal 13C-tracers that can accurately and precisely resolve all fluxes in a given metabolic network model (Antoniewicz, 2013a; Metallo et al., 2009; Schellenberger et al., 2012). Recently, several new approaches have been proposed to identify optimal tracers for labeling studies (Crown et al., 2012; Crown and Antoniewicz, 2012; Walther et al., 2013). It is now well recognized, however, that in most cases there will be no one best tracer for measuring fluxes in a given network model. Therefore, to improve flux precision a new 13C-MFA approach was developed based on combined analysis of multiple parallel labeling experiments, see Crown and Antoniewicz for a review (Crown and Antoniewicz, 2013a). This new approach has been termed COMPLETE-MFA (Leighty and Antoniewicz, 2013), short for complementary parallel labeling experiments technique for metabolic flux analysis. Studies have shown that COMPLETE-MFA can significantly improve not only flux precision but also flux observability in complex network models (Crown and Antoniewicz, 2013a). Parallel labeling experiments are also increasingly used for validation of metabolic network models, including in microbial (Au et al., 2014; Leighty and Antoniewicz, 2012), mammalian (Ahn and Antoniewicz, 2013; Sheikholeslami et al., 2014), and plant systems (Allen and Young, 2013; Alonso et al., 2007; Schwender et al., 2006).
In parallel labeling studies, the total number of parallel experiments will depend on the complexity of the system and the desired precision of fluxes. It is reasonable to expect anywhere from 2 to 4 experiments. Currently, the largest data set used for COMPLETE-MFA has consisted of 6 parallel experiments with six singly labeled glucose tracers in E. coli (Leighty and Antoniewicz, 2013). In this contribution, we report integrated analysis of 14 parallel labeling experiments, where we have combined six experiments from a previous study (Leighty and Antoniewicz, 2013) with eight new labeling experiments. In total, more than 1200 mass isotopomer measurements were used in this study to determine highly precise metabolic fluxes in E. coli. To our knowledge, an effort on such a massive scale has never been attempted before. In addition to several commonly used isotopic tracers such as [1,2-13C]glucose (He et al., 2014; Swarup et al., 2014) and mixtures of [1-13C]glucose and [U-13C]glucose (Antoniewicz et al., 2007c), several novel tracers were applied, including [2,3-13C]glucose, [4,5,6-13C]glucose, [2,3,4,5,6-13C]glucose and a mixture of [1-13C]glucose and [4,5,6-13C]glucose. These new tracers were selected using the EMU basis vector (EMU-BV) approach described in Crown and Antoniewicz (Crown et al., 2012; Crown and Antoniewicz, 2012). Overall, this study demonstrates that: 1) integrated flux analysis can be successfully conducted for large sets of experiments; 2) flux precision varies significantly for different isotopic tracers; 3) there is no single optimal tracer for the entire E. coli metabolic network model; and 4) parallel labeling experiments improve flux precision and increase the number of fluxes that can be resolved, especially exchange fluxes, which are difficult to estimate using single tracer experiments.
2. MATERIALS AND METHODS
2.1. Materials
Media and chemicals were purchased from Sigma-Aldrich (St. Louis, MO). [1-13C]Glucose (99 atom% 13C), [2,3-13C]glucose (99.5%), [4,5,6-13C]glucose (99.9%) and [U-13C]glucose (98.5%) were purchased from Cambridge Isotope Laboratories (Andover, MA); [1,2-13C]glucose (99.5%) and [2,3,4,5,6-13C]glucose (98.5%) were purchased from Sigma-Aldrich Isotec (St. Louis, MO). Glucose stock solutions (20 wt%) were prepared in distilled water. For experiments involving tracer mixtures, new glucose stock solutions were prepared by mixing the appropriate stock solutions at the desired ratio. Defined growth medium (M9 minimal medium) was used for all experiments (Leighty and Antoniewicz, 2012). All solutions were sterilized by filtration.
2.2. Strain and growth conditions
E. coli K-12 MG1655 (ATCC Cat. No. 700925, Manassas, VA) was used in this study. The inoculum for parallel experiments was grown from a single colony on an agar plate. First, the colony was suspended in 50 mL of M9 medium with 2.5 g/L glucose and grown overnight at 37°C in a shaker flask. The next day, approximately 1 mL of this culture, which was still in early exponential growth phase, was added to 50 mL of glucose-free M9 medium, which was then divided into eight tubes of 5 mL each. Next, glucose tracers were added to each tube from the eight stocks solutions that had been prepared: [1,2-13C]glucose; [2,3-13C]glucose; [4,5,6-13C]glucose; [2,3,4,5,6-13C]glucose; [1-13C] + [4,5,6-13C]glucose (1:1); [1-13C] + [U-13C]glucose (1:1); [1-13C] + [U-13C]glucose (4:1); and 20% [U-13C]glucose. The initial glucose concentration in all cultures was about 2.55 g/L. We estimated that about 0.05 g/L of unlabeled glucose was carried over from the inoculum to the cultures. The optical density (OD600) of the inoculated cultures was about 0.03. E. coli cells were then grown in parallel in aerated mini-bioreactors at 37°C, as described before (Leighty and Antoniewicz, 2013). A high precision multichannel peristaltic pump (Watson Marlow, Wilmington, MA) controlled the flow rate of air to the bioreactors, which was maintained at 5 mL/min. Gas flow rates were monitored by a digital flow-meter (Supelco, Veri-Flow 500).
