Abstract
Purpose
This study investigates the virulence and antimicrobial resistance in association with common clonal complexes (CCs) of enteroaggregative Escherichia coli (EAEC) isolated from Bangladesh. The aim was to determine whether specific CCs were more likely to be associated with putative virulence genes and/or antimicrobial resistance.
Methodology
The presence of 15 virulence genes (by PCR) and susceptibility to 18 antibiotics were determined for 151 EAEC isolated from cases and controls during an intestinal infectious disease study carried out between 2007–2011 in the rural setting of Mirzapur, Bangladesh (Kotloff KL, Blackwelder WC, Nasrin D, Nataro JP, Farag TH et al. Clin Infect Dis 2012;55:S232–S245). These data were then analysed in the context of previously determined serotypes and clonal complexes defined by multi-locus sequence typing.
Results
Overall there was no association between the presence of virulence or antimicrobial resistance genes in isolates of EAEC from cases versus controls. However, when stratified by clonal complex (CC) one CC associated with cases harboured more virulence factors (CC40) and one CC harboured more resistance genes (CC38) than the average. There was no direct link between the virulence gene content and antibiotic resistance. Strains within a single CC had variable virulence and resistance gene content indicating independent and multiple gene acquisitions over time.
Conclusion
In Bangladesh, there are multiple clonal complexes of EAEC harbouring a variety of virulence and resistance genes. The emergence of two of the most successful clones appeared to be linked to either increased virulence (CC40) or antimicrobial resistance (CC38), but increased resistance and virulence were not found in the same clonal complexes.
Keywords: enteroaggregative E. coli, EAEC, resistance, virulence, MLST, Bangladesh
Introduction
Enteroaggregative Escherichia coli (EAEC) have been linked to acute and persistent diarrhoea among children and adults in developing countries [1–6] where malnutrition contributes to increasing the severity of symptoms and infection can impair growth and development [7].
Strains of EAEC make a significant contribution to the burden of gastrointestinal disease in Bangladesh either as an important independent causal agent [8] or in combination with other pathogens [1, 9, 10]. In many countries, bacterial gastroenteritis is managed without recourse to antibiotics but in severe cases, high-risk patients and chronic persistent infections, specific therapy is warranted [11]. In Bangladesh, symptoms of EAEC infection are often severe or persistent and so antibiotic treatment is frequently recommended. With the exception of one report highlighting an extended spectrum beta-lactamase (ESBL)-producing strain of EAEC from a recurrent urinary tract infection [12], there is very little information published on resistance to antibiotics of EAEC in Bangladesh, or indeed globally.
The causal link of EAEC in relation to disease in the human population is not absolute [13]. Multiple studies have demonstrated the heterogeneity of EAEC with respect to both plasmid and chromosomal gene content [14, 15] and the association of single virulence factors with disease is confounded by the variation in genetic background of this group [15]. An alternative approach is to define sub-groups of EAEC, which are known to be present in multiple lineages of the E. coli population. Indeed multi-locus sequence typing (MLST) has been used to define sub-groups of EAEC associated, more or less, with disease [16], but few studies have analysed MLST sub-groups for virulence gene content.
There are a number of EAEC putative virulence factors, the majority of which are plasmid (pAA) borne [14, 17, 18]. The principal diagnostic target is aggR, which regulates the transcription of other EAEC plasmid- and chromosomally-encoded genes during the control of aggregative adherence [19, 20]. There are currently five known multiple aggregative adherence fimbriae (AAF) subunits associated with aggR positive EAEC: AAF/1, AAF/2, AAF/3, AAF/4 [21] and AAF/5 [22] encoded by the genes aggA, aafA, aag3a, aag4A and aaf5A respectively. Other plasmid-encoded EAEC genes include the anti-aggregative transporter (aat) responsible for transporting a protein called dispersin across the membrane [23], the dispersin protein encoded by aap [18], the enteroaggregative heat stable toxin 1 (EAST-1) encoded by astA [24] and the plasmid-encoded heat-liable cytotoxin encoded by pet [25]. Chromosomally encoded virulence factors include: the type VI secretion system on the aggR-activated island, aaiC [26]; the mucinase protein involved in colonisation encoded by pic [27]; the shigella enterotoxin 1 (ShET1) toxin, encoded by set1A and set1B [28]; the iron-repressible high-molecular-weight protein 2 encoded by irp2 [29] and the locus controlling intestinal epithelial adherence and toxigenic invasion tia [30].
