Abstract
We have isolated a new extremely thermophilic fast-growing Geobacillus strain that can efficiently utilize xylose, glucose, mannose and galactose for cell growth. When grown aerobically at 72 °C, Geobacillus LC300 has a growth rate of 2.15 h−1 on glucose and 1.52 h−1 on xylose (doubling time less than 30 minutes). The corresponding specific glucose and xylose utilization rates are 5.55 g/g/h and 5.24 g/g/h, respectively. As such, Geobacillus LC300 grows 3-times faster than E. coli on glucose and xylose, and has a specific xylose utilization rate that is 3-times higher than the best metabolically engineered organism to date. To gain more insight into the metabolism of Geobacillus LC300 its genome was sequenced using PacBio’s RS II single-molecule real-time (SMRT) sequencing platform and annotated using the RAST server. Based on the genome annotation and the measured biomass composition a core metabolic network model was constructed. To further demonstrate the biotechnological potential of this organism, Geobacillus LC300 was grown to high cell-densities in a fed-batch culture, where cells maintained a high xylose utilization rate under low dissolved oxygen concentrations. All of these characteristics make Geobacillus LC300 an attractive host for future metabolic engineering and biotechnology applications.
Keywords: Thermophile, genome analysis, metabolic network model, xylose metabolism, high-cell-density fermentation
1. INTRODUCTION
Thermophilic organisms are increasingly used as versatile hosts for a variety of metabolic engineering applications (Blumer-Schuette et al., 2008; Chang and Yao, 2011; Elleuche et al., 2014; Taylor et al., 2009). In theory, high growth temperatures can promote higher rates of feedstock conversion, reduce cooling costs in fermentations, facilitate the removal and recovery of products, and decrease chances of contaminations (Lin and Xu, 2013; Lynd, 1989). Moreover, many thermophiles readily ferment pentoses and hexoses, and even in some cases polymeric precursors and structurally complex polycarbohydrates such as cellulose and hemicellulose, an important characteristic for the development of next generation bioprocesses (Lynd, 1989; Taylor et al., 2009).
The genus Geobacillus includes a wide range of highly thermophilic bacilli that have attracted significant interest for their potential applications in biotechnology, e.g. for biofuel production (Cripps et al., 2009; Lin et al., 2014; Xiao et al., 2012), bioremediation (Feng et al., 2007), and as a source of thermostable enzymes (Anand et al., 2013; Canakci et al., 2012; Ezeji and Bahl, 2006; Ezeji et al., 2005; Verma et al., 2013). Geobacillus species are capable of growing at temperatures up to 70–80 °C (Coorevits et al., 2012; Shintani et al., 2014), and typically have optimal growth temperatures near 55–60 °C (Coorevits et al., 2012; Nazina et al., 2001; Xiao et al., 2012). Maximum growth rates are typically around 0.2–0.3 h−1 (Anand et al., 2013; Tang et al., 2009), although faster growing Geobacillus strains have also been described. For example, Marchant et al. (Marchant et al., 2002a) isolated several fast growing Geobacillus strains from soil samples with an optimal growth temperature of 70 °C and a doubling time of around 30 minutes in rich medium.
In this work, we have isolated a new aerobic extremely thermophilic fast-growing Geobacillus strain (termed strain LC300), that can efficiently utilize xylose, glucose and several other carbohydrates for cell growth. To gain more insight into the metabolism of this organism, its genome was sequenced and analyzed. Based on this information and the measured biomass composition a core metabolic network model was constructed for 13C-flux analysis studies. Finally, the biotechnological potential of Geobacillus LC300 was evaluated in high cell-density fed-batch fermentations.
2. MATERIALS AND METHODS
2.1. Materials
Media and chemicals were purchased from Sigma-Aldrich (St. Louis, MO). [U-13C]Xylose (99% 13C) was purchased from Isotec (St. Louis, MO). Wolfe’s minerals (Cat. No. MD-TMS) and Wolfe’s vitamins (Cat. No. MD-VS) were purchased from ATCC (Manassas, VA). Tris solution (1 mol/L) was purchased from Cellgro (Cat. No. 46-031-CM). Yeast extract was purchased from Fisher (Cat. No. BP-1422-500, lot 068366). Xylose stock solutions (20 wt%) and yeast extract stock solution (1 wt%) were prepared in distilled water. For growth of Geobacillus LC300, a previously described growth medium was used (Swarup et al., 2014). The medium contained (per liter of medium): 0.50 g K2HPO4, 0.30 g KH2PO4, 0.50 g NH4Cl, 0.50 g NaCl, 0.20 g MgCl2.6H2O, 0.04 g CaSO4.2H2O, 40 mL of 1 M Tris, 5 mL of Wolfe’s minerals, 5 mL of Wolfe’s vitamins, and 0.05 g/L of yeast extract. For growth of E. coli, M9 minimal medium was used. Xylose and other carbohydrates were added as indicated. All solutions were sterilized by filtration.
2.2. Strains
Geobacillus LC300 was isolated as a contaminant from a culture of Thermus thermophilus that was grown aerobically at 72 °C in medium containing 4 g/L xylose. The extremely fast growth rate observed in the culture suggested that the fast-growing strain was not T. thermophilus, but likely a contaminant species of a different genus altogether. A sample of the culture was streaked on a xylose-agar plate and incubated at 60 °C. Colonies appeared within 24 h. A single colony from the plate was suspended in 10 mL of medium with 2 g/L xylose and 0.05 g/L yeast extract. Fast growth on xylose was confirmed under aerobic growth conditions at 72 °C. Cells collected during the exponential growth phase were then re-suspended in medium containing 15% glycerol and frozen at −80°C. This frozen stock, termed LC300, was used for all subsequent experiments. For comparison, E. coli K-12 MG1655 (ATCC 700926) and Geobacillus stearothermophilus ATCC 12980 (the type strain for the genus Geobacillus) were also used in this study.
The origin of Geobacillus LC300 is uncertain at this time. The T. thermophilus culture from which Geobacillus LC300 was isolated was theoretically a pure culture. We believe that spores of Geobacillus LC300 may have accidentally contaminated one of the chemicals in the lab, or possibly some glass or plastic supplies. It is well known that Geobacillus strains can survive in harsh conditions (Marchant et al., 2002a; Marchant et al., 2002b), even in nutrient-limiting conditions of clean room environments (La Duc et al., 2007).
2.3. Growth conditions
All batch experiments in this study were performed in mini-bioreactors with a working volume of 10 mL, as described previously for growth of T. thermophilus and E. coli (Leighty and Antoniewicz, 2012; Swarup et al., 2014). E. coli was grown at 37 °C in M9 minimal medium with 30 g/L of xylose as the only carbon source. Geobacillus stearothermophilus and Geobacillus LC300 were grown at 68 °C and 72 °C, respectively, in growth medium described in section 2.1, supplemented with 30 g/L of xylose. Several additional growth experiments were also performed with 30 g/L of arabinose, glucose, mannose, or galactose. A high-precision multichannel peristaltic pump (Watson Marlow, Wilmington, MA) was used to control the air flow to the mini-bioreactors, which was set at 11.3 mL/min. Gas flow rates were monitored by a digital flow-meter (Supelco, Veri-Flow 500). Mixing in the mini-bioreactors was achieved through the rising gas bubbles and a constant temperature was maintained by placing the tubes in a heating block (Fisher Isotemp Digital Dry-Bath 125D) (Swarup et al., 2014). Samples were collected during the early to mid-exponential growth phase, when OD600 was less than 1.5.