2.3. Analytical methods
Samples were collected during the exponential growth phase to monitor cell growth and glucose uptake. Cell growth was monitored by measuring the optical density at 600nm (OD600) using a spectrophotometer (Eppendorf BioPhotometer). The OD600 values were converted to cell dry weight concentrations using a pre-determined OD600-dry cell weight relationship for E. coli (1.0 OD600 = 0.32 gDW/L). A molecular weight for dry biomass of 24.6 gDW/C-mol (CH1.8O0.5N0.2) was assumed (Antoniewicz et al., 2007c). After centrifugation, the supernatant was separated from the biomass pellet and glucose concentration was measured with a YSI 2700 biochemistry analyzer (YSI, Yellow Springs, OH). Acetate was measured by HPLC (Leighty and Antoniewicz, 2013).
2.4. Gas chromatography mass spectrometry
GC-MS analysis was performed on an Agilent 7890A GC system equipped with a DB-5MS capillary column (30 m, 0.25 mm i.d., 0.25 µm-phase thickness; Agilent J&W Scientific), connected to a Waters Quattro Micro Tandem Mass Spectrometer (GC-MS/MS) operating under ionization by electron impact (EI) at 70 eV. GC-MS analysis of tert-butyldimethylsilyl (TBDMS) derivatized proteinogenic amino acids was performed as described previously (Antoniewicz et al., 2007a). Labeling of glucose in the medium was confirmed by GC-MS analysis of the aldonitrile pentapropionate derivative of glucose (Antoniewicz et al., 2011). Mass isotopomer distributions were obtained by integration and corrected for natural isotope abundances (Fernandez et al., 1996).
2.5. Metabolic network model and flux analysis
13C-MFA was performed using the Metran software (Yoo et al., 2004), which is based on the elementary metabolite units (EMU) framework (Antoniewicz et al., 2007b; Young et al., 2008). The E. coli network model used for flux calculations was described previously by Leighty and Antoniewicz (Leighty and Antoniewicz, 2013), and is given in Supplementary Materials. The model includes all major metabolic pathways of central carbon metabolism, lumped amino acid biosynthesis pathways, and a lumped reaction for cell growth. In addition, the model accounts for the exchange of intracellular and atmospheric CO2, which dilutes labeling of certain intracellular metabolites (Leighty and Antoniewicz, 2012), and G-value parameters to describe fractional labeling of amino acids. As described previously (Antoniewicz et al., 2007c), the G-value represents the fraction of a metabolite produced from labeled glucose, while 1-G represents the fraction that is naturally labeled, due to the inoculum. By default, one G-value parameter was included for each measured amino acid in each data set. Reversible reactions were modeled as separate forward and backward fluxes. Net and exchange fluxes were calculated as follows: vnet = vf−vb; vexch = min(vf, vb). Exchange fluxes were then scaled to 0–100% as follows:
| [1] |
Metabolic fluxes were estimated by minimizing the variance-weighted sum of squared residuals (SSR) between the experimentally measured and model predicted extracellular rates and mass isotopomer distributions of amino acids using non-linear least-squares regression (Antoniewicz et al., 2006). For integrated analysis of parallel labeling experiments, all data sets were fitted simultaneously to a single flux model as described previously (Leighty and Antoniewicz, 2013). Flux estimation was repeated at least 10 times starting with random initial values for all fluxes to find a global solution. At convergence, accurate 68% and 95% confidence intervals were computed for all estimated fluxes by evaluating the sensitivity of the minimized SSR to flux variations (Antoniewicz et al., 2006). Standard deviations of fluxes were determined as follows:
| [2] |
2.6. Goodness-of-fit analysis
To determine the goodness-of-fit, 13C-MFA fitting results were subjected to a χ2-statistical test. In short, assuming that the model is correct and data are without gross measurement errors, the minimized SSR is a stochastic variable with a χ2-distribution (Antoniewicz et al., 2006). The number of degrees of freedom is equal to the number of fitted measurements n minus the number of estimated independent parameters p. The acceptable range of SSR values is between χ2α/2(n−p) and χ21−α/2(n−p), where α is a certain chosen threshold value, for example 0.05 for 95% confidence interval.
RESULTS
3.1. Parallel labeling experiments
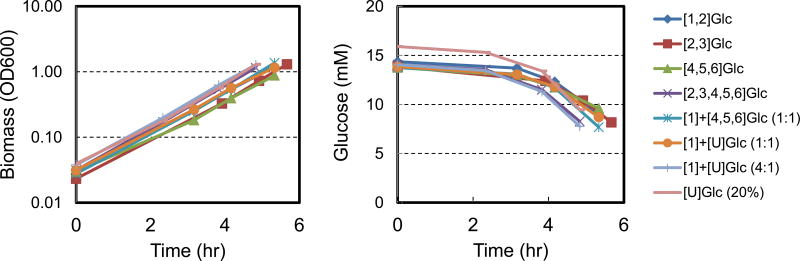
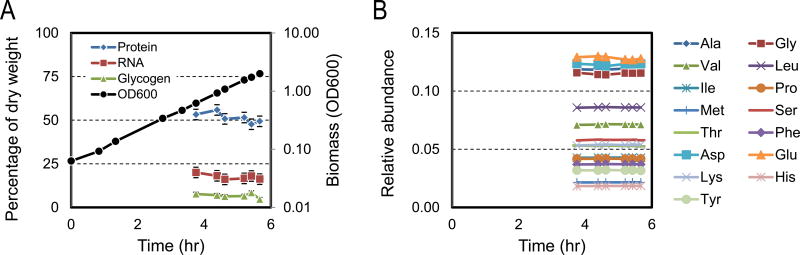
E. coli was grown aerobically in eight parallel batch cultures in mini-bioreactors (5 mL working volume) in M9 minimal medium with glucose as the only carbon source. Eight different isotopic tracers were applied in this study, including four widely used tracers: [1,2-13C]glucose, 20% [U-13C]glucose, and mixtures of [1-13C]glucose and [U-13C]glucose (1:1 and 4:1); and four novel tracers: [2,3-13C]glucose, [4,5,6-13C]glucose, [2,3,4,5,6-13C]glucose and a mixture of [1-13C]glucose and [4,5,6-13C]glucose (1:1) (Table 1). The growth rates and biomass yields for the eight cultures are shown in Table 2. Cell growth was very reproducible in the mini-bioreactors (Figure 1). The average specific growth rate was 0.72 ± 0.02 h−1 and the average biomass yield was 0.38 ± 0.02 gDW/g. The good consistency between the cultures suggests that the different glucose tracers did not influence macroscopic growth behavior of E. coli, as was also reported previous (Leighty and Antoniewicz, 2012; Leighty and Antoniewicz, 2013). The consistency also supports the assumption that E. coli was in a metabolically similar state in all cultures, which is an important assumption for conducting 13C-MFA across multiple data sets (Crown and Antoniewicz, 2013a). Another assumption of 13C-MFA is that biomass composition remains constant during cell growth. This was confirmed in a separate experiment under identical growth conditions, using the methods described by Long and Antoniewicz (Long and Antoniewicz, 2014b). The results are shown in Figure 2.