There are genetically related, clonal complexes (CCs) of EAEC with a defined ability to cause disease [15, 16]. In the Chattaway et al. [16] study, Bangladesh EAEC isolates were found in several of the EAEC complexes with the majority of strains falling into CC38, CC295, CC31 CC10 and CC40 but assessment of EAEC virulence genes or antibiotic resistance of these key complexes was not undertaken. Understanding pathogenic and resistant CCs of EAEC in Bangladesh can (1) help clinical management of these infections to prevent malnutrition and mortality and (2) have a baseline of data to assess that will enable further studies to assess the global impact of any major CCs. Here we report the virulence factors and antimicrobial resistance (AMR) profiles associated with these complexes. This is the first study analysing in detail EAEC isolates from the rural setting of Mirzapur, Bangladesh [31]. The aim of this study is to to seek an association between AMR, ability to cause disease and genetic background of EAEC strains from Bangladesh.
Methods
Bacterial strains and serotyping
One hundred and fifty-seven isolates of EAEC originally isolated between 2007–2011 from the rural setting of Mirzapur, Bangladesh using presence of the aat or aaiC gene by PCR as part of a case control study [32] were included in this study. Cases were defined as having acute onset of diarrhoea (≥3 abnormally loose stools in the previous 24 h) within 7 days of study enrolment. A control was defined as having no diarrhoea in the previous 7 days enrolled within the same community within 14 days of presentation of the index case [31].
The set comprised of 96 cases and 61 controls. Serotyping of the somatic and flagella antigens [33] was carried out on the heat stable lipopolysaccharide (LPS) (somatic or O) antigens and the flagellar (H) antigens. Strains that failed to produce LPS and could not be typed were termed ‘rough’ and those that did not react with any sera in the serotyping scheme were termed ‘O unidentifiable’ or ‘H unidentifiable’.
Multi-locus sequencing typing and genotyping
Extracted DNA was available for 151/157 EAEC Bangladesh isolate strains from a previous study which had previously defined their multi-locus sequence types [16]. The DNA from 151 EAEC isolates was screened by PCR for the plasmid encoded virulence genes: aat, aap, astA, aggR, affA, aafA, agg3A, agg4A, aaf5A and pet and chromosomal encoded genes: pic, Set1A, aaiC and irp2 (Table S1, available in the online Supplementary Material). The primers and probes for aaf5A were designed as part of this study FIM5_F 5′-GACTGGATTCTTCAGCTTAAATTAAG-3′, FIM_R 5′-TTCATTTGATGCTGGATTGA-3′, FIM5_P ‘GAGCCCGAGCCTGTACATAGATTTGT’. Products were amplified using the 7500 Fast Real-Time PCR System (Applied Biosystems) with amplification conditions: 95 °C for 5 min followed by 95 °C for 30 s, then 60 °C for 30 s and 72 °C for 10 s. Controls used were as follows: O42 (aafA, aat, aggR, pic, astA, set1A, aaiC, irp2, aap, pet), E099518 (aag3A), 8089 (agg4A), 601010 (tia), 900063 (aagA), 3036 (aaf5A). The total number of positive virulence genes was counted in each strain to give a virulence score.
Phylotyping (A, B1, B2, and D) was determined by PCR [34] for the sequence type (ST) complexes that contained 5 isolates or more.
A previous study [16] described an association of EAEC CCs with disease. Therefore, the ten main EAEC CCs in Bangladesh (CC10, 155, 165, 168, 295, 31, 38, 394, 40 and 720) were analysed for an association with virulence and resistance. Data were analysed in Stata version 13.1 (StataCorp, College Station, Texas). T-tests were used to compare means of continuous variables and the Chi-square test for categorical variables, such as associations between specific genes and the proportion of cases. Differences in virulence score according to complex were examined via multivariable linear regression.
Antimicrobial resistance typing
The antimicrobial drug susceptibilities of all 157 EAEC isolates were determined using the agar incorporation breakpoint method described in the British Society for Antimicrobial Chemotherapy guidelines [35, 36]. The concentrations of the antibiotics used for testing were: colistin (2 mg l−1), gentamicin (2 mg l−1), amikacin (8 mg l−1), streptomycin (8 and 16 mg l−1), tobramycin (2 mg l−1), ertapenem (0.064 and 0.5 mg l−1), cefoxitin (8 mg l−1), ceftazidime (0.5, 1 and 2 mg l−1), cefotaxime (0.25, 0.5 and 1 mg l−1), ceftiofur (1 mg l−1), cefpirome (1 mg l−1), chloramphenicol (8 and 16 mg l−1), trimethoprim (2 mg l−1), nalidixic acid (16 mg l−1), ciprofloxacin (0.064 and 0.5 mg l−1), sulfamethoxazole (256 mg l−1) and tetracycline (8 mg l−1).
