Abstract
Mechanisms underlying plant non-host resistance to Xanthomonas oryzae pv. oryzicola (Xoc), the pathogen causing rice leaf streak disease, are largely unknown. Cyclic nucleotide-gated ion channels (CNGCs) are calcium-permeable channels that are involved in various biological processes including plant resistance. In this study, functions of two tomato CNGC genes SlCNGC1 and SlCNGC14 in non-host resistance to Xoc were analyzed. Silencing of SlCNGC1 and SlCNGC14 in tomato significantly enhanced Xoc-induced hypersensitive response (HR) and non-host resistance, demonstrating that both SlCNGC1 and SlCNGC14 negatively regulate non-host resistance related HR and non-host resistance to Xoc in tomato. Silencing of SlCNGC1 and SlCNGC14 strikingly increased Xoc-induced callose deposition and strongly promoted both Xoc-induced and flg22-elicited H2O2, indicating that these two SlCNGCs repress callose deposition and ROS accumulation to attenuate non-host resistance and PAMP-triggered immunity (PTI). Importantly, silencing of SlCNGC1 and SlCNGC14 apparently compromised cytosolic Ca2+ accumulation, implying that SlCNGC1 and SlCNGC14 function as Ca2+ channels and negatively regulate non-host resistance and PTI-related responses through modulating cytosolic Ca2+ accumulation. SlCNGC14 seemed to play a stronger regulatory role in the non-host resistance and PTI compared to SlCNGC1. Our results reveal the contribution of CNGCs and probably also Ca2+ signaling pathway to non-host resistance and PTI.
Keywords: cyclic nucleotide-gated ion channel (CNGCs), Xanthomonas oryzae pv. oryzicola, PAMP-triggered immunity, non-host resistance, Ca2+ signaling
Introduction
Each pathogen has its own host range. Non-host resistance is triggered when a non-adapted pathogen attempts to infect a plant species outside of its host range. Thus, non-host resistance is widely occurring, durable and broad-spectrum to non-adapted pathogens and is highly potential to be exploited in crop resistance engineering (Schulze-Lefert and Panstruga, 2011; Senthil-Kumar and Mysore, 2013). It has been clear that plant non-host resistance utilizes both preformed and induced defense mechanisms and frequently elicites hypersensitive response (HR) (Mysore and Ryu, 2004; Senthil-Kumar and Mysore, 2013). The preformed mechanisms generally consist of plant physical and chemical barriers, including antibiotic compounds (Bednarek and Osbourn, 2009; Fan et al., 2011). The induced non-host resistance is elicited after the preformed defense is overcome. As observed for host resistance, pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) are often initiated in this layer of defense (Niks and Marcel, 2009; Senthil-Kumar and Mysore, 2013). Efforts to identifiy non-host resistance-related genes resulted in the finding that some genes, such as PLDδ, GOX, SGT1, and NHO1, contribute to both host and non-host resistance in Arabidopsis (Lu et al., 2001; Maimbo et al., 2010; Rojas et al., 2012; Pinosa et al., 2013). Generally speaking, the mechanisms underlying plant non-host resistance are far from well understood.
Xanthomonas oryzae pv. oryzae (Xoo) and X. oryzae pv. oryzicola (Xoc) are two important pathovars of X. oryzae, causing bacterial blight and leaf streak diseases, respectively, in rice (Oryza sativa), which is a staple food in many countries and a model plant for cereal biology (Nino-Liu et al., 2006). It is notable that these two pathovars utilize distinct mechanisms to infect rice leaves. Xoo is a vascular pathogen that enters rice leaves via the hydathodes, while Xoc penetrates rice leaves mainly through stomata or wound sites and colonizes intercellular spaces in the mesophyll (Nino-Liu et al., 2006). During infection of their plant hosts, many strains secrete transcription activator-like (TAL) effectors, which enter the host cell nucleus and activate specific corresponding host genes at effector binding elements (EBEs) in the promoter (Boch et al., 2009; Moscou and Bogdanove, 2009). It has been established that host resistance to Xoo is mostly related to the action of TAL effectors, either by polymorphisms that prevent the induction of susceptibility (S) genes or by executor resistance (R) genes with EBEs embedded in their promoter, and that induce cell death and resistance (Bogdanove et al., 2010; Zhang et al., 2015). Xoc is known to suppress host resistance, and no host R gene has been identified against it (Cai et al., 2017). Compared with the host resistance (Zhang and Wang, 2013), non-host resistance to Xoo and Xoc is much less studied. We have previously identified seven genes required for non-host resistance to Xoo in Nicotiana benthamiana. Among them are a calreticulin and a peroxidase, indicating that oxidative burst and calcium-dependent signaling pathways play an important role in non-host resistance to Xoo (Li et al., 2012). However, molecular mechanisms underlying non-host resistance to Xoc remain largely unknown. Whether the oxidative burst and calcium signaling pathway contribute to the non-host resistance to Xoc as to Xoo awaits examination.
The cyclic nucleotide-gated ion channels (CNGCs) are suggested to be one of the important pathways for conducting Ca2+ ions in signaling transduction (Talke et al., 2003). They are ligand-gated Ca2+-permeable divalent cation-selective channels that are often localized in plasma membrane, presumptively are activated by direct binding of cyclic nucleotides and are complexly regulated by binding of calmodulin (CaM) to the CaM Binding (CaMB) domain (Chin et al., 2009; Ma et al., 2009; Wang et al., 2013; Gao et al., 2014; DeFalco et al., 2016a,b; Fischer et al., 2017). The plant CNGCs are involved in numerous biological functions varying from plant development and stress tolerance to disease resistance (Kaplan et al., 2007; Qi et al., 2010; Ma and Berkowitz, 2011; Moeder et al., 2011). CNGCs are well conserved among plant species, comprising 20 members in Arabidopsis and 18 members in tomato (Saand et al., 2015a,b). Earlier studies revealed that AtCNGC2, AtCNGC4, AtCNGC11, and AtCNGC12 and their homologs play an important role in disease resistance against various pathogens (Yu et al., 1998; Clough et al., 2000; Balagué et al., 2003; Jurkowski et al., 2004; Yoshioka et al., 2006; Ali et al., 2007; Ma and Berkowitz, 2011; Chin et al., 2013; Fortuna et al., 2015; Saand et al., 2015a). Later, other CNGC members such as SlCNGC1 and SlCNGC14 were also found to contribute to disease resistance (Saand et al., 2015a,b). We found that SlCNGC1 and SlCNGC14 play a negative role in non-host resistance to Xoo in tomato (Saand et al., 2015b). However, whether these SlCNGCs indeed encode functional CNGC channels and whether and how they function in non-host resistance to Xoc in tomato remains further study.
