Abstract
The alphaherpesvirus simian varicella virus (SVV) causes varicella and zoster in nonhuman primates. Herpesviruses evolved elaborate mechanisms to escape host immunity, but the immune evasion strategies employed by SVV remain ill-defined. We analysed whether SVV impairs the cellular response to key antiviral cytokine interferon-γ (IFNγ). SVV infection inhibited the expression of IFNγ-induced genes like C-X-C motif chemokine 10 and interferon regulatory factor 1. Phosphorylation and nuclear translocation of the signal transducer and activator of transcription 1 (STAT1) was blocked in SVV-infected cells, which did not involve cellular and viral phosphatases. SVV infection did not downregulate IFNγ receptor α and β chain expression on the cell surface. Instead, STAT1, Janus tyrosine kinases 1 (JAK1) and JAK2 protein levels were significantly decreased in SVV-infected cells. Collectively, these results demonstrate that SVV targets three proteins in the IFNγ signal transduction pathway to escape the antiviral effects of IFNγ.
Keywords: simian varicella virus, interferon-γ, STAT1, JAK1, JAK2
Simian varicella virus (SVV) is a neurotropic alphaherpesvirus that naturally infects Old World monkeys [1, 2]. Primary SVV infection causes a generalized vesicular skin rash (varicella), after which the virus establishes a lifelong latent infection in sensory neurons of the dorsal root and trigeminal ganglia [3, 4]. Upon stress or immune suppression, SVV reactivates to cause a dermatome-restricted skin rash referred to as herpes zoster [5, 6]. To persist in the host, herpesviruses have developed an impressive arsenal of defence mechanisms to counteract innate and adaptive immune responses [7, 8]. However, the immune evasion strategies employed by SVV are largely unknown.
The archetypal antiviral response is mediated by interferons (IFNs) [9]. IFNs are categorized into types I and II and more recently identified type III IFNs that signal through distinct receptors [10]. IFN gamma (IFNγ) is the sole member of type II IFNs and is mainly produced by natural killer (NK) cells and T-cells [11, 12]. The IFNγ receptor complex (IFNγR) consists of two ligand-binding IFNγRα chains that are associated with Janus tyrosine kinase 1 (JAK1) and two signal-transducing IFNγRβ chains associated with JAK2 [11, 12]. Binding of IFNγ to its receptor activates JAK2 to autophosphorylate, and to transphosphorylate and activate JAK1. Activated JAK1 phosphorylates the IFNγRα chain to create a docking site for signal transducer and activator of transcription 1 (STAT1) binding and phosphorylation, after which phosphorylated STAT1 (pSTAT1) dissociates from the IFNγR and forms homodimers that translocate to the nucleus and induce the expression of a broad range of IFNγ-stimulated genes (ISGs) [11, 12]. Collectively, ISGs inhibit cell proliferation and stimulate pathogen recognition, antigen processing/presentation and apoptosis [11, 12].
The IFNγ signalling cascade is broadly targeted by human herpesviruses. Herpes simplex virus type 1 (HSV-1) post-transcriptionally modifies and reduces IFNγRα protein levels, and downregulates JAK1 expression in infected cells [13–15]. Epstein–Barr virus (EBV) and human cytomegalovirus (HCMV) target IFNγRα and JAK1 proteins [16, 17]. Varicella-zoster virus (VZV), the human homologue of SVV, downregulates STAT1 and JAK2 protein levels in virus-infected cells [18]. In this study, we investigated whether the nonhuman primate alphaherpesvirus SVV affects the IFNγ signalling pathway and determined the mechanisms.