Table 1.
Overview of isotopic labeling experiments used in this study.
| Tracer experiment | Abbreviation | Reference |
|---|---|---|
| 97.5% [1,2-13C]glucose | [1,2]Glc | This study |
| 97.5% [2,3-13C]glucose | [2,3]Glc | This study |
| 97.5% [4,5,6-13C]glucose | [4,5,6]Glc | This study |
| 97.5% [2,3,4,5,6-13C]glucose | [2,3,4,5,6]Glc | This study |
| 47.5% [1-13C]glucose + 50.0 % [4,5,6-13C]glucose | [1]+[4,5,6]Glc (1:1) | This study |
| 47.5% [1-13C]glucose + 50.0 % [U-13C]glucose | [1]+[U]Glc (1:1) | This study |
| 77.0% [1-13C]glucose + 20.5 % [U-13C]glucose | [1]+[U]Glc (4:1) | This study |
| 20.5 % [U-13C]glucose | [U]Glc (20%) | This study |
| 100 % [1-13C]glucose | [1]Glc | Leighty (2013) |
| 100 % [2-13C]glucose | [2]Glc | Leighty (2013) |
| 100 % [3-13C]glucose | [3]Glc | Leighty (2013) |
| 100 % [4-13C]glucose | [4]Glc | Leighty (2013) |
| 100 % [5-13C]glucose | [5]Glc | Leighty (2013) |
| 100 % [6-13C]glucose | [6]Glc | Leighty (2013) |
Table 2.
Growth characteristics of E. coli grown in eight parallel batch cultures with different isotopic tracers.
| Tracer experiment | Growth rate (h−1) |
Biomass yield (g/g) |
|---|---|---|
| [1,2]Glc | 0.71 | 0.39 |
| [2,3]Glc | 0.70 | 0.40 |
| [4,5,6]Glc | 0.73 | 0.36 |
| [2,3,4,5,6]Glc | 0.75 | 0.35 |
| [1]+[4,5,6]Glc (1:1) | 0.73 | 0.39 |
| [1]+[U]Glc (1:1) | 0.68 | 0.39 |
| [1]+[U]Glc (4:1) | 0.73 | 0.38 |
| [U]Glc (20%) | 0.72 | 0.37 |
Figure 1.
Cell growth and glucose consumption for E. coli in eight parallel batch cultures with different isotopic tracers.
Figure 2.
Validation of constant biomass composition of E. coli grown in batch culture. (A) Cell growth and relative amounts of protein, RNA and glycogen in E. coli biomass. (B) Relative abundance of amino acids in biomass proteins.
3.2. 13C-MFA and COMPLETE-MFA
During the exponential growth phase biomass samples were collected for isotopomer analysis. Mass isotopomer distributions of protein-bound amino acids were measured for all experiments using GC-MS. In total, 14 amino acid fragments were obtained for each experiment, totaling 1246 mass isotopomer abundance measurements for the eight experiments. All data is provided in Supplementary Materials. The data from these eight tracer experiments were then combined with data from a previous study where six singly-labeled glucose tracers had been used under the same experimental conditions (Leighty and Antoniewicz). Thus, in total we had 14 parallel labeling experiments for conducting combined 13C-MFA (i.e. COMPLETE-MFA) and comparing the performance of the different isotopic tracers.
First, 13C-MFA was conducted for each tracer experiment individually, i.e. 14 flux maps were generated, one for each data set. For all 13C-MFA calculations, the precision of isotopomer measurements was assumed to be 0.4 mol%. The measured acetate yield of 70±5 mol acetate produced per 100 mol of glucose consumed was also used as an external constraint. Glucose uptake was fixed at 100. Biomass yield was not used as a constraint in flux calculations. The minimized sum of squared residuals (SSR) values for the individual 13C-MFA fits are shown in Table 3, together with the number of redundant measurements and statistically acceptable SSR values. For all 14 data sets a statistically acceptable fit was obtained. Next, COMPLETE-MFA was performed by fitting all 14 data sets simultaneously to a single flux model. For this combined analysis, a total of 811 mass isotopomer measurements (counting only non-zero mass isotopomers after correction for natural isotope abundances) and 2 external flux constraints (i.e. glucose and acetate) were used to estimate 10 net free fluxes, 9 exchange fluxes, and 129 G-value parameters. Thus, we had a total of 665 redundant measurements. The SSR value of 690 was lower than the maximum statistically acceptable SSR value of 738 at 95% confidence level, indicating a statistically acceptable fit. To our best knowledge, this is the first time that such a large number of isotopic labeling experiments have been successfully integrated for 13C-MFA. At convergence, accurate confidence intervals were calculated for the fluxes. The flux results for all 14 individual fits and for the COMPLETE-MFA fit are provided in Supplementary Materials, including 95% confidence intervals for all fluxes.