Our data in this study strongly indicate that SlCNGC1 and SlCNGC14 function as Ca2+ channels and negatively regulate tomato non-host resistance to Xoc through modulating ROS accumulation and callose deposition. Our results provide evidence for the contribution of CNGC-mediated Ca2+ signaling pathway to non-host resistance.
Materials and Methods
Plant Growth and Inoculation
Tomato (cv. Money Maker) plants were grown in growth room at 21°C with16 h light/8 h dark photoperiod. For disease resistance evaluation analyses, tomato plants were inoculated with X. oryzae pv. oryzicola (Xoc). After single colony propagation culture in NA medium, the Xoc bacterial cells were collected and resuspended in ddH2O to 1 × 108 cfu mL-1. The bacterial suspension was then infiltrated into leaves with sterilized needleless syringe. The infiltration zones were marked immediately after infiltration. The inoculated plants were grown in growth room at 26°C with16 h light/8 h dark photoperiod.
Gene Silencing Analyses
The virus-induced gene silencing (VIGS) target fragments of SlCNGC1 (Solyc01g095770.2) and SlCNGC14 (Solyc03g114110.2) were amplified by RT-PCR with gene-specific primers (Supplementary Table S1) and ligated into the TRV-based VIGS vector pYL156, which was subsequently electroporated into Agrobacterium tumefaciens strain GV3101 for VIGS analyses. VIGS analyses were conducted with vacuum infiltration delivery approach as described using recombinant pYL156 with insertion of an eGFP fragment instead of an empty pYL156 as control to alleviate viral symptom (Saand et al., 2015a). At about 3 weeks post agro-infiltration, plants were inoculated with Xoc as described above.
Detection of Callose Deposition
Callose deposition were stained with aniline blue and visualized in fluorescence microscope. Briefly, the collected leaves were washed twice with ddH2O and ethanol, respectively, and cleared in acetic acid-ethanol (1:3) for 4 h. After washed twice with ddH2O, the leaves were incubated in aniline blue solution [150 mM KH2PO4, 0.1% (w/v) aniline blue, pH 9.5] for 1 h. The stained leaves were washed with ddH2O and observed in 30% glycerol by fluorescence microscopy.
Bacterial Number Counting Assays
Bacterial numbers in Xoc-infiltrated leaves of silenced plants were determined as previously reported (Li et al., 2012).
Detection of ROS
Xoc-inoculated leaves of tomato plants were detached and stained with 3,3-diamino benzidine hydrochloride (DAB) (1mg/mL) as described (Li et al., 2015). Quantitative determination of PAMP-elicited H2O2 was conducted using a luminol-based approach (Saand et al., 2015a). For each experiment, six tomato leaf disks of 3 mm at diameter from three plants were dipped in distilled water in the light over night. The leaf disks were then transferred into 50 μL of distilled water in a 96-well plate. After addion of the same volume mixture which contains 200 nM luminol (Sigma-Aldrich), 20 μg of horseradish peroxidase and 200 nM flg22, H2O2 were measured for 35 min as luminescence using a Microplate Luminometer (Titertek Berthold, Germany).
Calcium Assay
Transient increase of cytosolic Ca2+ concentration was monitored in the Aequarin transgenic tomato lines. For each experiment, six leaf disks were punched from three plants and vacuum-infiltrated in 12.5 μM coelenterazin h (Sigma) on a 96-well plate for 1 min and then set for 2 h at room temperature. Before measurement, the coelenterazin solution was gently removed and 200 μL of PAMP solution (100 nM flg22) was added to the wells. Luminescence was measured using a Microplate Luminometer (Titertek Berthold, Germany).
Gene Expression Analyses by qRT-PCR
Five defense-related Ca2+ signaling genes SlCDPK10 (Solyc11g018610.1), SlCAMTA3 (Solyc04g056270.2), SlCBP60G (Solyc01g100240.2), SlCAM2 (Solyc10g081170.1) and SlCAM6 (Solyc03g098050.2) were subjected to gene expression analyses (Zhao et al., 2013; Rahman et al., 2016b; Wang et al., 2016). Total RNA was isolated by Trizol (TaKaRa, Japan) extraction according to the manufacture’s instructions. RNAs were treated with DNase I (TaKaRa, Japan) and then reverse transcribed. Quantitative real time PCR (qRT-PCR) was performed as previously described using the StepOne Real-Time PCR system (Applied Biosystems, United States) with SYBR Green PCR Master Mix (TaKaRa, Japan) (Saand et al., 2015a). 18s rDNA was used as internal control. The accession number and the specific primers for amplification of the analyzed genes were listed (Supplementary Table S1). The expression of target genes relative to control was calculated based on a value of 2-ΔΔCt as recommended by the manufacturer. Meanwhile, semi-qRT-PCR for these genes were conducted in parallel and obtained PCR products were analyzed by agarose gel electrophoresis.
Statistical Analyses of Data
All experiments were conducted three times independently. The quantitative measurement data were statistically analyzed using SPSS software and represent means ± standard error. Significant difference between mean values was determined with DMRT (p < 0.05).
Results
Silencing of SlCNGC1 and SlCNGC14 Enhanced Xoc-Induced Hypersensitive Response and Non-host Resistance
In order to dissect the function of SlCNGC1 and SlCNGC14 in Xoc-induced HR and non-host resistance, we used TRV-based VIGS vector to perform VIGS analyses for these genes. Transcripts of SlCNGC1 and SlCNGC14 genes accumulated to only about 20% of the eGFP silencing control (Figure 1A), indicating that SlCNGC1 and SlCNGC14 genes had been effictiently silenced in these plants. Inoculation assays with Xoc in these silencing plants demonstrated that SlCNGC1- and SlCNGC14-silenced leaves showed more severe Xoc-induced HR than the eGFP control leaves. SlCNGC1- and SlCNGC14-silenced leaves showed obvious HR necrosis at 3 h post Xoc inoculation, and exhibited strong HR necrosis at 9 hpi, especially SlCNGC14-silenced leaves, which had turned into desiccative, while the eGFP control plants displayed barely visible and weak HR necrosis at 3 and 9 hpi, respectively (Figure 1B). This result indicated that silencing of SlCNGC1 and SlCNGC14 enhanced Xoc-induced HR.