A fed-batch culture of Geobacillus LC300 was also performed in a 3-L benchtop fermentor with a working volume of 1.5 L (New Brunswick Scientific BioFlow/ CelliGen 115, New Brunswick, NJ). For this culture, the medium described in section 2.1 was supplemented with 5.2 g/L of xylose, 0.36 g/L of NH4Cl, and 1.0 g/L of yeast extract. The pH during the culture was controlled at 6.5 by addition of 1 N NaOH, and the temperature was controlled at 70 °C. The dissolved oxygen concentration was controlled at 1.6 mg/L by automatic control of the agitator speed (between 400 rpm and 1000 rpm). A constant gas flow rate of 4 L/min was maintained. The inoculum for the fed-batch culture was grown in one of the mini-bioreactors described above to an OD600 of about 0.5. Next, 10 mL of this culture was used to inoculate the 1.5 L benchtop bioreactor. The initial OD600 was about 0.003. After 2.7 hours of batch culture, xylose feed was initiated. The feed medium contained (per liter of feed medium): 36.8 g xylose, 6.1 g NH4Cl, 7.4 g yeast extract, 20 mL of Wolfe’s minerals, and 20 mL of Wolfe’s vitamins. The feed rate was maintained constant at 100 mL/min.
2.4. Analytical methods
Biomass concentration was determined by measuring the optical density at 600 nm (OD600) using a spectrophotometer (Eppendorf BioPhotometer). The OD600 values were converted to cell dry weight concentrations using the pre-determined OD600-dry cell weight relationships: 1.0 OD600 = 0.27 gDW/L for Geobacillus LC300; and 1.0 OD600 = 0.31 gDW/L for E. coli. Xylose concentration was determined by isotope ratio analysis using [U-13C]xylose as internal standard (Ahn and Antoniewicz, 2013). Briefly, 50 µL of supernatant was combined with 50 µL of a 3.5 g/L [U-13C]xylose solution. The mixture was then evaporated and derivatized using the aldonirile propionate derivatization method described by Antoniewicz et al. (Antoniewicz et al., 2011). The ion fragment at m/z 173 was measured by GC-MS, which contains the last two carbon atoms of xylose.
2.5. Biomass composition analysis
The biomass composition of Geobacillus LC300 was determined by GC-MS analysis, as described previously (Long and Antoniewicz, 2014b). Specifically, weight percentages of protein, RNA, glycogen and lipids (wt% of dry biomass) were measured. Constant values were assumed for weight percentages of DNA (3 wt%), and ions and metabolites (3 wt%). The remaining fraction was assumed to be cell wall components, e.g. peptidoglycan and teichoic acids. For fatty acid quantification, [U-13C]palmitate was used as the internal standard. For cell wall components, the composition reported for the closely related B. subtilis was assumed (Coorevits et al., 2012; Dauner and Sauer, 2001; Nazina et al., 2001).
2.6. Gas chromatography mass spectrometry
GC-MS analysis was performed on an Agilent 7890A GC system equipped with a DB-5MS capillary column (30 m, 0.25 mm i.d., 0.25 µm-phase thickness; Agilent J&W Scientific), connected to a Waters Quattro Micro Tandem Mass Spectrometer (GC-MS/MS) operating under ionization by electron impact (EI) at 70 eV. Mass isotopomer distributions were obtained by integration selected ion chromatograms (Antoniewicz et al., 2007), and corrected for natural isotope abundances (Fernandez et al., 1996).
2.7. Genome sequencing, assembly and annotation
High molecular weight (HMW) genomic DNA from Geobacillus LC300 was isolated using Qiagen’s Genomic-tip 20/G, following the protocol by the manufacturer. After extraction, HMW DNA (>23 Kb) was confirmed using agarose gel electrophoresis with λ-DNA digested with HindIII as the marker and fragment analyzer. The isolated genomic DNA was then sequenced using PacBio’s RSII third generation DNA sequencing system, known as SMRT (Single Molecule, Real-Time) sequencing, at the University of Delaware Sequencing and Genotyping Center. Assembly was performed using HGAP 2.0 (hierarchical genome assembly process) in PacBio’s SMRT portal. The assembled two contigs of 3.49 Mb (>150× coverage, LC300 chromosome) and 38.4 Kb (>300× coverage, pGt35 plasmid) were subsequently annotated using the RAST server (Aziz et al., 2008). To perform an unbiased annotation, the genus and species name were declared as “unknown” and annotation was performed with the default RAST annotation settings.
The nucleotide sequences of Geobacillus LC300 chromosome and pGt35 plasmid were deposited in the NCBI GenBank database under accession numbers CP008903 and CP008904, respectively. The deposited sequences were annotated by NCBI’s Prokaryotic Genome Automatic Annotation Pipeline (PGAAP).
3. RESULTS AND DISCUSSION
3.1. General genome features
The genome of strain LC300 was sequenced using PacBio’s RSII third generation DNA sequencing system, known as SMRT (Single Molecule, Real-Time) sequencing. Analysis of 16S rDNA indicated that the strain belonged to the genus Geobacillus. High similarity of 16S rDNA gene sequence was found with that of G. stearothermophilus (99.7%), the type species of this genus (Coorevits et al., 2012; Nazina et al., 2001), G. thermocatenulatus (98.9%), G. kaustophilus (98.7%), G. thermoleovorans (98.5%), G. thermodenitrificans (98.5%), and to a lesser extent G. thermoglucosidasius (96.7%).
The genome sequence was then annotated using the RAST server (Rapid Annotations using Subsystems Technology) (Aziz et al., 2008). The annotation generated by the RAST server is included in Supplementary Materials. The genome of Geobacillus LC300 is composed of a 3,494,216-bp chromosome and a 38,364-bp plasmid, designated pGt35, with mean GC contents of 52.2% and 46.8%, respectively (Table 1). On the chromosome, 3,988 protein-coding sequences (CDSs) were identified, 10 rRNA operons and 88 tRNA genes for all 20 amino acids, covering 86.9% of the chromosome. Putative biological roles were assigned to 2,630 CDSs (66%). The remaining 1,358 CDSs (34%) were classified as hypothetical proteins of unknown function. On the plasmid, 51 CDSs were identified, covering 67.6% of the plasmid, and putative biological roles were assigned to 26 CDSs (51%).
Table 1.