Table 3.
Goodness-of-fit analysis for individual 13C-MFA fits and for combined analysis of all 14 data sets using COMPLETE-MFA.
| Data set | No. of fitted measurements* |
No. of estimated net free fluxes, exchange fluxes, G-values |
No. of redundant measurements** |
Maximum acceptable SSR*** |
SSR (contribution to SSR in COMPLETE- MFA) |
|---|---|---|---|---|---|
| [1,2]Glc | 66 | 8, 5, 10 | 43 | 63 | 11 (45) |
| [2,3]Glc | 66 | 10, 3, 10 | 43 | 63 | 8 (47) |
| [4,5,6]Glc | 67 | 7, 4, 10 | 46 | 67 | 32 (85) |
| [2,3,4,5,6]Glc | 67 | 9, 5, 10 | 43 | 63 | 31 (71) |
| [1]+[4,5,6]Glc (1:1) | 66 | 7, 4, 9 | 46 | 67 | 11 (35) |
| [1]+[U]Glc (1:1) | 67 | 7, 5, 9 | 46 | 67 | 10 (30) |
| [1]+[U]Glc (4:1) | 67 | 7, 6, 9 | 45 | 65 | 33 (61) |
| [U]Glc (20%) | 67 | 5, 5, 9 | 48 | 69 | 54 (77) |
| [1]Glc | 56 | 7, 6, 8 | 35 | 53 | 6 (54) |
| [2]Glc | 53 | 9, 4, 10 | 30 | 47 | 8 (30) |
| [3]Glc | 51 | 4, 5, 5 | 37 | 56 | 5 (23) |
| [4]Glc | 42 | 2, 2, 0 | 38 | 57 | 15 (20) |
| [5]Glc | 51 | 10, 5, 10 | 26 | 42 | 14 (53) |
| [6]Glc | 53 | 7, 2, 8 | 36 | 54 | 13 (58) |
| COMPLETE-MFA | 813 | 10, 9, 129 | 665 | 738 | 690 |
Number of fitted measurements includes mass isotopomers that were non-zero after correction for natural isotope abundances and external flux constraints (i.e. glucose and acetate).
The number of redundant measurements equals the number of fitted measurements minus the number of estimated parameters, i.e. net free fluxes, exchange fluxes, and G-values.
Maximum acceptable SSR value at 95% confidence level
3.3. Comparison of flux resolution for different tracers
The flux values estimated in this study were in good agreement with the results obtained in our previous study (Leighty and Antoniewicz, 2013), where six singly labeled glucose tracers had been used for combined flux analysis. Therefore, we will not focus on describing the flux results, as these have been described in detail previously (Leighty and Antoniewicz, 2013), but instead we will focus on comparing the performance of the different isotopic tracers relative to each other, first based on the number of estimated fluxes (i.e. flux observability), and then based on flux precision.
Table 3 shows the number of estimated net free fluxes, exchange fluxes and G-values for each isotopic tracer. Here, a flux was considered estimated if it had a well-defined local minimum at the optimal solution (based on the sensitivity of SSR to flux variations), and the width of the 95% confidence interval was not greater than 100% of the estimated flux value. As summarized in Table 3, only two of the fourteen tracers, [2,3]Glc and [5]Glc, were able to estimate all 10 net free fluxes in the model. Two tracers, [2,3,4,5,6]Glc and [2]Glc, estimated 9 free fluxes, and [1,2]Glc estimated 8 net free fluxes. Six other tracers estimated 7 out of 10 net free fluxes. The poorest performances in terms of flux resolution were for 20% [U]Glc with 5 net free fluxes estimated, [3]Glc with 4 net free fluxes estimated, and [4]Glc with only 2 net free fluxes estimated. As indicated in the previous section, COMPLETE-MFA estimated all 10 net free fluxes in the model. It is worth noting that the two best performing tracers, [2,3]Glc and [5]Glc, had not been previously suggested as optimal tracers for 13C-MFA to our knowledge.