Isolates were screened by multiplex PCR for the presence of CTX-M, AmpC, TEM, SHV, VEB, PER and GES beta-lactamase genes and ESBL production was confirmed by double-disc synergy test. Group 9 CTX-M genes were identified to allele level by sequencing where possible [37]. Group 1 CTX-M alleles and their upstream genetic environments were investigated by PCR and sequencing using primers specific for ISEcp1-like and IS26-like elements [37].
Results
Virulence of EAEC cases and controls versus clonal complexes
Virulence gene content between cases and controls was heterogeneous (Tables S2 and S3) and none of the individual genes were significantly associated with either cases or controls. However, the average virulence score (total number of virulence genes present) was higher in cases (7.3) than controls (6.2) (Table 1) (P-value=0.027). The chromosomal gene irp2 was higher in cases (70 %) than controls (58 %) but did not reach significance (P=0.107). There was relatively low power to detect differences in individual genes with the sample size collected in this study as only differences of 20 % or more between cases and controls would approach 80 % power. Therefore modest differences could not be ruled out (10–20 %) in the proportion of genes between cases and controls.
Table 1. Percentage of virulence gene in EAEC from cases and controls.
| aggR | aat | aap | aggA | aafA | agg3A | agg4A | aaf5A | astA | pet | pic | setA | irp2 | tia | aaiC | Virulence score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case (93) | 72 % (67) | 80 % (74) | 86 % (80) | 14 % (13) | 22 % (20) | 17 % (16) | 15 % (14) | 18 % (17) | 43 % (40) | 22 .% (21) | 44 % (41) | 45 % (42) | 70 % (66) | 37 % (34) | 45 % (42) | 7.3 |
| Control (58) | 67 % (40) | 83 % (50) | 83 % (50) | 17 % (10) | 22 % (13) | 13 % (8) | 17 % (10) | 13 % (8) | 47 % (28) | 23 % (14) | 48 % (29) | 40 % (24) | 58 % (35) | 37 % (22) | 56 % (33) | 6.2 |
| Probability | 0.479 | 0.562 | 0.65 | 0.65 | 0.981 | 0.52 | 0.811 | 0.419 | 0.657 | 0.914 | 0.607 | 0.529 | 0.107 | 0.989 | 0.196 | 0.027 |
Table showing the content of each gene in association with cases and controls from 151 EAEC from Bangladesh.
The virulence genes aggR, aat, aap, AAF1-4, astA, pet, pic, setA, ipr2 and tia were significantly associated with the common EAEC CCs [16]. There was no statistical significant association with the common EAEC CCs for aaf5A and aaiC (Table S4). Comparison of virulence scores between EAEC CCs revealed that isolates from CC40 and CC295 harboured more virulence factors than other CCs (Table 2). Within CCs, there was no significant association between fimbrial type and whether the strain was from a case or control. ST165 strains (cases only) were positive for agg3A and all ST40 strains (both cases and controls) were positive for aafA.
Table 2. Mean virulence score (and association with disease) of EAEC complexes.
| Clonal complex | Phylogroup | Sample size | Mean of virulence score | Association of EAEC CC with disease* |
|---|---|---|---|---|
| 10 | A | 18 | 6.8 | 0.01 |
| 155 | B1 | 8 | 6.6 | 0.2 |
| 165 | A | 5 | 4.6 | 0.3 |
| 168 | A | 5 | 6.8 | 0.2 |
| 295 | B1 | 20 | 9.2 | 0.2 |
| 31 | D | 11 | 7.7 | Associated with controls=P0.005 |
| 38 | D | 21 | 6.3 | 0.3 |
| 394 | D | 7 | 5.6 | 0.56 |
| 40 | B1 | 10 | 10.4 | 0.03 |
*Taken from Chattaway et al. study [16].
Mean of virulence scores per EAEC Clonal complex (CC) from Bangladesh with 5 or more samples. CC40 and 295 are the most virulent in terms of average virulent gene content.