FIGURE 1.
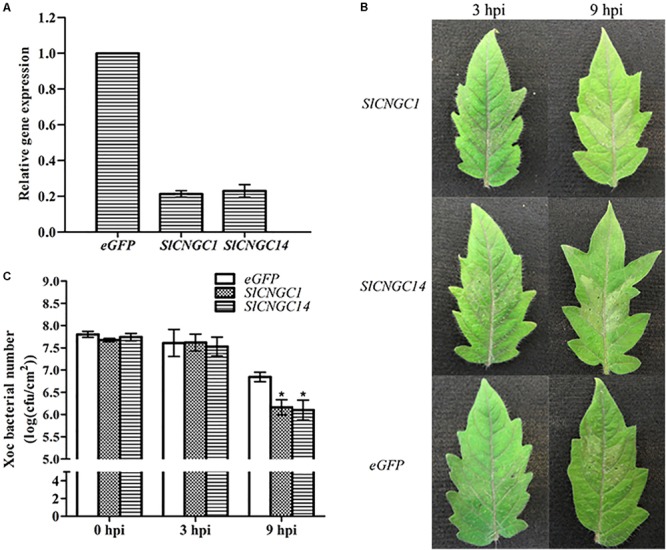
Silencing of SlCNGC1 and SlCNGC14 enhanced HR and non-host resistance to Xoc in tomato. (A) Silencing efficiency analysis. Plants infiltrated with Agrobacterium suspensions carrying an eGFP-control vector served as control plants. Accumulation level of SlCNGC1 and SlCNGC14 transcript in VIGS-treated plants and the eGFP-control plants was detected by qRT-PCR analyses. (B) Xoc-induced hypersensitive response (HR). Photographs were taken at 3 and 9 hpi. (C) Xoc bacterial number counting. Xoc bacterial numbers were counted in leaf areas that were inoculated with bacterium at 3 and 9 hpi. At least five leaves were tested for each treatment, and the data represent means ± standard error (SE). Significant differences between treatments and the control are indicated by an asterisk (p < 0.05).
We further counted Xoc bacterial number in the infiltrated leaf areas. The result showed that compared with the eGFP controls, Xoc bacterial number in SlCNGC1- and SlCNGC14-silenced plants decreased significantly by 0.7 orders of magnitude at 9 hpi (Figure 1C), indicating that silencing of SlCNGC1 and SlCNGC14 enhanced non-host resistance to Xoc.
Collectively, these results implied that both SlCNGC1 and SlCNGC14 might negatively regulate non-host resistance related HR cell death and non-host resistance to Xoc in tomato.
Silencing of SlCNGC1 and SlCNGC14 Increased Xoc-Induced Callose Deposition
To probe the mechanisms underlying SlCNGC1- and SlCNGC14-dependent regulation of non-host resistance against Xoc, effect of these CNGC genes on callose deposition upon pathogen infection was examined. Callose deposition was visible in the aniline blue-stained leaves under the fluorescence microscopy. Microscopic observation result showed that SlCNGC1- and SlCNGC14-silenced leaves deposited much more callose than eGFP control leaves (Figure 2A). Further quantification revealed that callose deposits in SlCNGC1- and SlCNGC14-silenced leaves was 5.9- and 7.3-fold respectively, as many as in the eGFP control leaves at 4 hpi (Figure 2B). This result demonstrates that SlCNGC1 and SlCNGC14 repress the callose deposition upon pathogen infection to alleviate non-host resistance to Xoc in tomato.
FIGURE 2.
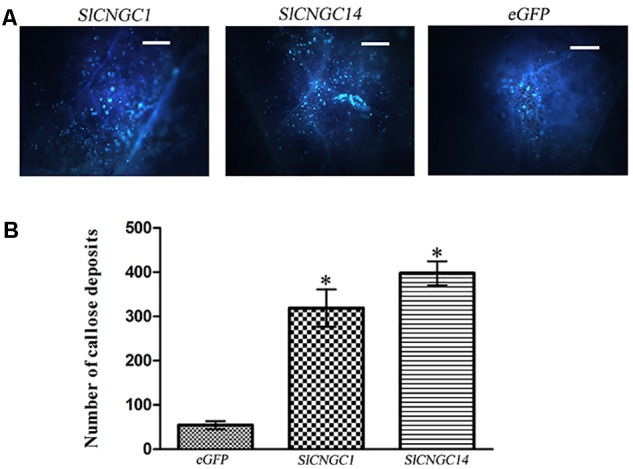
Silencing of SlCNGC1 and SlCNGC14 increased Xoc-induced callose deposition. Leaves inoculated with Xoc at 4 hpi were stained with aniline blue and visualized in fluorescence microscope. A typical staining result (A) and its quantification result (B) were shown. Significant differences between treatments and the control are indicated by an asterisk (p < 0.05).
Silencing of SlCNGC1 and SlCNGC14 Promoted ROS Accumulation
Reactive oxygen species (ROS) is indispensable to X. oryzae pv. oryzae (Xoo)-induced HR and non-host resistance (Li et al., 2015). Thus, we analyzed the effect of SlCNGC1 and SlCNGC14 on the production of hydrogen peroxide (H2O2), one of the primary species of ROS. DAB staining result demonstrated that infiltration areas of SlCNGC1- and SlCNGC14-silenced leaves showed significantly stronger DAB staining than those of control (Figure 3A), implying that silencing of SlCNGC1 and SlCNGC14 increased the production of H2O2.
FIGURE 3.