General features of the genome of Geobacillus LC300.
| Feature | Chromosome | Plasmid pGt35 |
|---|---|---|
| Length (bp) | 3,494,216 | 38,364 |
| GC content (%) | 52.2 | 46.8 |
| Coding sequence (%) | 86.9 | 67.6 |
|
| ||
| Predicted CDSs | 3988 | 51 |
| With assigned function | 2630 (66%) | 26 (51%) |
| Hypothetical protein | 1358 (34%) | 25 (49%) |
| With assigned EC number | 946 | 3 |
|
| ||
| RNAs | ||
| rRNA operons | 10 | - |
| tRNAs | 88 | - |
The genome length, mean GC content, and number of predicted CDSs of Geobacillus LC300 (3.49 Mbp, 52.2% GC, 3988 CDS) are similar to those reported for other Geobacillus species, notably Geobacillus sp. JF8 (3.49 Mbp, 52.0% GC, 3584 CDS) (Shintani et al., 2014), G. thermoleovorans (3.60 Mbp, 52.3% GC, 3887 CDS) (Muhd Sakaff et al., 2012), G. kaustophilus (3.54 Mbp, 52.1% GC, 3498 CDS) (Takami et al., 2004), G. stearothermophilus (3.27 Mbp, 52.6% GC, 3042 CDS) (Rozanov et al., 2014), and to a lesser extent G. thermodenitrificans (3.55 Mbp, 49.0% GC, 3444 CDS) (Feng et al., 2007) and G. thermoglucosidasius (3.75 Mbp, 43.9% GC, 3443 CDS) (Zhao et al., 2012). The pGt3 plasmid of Geobacillus LC300 shows similarity with plasmids pHTA426 from G. kaustophilus (97% identity, 37% coverage, BLASTN), pBt40 from Geobacillus sp. JF8 (97% identity, 35% coverage), and pGS18 from G. stearothermophilus (98% identity, 26% coverage).
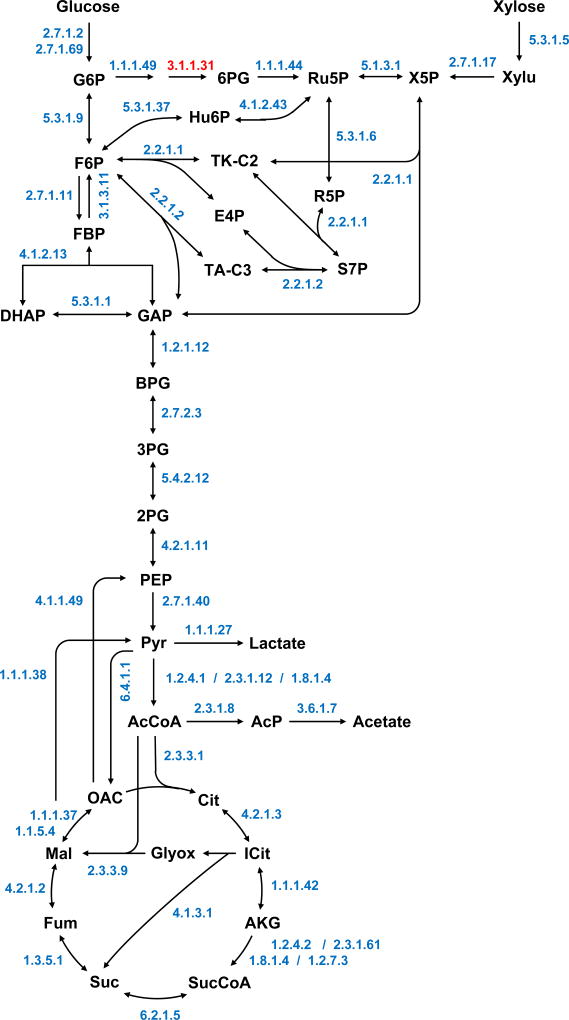
3.2. General metabolism
The Geobacillus LC300 genome encodes many common central metabolic pathways, including glycolysis, pentose phosphate pathway, tricarboxylic acid (TCA) cycle, glyoxylate shunt, and the ribulose monophosphate pathway (RuMP), but not Entner–Doudoroff pathway (Figure 1). No gene encoding for 6-phosphogluconolactonase (EC 3.1.1.31) of the oxidative pentose phosphate pathway was identified. However, this reaction is also known to proceed spontaneously (Kupor and Fraenkel, 1969; Thomason et al., 2004; Zimenkov et al., 2005), so the oxidative pentose phosphate pathway was considered complete. Enzymes for anaplerosis (pyruvate carboxylase, EC 6.4.1.1), cataplerosis (NAD-dependent malic enzyme, EC 1.1.38) and gluconeogenesis (phosphoenolpyruvate carboxykinase, EC 4.1.1.49; and fructose-bisphosphatase, EC 3.1.3.11) were also identified. No genes encoding for transhydrogenases were found. Several genes encoding typical acid fermentation pathways were identified, including phosphate acetyltransferase (EC 2.3.1.8) and acetate kinase (EC 2.7.2.1) for acetate production, and lactate dehydrogenase (EC 1.1.1.27) for lactate production (Figure 1).
Figure 1.
Central metabolic pathways of Geobacillus LC300 based on the genome annotation. EC numbers of identified enzymes are shown. No gene encoding for 6-phosphogluconolactonase (EC 3.1.1.31) was identified. However, this reaction is also known to proceed spontaneously, so the oxidative pentose phosphate pathway was considered complete.
In batch growth experiments, Geobacillus LC300 was found to be a strict aerobe. No growth was observed under anaerobic growth conditions. In contrast to G. thermoglucosidasius, no genes homologous to pyruvate formate lyase (PFL, EC 2.3.1.54) were identified in the Geobacillus LC300 genome. G. thermoglucosidasius grows anaerobically by using PFL instead of pyruvate dehydrogenase (PDH, EC 1.2.4.1) to convert pyruvate to acetyl-CoA (Loftie-Eaton et al., 2013; Tang et al., 2009). Moreover, in contrast to G. thermodenitrificans (Feng et al., 2007), no genes encoding pyruvate-ferredoxin oxidoreductase (PFOR, EC 1.2.7.1) were identified in the Geobacillus LC300 genome.
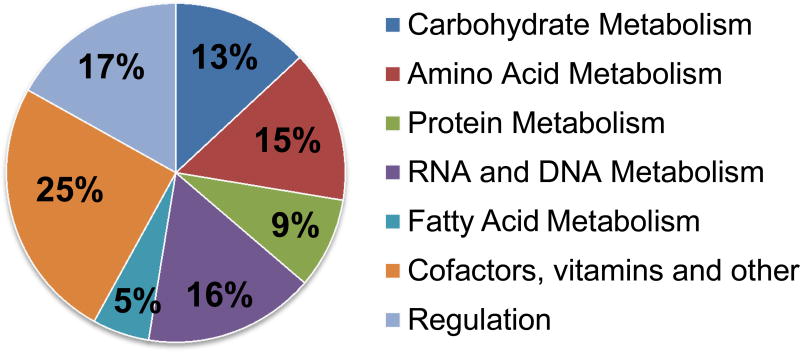
Biosynthetic pathways for purine and pyrimidine nucleotides, fatty acids, and all 20 amino acids were identified, with the exception of phosphoserine phosphatase enzyme (serB, EC 3.1.3.3) in the serine biosynthesis pathway. To date, no phosphoserine phosphatase gene has been identified in any of the sequenced Geobacillus genomes, possibly suggesting the presence of a non-orthologous gene. In E. coli, it was recently shown that deletion of serB can be rescued by several other phosphatase enzymes, including phosphoglycolate phosphatase (gph, EC 3.1.3.18), histidinol phosphate phosphatase (hisB, EC 3.1.3.15), and ytjC, a predicted bisphosphatase (Patrick et al., 2007). In the Geobacillus LC300 genome, several related histidinol phosphate phosphatases and several hypothetical phosphatases were identified. Biosynthetic pathways for many cofactors and vitamins were also identified, including for vitamin B12 (cobalamin), as well as catabolic pathways for most amino acids (Figure 2).