Phylogrouping showed that CCs were evenly distributed among three groups (A, B1, D) although no CC was associated with group B2. Isolates from group B1 had a higher average virulence score (8.7) than isolates from group A (6.1) and D (6.5) (Table 2) but there was no obvious association with disease from the case control data. However, analysing the groups by sequence type revealed that B1 could be further divided into three groups: CC155, 295 and 40 (Table 2). Comparison with the case control data showed that CC40 was also associated with disease (case control study P=0.03 [16]) and was the sequence type that harboured the most virulence factors (10.4, Table 2). Thus CC40 has been shown by two independent methods to be a virulent subtype of EAEC.
Resistance of EAEC clonal complexes
The phenotypic antimicrobial typing results showed a high incidence of multidrug resistance (MDR) in this dataset (defined as resistant to three or more antibiotic classes). MDR was identified in 119 (75.8 %) of 157 isolates. Thirty-two (20.4 %) of isolates were resistant to the third-generation cephalosporins (ceftazidime 0.25 mg l−1 and cefotaxime 1 mg l−1) and 120 (75.9 %) were resistant to ampicillin (8 mg l−1). One hundred and ten (70.2 %) exhibited reduced susceptbility (0.064 mg l−1 ciprofloxacin) and 21 were resistant (0.5 mg l−1 ciprofloxacin) to the quinolones. The isolates were also resistant to streptomycin 16 mg l−1 (n=49; 31 %), trimethoprim 2 mg l−1 (n=88; 56 %) and tetracycline 8 mg l−1 (n=70; 44 %) (Table S5). Analysis of resistance gene content by CC indicated multiple drug resistance of strains within these complexes (Table 3). Of the 32 presumptive ESBL isolates, 28 had double-disk (DD) synergy and resistance was encoded by blaCTX-M (blaCTX-M-9, n=17; blaCTX-M-15, n=11) which were identified as the sole mechanism explaining third-generation cephalosporin resistance.
Table 3. Number of resistant isolates from each complex in the dataset.
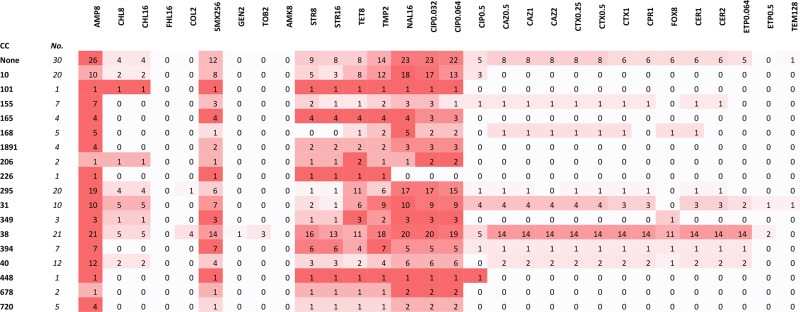 |
Table summarising the number of resistant isolates from each main clonal complex (CC) in the Bangladesh dataset. No=number of isolates in that complex that were resistant against the antibiotic. Colour Key=For each CC per row, the more prevalence the resistance within that complex (i.e. the greatest number of isolates per antibiotic), the darker the red.
Antibiotics: COL (colistin, 2 mg l−1), GEN (gentamicin, 2 mg l−1), AMK (amikacin, 8 mg l−1), STR (streptomycin, 8 and 16 mg l−1 respectively), TOB (tobramycin, 2 mg l−1), AMP (ampicillin, 8 mg l−1), ETP1 and ETP2 (ertapenem, 0.064 and 0.5 mg l−1 respectively), FOX (cefoxtin, 8 mg l−1), TAZ 0.5, TAZ 1 and TAZ2 (cefazidime 0.5, 1 and 2 mg l−1 respectively), CTX 0.25, CTX 1 and CTX 2 (cefotaxime 0.25, 0.5 and 1 mg l−1 respectively), CER 1 (ceftiofur, 1 mg l−1), CPR (cefpirome, 1 mg l−1) CHL1 and CHL2 (chloramphenicols, 8 and 16 mg l−1 respectively), TMP (trimethoprim, 2 mg l−1), NAL (nalidixic acid, 16 mg l−1), CIP 0.32, CIP 1 and CIP2 (ciprofloxacin 0.032, 0.064 and 0.5 mg l−1 respectively), SMX (sulfamethoxazole, 256 mg l−1), TET (tetracycline, 8 mg l−1).