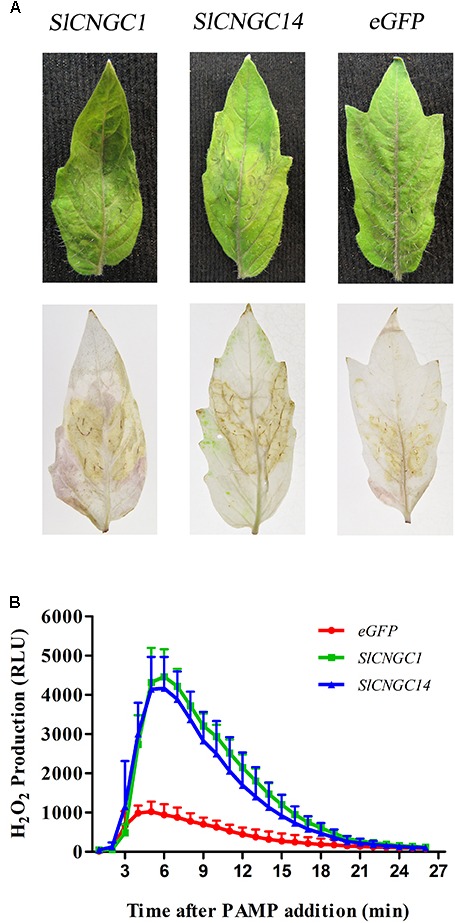
Silencing of SlCNGC1 and SlCNGC14 promoted Xoc-induced and flg22-elicited ROS accumulation. (A) Xoc-induced ROS assay. Hydrogen peroxide (H2O2) accumulation at 4 hpi was detected through Diaminobenzidine (DAB) staining analysis. The Xoc-infriltrated leaves before (Up panel) and after (Down panel) staining analyses were presented. (B) flg22-elicited ROS assay. Production of H2O2 induced by 100 nM flg22 was detected using a luminol-based assay in leaf disks of SlCNGC1-, SlCNGC14-silenced plants and the eGFP non-silenced control plants. Data are shown as relative luminol units (RLUs) and represent the mean ± SE of three independent experiments.
Effect of SlCNGC1 and SlCNGC14 on the PAMP (flg22)-elicited H2O2 accumulation was further examined using leaf disk assays and was indicated as relative luminescence (RLU). The luminol-based assay showed that flg22-induced H2O2 in SlCNGC1- and SlCNGC14-silenced leaves culminated to over 4100 RLU, while that in eGFP control leaves was peaked only to 940 RLU (Figure 3B), demonstrating that silencing of SlCNGC1 and SlCNGC14 promoted the production of flg22-elicited H2O2.
Together, these results suggest that SlCNGC1 and SlCNGC14 supress the ROS accumulation to attenuate non-host resistance and PTI.
Silencing of SlCNGC1 and SlCNGC14 Compromised Cytosol Ca2+ Influx
Some Arabidopsis CNGCs have been proved to be functional Ca2+ channels (Gao et al., 2014; Wang et al., 2017). To examine whether SlCNGC1 and SlCNGC14 function as Ca2+ channels, effect of silencing of these genes on accumulation of cytosolic Ca2+ elicited by the PAMP flg22 was monitored through leaf disk assays using aequorin transgenic tomato lines. In eGFP control aequorin transgenic plants, flg22-triggered Ca2+ increased rapidly and peaked to 192 RLU, while those in SlCNGC1- and SlCNGC14-silenced aequorin transgenic plants strongly decreased with the peak value drop to only 97 and 39 RLU, respectively (Figure 4). This result indicates that SlCNGC1 and SlCNGC14 function as Ca2+ channels and negatively regulate non-host resistance and PTI through modulating cytosolic Ca2+ accumulation.
FIGURE 4.
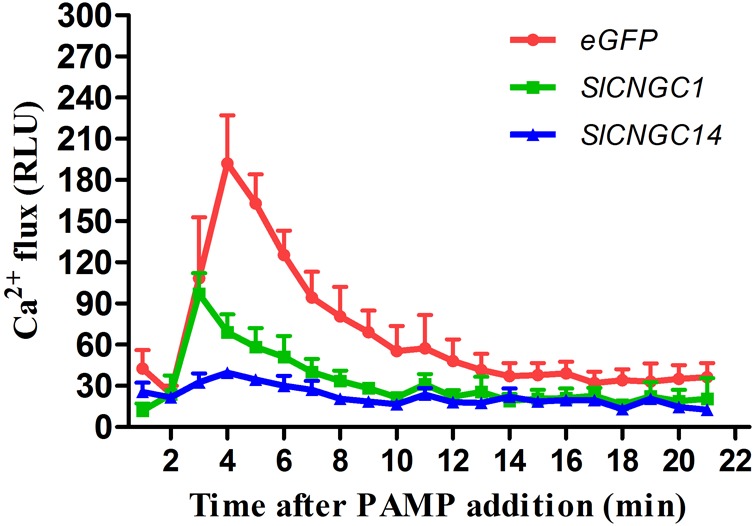
Silencing of SlCNGC1 and SlCNGC14 compromised cytosolic Ca2+ accumulation. Production of Ca2+ flux elicited by 100 nM flg22 was detected using a luminescence-based assay in leaf disks of SlCNGC1-, SlCNGC14-silenced plants and the eGFP non-silenced control plants of the aequorin transgenic tomato lines. Data are shown as relative luminescence units (RLU) and represent the mean ± SE of three independent experiments.
Silencing of SlCNGC1 and SlCNGC14 Altered Expression of Defense-Related Ca2+ Signaling Genes
To obtain a clue to understand how SlCNGC1 and SlCNGC14 genes, as Ca2+ channel genes, regulate plant disease resistance, we examined effect of silencing of these genes on expression of a set of defense-related Ca2+ signaling genes to probe the possibility of their involvement in SlCNGC1- and SlCNGC14-mediated resistance regulation. The genes under this expression analysis included two CaM genes SlCaM2 and SlCaM6, a tomato homolog (SlCDPK10) of Arabidopsis calcium-dependent protein kinase gene AtCDPK11, and the tomato homologs (SlCBP60g and SlCAMTA3) of two Arabidopsis CaM-binding transcription factors AtCBP60g and AtCAMTA3. All these genes play an important role in regulating disease resistance (Du et al., 2009; Wang et al., 2009, 2011; Boudsocq et al., 2010; Zhao et al., 2013; Sun et al., 2015; Rahman et al., 2016a,b; Wang et al., 2016). Result of qRT-PCR showed that at 4 h after inoculation with Xoc, SlCNGC1- and SlCNGC14-silenced leaves reduced expression of the negative defense regulatory gene SlCAMTA3 by 2.5-fold, and generally increased expression of the positive defense regulatory genes SlCaM6, SlCDPK10, and SlCBP60g to different extent, which was much higher in SlCNGC14-silenced leaves than SlCNGC1-silenced leaves. Expression of SlCaM6, SlCDPK10 and SlCBP60g genes in SlCNGC1-silenced leaves was 4.3-, 1.5- and 2.5-fold, respectively, as high as that in the eGFP controls, while these folds were 34.0, 4.5, and 9.1 for that in SlCNGC14-silenced leaves (Figure 5). This result implies that SlCNGC1 and SlCNGC14 might modulate the expression of these Ca2+ signaling genes.