Figure 2.
Subsystem distribution of Geobacillus LC300 based on RAST server annotation.
The Geobacillus LC300 genome encodes a large number of transporters for uptake of carbohydrates, amino acids and peptides. Of the 118 ORFs belonging to the ATP-binding cassette (ABC) substrate transporter family, 22 were predicted to be involved in carbohydrate transport (with predicted specificities for xylose, ribose and maltose), 29 for amino acid transport (among these, a large number for branched chain amino acids and methionine), and 16 for peptide transport. Additionally, 12 ORFs belonging to the phosphotransferase transport system (PTS) were identified, with predicted specificities for glucose, fructose, sucrose, cellobiose, mannitol, N-acetylmuramic acid and N-acetylglucosamine.
3.3. Oxidative phosphorylation and energy metabolism
Geobacillus LC300 generates energy largely by oxygen respiration (see also accompanying paper, (Cordova and Antoniewicz, 2015)). Reducing equivalents are fed into the membrane-bound oxidative respiratory chain via NADH dehydrogenases and succinate dehydrogenase. Geobacillus LC300 genome encodes three NADH dehydrogenases: a Complex I-like NADH dehydrogenase (H+ translocating) (nuoA-D, nuoH-N, EC 1.6.5.3, NADH + Q → QH2 + 4 H+out), and two type II NADH dehydrogenases (ndh, EC 1.6.99.3, NADH + Q → QH2). The LC300 genome also encodes a succinate dehydrogenase complex (EC 1.3.5.1, Fum → Suc + FADH2, FADH2 + Q → QH2) and a cytochrome c reductase complex. Gene clusters for four terminal cytochrome oxidases were identified: cytochrome aa3-type quinol oxidase (QoxABCD, H+/e− = 2), cytochrome bd-type quinol oxidase (CydAB, H+/e− = 1), cytochrome caa3-type cytochrome c oxidase (CoxABCD, H+/e− = 2), and cytochrome b(O/a)3-type cytochrome c oxidase (H+/e− = 2). These enzymes act as the terminal oxidases in the aerobic respiratory chain of Geobacillus LC300. ATP is synthesized by an ATP synthase of the F-type. Assuming a H+/ATP ratio of 4 for ATP synthase, the expected (P/O) ratio for oxidation of NADH ranges between 0.5 and 2, and between 0.5 and 1 for oxidation of FADH2.
3.4. Growth characteristics of Geobacillus LC300
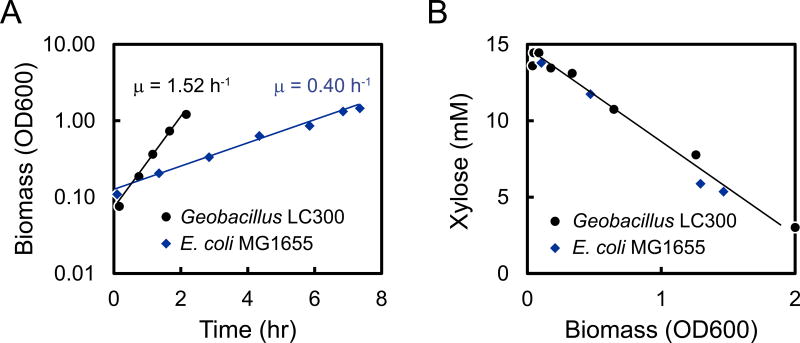
A unique characteristic of Geobacillus strain LC300 is its very fast growth rate on xylose. To determine the maximum specific growth rate, cells were grown aerobically at 72 °C in batch culture in medium containing 30 g/L xylose. Geobacillus LC300 grew exponentially with an average specific growth rate of 1.52 h−1 (Figure 3A). To our knowledge, this is the fastest growth rate reported for any organism to date on xylose. For comparison, wild-type E. coli MG1655 was also grown aerobically in M9 minimal medium with 30 g/L xylose at 37 °C. E. coli grew exponentially with an average specific growth rate of 0.40 h−1 (Figure 3A), thus almost 4-times slower than Geobacillus LC300. To quantify biomass yield on xylose, cells were grown on medium containing 2 g/L xylose (Figure 3B). The biomass yield on xylose was very similar for Geobacillus LC300 and E. coli MG1655, about 0.29 g/g for Geobacillus LC300 and 0.31 g/g for E. coli. However, due to the faster growth rate, Geobacillus LC300 had a 4-fold higher specific xylose utilization rate of 5.24 g/g/h, compared to 1.29 g/g/h for E. coli.
Figure 3.
Determination of specific growth rate and biomass yield for Geobacillus LC300 grown at 72 °C and E. coli MG1655 grown at 37 °C with xylose as the main carbon source. (A) Cells were grown in medium containing 30 g/L of xylose. Exponential growth was observed for both strains. (B) Cells were grown in medium containing 2 g/L of xylose. The biomass yield was similar for both strains.
Table 2 compares the performance of Geobacillus LC300 to other mesophilic and thermophilic organisms that have been selected or engineered for xylose utilization. Tang et al. (Tang et al., 2009) reported growth rates of 0.27 h−1 and 0.15 h−1 for Geobacillus thermoglucosidasius M10EXG grown on 10 g/L xylose under aerobic and anaerobic conditions, respectively, and reported specific xylose utilization rates of 1.17 g/g/h and 1.50 g/g/h, respectively. Gonzalez et al. (Gonzalez et al., 2002) reported a specific growth rate of 0.19 h−1 and xylose utilization rate of 1.58 g/g/h for E. coli KO11 grown anaerobically on 100 g/L xylose. Recently, the Stephanopoulos lab developed several efficient yeast strains for xylose utilization. S. cerevisiae strain H131-A3-AL is reported to have maximum growth rates of 0.23 h−1 and 0.20 h−1 under aerobic and anaerobic conditions, respectively, and specific xylose utilization rates of 1.50 g/g/h and 1.87 g/g/h, respectively (Wasylenko and Stephanopoulos, 2014; Zhou et al., 2012). Thus, compared to these strains, Geobacillus LC300 has a xylose utilization rate that is 3-times higher than the best metabolically engineered S. cerevisiae strain to date, and grows 3-times faster than E. coli on xylose under its optimal growth conditions.
Table 2.