Discussion
This study showed that isolates of EAEC belonging to CC295 and 40 were associated with the highest virulence factor score, whereas ST165 and CC394 had the lowest virulence factor scores. This supports previous evidence from case control studies that highlighted CC40 as a potentially virulent group of EAEC [16, 38]. In previous studies, it was not always possible to correlate the presence of individual EAEC virulence factors with disease [39, 40] but this study showed an association between the presence of combined EAEC virulence factors with disease (P=0.027; virulence score in case versus control). While the individual virulence genes were not independently associated with the ability to cause disease (hence the non-significance association of individual gene with cases), the number of virulence genes present may increase the pathogenic potential of the strain. The heterogeneity of virulence profiles (Tables 1 and 2) suggests multiple acquisition events of different virulence genes. Evolutionary analysis of the different EAEC complexes have indicated that some clonal complexes have evolved predominantly by recombination (such as CC38) rather than mutation (such as CC10) [41]. Clonal complexes that can successfully encounter multiple recombination events can easily acquire mobile genetic elements without impact to survival of the organism. Although the functions of many of the genes described here have been published, a comprehensive understanding of all the gene functions and how they interact with other genes has yet to be fully elucidated.
We also considered the possibility that the presence of multiple virulence factors may be associated with a specific genetic background (as defined by EAEC CC). ST40 has been previously linked to a household outbreak of EAEC O111:H21 encoding the stx2 gene in Northern Ireland providing further evidence that CC40 has the capacity to acquire and maintain a repertoire of virulence genes [38].
The chromosomal marker aaiC is located on the AAI pathogenicity island that encodes a type VI secretion system regulated by the aggR activator [42]. The gene aaiC is an important diagnostic marker for EAEC and is now used alongside aat or aggR to detect EAEC. It has been postulated that the presence of the AAI operon may be associated with increased pathogenicity irrespective of other virulence factors [26]. However, as with previous studies [3, 14, 15, 43], this study showed no statistical association with the presence of aaiC in cases versus controls [3, 10, 14, 15, 43, 44].
Previous studies have highlighted other main EAEC complexes to be pathogenic including the EAEC CC38 associated with extra-intestinal infection, which had been previously shown to have an average virulence score of 9 (including extra-intestinal virulence markers) [41] as did CC10, also statistically shown to be associated with disease in adults [16] and in children [15]. Since data from this study has shown that virulence content is not directly proportional to association with disease, it is suggested that the content of known virulence genes for EAEC plays a role in the ability to cause disease but also in biological success and therefore advantageous to retain these genes as CC evolve. This may explain why the virulence scores of strains within the same complexes vary (Table 2).
CC38 (phylogroup D) is the most resistant of all of the CC in this study (Table 3). CC38 has adapted to both gastrointestinal and extra-intestinal environments [15, 16, 41]. The ability to colonise multiple niches may provide an increased opportunity to acquire resistance genes from a wide variety of bacterial species. Recombination events are a major contributory factor in the evolutionary history of CC38 [16] and may facilitate its ability to adapt to retaining multiple resistance mechanisms. CC10 is one of the largest CC in this study and an ancestral group of multiple pathotypes of E. coli. In CC10, mutation rather than recombination has had a higher impact on the evolution of this group [15, 41] and although the ability of CC10 to naturally evolve over time has enabled this group to evolve into multiple pathotypes of E. coli, this study showed that CC10 was less resistant than CC38 (Table 3). The results of this study showed that some CC are more resistant than others and that strains within a complex are not consistently resistant against the same panel of antimicrobials indicating independent acquisition of resistance mechanisms within a CC. As with the heterogeneous virulence score and there was no direct link between resistance with virulence in any of the CC.
Multidrug resistance was identified in over 75 % of the isolates including all of the main EAEC CCs (Tables 3 and S5). There is little Bangladesh EAEC resistance data available for comparison but the Talukdar et al. study screening E. coli resistance from household water supply showed 36 % of MDR of the E. coli strains [45]. Although the MDR EAEC strains from this study are relatively high, only ETEC and EPEC pathotypes were detected in this study, and additional studies in Bangladesh of EAEC resistance would be required for a fairer comparison. The widespread use of antibiotics has been linked with the selection of resistance mechanisms in pathogenic and non-pathogenic isolates of E. coli [46]. Southeast Asia has been identified as a hotspot for the emergence of MDR in both extra-intestinal bacteria and gastrointestinal pathogens [47]. Despite this fact, the level of MDR detected in the set of EAEC from Bangladesh is high, and includes a high percentage of low level quinolone resistance and ESBL-producing strains. Ciprofloxacin is often used to treat gastrointestinal infections so increasing resistance in these isolates is of concern [48]. Furthermore, it is generally accepted that gut pathogens and gut commensals act as a reservoir of resistance genes that may be acquired by extra-intestinal pathogens associated with life-threatening conditions, such as septicaemia and pneumonia [49].