FIGURE 5.
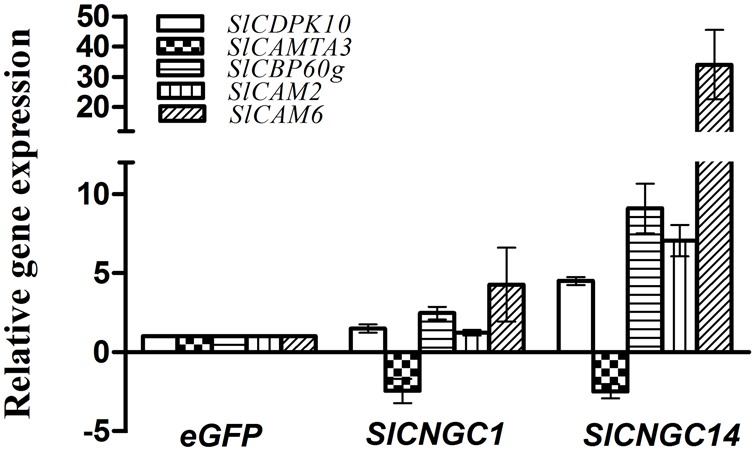
Silencing of SlCNGC1 and SlCNGC14 altered expression of defense-related Ca2+ signaling genes. Expression of defense-related Ca2+ signaling genes SlCAM2, SlCAM6, SlCDPK10, SlCAMTA3, and SlCBP60g were examined for SlCNGC1-, SlCNGC14-silenced plants relative to the eGFP non-silenced control plants after inoculation with Xoc at 4 hpi by qRT-PCR with 18s rDNA gene serving as a loading control gene. Data represent the mean ± SE of three independent experiments.
Discussion
We previously found that two tomato CNGC genes SlCNGC1 and SlCNGC14 play an important role in tomato HR elicited by the rice bacterial blight pathogen X. oryzae pv. oryzae (Xoo) (Saand et al., 2015a). Here, we further reveal that these two CNGC genes likely encode functional Ca2+ channels and negatively affect tomato non-host resistance to the rice bacterial leaf streak pathogen Xoc, through modulating ROS accumulation and callose deposition. The role of Ca2+ signaling pathway in non-host resistance to Xoc is previously unknown. Our results provide evidence for the contribution of CNGC-mediated Ca2+ signaling pathway to this non-host resistance.
In plants, only a few CNGCs such as AtCNGC2 and AtCNGC18 have been proved to function as Ca2+ channels (Gao et al., 2014; Wang et al., 2017). Whether the tomato CNGC genes encode functional Ca2+ channels remain to be verified. In this study, we demonstrated that silencing of SlCNGC1 and SlCNGC14 significantly reduced or almost abolished cytosolic Ca2+ accumulation (Figure 4). Thus, SlCNGC1 and SlCNGC14 likely function as Ca2+ channels. Further electrophysiological studies are required to provide more evidences to verify it.
It is recently reported that Arabidopsis cngc2 mutant overaccumulates Ca2+ in apoplastic compartment and increased HR and resistance (Wang et al., 2017). Similarly, in this study, we found that silencing of SlCNGC1 and SlCNGC14 significantly reduced cytosolic Ca2+ accumulation and enhanced HR and non-host resistance to Xoc and PTI. Whether silencing of SlCNGC1 and SlCNGC14 results in overaccumulation of Ca2+ in apoplastic compartment as observed in Atcngc2 mutant (Wang et al., 2017) requires further confirmation.
ROS is the well-known master signal in plant immunity and is indispensable to Xoo-induced HR and non-host resistance (Li et al., 2015). Thus, it is considerable that regulation of ROS accumulation represents one of the essential mechanisms underlying SlCNGC1- and SlCNGC14-dependent regulation of HR and non-host resistance to Xoc and PTI. How SlCNGC1 and SlCNGC14 negatively regulate ROS accumulation remains an intriguing question to be addressed. NADPH oxidase encoded by RBOH genes is the key enzyme to generate ROS during plant-pathogen interactions. Interestingly, Ca2+ affects the activity of this enzyme which contains EF-hands to bind Ca2+. Moreover, AtCPK28, a calcium-dependent protein kinase gene, negatively regulate ROS accumulation during PTI (Monaghan et al., 2014). In this context, it is notable that silencing of SlCNGC1 and SlCNGC14 strongly represses cytosolic Ca2+ accumulation and expression of a set of denfense-related Ca2+ signaling genes including SlCaM6, SlCDPK10 and SlCBP60g (Figures 4, 5). Whether and how these changes affect NADPH oxidase thereby modulate ROS accumulation deserves further investigation.
SlCNGC1 and SlCNGC14 belong to group I and group III of tomato CNGC family. They function similarly in negative modulation of tomato HR and non-host resistance to Xoc. Both of them suppress ROS accumulation and callose deposition and alter expression of a same set of defense-related Ca2+ signaling genes including SlCaM6, SlCDPK10 and SlCBP60g. However, the degree of effect in resistance responses by silencing SlCNGC1 and SlCNGC14 varies (Figures 1–5), indicating the difference of their involvement in this non-host resistance although it can be the consequence of difference of silencing degree. In animal, CNGC often forms homo- or hetero-oligomer to function (Kaupp and Seifert, 2002). Furthermore, some plant CNGC isoforms, such as AtCNGC2 and AtCNGC4, have been reported to be able to homo- or hetero-complex in planta (Chin et al., 2013). In this context, whether SlCNGC1 and SlCNGC14 form oligomer to regulate HR and plant immunity is worth further clarifying. Additionally, cellular localization of plant CNGCs other than long expected plasma membrane have been reported recently (DeFalco et al., 2016b). Whether the cellular localization and accumulation of SlCNGC1 and SlCNGC14 differ awaits further study.