Comparison of strains grown in batch culture on medium with xylose as the main carbon source
| Organism | Growth rate (h−1) |
Biomass yield (g/g) |
Xylose utilization rate (g/g/h) |
Temp. (°C) |
Growth conditions | Reference |
|---|---|---|---|---|---|---|
| Geobacillus LC300 | 1.52 | 0.29 | 5.24 | 72 | Aerobic, 30 g/L xylose | This study |
| Geobacillus thermoglucosidasius M10EXG | 0.27 | 0.23 | 1.17 | 60 | Aerobic, 10 g/L xylose | Tang, 2009 |
| Geobacillus thermoglucosidasius M10EXG | 0.15 | 0.10 | 1.50 | 60 | Anaerobic, 10 g/L xylose | Tang, 2009 |
| Escherichia coli MG1655 | 0.40 | 0.31 | 1.29 | 37 | Aerobic, 30 g/L xylose | This study |
| Escherichia coli KO11 | 0.19 | NR | 1.58 | 35 | Anaerobic, 100 g/L xylose | Gonzalez, 2002 |
| Saccharomyces cerevisiae H131-A3-AL | 0.23 | NR | 1.50 | 30 | Aerobic, 20 g/L xylose | Wasylenko, 2014 |
| Saccharomyces cerevisiae H131-A3-AL | 0.20 | 0.06 | 1.87 | 30 | Anaerobic, 50 g/L xylose | Zhou, 2012 |
NR = not reported
Based on the genome sequence analysis, Geobacillus LC300 was found to be closely related to G. stearothermophilus, i.e. 99.7% 16S rDNA gene sequence similarity (see section 3.1). It has been suggested that organisms with such high similarity in their 16S rDNA sequences are likely to be members of the same species (Goebel and Stackebrandt, 1994). However, we found several major physiological differences between Geobacillus LC300 and G. stearothermophilus, as summarized in Table 3. First, we found that G. stearothermophilus was unable to grow on xylose. In batch culture, G. stearothermophilus grew only on glucose and mannose, whereas Geobacillus LC300 was able to grow on xylose, glucose, mannose, and galactose. The growth rates of Geobacillus LC300 on glucose and mannose (2.15 and 1.15 h−1, respectively) were more than 2-fold higher compared to G. stearothermophilus (0.82 and 0.39 h−1, respectively). Furthermore, the optimal growth temperature of Geobacillus LC300 (Toptimal = 72 °C) was significantly higher than G. stearothermophilus (Toptimal = 65–68 °C). Based on these physiological differences, we believe that Geobacillus LC300 should be classified as a new species of Geobacillus and not as a strain of G. stearothermophilus.
Table 3.
Maximum growth rates (h−1) of Geobacillus LC300 and Geobacillus stearothermophilus grown aerobically on different C6 and C5 sugars.
| Sugar |
Geobacillus LC300 |
Geobacillus stearothermophilus |
|---|---|---|
| Glucose | 2.15 | 0.82 |
| Galactose | 0.83 | No growth |
| Mannose | 1.15 | 0.39 |
| Xylose | 1.52 | No growth |
| Arabinose | No growth | No growth |
The growth medium contained 30 g/L of the respective sugar and 0.05 g/L of yeast extract. Geobacillus LC300 was grown at 72°C. Geobacillus stearothermophilus was grown at 68°C.
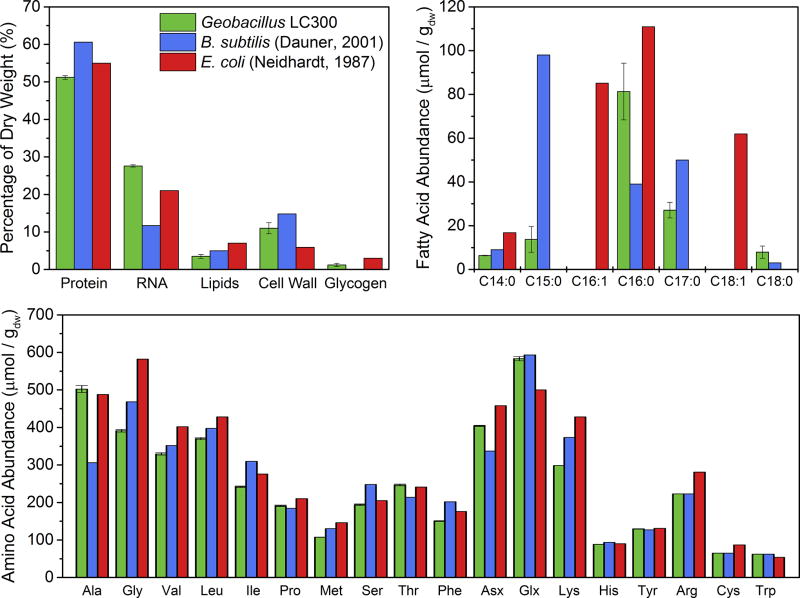
3.5. Biomass composition analysis
Detailed knowledge of biomass composition is needed to construct metabolic network models for metabolic flux analysis studies (Antoniewicz, 2015; Antoniewicz et al., 2006; Long and Antoniewicz, 2014b). Figure 4 shows the measured biomass composition of Geobacillus LC300. For comparison, the biomass compositions of B. subtilis and E. coli are also shown (Dauner and Sauer, 2001; Neidhardt, 1987). Characteristic of Geobacillus LC300 is its high RNA content (28 wt% of dry biomass), compared to B. subtilis (12 wt%) and E. coli (21 wt%). It is well known that high RNA content often correlates with high growth rate (Dauner and Sauer, 2001). For example, the high RNA content of Geobacillus LC300 matches well with the RNA content reported for fast growing E. coli (Stephanopoulos et al., 1998). The protein content of Geobacillus LC300 (51 wt% of dry biomass) was slightly lower than reported for B. subtilis (60 wt%) and E. coli (55 wt%), likely due to the relatively larger RNA fraction in Geobacillus LC300. The lipid and cell wall fractions were also slightly lower. Lastly, only a negligible amount of glycogen (1 wt%) was measured in Geobacillus LC300.
Figure 4.
Measured biomass composition of Geobacillus LC300, compared to B. subtilis (Dauner and Sauer, 2001) and E. coli (Neidhardt, 1987).
The amino acid composition of biomass proteins in Geobacillus LC300 was similar to that reported previously for B. subtilis and E. coli (Figure 4). Compared to B. subtilis, Geobacillus LC300 contains a relatively larger fraction of alanine, threonine and aspartate/aspargine; and compared to E. coli, Geobacillus LC300 contains a relatively larger fraction of alanine, threonine and glutamate/glutamine. The dominant cellular fatty acids of Geobacillus LC300 are saturated C16:0 and C17:0 fatty acids (including branched-chain iso- and anteiso-), with smaller amounts of saturated C14:0, C15:0 and C18:0 fatty acids. A similar fatty acid profile was recently reported for Geobacillus thermoglucosidasius M10EXG (Tang et al., 2009).