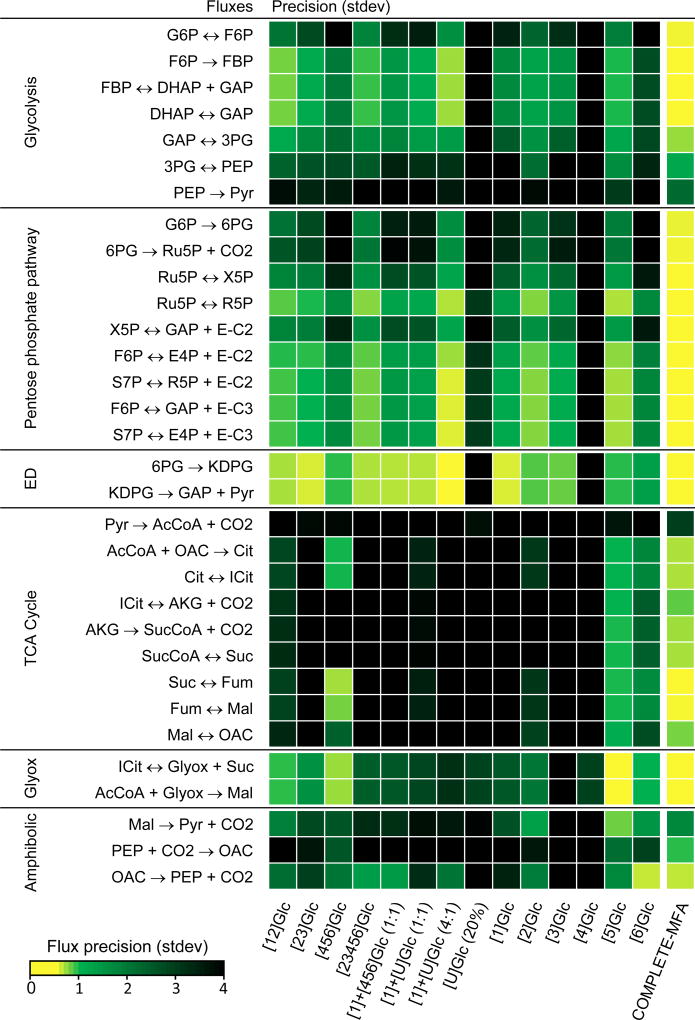
In our E. coli model 22 reactions were considered reversible. As shown in Table 3, however, for the majority of these reversible reactions no exchange flux could be determined. Even with COMPLETE-MFA, only 9 out of 22 exchange fluxes could be estimated. Of these, two reactions were determined to be almost completely equilibrated: fumarase (v27exch = 98% ± 0.1%) and malate dehydrogenase (v28exch = 100% ± 0.3%), and three reactions were highly reversible: phosphoglucose isomerate (v2exch = 75% ± 8%), transaldolase (v16exch = 80% ± 6%) and isocitrate dehydrogenase (v23exch = 80% ± 11%). Two reactions were determined to be moderately reversible: transketolase (v14exch = 13% ± 3%) and serine hydroxymethyltransferase (v44, 18% ± 1), and two reactions were determined to be nearly irreversible: isocitrate lyase (v29exch = 0% ± 0.2%) and glycine cleavage to CO2 (v45exch = 0% ± 0.3%).
Of the 14 tracers, the best performance for the estimation of exchange fluxes was for [1]Glc and [1]+[U]Glc (4:1) with 6 exchange fluxes estimated each. Six other tracers estimated 5 exchange fluxes each, [1,2]Glc, [2,3,4,5,6]Glc, [1]+[U]Glc (1:1), 20% [U]Glc, [3]Glc, and [5]Glc. The poorest performance was observed for [2,3]Glc with 3 exchange fluxes estimated, and [4]Glc and [6]Glc with only 2 exchange fluxes estimated. Note that [2,3]Glc, which was one of the two best performing tracers for the estimation of net free fluxes, performed rather poorly for the estimation of exchange fluxes, while the other optimal tracer, [5]Glc, performed well for both net and exchange fluxes. The fact that less than half of the exchange fluxes could be determined even with the massive amount of labeling data used in COMPLETE-MFA illustrates the general difficulty in determining exchange fluxes with 13C-MFA.
3.4. Comparison of flux precision for different tracers
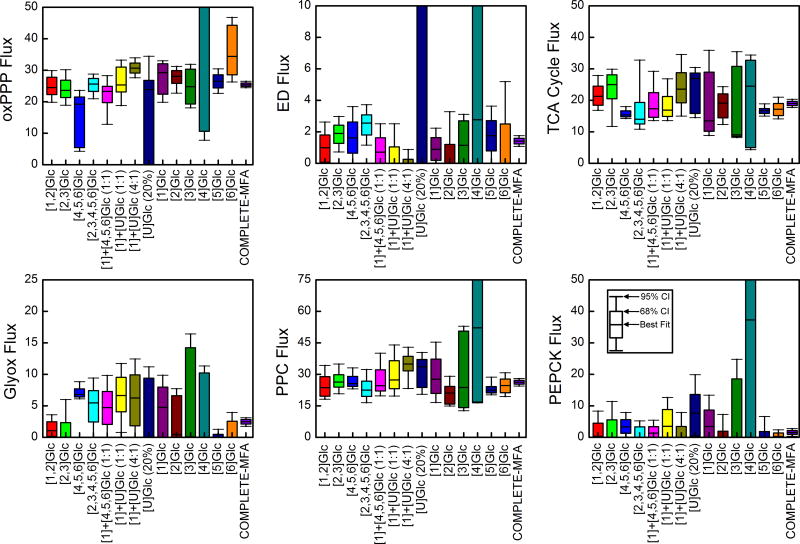
Confidence intervals of six representative fluxes in central carbon metabolism are shown in Figure 3. In each plot, the box and whiskers represent 68% and 95% confidence intervals, respectively. Figure 3 illustrates that there was no single optimal isotopic tracer for the estimation of all fluxes in the E. coli model. As an example, the widely used isotopic tracer mixture [1]+[U]Glc (4:1) performed well for glycolysis, oxidative pentose phosphate pathway and the Entner-Doudoroff pathway; however, it’s performance was poor for TCA cycle and anaplerotic fluxes. The opposite was true for [6]Glc. While this tracer performed well for fluxes in the lower part of metabolism, it was not optimal for estimation of fluxes in the upper part of central metabolism.
Figure 3.
Flux confidence intervals obtained from 13C-MFA of individual tracer experiments and for combined 13C-MFA of all 14 tracer experiments (i.e. COMPLETE-MFA). The 68% and 95% confidence intervals are shown for six representative metabolic fluxes in central metabolism: oxidative pentose phosphate pathway (v10, oxPPP); Entner–Doudoroff pathway (v18, ED); citrate synthase (v21, TCA cycle); glyoxylate shunt (v29, Glyox); phosphoenolpyruvate carboxylase (v33, PPC); and phosphoenolpyruvate carboxykinase (v34, PEPCK). COMPLETE-MFA produced the smallest confidence intervals for all fluxes.