Roles of CNGCs in plant disease resistance seem to be complex. Results from previous studies using Ca2+ channel blockers indicated that cytosolic Ca2+ elevation in response to PAMPs such as flg22 is required for PAMP induced defense resposnes including ROS burst (Jeworutzki et al., 2010; Ranf et al., 2011; Segonzac et al., 2011), suggesting a positive role of the possibly involved CNGCs in PTI defenses. However, CNGC genes likely play different roles in various types of resistance. For instance, T-DNA insertion knockout mutants for AtCNGC11 and AtCNGC12 exhibit unaffected flg22 induced PTI responses but show a partial breakdown of resistance against avirulent, but not virulent Hyaloperonospora arabidopsidis and Pseudomonas syringae, indicating that both AtCNGC11 and AtCNGC12 might be not involved in PTI but act as positive regulators of R gene-mediated resistance responses (Yoshioka et al., 2006; Moeder et al., 2011). Differently, null mutants for AtCNGC2 and AtCNGC4 display impaired HR cell death, while maintaining resistance against avirulent pathogens, exhibiting enhanced broad-spectrum resistance against virulent pathogens, elevated levels of SA, and constitutive expression of PR genes (Yu et al., 1998; Clough et al., 2000; Yu et al., 2000; Balagué et al., 2003; Jurkowski et al., 2004; Genger et al., 2008), and thus these AtCNGCs seem to play positive role in HR while have negative role in R gene-mediated resistance and basal resistance. Additionally, AtCNGC2 was reported to positively contribute to Pep-elicited immunity (Qi et al., 2010; Ma et al., 2012, 2013) and LPS-triggered Ca2+ influx that led to nitric oxide production and consequent HR formation (Ali et al., 2007; Ma et al., 2007), but might be not involved in flg22-triggered immunity (Jeworutzki et al., 2010). In this study, we found the negative role of SlCNGC1 and SlCNGC14 in Xoc-induced HR and non-host resistance, adding further complexity to the function of plant CNGCs. It is likely that different members of CNGC family might have different roles in resistance and at least some CNGCs are multifunctional and differentially regulate various types of resistance against different pathogens. SlCNGC1 and SlCNGC14, which exhibit similar function in Xoc-induced HR and non-host resistance, belong to different groups of CNGCs, with SlCNGC1 to group I as AtCNGC11 and AtCNGC12, while SlCNGC14 to group III, indicating that the function of CNGCs is not group-dependent, as we suggested previously (Saand et al., 2015b). Mechanisms underlying plant CNGC-mediated resistance againt various pathogens await further elucidation.
Conclusion
Gene silencing analyses demonstrated that SlCNGC1 and SlCNGC14 negatively regulate non-host resistance related HR cell death and non-host resistance to X. oryzae pv. oryzicola (Xoc) in tomato. These two SlCNGCs repress callose deposition and ROS accumulation to alleviate non-host resistance and likely also PTI. Silencing of SlCNGC1 and SlCNGC14 strongly reduced cytosolic Ca2+ accumulation, suggesting that SlCNGC1 and SlCNGC14 function as Ca2+ channels and negatively regulate non-host resistance and PTI through modulating cytosolic Ca2+ accumulation. SlCNGC14 likely played a stronger regulatory role in the non-host resistance and PTI than SlCNGC1. Our results reveal the contribution of CNGC-mediated Ca2+ signaling pathway to non-host resistance and PTI.
Author Contributions
The project was coordinated by X-ZC. X-RZ conducted the gene silencing analyses. X-RZ and Y-PX carried out the gene expression, designed and analyzed all statistical data. X-ZC conceived of the study, and participated in its design and coordination. X-ZC and X-RZ prepared the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Prof. Gerald Alan Berkowitz, Department of Plant Science and Landscape Architecture, Agricultural Biotechnology Laboratory, University of Connecticut, United States, for providing seeds of aequorin transgenic tomato lines.
Footnotes
Funding. This work was financially supported by grants from the Zhejiang Provincial Natural Science Foundation of China (No. LZ18C140002), the Genetically Modified Organisms Breeding Major Projects (No. 2014ZX0800905B), the National Natural Science Foundation of China (No. 31672014), and the National Key Research and Development Project (No. 2017YFD0200602).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00285/full#supplementary-material
References
- Ali R., Ma W., Lemtiri-Chlieh F., Tsaltas D., Leng Q., von Bodman S., et al. (2007). Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19 1081–1095. 10.1105/tpc.106.045096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagué C., Lin B., Alcon C., Flottes C., Malmström M., Köhler C., et al. (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide–gated channel ion channel family. Plant Cell 15 365–379. 10.1105/tpc.006999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P., Osbourn A. (2009). Plant-microbe interactions: chemical diversity in plant defense. Science 324 746–748. 10.1126/science.1171661 [DOI] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326 1509–1512. 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]
- Bogdanove A. J., Schornack S., Lahaye T. (2010). TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13 394–401. 10.1016/j.pbi.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M. R., Mccormack M., Lee H., Shan L., He P.et al. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464 418–422. 10.1038/nature08794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Cao Y., Xu Z., Ma W., Zakria M., Zou L., et al. (2017). A transcription activator-like effector Tal7 of Xanthomonas oryzae pv. oryzicola activates rice gene Os09g29100 to suppress rice immunity. Sci. Rep. 7:5089. 10.1038/s41598-017-04800-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K., DeFalco T. A., Moeder W., Yoshioka K. (2013). The Arabidopsis cyclic nucleotide-gated ion channels AtCNGC2 and AtCNGC4 work in the same signaling pathway to regulate pathogen defense and floral transition. Plant Physiol. 163 611–624. 10.1104/pp.113.225680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K., Moeder W., Yoshioka K. (2009). Biological roles of cyclic-nucleotide-gated ion channels in plants: what we know and don’t know about this 20 member ion channel family. Botany 87 668–677. 10.1139/B08-147 [DOI] [Google Scholar]