3.5. Metabolic network model construction for metabolic flux analysis
To facilitate quantitative studies of Geobacillus LC300, a core metabolic network model was constructed for 13C-metabolic flux analysis (13C-MFA) and dynamic metabolic flux analysis (Antoniewicz, 2013a; Antoniewicz, 2013b). The model is provided in Supplementary Materials. It includes all metabolic pathways in central carbon metabolism that were identified, including glycolysis, pentose phosphate pathway, TCA cycle, glyoxylate shunt, anaplorotic and cataplerotic reactions, as well as lumped amino acid biosynthesis pathways, and a lumped cell growth reaction based on the measured biomass composition. The model also accounts for exchange of intracellular and atmospheric CO2, which can dilute labeling of intracellular metabolites via carboxylation reactions (Au et al., 2014; Leighty and Antoniewicz, 2012). Overall, the model contains 73 reactions, 63 balanced intracellular metabolites, and 9 external metabolites (glucose, xylose, acetate, lactate, CO2, O2, NH3, SO42−, and biomass). Carbon atom transitions were manually assigned for all reactions. This model was used in the accompanying paper to elucidate xylose metabolism in Geobacillus LC300 in detail (Cordova and Antoniewicz, 2015).
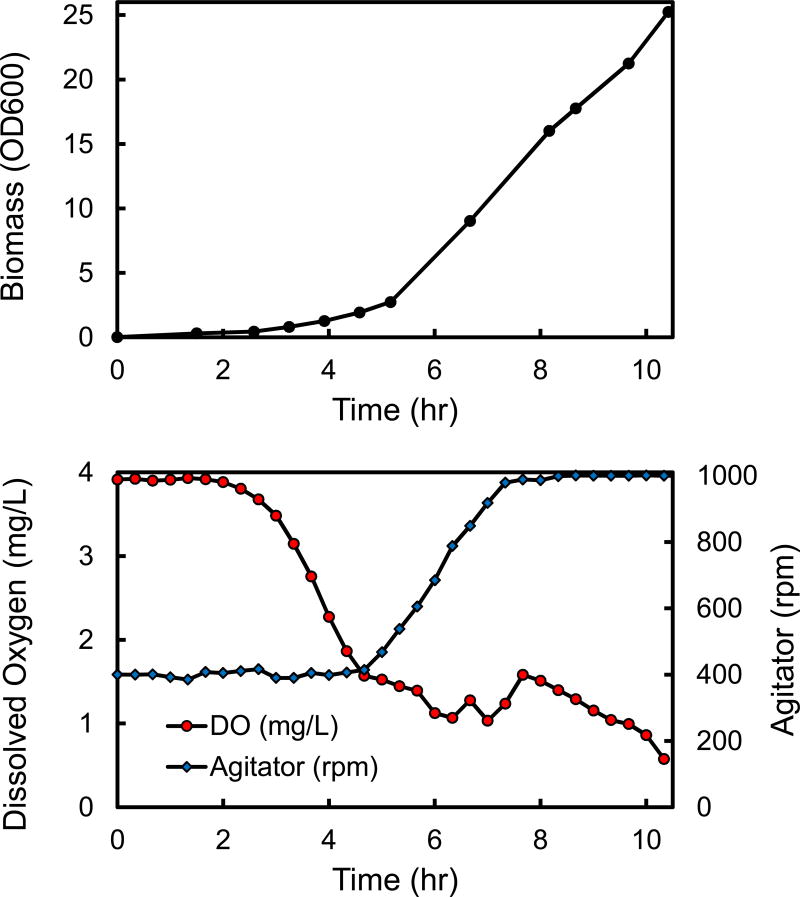
3.6. High cell-density fed-batch culture
From a biotechnological perspective, fast growth at high cell-densities and at low dissolved oxygen concentrations is desirable. To evaluate the biotechnological potential Geobacillus LC300, a fed-batch fermentation was performed at 70 °C with xylose as the main carbon source (Figure 5). A 1.5-L benchtop fermentor was inoculated with Geobacillus LC300 at an OD600 of 0.003. After 2.7 hrs of batch culture, xylose feed was initiated (3.7 gram of xylose added per hour). During the feeding phase, the optical density (OD600) increased linearly from 2.7 at 5.2 hr to 25.2 at 10.4 hr (Figure 5). At the same time, the dissolved oxygen concentration ranged between 0.5 and 1.5 mg/L. These results demonstrate that Geobacillus LC300 can achieve high cell-densities and maintain a high rate of xylose utilization under low dissolved oxygen conditions, thus making it an attractive host for future metabolic engineering and biotechnology applications.
Figure 5.
Fed-batch culture of Geobacillus LC300 grown on xylose as the main carbon source at 70 °C. After 2.7 hours of batch growth, xylose feed was initiated. The set point for dissolved oxygen concentration was 1.6 mg/L. The agitator speed was controlled between 400 rpm and 1000 rpm.
4. CONCLUSIONS
In this work, we have described a new thermophilic Geobacillus strain that can efficiently utilize xylose, glucose, mannose and galactose for cell growth. The Geobacillus LC300 genome provides an excellent resource for future studies of this organism. The extremely fast growth rate of Geobacillus LC300 on multiple carbon sources suggests the presence of highly efficient transporters and enzymes (Liu et al., 2014). Xylose isomerase and xylulokinase are often cited as the rate limiting steps in xylose utilization (Zhou et al., 2012). In future work, it would be interesting to characterize the kinetic properties of these enzymes, as well as other key transporters and catabolic and anabolic enzymes. Codon-optimized enzymes from Geobacillus LC300 could also be overexpressed in other thermophiles with similar growth temperatures to improve specific bioconversion rates.
The reconstructed metabolic network model provides a solid foundation for future 13C-flux analysis of this and other related organisms. Flux analysis studies can yield important insights into cellular metabolism that cannot be obtained from genome analysis alone (Crown and Antoniewicz, 2013b; He et al., 2014; Long and Antoniewicz, 2014a). In the accompanying paper (Cordova and Antoniewicz, 2015), a detailed characterization of xylose metabolism of Geobacillus LC300 is described using the developed model and the recently established COMPLETE-MFA methodology (Crown and Antoniewicz, 2013a; Leighty and Antoniewicz, 2013).
Supplementary Material
Acknowledgments
This work was supported by NSF-MCB-1120684 grant.