Table 4 lists the top five tracers for each of the six fluxes shown in Figure 3. As expected, COMPLETE-MFA produced the smallest confidence intervals is all cases. The improvement in flux precision relative to the best single tracer was significant in many cases. For example, for the oxidative pentose phosphate pathway flux, COMPLETE-MFA improved the flux precision by 3-fold compared to the best single tracer [1]+[U]Glc (4:1), while a 2-fold improvement was observed for phosphoenolpyurvate carboxylase flux compared to the best single tracer [5]Glc. For the TCA cycle, a more modest 1.5-fold improvement was observed compared to the best single tracer [4,5,6]Glc. At the same time, it is important to note that for several fluxes in the model the improvement in flux precision was rather small. For example, for Entner-Doudoroff flux, COMPLETE-MFA performed only slightly better than the best single tracer [1]+[U]Glc (4:1), while for the glyoxylate shunt flux no improvement in flux precision was observed compared to the best single tracer [5]Glc. The same was true for the phosphoenolpyruvate carboxykinase flux, where [6]Glc and COMPLETE-MFA both produced the same flux precision. Figure 4 provides a visual overview of the flux precisions for all fluxes in central metabolism and for all 14 tracers.
Table 4.
Comparison of flux precision for different isotopic tracers and COMPLETE-MFA. Tracers that produced the smallest 95% confidence intervals for the fluxes shown in Figure 4 are listed.
| Oxidative pentose phosphate pathway (v10) | Entner–Doudoroff pathway (v18) | ||
|
|
|
||
| COMPLETE-MFA | 2.1 | COMPLETE-MFA | 0.7 |
| [1]+[U]Glc (4:1) | 6.3 | [1]+[U]Glc (4:1) | 0.9 |
| [5]Glc | 7.7 | [1]Glc | 2.2 |
| [2,3,4,5,6]Glc | 7.8 | [2,3]Glc | 2.3 |
| [2]Glc | 8.5 | [1]+[4,5,6]Glc (1:1) | 2.5 |
| TCA cycle (v21) | Glyoxylate shunt (v29) | ||
|
|
|
||
| COMPLETE-MFA | 2.6 | COMPLETE-MFA | 1.3 |
| [4,5,6]Glc | 3.8 | [5]Glc | 1.3 |
| [5]Glc | 3.9 | [4,5,6]Glc | 2.7 |
| [6]Glc | 6.9 | [1,2]Glc | 3.6 |
| [1,2]Glc | 11.1 | [6]Glc | 3.9 |
| Phosphoenolpyruvate carboxylase (v33) | Phosphoenolpyruvate carboxykinase (v34) | ||
|
|
|
||
| COMPLETE-MFA | 3.6 | COMPLETE-MFA | 2.4 |
| [5]Glc | 8.4 | [6]Glc | 2.4 |
| [4,5,6]Glc | 10.0 | [2,3,4,5,6]Glc | 5.2 |
| [6]Glc | 11.3 | [1]+[4,5,6]Glc (1:1) | 5.4 |
| [2,3]Glc | 14.2 | [5]Glc | 6.6 |
The numbers in this table denote the width of the 95% confidence interval, i.e. (95% CI upper bound) – (95% CI lower bound).
Figure 4.
Precision of central carbon metabolic fluxes for 14 individual tracer experiments and for combined 13C-MFA of all tracer experiments (i.e. COMPLETE-MFA). COMPLETE-MFA produced high precision for fluxes in central metabolism.
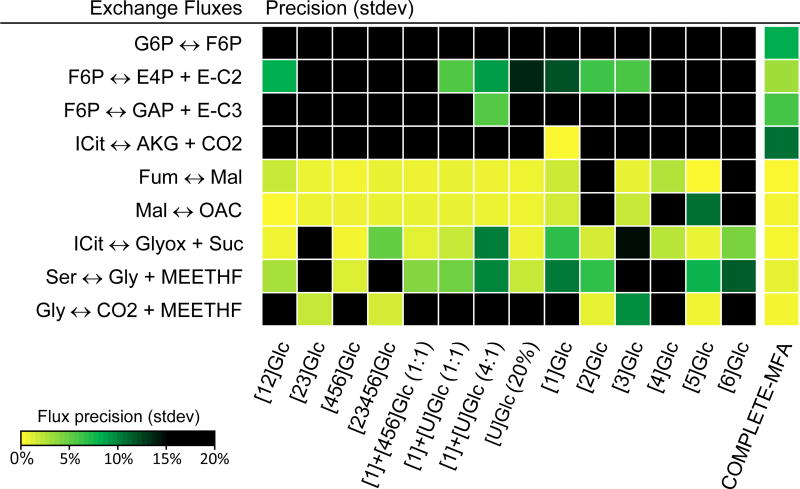
As described above, COMPLETE-MFA performed significantly better than 13C-MFA in the resolution of exchange fluxes. This is also illustrated in Figure 5, which displays flux precisions for the observable exchange fluxes and for all 14 tracers. We found that while many tracers could estimate exchange fluxes for fumarase, malate dehydrogenase, isocitrate lyase and serine hydroxymethyltransferase, only about half of the tracers were able to estimate exchange fluxes for transketolase and glycine cleavage. For transaldolase and isocitrate dehydrogenase, only one tracer provided a well resolved exchange flux. Finally, none of the 14 tracers could determine the exchange flux for phosphoglucose isomerase. This was only possible with COMPLETE-MFA.
Figure 5.
Precision of exchange fluxes in the model for 14 individual tracer experiments and for combined 13C-MFA of all tracer experiments (i.e. COMPLETE-MFA). With COMPLETE-MFA, 9 out of 22 exchange fluxes in the model could be determined.