In conclusion we believe that the definition of the EAEC group of bacteria includes a mixture of commensal bacteria, opportunistic pathogens and primary pathogens. Only by defining the genetic basis of virulence and biological success in these three distinct groups of EAEC will we be able to understand the association with gastrointestinal disease. This study shows that it is possible to define pathogenic and MDR CCs of EAEC and that EAEC CC40 is likely to represent a true pathogen.
Funding information
This study was supported by Gastrointestinal Bacteria Reference Unit, Public Health England, International Centre for Diarrhoeal Disease Research, Bangladesh, Society for Applied Microbiology (Supervisor fund) and Society for General Microbiology (Presidents fund). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
Acknowledgements to the scientists involved in the case control studies, diagnostic and reference laboratories. Thank you to Dawn Hedges and Amy Gentle for Serotyping and Yoshini Taylor and Vivienne DoNascimento for identification at the Gastrointestinal Bacteria Reference Unit (GBRU), PHE, and Benjamin Evens (UEA) for proof reading the article. Thank you to Mark Achtman, University of Warwick, UK and his group for development, management and access and to the public database as well as the scientists who have submitted their data on multiple studies.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
All data was anonymized for the study and ethics approval was not required.
Supplementary Data
Footnotes
Abbreviations: CC, clonal complex; EAEC, enteroaggregative Escherichia coli; ESBL, extended spectrum beta-lactamase; MDR, multidrug resistance.
Five supplementary tables are available with the online Supplementary Material.
References
- 1.Albert MJ, Faruque AS, Faruque SM, Sack RB, Mahalanabis D. Case-control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. J Clin Microbiol. 1999;37:3458–3464. doi: 10.1128/jcm.37.11.3458-3464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araujo JM, Tabarelli GF, Aranda KR, Fabbricotti SH, Fagundes-Neto U, et al. Typical enteroaggregative and atypical enteropathogenic types of Escherichia coli are the most prevalent diarrhea-associated pathotypes among Brazilian children. J Clin Microbiol. 2007;45:3396–3399. doi: 10.1128/JCM.00084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boisen N, Scheutz F, Rasko DA, Redman JC, Persson S, et al. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J Infect Dis. 2012;205:431–444. doi: 10.1093/infdis/jir757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cravioto A, Tello A, Navarro A, Ruiz J, Villafán H, et al. Association of Escherichia coli HEp-2 adherence patterns with type and duration of diarrhoea. Lancet. 1991;337:262–264. doi: 10.1016/0140-6736(91)90868-P. [DOI] [PubMed] [Google Scholar]
- 5.Okeke IN, Lamikanra A, Czeczulin J, Dubovsky F, Kaper JB, et al. Heterogeneous virulence of enteroaggregative Escherichia coli strains isolated from children in Southwest Nigeria. J Infect Dis. 2000;181:252–260. doi: 10.1086/315204. [DOI] [PubMed] [Google Scholar]
- 6.Okeke IN, Nataro JP. Enteroaggregative Escherichia coli. Lancet Infect Dis. 2001;1:304–313. doi: 10.1016/S1473-3099(01)00144-X. [DOI] [PubMed] [Google Scholar]
- 7.Roche JK, Cabel A, Sevilleja J, Nataro J, Guerrant RL. Enteroaggregative Escherichia coli (EAEC) impairs growth while malnutrition worsens EAEC infection: a novel murine model of the infection malnutrition cycle. J Infect Dis. 2010;202:506–514. doi: 10.1086/654894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Kabir F, Manneh J, Lertsethtakarn P, Begum S, et al. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis. 2014;14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]
- 9.Haider K, Faruque SM, Shahid NS, Albert MJ, Nahar S, et al. Enteroaggregative Escherichia coli infections in Bangladeshi children: clinical and microbiological features. J Diarrhoeal Dis Res. 1991;9:318–322. [PubMed] [Google Scholar]
- 10.Taniuchi M, Sobuz SU, Begum S, Platts-Mills JA, Liu J, et al. Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods. J Infect Dis. 2013;208:1794–1802. doi: 10.1093/infdis/jit507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Vaziri H. Treatment of chronic diarrhoea. Best Pract Res Clin Gastroenterol. 2012;26:677–687. doi: 10.1016/j.bpg.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Ahmed D, Sultana N, Hossain A, Sadique T, Islam N, et al. Recurrent urinary tract infection due to co‐infection with extended spectrum β‐lactamase‐producer uropathogenic Escherichia coli and enteroaggregative E. coli. JMM Case Rep. 2014;1 doi: 10.1099/jmmcr.0.001404. [DOI] [Google Scholar]