References
- Ahn WS, Antoniewicz MR. Parallel labeling experiments with [1,2-(13)C]glucose and [U-(13)C]glutamine provide new insights into CHO cell metabolism. Metab Eng. 2013;15:34–47. doi: 10.1016/j.ymben.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Anand A, Kumar V, Satyanarayana T. Characteristics of thermostable endoxylanase and beta-xylosidase of the extremely thermophilic bacterium Geobacillus thermodenitrificans TSAA1 and its applicability in generating xylooligosaccharides and xylose from agro-residues. Extremophiles : life under extreme conditions. 2013;17:357–66. doi: 10.1007/s00792-013-0524-x. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR. 13C metabolic flux analysis: optimal design of isotopic labeling experiments. Curr Opin Biotechnol. 2013a;24:1116–21. doi: 10.1016/j.copbio.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR. Dynamic metabolic flux analysis--tools for probing transient states of metabolic networks. Curr Opin Biotechnol. 2013b;24:973–8. doi: 10.1016/j.copbio.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR. Methods and advances in metabolic flux analysis: a mini-review. J Ind Microbiol Biotechnol. 2015;42:317–25. doi: 10.1007/s10295-015-1585-x. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal Chem. 2007;79:7554–9. doi: 10.1021/ac0708893. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Measuring deuterium enrichment of glucose hydrogen atoms by gas chromatography/mass spectrometry. Anal Chem. 2011;83:3211–6. doi: 10.1021/ac200012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Stephanopoulos G, Kelleher JK. Evaluation of regression models in metabolic physiology: predicting fluxes from isotopic data without knowledge of the pathway. Metabolomics. 2006;2:41–52. doi: 10.1007/s11306-006-0018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au J, Choi J, Jones SW, Venkataramanan KP, Antoniewicz MR. Parallel labeling experiments validate Clostridium acetobutylicum metabolic network model for C metabolic flux analysis. Metab Eng. 2014;26:23–33. doi: 10.1016/j.ymben.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr Opin Biotechnol. 2008;19:210–7. doi: 10.1016/j.copbio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Canakci S, Cevher Z, Inan K, Tokgoz M, Bahar F, Kacagan M, Sal FA, Belduz AO. Cloning, purification and characterization of an alkali-stable endoxylanase from thermophilic Geobacillus sp. 71. World journal of microbiology & biotechnology. 2012;28:1981–8. doi: 10.1007/s11274-011-1000-3. [DOI] [PubMed] [Google Scholar]
- Chang T, Yao S. Thermophilic, lignocellulolytic bacteria for ethanol production: current state and perspectives. Appl Microbiol Biotechnol. 2011;92:13–27. doi: 10.1007/s00253-011-3456-3. [DOI] [PubMed] [Google Scholar]
- Coorevits A, Dinsdale AE, Halket G, Lebbe L, De Vos P, Van Landschoot A, Logan NA. Taxonomic revision of the genus Geobacillus: emendation of Geobacillus, G. stearothermophilus, G. jurassicus, G. toebii, G. thermodenitrificans and G. thermoglucosidans (nom. corrig., formerly 'thermoglucosidasius'); transfer of Bacillus thermantarcticus to the genus as G. thermantarcticus comb. nov.; proposal of Caldibacillus debilis gen. nov., comb. nov.; transfer of G. tepidamans to Anoxybacillus as A. tepidamans comb. nov.; and proposal of Anoxybacillus caldiproteolyticus sp. nov. International journal of systematic and evolutionary microbiology. 2012;62:1470–85. doi: 10.1099/ijs.0.030346-0. [DOI] [PubMed] [Google Scholar]
- Cordova LT, Antoniewicz MR. 13C Metabolic flux analysis of the extremely thermophilic, fast growing, xylose-utilizing Geobacillus strain LC300. Metab Eng. 2015 doi: 10.1016/j.ymben.2015.06.004. [DOI] [PubMed] [Google Scholar]
- Cripps RE, Eley K, Leak DJ, Rudd B, Taylor M, Todd M, Boakes S, Martin S, Atkinson T. Metabolic engineering of Geobacillus thermoglucosidasius for high yield ethanol production. Metab Eng. 2009;11:398–408. doi: 10.1016/j.ymben.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Crown SB, Antoniewicz MR. Parallel labeling experiments and metabolic flux analysis: Past, present and future methodologies. Metab Eng. 2013a;16:21–32. doi: 10.1016/j.ymben.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Antoniewicz MR. Publishing 13C metabolic flux analysis studies: a review and future perspectives. Metab Eng. 2013b;20:42–8. doi: 10.1016/j.ymben.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauner M, Sauer U. Stoichiometric growth model for riboflavin-producing Bacillus subtilis. Biotechnol Bioeng. 2001;76:132–43. doi: 10.1002/bit.1153. [DOI] [PubMed] [Google Scholar]
- Elleuche S, Schroder C, Sahm K, Antranikian G. Extremozymes--biocatalysts with unique properties from extremophilic microorganisms. Curr Opin Biotechnol. 2014;29:116–23. doi: 10.1016/j.copbio.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Ezeji TC, Bahl H. Purification, characterization, and synergistic action of phytate-resistant alpha-amylase and alpha-glucosidase from Geobacillus thermodenitrificans HRO10. J Biotechnol. 2006;125:27–38. doi: 10.1016/j.jbiotec.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Ezeji TC, Wolf A, Bahl H. Isolation, characterization, and identification of Geobacillus thermodenitrificans HRO10, an alpha-amylase and alpha-glucosidase producing thermophile. Canadian journal of microbiology. 2005;51:685–93. doi: 10.1139/w05-054. [DOI] [PubMed] [Google Scholar]
- Feng L, Wang W, Cheng J, Ren Y, Zhao G, Gao C, Tang Y, Liu X, Han W, Peng X, Liu R, Wang L. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc Natl Acad Sci U S A. 2007;104:5602–7. doi: 10.1073/pnas.0609650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–62. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Goebel BM, Stackebrandt E. Cultural and phylogenetic analysis of mixed microbial populations found in natural and commercial bioleaching environments. Appl Environ Microbiol. 1994;60:1614–21. doi: 10.1128/aem.60.5.1614-1621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R, Tao H, Shanmugam KT, York SW, Ingram LO. Global gene expression differences associated with changes in glycolytic flux and growth rate in Escherichia coli during the fermentation of glucose and xylose. Biotechnol Prog. 2002;18:6–20. doi: 10.1021/bp010121i. [DOI] [PubMed] [Google Scholar]