4. DISCUSSION AND CONCLUSIONS
In this work, we have tested the limits of COMPLETE-MFA. We have conducted eight 13C-tracer experiments with E. coli using four common tracers and four novel tracers. Data from these experiments was combined with data from a previous study where an additional six tracers had been used (Leighty and Antoniewicz, 2013). With the 14 data sets in total, 13C-MFA was performed for each data set individually, followed by integrated analysis of all experiments. The four novel isotopic tracers used in this study performed well compared to the commonly used isotopic tracers. For example, [4,5,6-13C]glucose produced the best precision for the TCA cycle flux, while [2,3-13C]glucose was one of only two tracers that could successfully estimate all 10 net free fluxes in the model. Of the commonly used isotopic tracers, the 4:1 mixture of [1-13C]glucose and [U-13C]glucose performed optimally for fluxes in the upper part of central metabolism, i.e. glycolysis, pentose phosphate pathway, and Entner-Doudoroff pathway. However, this tracer mixture performed rather poorly for TCA cycle fluxes and anaplerotic reactions.
Overall, no single isotopic tracers was identified that performed optimally for all fluxes in central carbon metabolism. Of the 14 isotopic tracers evaluated in this study, the best single tracer was perhaps [5-13C]glucose. It produced good flux precision for many of the key fluxes in central metabolism (Table 4), while also allowing a large number of exchange fluxes to be determined (Figure 5).
Despite the successful integration of 14 parallel labeling experiments in this work, parallel labeling studies of this magnitude are not practical. It can be experimentally difficult and cost-prohibitive to conduct this many tracer experiments on a routine basis. This study can be viewed as an important validation of the COMPLETE-MFA methodology and the starting point for identifying smaller sets of parallel labeling experiments that can still produce high flux resolution while minimizing experimental and analytical cost and effort. Also of interest will be determining the marginal return of performing additional parallel labeling experiments, i.e. at what point does running an additional experiment not improve flux precision beyond some threshold value? To probe these questions, novel theories and algorithms will be needed that allow more rational and cost-effective design of parallel labeling studies (Crown et al., 2012; Crown and Antoniewicz, 2012).
Lastly, it is important to note that the results obtained in this study were derived for GC-MS measurements of protein-bound amino acids as the labeling input. Some of the results might have been different if different isotopic measurements had been used, for example NMR or tandem mass spectrometry data (Antoniewicz, 2013b; Choi and Antoniewicz, 2011; Choi et al., 2012; Masakapalli et al., 2014), or even if intracellular metabolite labeling was measured (Millard et al., 2014). Therefore, when designing labeling studies, it is important to consider not only the choice of isotopic tracers but also the choice of labeling measurements (Chang et al., 2008).
Supplementary Material
Acknowledgments
This work was supported by the NSF CAREER Award (CBET-1054120) and NSF Graduate Fellowship to SBC.
References
- Ahn WS, Antoniewicz MR. Parallel labeling experiments with [1,2-(13)C]glucose and [U-(13)C]glutamine provide new insights into CHO cell metabolism. Metab Eng. 2013;15:34–47. doi: 10.1016/j.ymben.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Allen DK, Young JD. Carbon and nitrogen provisions alter the metabolic flux in developing soybean embryos. Plant Physiol. 2013;161:1458–75. doi: 10.1104/pp.112.203299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso AP, Raymond P, Hernould M, Rondeau-Mouro C, de Graaf A, Chourey P, Lahaye M, Shachar-Hill Y, Rolin D, Dieuaide-Noubhani M. A metabolic flux analysis to study the role of sucrose synthase in the regulation of the carbon partitioning in central metabolism in maize root tips. Metab Eng. 2007;9:419–32. doi: 10.1016/j.ymben.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR. 13C metabolic flux analysis: optimal design of isotopic labeling experiments. Curr Opin Biotechnol. 2013a;24:1116–21. doi: 10.1016/j.copbio.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR. Tandem mass spectrometry for measuring stable-isotope labeling. Curr Opin Biotechnol. 2013b;24:48–53. doi: 10.1016/j.copbio.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–37. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal Chem. 2007a;79:7554–9. doi: 10.1021/ac0708893. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab Eng. 2007b;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Measuring deuterium enrichment of glucose hydrogen atoms by gas chromatography/mass spectrometry. Anal Chem. 2011;83:3211–6. doi: 10.1021/ac200012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kraynie DF, Laffend LA, Gonzalez-Lergier J, Kelleher JK, Stephanopoulos G. Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol. Metab Eng. 2007c;9:277–92. doi: 10.1016/j.ymben.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au J, Choi J, Jones SW, Venkataramanan KP, Antoniewicz MR. Parallel labeling experiments validate Clostridium acetobutylicum metabolic network model for C metabolic flux analysis. Metab Eng. 2014;26:23–33. doi: 10.1016/j.ymben.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Chang Y, Suthers PF, Maranas CD. Identification of optimal measurement sets for complete flux elucidation in metabolic flux analysis experiments. Biotechnol Bioeng. 2008;100:1039–49. doi: 10.1002/bit.21926. [DOI] [PubMed] [Google Scholar]