- 13.Chattaway MA, Harris R, Jenkins C, Tam C, Coia JE, et al. Investigating the link between the presence of enteroaggregative Escherichia coli and infectious intestinal disease in the United Kingdom, 1993 to 1996 and 2008 to 2009. Euro Surveill. 2013;18:20582. doi: 10.2807/1560-7917.ES2013.18.37.20582. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins C, Chart H, Willshaw GA, Cheasty T, Smith HR. Genotyping of enteroaggregative Escherichia coli and identification of target genes for the detection of both typical and atypical strains. Diagn Microbiol Infect Dis. 2006;55:13–19. doi: 10.1016/j.diagmicrobio.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Okeke IN, Wallace-Gadsden F, Simons HR, Matthews N, Labar AS, et al. Multi-locus sequence typing of enteroaggregative Escherichia coli isolates from Nigerian children uncovers multiple lineages. PLoS One. 2010;5:e14093. doi: 10.1371/journal.pone.0014093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chattaway MA, Jenkins C, Rajendram D, Cravioto A, Talukder KA, et al. Enteroaggregative Escherichia coli have evolved independently as distinct complexes within the E. coli population with varying ability to cause disease. PLoS One. 2014;9:e112967. doi: 10.1371/journal.pone.0112967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerna JF, Nataro JP, Estrada-Garcia T. Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. J Clin Microbiol. 2003;41:2138–2140. doi: 10.1128/JCM.41.5.2138-2140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piva IC, Pereira AL, Ferraz LR, Silva RS, Vieira AC, et al. Virulence markers of enteroaggregative Escherichia coli isolated from children and adults with diarrhea in Brasília, Brazil. J Clin Microbiol. 2003;41:1827–1832. doi: 10.1128/JCM.41.5.1827-1832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nataro JP, Yikang D, Yingkang D, Walker K. AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli. J Bacteriol. 1994;176:4691–4699. doi: 10.1128/jb.176.15.4691-4699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boisen N, Ruiz-Perez F, Scheutz F, Krogfelt KA, Nataro JP. Short report: high prevalence of serine protease autotransporter cytotoxins among strains of enteroaggregative Escherichia coli. Am J Trop Med Hyg. 2009;80:294–301. [PMC free article] [PubMed] [Google Scholar]
- 22.Prager R, Lang C, Aurass P, Fruth A, Tietze E, et al. Two novel EHEC/EAEC hybrid strains isolated from human infections. PLoS One. 2014;9:e95379. doi: 10.1371/journal.pone.0095379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishi J, Sheikh J, Mizuguchi K, Luisi B, Burland V, et al. The export of coat protein from enteroaggregative Escherichia coli by a specific ATP-binding cassette transporter system. J Biol Chem. 2003;278:45680–45689. doi: 10.1074/jbc.M306413200. [DOI] [PubMed] [Google Scholar]
- 24.Savarino SJ, Fasano A, Robertson DC, Levine MM. Enteroaggregative Escherichia coli elaborate a heat-stable enterotoxin demonstrable in an in vitro rabbit intestinal model. J Clin Invest. 1991;87:1450–1455. doi: 10.1172/JCI115151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheikh J, Czeczulin JR, Harrington S, Hicks S, Henderson IR, et al. A novel dispersin protein in enteroaggregative Escherichia coli. J Clin Invest. 2002;110:1329–1337. doi: 10.1172/JCI16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dallman TJ, Chattaway MA, Cowley LA, Doumith M, Tewolde R, et al. An investigation of the diversity of strains of enteroaggregative Escherichia coli isolated from cases associated with a large multi-pathogen foodborne outbreak in the UK. PLoS One. 2014;9:e98103. doi: 10.1371/journal.pone.0098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP. Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect Immun. 1999;67:5587–5596. doi: 10.1128/iai.67.11.5587-5596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fasano A, Noriega FR, Maneval DR, Chanasongcram S, Russell R, et al. Shigella enterotoxin 1: an enterotoxin of Shigella flexneri 2a active in rabbit small intestine in vivo and in vitro. J Clin Invest. 