- He L, Xiao Y, Gebreselassie N, Zhang F, Antoniewicz MR, Tang YJ, Peng L. Central metabolic responses to the overproduction of fatty acids in Escherichia coli based on 13C-metabolic flux analysis. Biotechnol Bioeng. 2014;111:575–585. doi: 10.1002/bit.25124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupor SR, Fraenkel DG. 6-phosphogluconolactonase mutants of Escherichia coli and a maltose blue gene. J Bacteriol. 1969;100:1296–301. doi: 10.1128/jb.100.3.1296-1301.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Duc MT, Dekas A, Osman S, Moissl C, Newcombe D, Venkateswaran K. Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean room environments. Appl Environ Microbiol. 2007;73:2600–11. doi: 10.1128/AEM.03007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighty RW, Antoniewicz MR. Parallel labeling experiments with [U-13C]glucose validate E. coli metabolic network model for 13C metabolic flux analysis. Metab Eng. 2012;14:533–41. doi: 10.1016/j.ymben.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Leighty RW, Antoniewicz MR. COMPLETE-MFA: complementary parallel labeling experiments technique for metabolic flux analysis. Metab Eng. 2013;20:49–55. doi: 10.1016/j.ymben.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Lin L, Xu J. Dissecting and engineering metabolic and regulatory networks of thermophilic bacteria for biofuel production. Biotechnol Adv. 2013;31:827–37. doi: 10.1016/j.biotechadv.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Lin PP, Rabe KS, Takasumi JL, Kadisch M, Arnold FH, Liao JC. Isobutanol production at elevated temperatures in thermophilic Geobacillus thermoglucosidasius. Metab Eng. 2014;24:1–8. doi: 10.1016/j.ymben.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Liu JK, O'Brien EJ, Lerman JA, Zengler K, Palsson BO, Feist AM. Reconstruction and modeling protein translocation and compartmentalization in Escherichia coli at the genome-scale. BMC Syst Biol. 2014;8:110. doi: 10.1186/s12918-014-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftie-Eaton W, Taylor M, Horne K, Tuffin MI, Burton SG, Cowan DA. Balancing redox cofactor generation and ATP synthesis: key microaerobic responses in thermophilic fermentations. Biotechnol Bioeng. 2013;110:1057–65. doi: 10.1002/bit.24774. [DOI] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR. Metabolic flux analysis of Escherichia coli knockouts: lessons from the Keio collection and future outlook. Curr Opin Biotechnol. 2014a;28:127–33. doi: 10.1016/j.copbio.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR. Quantifying biomass composition by gas chromatography/mass spectrometry. Anal Chem. 2014b;86:9423–7. doi: 10.1021/ac502734e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd LR. In: Production of ethanol from lignocellulosic materials using thermophilic bacteria: Critical evaluation of potential and review. Fiechter A, editor. Springer-Verlag; Heidelberg: 1989. pp. 1–52. [Google Scholar]
- Marchant R, Banat IM, Rahman TJ, Berzano M. The frequency and characteristics of highly thermophilic bacteria in cool soil environments. Environmental microbiology. 2002a;4:595–602. doi: 10.1046/j.1462-2920.2002.00344.x. [DOI] [PubMed] [Google Scholar]
- Marchant R, Banat IM, Rahman TJ, Berzano M. What are high-temperature bacteria doing in cold environments? Trends Microbiol. 2002b;10:120–1. doi: 10.1016/s0966-842x(02)02311-9. [DOI] [PubMed] [Google Scholar]
- Muhd Sakaff MK, Abdul Rahman AY, Saito JA, Hou S, Alam M. Complete genome sequence of the thermophilic bacterium Geobacillus thermoleovorans CCB_US3_UF5. J Bacteriol. 2012;194:1239. doi: 10.1128/JB.06580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazina TN, Tourova TP, Poltaraus AB, Novikova EV, Grigoryan AA, Ivanova AE, Lysenko AM, Petrunyaka VV, Osipov GA, Belyaev SS, Ivanov MV. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. International journal of systematic and evolutionary microbiology. 2001;51:433–46. doi: 10.1099/00207713-51-2-433. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC. Escherichia Coli and Salmonella Typhimurium. ASM Press; 1987. [Google Scholar]
- Patrick WM, Quandt EM, Swartzlander DB, Matsumura I. Multicopy suppression underpins metabolic evolvability. Mol Biol Evol. 2007;24:2716–22. doi: 10.1093/molbev/msm204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanov AS, Logacheva MD, Peltek SE. Draft Genome Sequences of Geobacillus stearothermophilus Strains 22 and 53, Isolated from the Garga Hot Spring in the Barguzin River Valley of the Russian Federation. Genome announcements. 2014;2 doi: 10.1128/genomeA.01205-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani M, Ohtsubo Y, Fukuda K, Hosoyama A, Ohji S, Yamazoe A, Fujita N, Nagata Y, Tsuda M, Hatta T, Kimbara K. Complete Genome Sequence of the Thermophilic Polychlorinated Biphenyl Degrader Geobacillus sp. Strain JF8 (NBRC 109937) Genome announcements. 2014;2 doi: 10.1128/genomeA.01213-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos GN, Aristidou AA, Nielsen J. Metabolic Engineering: Principles and Methodologies. Academic Press; 1998. [Google Scholar]
- Swarup A, Lu J, DeWoody KC, Antoniewicz MR. Metabolic network reconstruction, growth characterization and 13C-metabolic flux analysis of the extremophile Thermus thermophilus HB8. Metab Eng. 2014;24:173–80. doi: 10.1016/j.ymben.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Takami H, Takaki Y, Chee GJ, Nishi S, Shimamura S, Suzuki H, Matsui S, Uchiyama I. Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Res. 2004;32:6292–303. doi: 10.1093/nar/gkh970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YJ, Sapra R, Joyner D, Hazen TC, Myers S, Reichmuth D, Blanch H, Keasling JD. Analysis of metabolic pathways and fluxes in a newly discovered thermophilic and ethanol-tolerant Geobacillus strain. Biotechnol Bioeng. 2009;102:1377–86. doi: 10.1002/bit.22181. [DOI] [PubMed] [Google Scholar]
- Taylor MP, Eley KL, Martin S, Tuffin MI, Burton SG, Cowan DA. Thermophilic ethanologenesis: future prospects for second-generation bioethanol production. Trends Biotechnol. 2009;27:398–405. doi: 10.1016/j.tibtech.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Thomason LC, Court DL, Datta AR, Khanna R, Rosner JL. Identification of the Escherichia coli K-12 ybhE gene as pgl, encoding 6-phosphogluconolactonase. J Bacteriol. 2004;186:8248–53. doi: 10.1128/JB.186.24.8248-8253.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Anand A, Satyanarayana T. Thermostable and alkalistable endoxylanase of the extremely thermophilic bacterium Geobacillus thermodenitrificans TSAA1: cloning, expression, characteristics and its applicability in generating xylooligosaccharides and fermentable sugars. Applied biochemistry and biotechnology. 2013;170:119–30. doi: 10.1007/s12010-013-0174-6. [DOI] [PubMed] [Google Scholar]
- Wasylenko TM, Stephanopoulos G. Metabolomic and C-metabolic flux analysis of a xylose-consuming Saccharomyces cerevisiae strain expressing xylose isomerase. Biotechnol Bioeng. 2014 doi: 10.1002/bit.25447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Wang X, Huang Y, Huo F, Zhu X, Xi L, Lu JR. Thermophilic fermentation of acetoin and 2,3-butanediol by a novel Geobacillus strain. Biotechnology for biofuels. 2012;5:88. doi: 10.1186/1754-6834-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Caspers MP, Abee T, Siezen RJ, Kort R. Complete genome sequence of Geobacillus thermoglucosidans TNO-09.020, a thermophilic sporeformer associated with a dairy-processing environment. J Bacteriol. 2012;194:4118. doi: 10.1128/JB.00318-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Cheng JS, Wang BL, Fink GR, Stephanopoulos G. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng. 2012;14:611–22. doi: 10.1016/j.ymben.2012.07.011. [DOI] [PubMed] [Google Scholar]
- Zimenkov D, Gulevich A, Skorokhodova A, Biriukova I, Kozlov Y, Mashko S. Escherichia coli ORF ybhE is pgl gene encoding 6-phosphogluconolactonase (EC 3.1.1.31) that has no homology with known 6PGLs from other organisms. FEMS Microbiol Lett. 2005;244:275–80. doi: 10.1016/j.femsle.2005.01.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.