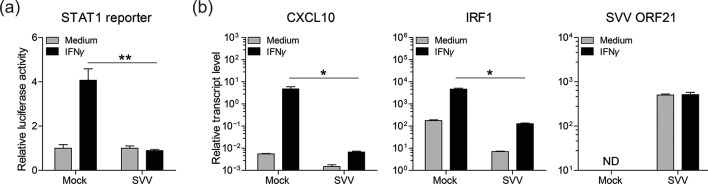
To determine whether SVV infection impairs IFNγ signalling, we analysed IFNγ-induced STAT1-dependent transcription in SVV-infected luciferase reporter cells. African green monkey kidney epithelial (BSC-1) cells were transiently transfected with a plasmid encoding firefly luciferase under the control of a STAT1-responsive promotor [19, 20] and a plasmid constitutively expressing renilla luciferase. Forty-eight hours post-transfection, the transfected cells were mock- or SVV-infected by co-cultivation with SVV-infected BSC-1 cells (showing ≥70 % cytopathic effect) at ratio of one SVV-infected cell to eight transfected cells for 72 h. Subsequently, cells were cultured with or without recombinant human IFNγ (1000 U ml−1) for an additional 6 h, after which luciferase activity was measured. While robust IFNγ-induced STAT1 reporter activity was detected in mock-infected cells, only a minimal STAT1 response was observed after IFNγ treatment of SVV-infected cells (Fig. 1a). Next, we analysed the expression of the ISGs’ C-X-C motif chemokine 10 (CXCL10) and interferon regulatory factor 1 (IRF1) [12] upon IFNγ treatment of mock- or SVV-infected BSC-1 cells. At 72 h p.i. (8 : 1 ratio of uninfected to SVV-infected cells), fresh medium with or without 1000 U ml−1 recombinant human IFNγ (Peprotech) was added to the cells. Twenty-four hours later, the CXCL10, IRF1, SVV open reading frame 21 (ORF21) and cellular control gene oncostatin-M transcript levels were determined by real-time PCR as described [21, 22]. IFNγ treatment induced on average an 853-fold increase in CXCL10 and a 27-fold increase in IRF1 transcript levels in mock-infected cells (Fig. 1). Notably, SVV infection significantly reduced IFNγ-induced CXCL10 and IRF1 expression in BSC-1 cells (average 4.9-fold and 18-fold increases, respectively). Moreover, IFNγ treatment did not reduce the expression levels of the viral early transcript ORF21 in SVV-infected cells. These findings suggest that SVV infection inhibits IFNγ-induced expression of ISGs like CXCL10 and IRF1.
Fig. 1.
SVV inhibits the expression of IFNγ-stimulated genes. (a) BSC-1 cells were transiently transfected to express firefly luciferase under a STAT1-responsive promotor [19, 20] and renilla luciferase under a constitutively active SV40 promotor. The next day, cells were mock- or SVV-infected for 72 h and subsequently cultured with or without recombinant human IFNγ (1000 U ml−1) for an additional 6 h, after which luciferase activity was measured. The relative STAT1 promotor activity of IFNγ-stimulated cells compared to medium control cells, after normalization to renilla luciferase activity, is shown. Bars represent average ± standard error of the mean (SEM) of three replicates measured in duplicate. **P<0.01 by unpaired Student’s t-test. (b) BSC-1 cells were mock- or SVV-infected for 72 h and subsequently cultured with or without recombinant human IFNγ (1000 U ml−1) for an additional 24 h. The transcript levels of C-X-C motif chemokine 10 (CXCL10), interferon regulatory factor 1 (IRF1) and SVV open reading frame 21 (ORF21) were determined in cells by real-time PCR. Relative transcript levels were normalized to the cellular oncostatin-M control transcript and expressed as log10 values. Bars represent average ± sem of two experiments, each performed in duplicate. nd, not detected. *P<0.05 by the Mann–Whitney U test.
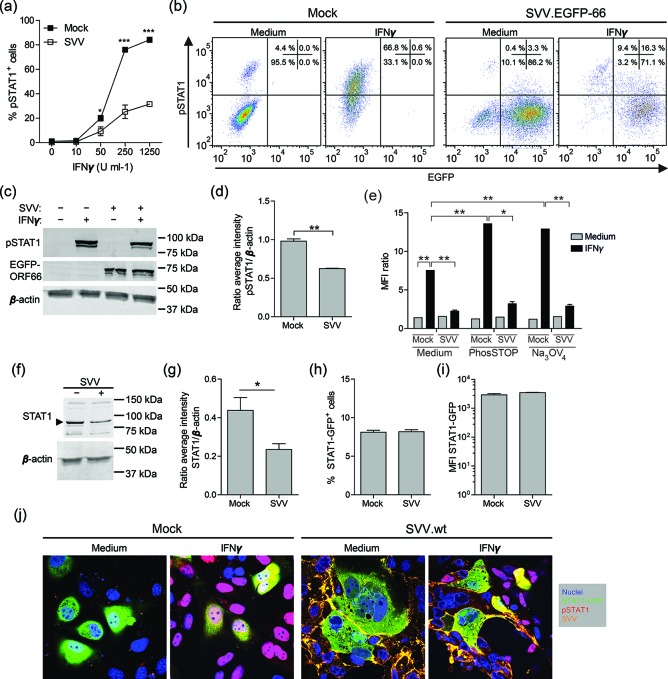