- Clough S. J., Fengler K. A., Yu I. C., Lippok B., Smith R. K., Jr., Bent A. F. (2000). The Arabidopsis dnd1 ”defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. U.S.A. 97 9323–9328. 10.1073/pnas.150005697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T. A., Marshall C. B., Munro K., Kang H. G., Moeder W., Ikura M., et al. (2016a). Multiple calmodulin-binding sites positively and negatively regulate Arabidopsis CYCLIC NUCLEOTIDE-GATED CHANNEL12. Plant Cell 28 1738–1751. 10.1105/tpc.15.00870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T. A., Moeder W., Yoshioka K. (2016b). Opening the gates: insights into cyclic nucleotide-gated channel-mediated signaling. Trends Plant Sci. 21 903–906. 10.1016/j.tplants.2016.08.011 [DOI] [PubMed] [Google Scholar]
- Du L., Ali G. S., Simons K. A., Hou J., Yang T., Reddy A. S.et al. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457 1154–1158. 10.1038/nature07612 [DOI] [PubMed] [Google Scholar]
- Fan J., Crooks C., Creissen G., Hill L., Fairhurst S., Doerner P., et al. (2011). Pseudomonas sax genes overcome aliphatic isothiocyanate–mediated non-host resistance in Arabidopsis. Science 331 1185–1188. 10.1126/science.1201477 [DOI] [PubMed] [Google Scholar]
- Fischer C., DeFalco T. A., Karia P., Snedden W. A., Moeder W., Yoshioka K., et al. (2017). Calmodulin as a Ca2+-sensing subunit of Arabidopsis cyclic nucleotide-gated channel complexes. Plant Cell Physiol. 58 1208–1221. 10.1093/pcp/pcx052 [DOI] [PubMed] [Google Scholar]
- Fortuna A., Lee J., Ung H., Chin K., Moeder W., Yoshioka K. (2015). Crossroads of stress responses, development and flowering regulation–the multiple roles of Cyclic Nucleotide Gated Ion Channel 2. Plant Signal. Behav. 10:e989758. 10.4161/15592324.2014.989758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q. F., Fei C. F., Dong J. Y., Gu L. L., Wang Y. F. (2014). Arabidopsis CNGC18 is a Ca2+-permeable channel. Mol. Plant 7 739–743. 10.1093/mp/sst174 [DOI] [PubMed] [Google Scholar]
- Genger R. K., Jurkowski G. I., McDowell J. M., Lu H., Jung H. W., Greenberg J. T., et al. (2008). Signaling pathways that regulate the enhanced disease resistance of Arabidopsis ‘defense, no death’ mutants. Mol. Plant Microbe Interact. 10 1285–1296. 10.1094/MPMI-21-10-1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeworutzki E., Roelfsema M. R., Anschutz U., Krol E., Elzenga J. T., Felix G., et al. (2010). Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca2+-associated opening of plasma membrane anion channels. Plant J. 62 367–378. 10.1111/j.1365-313X.2010.04155.x [DOI] [PubMed] [Google Scholar]
- Jurkowski G. I., Smith R. K., Jr., Yu I. C., Ham J. H., Sharma S. B., Klessig D. F., et al. (2004). Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol. Plant Microbe Interact. 17 511–520. 10.1094/MPMI.2004.17.5.511 [DOI] [PubMed] [Google Scholar]
- Kaplan B., Sherman T., Fromm H. (2007). Cyclic nucleotide-gated channels in plants. FEBS Lett. 581 2237–2246. 10.1016/j.febslet.2007.02.017 [DOI] [PubMed] [Google Scholar]
- Kaupp U. B., Seifert R. (2002). Cyclic nucleotide-gated ion channels. Physiol. Rev. 82 769–824. 10.1146/annurev.cellbio.19.110701.154854 [DOI] [PubMed] [Google Scholar]
- Li W., Xu Y. P., Yang J., Chen G. Y., Cai X. Z. (2015). Hydrogen peroxide is indispensable to Xanthomonas oryzae pv. oryzae-induced hypersensitive response and nonhost resistance in Nicotiana benthamiana. Australas. Plant Pathol. 44 611–617. 10.1007/s13313-015-0376-1 [DOI] [Google Scholar]
- Li W., Xu Y. P., Zhang Z. X., Cao W. Y., Li F., Zhou X., et al. (2012). Identification of genes required for nonhost resistance to Xanthomonas oryzae pv. oryzae reveals novel signaling components. PLoS One 7:e42796. 10.1371/journal.pone.0042796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Tang X., Zhou J. M. (2001). Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. Plant Cell 13 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Berkowitz G. A. (2011). Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol. 190 566–572. 10.1111/j.1469-8137.2010.03577.x [DOI] [PubMed] [Google Scholar]
- Ma W., Qi Z., Smigel A., Walker R. K., Verma R., Berkowitz G. A. (2009). Ca2+, cAMP, and transduction of non-self perception during plant immune responses. Proc. Natl. Acad. Sci. U.S.A. 106 20995–21000. 10.1073/pnas.0905831106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Yoshioka K., Berkowitz G. (2007). Cyclic nucleotide gated channels and Ca2+-mediated signal transduction during plant innate immune response to pathogens. Plant Signal. Behav. 2 548–550. PMID: 19704555; PMCID: PMC2634365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Walker R. K., Zhao Y., Berkowitz G. A. (2012). Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 109 19852–19857. 10.1073/pnas.1205448109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhao Y., Walker R. K., Berkowitz G. A. (2013). Molecular steps in the immune signaling pathway evoked by plant elicitor peptides: Ca2+-dependent protein kinases, nitric oxide, and reactive oxygen species are downstream from the early Ca2+ signal. Plant Physiol. 163 1459–1471. 10.1104/pp.113.226068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimbo M., Ohnishi K., Hikichi Y., Yoshioka H., Kiba A. (2010). S-glycoprotein-like protein regulates defense responses in Nicotiana plants against Ralstonia solanacearum. Plant Physiol. 152 2023–2035. 10.1104/pp.109.148189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder W., Urquhart W., Ung H., Yoshioka K. (2011). The role of cyclic nucleotide-gated ion channels in plant immunity. Mol. Plant 4 442–452. 10.1093/mp/ssr018 [DOI] [PubMed] [Google Scholar]
- Monaghan J., Matschi S., Shorinola O., Rovenich H., Matei A., Segonzac C., et al. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16 605–615. 10.1016/j.chom.2014.10.007 [DOI] [PubMed] [Google Scholar]
- Moscou M. J., Bogdanove A. J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326:1501. 10.1126/science.1178817 [DOI] [PubMed] [Google Scholar]