1995;95:2853–2861. doi: 10.1172/JCI117991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the "high-pathogenicity island" of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleckenstein JM, Kopecko DJ, Warren RL, Elsinghorst EA. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect Immun. 1996;64:2256–2265. doi: 10.1128/iai.64.6.2256-2265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotloff KL, Blackwelder WC, Nasrin D, Nataro JP, Farag TH, et al. The Global Enteric Multicenter Study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin Infect Dis. 2012;55:S232–S245. doi: 10.1093/cid/cis753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 33.Gross RJ, Rowe B. Serotyping of Escherichia coli. In: Sussman M, editor. The Virulence of Escherichia coli. Cambridge: CambridgeUniversity Press; 1985. pp. 345–360. (editor) [Google Scholar]
- 34.Doumith M, Day MJ, Hope R, Wain J, Woodford N. Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J Clin Microbiol. 2012;50:3108–3110. doi: 10.1128/JCM.01468-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrews JM, BSAC Working Party on Susceptibility Testing FT BSAC standardized disc susceptibility testing method. J Antimicrob Chemother. 2001;48:43–57. doi: 10.1093/jac/48.suppl_1.43. [DOI] [PubMed] [Google Scholar]
- 36.Howe RA, Andrews JM, BSAC Working Party on Susceptibility Testing BSAC standardized disc susceptibility testing method (version 11) J Antimicrob Chemother. 2012;67:2783–2784. doi: 10.1093/jac/dks391. [DOI] [PubMed] [Google Scholar]
- 37.Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother. 2006;57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 38.Dallman T, Smith GP, O'Brien B, Chattaway MA, Finlay D, et al. Characterization of a verocytotoxin-producing enteroaggregative Escherichia coli serogroup O111:H21 strain associated with a household outbreak in Northern Ireland. J Clin Microbiol. 2012;50:4116–4119. doi: 10.1128/JCM.02047-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bafandeh S, Haghi F, Zeighami H. Prevalence and virulence characteristics of enteroaggregative Escherichia coli in a case-control study among patients from Iran. J Med Microbiol. 2015;64:519–524. doi: 10.1099/jmm.0.000055. [DOI] [PubMed] [Google Scholar]
- 40.Vila J, Vargas M, Henderson IR, Gascón J, Nataro JP. Enteroaggregative Escherichia coli virulence factors in traveler's diarrhea strains. J Infect Dis. 2000;182:1780–1783. doi: 10.1086/317617. [DOI] [PubMed] [Google Scholar]
- 41.Chattaway MA, Jenkins C, Ciesielczuk H, Day M, Donascimento V, et al. Evidence of evolving extraintestinal enteroaggregative Escherichia coli ST38 clone. Emerg Infect Dis. 2014;20:1935–1937. doi: 10.3201/eid2011.131845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:1267–1282. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 43.Lima IF, Boisen N, Quetz JS, Havt A, de Carvalho EB, et al. Prevalence of enteroaggregative Escherichia coli and its virulence-related genes in a case-control study among children from North-Eastern Brazil. J Med Microbiol. 2013;62:683–693. doi: 10.1099/jmm.0.054262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dutta S, Guin S, Ghosh S, Pazhani GP, Rajendran K, et al. Trends in the prevalence of diarrheagenic Escherichia coli among hospitalized diarrheal patients in Kolkata, India. PLoS One. 2013;8:e56068. doi: 10.1371/journal.pone.0056068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talukdar PK, Rahman M, Rahman M, Nabi A, Islam Z, et al. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One. 2013;8:e61090. doi: 10.1371/journal.pone.0061090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livermore DM, Winstanley TG, Shannon KP. Interpretative reading: recognizing the unusual and inferring resistance mechanisms from resistance phenotypes. J Antimicrob Chemother. 2001;48:87–102. doi: 10.1093/jac/48.suppl_1.87. [DOI] [PubMed] [Google Scholar]
- 47.Molton JS, Tambyah PA, Ang BS, Ling ML, Fisher DA. The global spread of healthcare-associated multidrug-resistant bacteria: a perspective from Asia. Clin Infect Dis. 2013;56:1310–1318. doi: 10.1093/cid/cit020. [DOI] [PubMed] [Google Scholar]
- 48.Steffen R, Hill DR, Dupont HL. Traveler's diarrhea: a clinical review. JAMA. 2015;313:71–80. doi: 10.1001/jama.2014.17006. [DOI] [PubMed] [Google Scholar]
- 49.Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