- Choi J, Antoniewicz MR. Tandem mass spectrometry: a novel approach for metabolic flux analysis. Metab Eng. 2011;13:225–33. doi: 10.1016/j.ymben.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Choi J, Grossbach MT, Antoniewicz MR. Measuring complete isotopomer distribution of aspartate using gas chromatography/tandem mass spectrometry. Anal Chem. 2012;84:4628–32. doi: 10.1021/ac300611n. [DOI] [PubMed] [Google Scholar]
- Crown SB, Ahn WS, Antoniewicz MR. Rational design of (1)(3)C-labeling experiments for metabolic flux analysis in mammalian cells. BMC Syst Biol. 2012;6:43. doi: 10.1186/1752-0509-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Antoniewicz MR. Selection of tracers for 13C-metabolic flux analysis using elementary metabolite units (EMU) basis vector methodology. Metab Eng. 2012;14:150–61. doi: 10.1016/j.ymben.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Antoniewicz MR. Parallel labeling experiments and metabolic flux analysis: Past, present and future methodologies. Metab Eng. 2013a;16:21–32. doi: 10.1016/j.ymben.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Antoniewicz MR. Publishing 13C metabolic flux analysis studies: a review and future perspectives. Metab Eng. 2013b;20:42–8. doi: 10.1016/j.ymben.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–62. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- He L, Xiao Y, Gebreselassie N, Zhang F, Antoniewicz MR, Tang YJ, Peng L. Central metabolic responses to the overproduction of fatty acids in Escherichia coli based on 13C-metabolic flux analysis. Biotechnol Bioeng. 2014;111:575–585. doi: 10.1002/bit.25124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller K, Metallo CM. Profiling metabolic networks to study cancer metabolism. Curr Opin Biotechnol. 2013 doi: 10.1016/j.copbio.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Leighty RW, Antoniewicz MR. Parallel labeling experiments with [U-13C]glucose validate E. coli metabolic network model for 13C metabolic flux analysis. Metab Eng. 2012;14:533–41. doi: 10.1016/j.ymben.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Leighty RW, Antoniewicz MR. COMPLETE-MFA: complementary parallel labeling experiments technique for metabolic flux analysis. Metab Eng. 2013;20:49–55. doi: 10.1016/j.ymben.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR. Metabolic flux analysis of Escherichia coli knockouts: lessons from the Keio collection and future outlook. Curr Opin Biotechnol. 2014a;28:127–33. doi: 10.1016/j.copbio.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR. Quantifying biomass composition by gas chromatography/mass spectrometry. Anal Chem. 2014b;86:9423–7. doi: 10.1021/ac502734e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masakapalli SK, Ratcliffe RG, Williams TC. Quantification of (1)(3)C enrichments and isotopomer abundances for metabolic flux analysis using 1D NMR spectroscopy. Methods Mol Biol. 2014;1090:73–86. doi: 10.1007/978-1-62703-688-7_5. [DOI] [PubMed] [Google Scholar]
- Metallo CM, Walther JL, Stephanopoulos G. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J Biotechnol. 2009;144:167–74. doi: 10.1016/j.jbiotec.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard P, Massou S, Wittmann C, Portais JC, Letisse F. Sampling of intracellular metabolites for stationary and non-stationary C metabolic flux analysis in Escherichia coli. Anal Biochem. 2014 doi: 10.1016/j.ab.2014.07.026. [DOI] [PubMed] [Google Scholar]
- Schellenberger J, Zielinski DC, Choi W, Madireddi S, Portnoy V, Scott DA, Reed JL, Osterman AL, Palsson B. Predicting outcomes of steady-state (1)(3)C isotope tracing experiments using Monte Carlo sampling. BMC Syst Biol. 2012;6:9. doi: 10.1186/1752-0509-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K, Carlsen M, Nielsen J, Villadsen J. Modeling isotopomer distributions in biochemical networks using isotopomer mapping matrices. Biotechnol Bioeng. 1997;55:831–40. doi: 10.1002/(SICI)1097-0290(19970920)55:6<831::AID-BIT2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Schwender J, Shachar-Hill Y, Ohlrogge JB. Mitochondrial metabolism in developing embryos of Brassica napus. J Biol Chem. 2006;281:34040–7. doi: 10.1074/jbc.M606266200. [DOI] [PubMed] [Google Scholar]
- Sheikholeslami Z, Jolicoeur M, Henry O. Elucidating the effects of postinduction glutamine feeding on the growth and productivity of CHO cells. Biotechnol Prog. 2014;30:535–46. doi: 10.1002/btpr.1907. [DOI] [PubMed] [Google Scholar]
- Suthers PF, Chang YJ, Maranas CD. Improved computational performance of MFA using elementary metabolite units and flux coupling. Metab Eng. 2010;12:123–8. doi: 10.1016/j.ymben.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Swarup A, Lu J, DeWoody KC, Antoniewicz MR. Metabolic network reconstruction, growth characterization and 13C-metabolic flux analysis of the extremophile Thermus thermophilus HB8. Metab Eng. 2014;24:173–80. doi: 10.1016/j.ymben.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Toya Y, Shimizu H. Flux analysis and metabolomics for systematic metabolic engineering of microorganisms. Biotechnol Adv. 2013;31:818–26. doi: 10.1016/j.biotechadv.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Walther JL, Metallo CM, Zhang J, Stephanopoulos G. Optimization of (13)C isotopic tracers for metabolic flux analysis in mammalian cells. Metab Eng. 2013 doi: 10.1016/j.ymben.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiechert W, Mollney M, Petersen S, de Graaf AA. A universal framework for 13C metabolic flux analysis. Metab Eng. 2001;3:265–83. doi: 10.1006/mben.2001.0188. [DOI] [PubMed] [Google Scholar]
- Yoo H, Stephanopoulos G, Kelleher JK. Quantifying carbon sources for de novo lipogenesis in wild-type and IRS-1 knockout brown adipocytes. J Lipid Res. 2004;45:1324–32. doi: 10.1194/jlr.M400031-JLR200. [DOI] [PubMed] [Google Scholar]
- Young JD. Metabolic flux rewiring in mammalian cell cultures. Curr Opin Biotechnol. 2013;24:1108–15. doi: 10.1016/j.copbio.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol Bioeng. 2008;99:686–99. doi: 10.1002/bit.21632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.