- Mysore K. S., Ryu C. M. (2004). Nonhost resistance: How much do we know? Trends Plant Sci. 9 97–104. 10.1016/j.tplants.2003.12.005 [DOI] [PubMed] [Google Scholar]
- Niks R. E., Marcel T. C. (2009). Nonhost and basal resistance: How to explain specificity? New Phytol. 182 817–828. 10.1111/j.1469-8137.2009.02849.x [DOI] [PubMed] [Google Scholar]
- Nino-Liu D. O., Ronald P. C., Bogdanove A. (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7 303–324. 10.1111/j.1364-3703.2006.00344.x [DOI] [PubMed] [Google Scholar]
- Pinosa F., Buhot N., Kwaaitaal M., Fahlberg P., Thordal-Christensen H., Ellerström M., et al. (2013). Arabidopsis phospholipase Dδ is involved in basal defense and nonhost resistance to powdery mildew fungi. Plant Physiol. 163 896–906. 10.1104/pp.113.223503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Verma R., Gehring C., Yamaguchi Y., Zhao Y., Ryan C. A., et al. (2010). Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. U.S.A. 107 21193–21198. 10.1073/pnas.1000191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman H., Xu Y. P., Zhang X. R., Cai X. Z. (2016a). Brassica napus genome possesses extraordinary high number of CAMTA genes and CAMTA3 contributes to PAMP-triggered immunity and resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 7:581. 10.3389/fpls.2016.00581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman H., Yang J., Xu Y. P., Munyampundu J. P., Cai X. Z. (2016b). Phylogeny of plant CAMTAs and role of AtCAMTAs in nonhost resistance to Xanthomonas oryzae pv. oryzae. Front. Plant Sci. 7:117. 10.3389/fpls.2016.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S., Eschen-Lippold L., Pecher P., Lee J., Scheel D. (2011). Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 68 100–113. 10.1111/j.1365-313X.2011.04671.x [DOI] [PubMed] [Google Scholar]
- Rojas C. M., Senthil-Kumar M., Wang K., Ryu C. M., Kaundal A., Mysore K. S. (2012). Glycolate oxidase modulates reactive oxygen species–mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24 336–352. 10.1105/tpc.111.093245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saand M. A., Xu Y. P., Li W., Wang J. P., Cai X. Z. (2015a). Cyclic nucleotide gated channel gene family in tomato: genome-wide identification and functional analyses in disease resistance. Front. Plant Sci. 6:303. 10.3389/fpls.2015.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saand M. A., Xu Y. P., Munyampundu J. P., Li W., Zhang X. R., Cai X. Z. (2015b). Phylogeny and evolution of plant cyclic nucleotide-gated ion channel (CNGC) gene family and functional analyses of tomato CNGCs. DNA Res. 22 471–483. 10.1093/dnares/dsv029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P., Panstruga R. (2011). A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16 117–125. 10.1016/j.tplants.2011.01.001 [DOI] [PubMed] [Google Scholar]
- Segonzac C., Feike D., Gimenez-Ibanez S., Hann D. R., Zipfel C., Rathjen J. P. (2011). Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol. 156 687–699. 10.1104/pp.110.171249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M., Mysore K. S. (2013). Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu. Rev. Phytopathol. 51 407–427. 10.1146/annurev-phyto-082712-102319 [DOI] [PubMed] [Google Scholar]
- Sun T., Zhang Y., Li Y., Zhang Q., Ding Y., Zhang Y. (2015). ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 6:10159. 10.1038/ncomms10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talke I. N., Blaudez D., Maathuis F. J., Sanders D. (2003). CNGCs: prime targets of plant cyclic nucleotide signalling? Trends Plant Sci. 8 286–293. 10.1016/S1360-1385(03)00099-2 [DOI] [PubMed] [Google Scholar]
- Wang J. P., Xu Y. P., Munyampundu J. P., Liu T. Y., Cai X. Z. (2016). Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: genome-wide identification and functional analyses in disease resistance. Mol. Genet. Genomics 291 661–676. 10.1007/s00438-015-1137-0 [DOI] [PubMed] [Google Scholar]
- Wang L., Tsuda K., Sato M., Cohen J. D., Katagiri F., Glazebrook J. (2009). Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog. 5:e1000301. 10.1371/journal.ppat.1000301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Tsuda K., Truman W., Sato M., Nguyen L. V., Katagiri F., et al. (2011). CBP60g and SARD1 play partially redundant, critical roles in salicylic acid signaling. Plant J. 67 1029–1041. 10.1111/j.1365-313X.2011.04655.x [DOI] [PubMed] [Google Scholar]
- Wang Y., Kang Y., Ma C., Miao R., Wu C., Long Y., et al. (2017). CNGC2 is a Ca2+ influx channel that prevents accumulation of apoplastic Ca2+ in the leaf. Plant Physiol. 173 1342–1354. 10.1104/pp.16.01222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. F., Munemasa S., Nishimura N., Ren H. M., Robert N., Han M., et al. (2013). Identification of cyclic GMP-activated nonselective Ca2+-permeable cation channels and associated CNGC5 and CNGC6 genes in Arabidopsis guard cells. Plant Physiol. 163 578–590. 10.1104/pp.113.225045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K., Moeder W., Kang H. G., Kachroo P., Masmoudi K., Berkowitz G., et al. (2006). The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18 747–763. 10.1105/tpc.105.038786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I. C., Fengler K. A., Clough S. J., Bent A. F. (2000). Identification of Arabidopsis mutants exhibiting an altered hypersensitive response in gene-for-gene disease resistance. Mol. Plant Microbe Interact. 13 227–286. 10.1094/MPMI.2000.13.3.277 [DOI] [PubMed] [Google Scholar]
- Yu I. C., Parker J., Bent A. F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. U.S.A. 95 7819–7824. PMID: 9636234; PMCID: PMC22769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang S. (2013). Rice versus Xanthomonas oryzae pv. oryzae: a unique pathosystem. Curr. Opin. Plant Biol. 16 188–195. 10.1016/j.pbi.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Zhang J., Yin Z., White F. (2015). TAL effectors and the executor R genes. Front. Plant Sci. 6:641. 10.3389/fpls.2015.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Liu W., Xu Y. P., Cao J. Y., Braam J., Cai X. Z. (2013). Genome-wide identification and functional analyses of calmodulin genes in Solanaceous species. BMC Plant Biol. 13:70. 10.1186/1471-2229-13-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