To investigate whether SVV infection interferes with IFNγ signalling at the level of STAT1 phosphorylation, mock- and SVV-infected BSC-1 cells (72 h p.i.; 8 : 1 ratio of uninfected to SVV-infected cells) were treated with the indicated amounts of IFNγ for 20 min at 37 °C and phosphorylated STAT1 was measured by flow cytometry (mouse anti-pY701-STAT1, clone 4a; BD Biosciences). While >50 U ml−1 IFNγ readily induced pSTAT1 expression in mock-infected BSC-1 cells, SVV infection significantly reduced the frequency of pSTAT1-expressing BSC-1 cells in an IFNγ dose-dependent manner (Fig. 2a). The reduced pSTAT1 levels detected in SVV-infected cells could result from an incomplete blockade of IFNγ signalling or be due to residual uninfected cells in the infected cell cultures. To distinguish between the two possibilities, we generated recombinant SVV expressing enhanced green fluorescent protein fused to the N-terminus of ORF66 (SVV.EGFP-66) using red recombination with a bacmid containing the complete SVV genome (Mahalingam et al., manuscript in preparation) [23]. Mock- and SVV.EGFP-66−infected BSC-1 cells (72 h p.i.; 8 : 1 ratio of uninfected to SVV-infected cells) were stimulated with and without 1000 U ml−1 IFNγ and subsequently analysed for pSTAT1 and EGFP expression by flow cytometry. Infection with SVV.EGFP-66 inhibited STAT1 phosphorylation in EGFP-positive SVV-infected cells, but not in EGFP-negative uninfected cells (Fig. 2b). Finally, lysates from mock- and SVV.EGFP-66−infected cells (72 h p.i.; 8 : 1 ratio of uninfected to SVV-infected cells) stimulated with and without IFNγ were analysed for pSTAT1 (mouse anti-pY701-STAT1; clone ST1P-11A5, ThermoFisher Scientific) and β-actin (mouse anti-β-actin; clone C4; Santa Cruz Biotechnology) by Western blotting. Consistent with the flow cytometric results (Fig. 2a, b), reduced pSTAT1 levels were detected in IFNγ-stimulated SVV.EGFP-66−infected cells compared to IFNγ-stimulated mock-infected cells (on average 35 % reduction; Fig. 2c, d). Thus, SVV infection reduced IFNγ-induced STAT1 phosphorylation in BSC-1 cells.
Fig. 2.
SVV inhibits IFNγ-induced phosphorylation of STAT1. (a) BSC-1 cells were mock- or SVV-infected and 72 h later (72 h p.i.), stimulated with the indicated dose of IFNγ for 20 min and finally analysed for intracellular phosphorylated STAT1 (pSTAT1) expression by flow cytometry. The frequency of pSTAT1-positive mock- or SVV-infected BSC-1 cells is shown. Data represent average values ± sem from two independent experiments. *P<0.05, ***P<0.001 by two-way ANOVA test with Bonferroni correction. (b) Flow cytometric analysis of intracellular pSTAT1 and EGFP expression by BSC-1 cells infected with SVV expressing EGFP-ORF66 fusion protein (SVV.EGFP-66) (72 h p.i.), and control mock-infected BSC-1 cells, upon incubation with and without 1000 U ml−1 IFNγ for 20 min. Quadrants were set based on isotype control stained cells and the frequencies of cells within each quadrant are indicated. (c) Mock- and SVV.EGFP-66−infected BSC-1 cells were incubated with and without 1000 U ml−1 IFNγ for 20 min and total cell lysates were analysed by Western blotting for pSTAT1 (two bands, STAT1α=91 kDa and STAT1β=84 kDa), EGFP and β-actin protein expression. (d) The signal intensity of pSTAT1 and β-actin was quantified and the average ratio pSTAT1/β-actin from three experiments is shown. (e) Mock- and SVV.EGFP-66−infected BSC-1 cells (72 h p.i.) were stimulated with medium or 1000 U ml−1 IFNγ for 20 min in the presence or absence of phosphatase inhibitors phosSTOP (Roche) and sodium orthovanadate (Na3OV4). Intracellular phosphorylated STAT1 (pSTAT1) expression was determined by flow cytometry and presented as the median fluorescence intensity (MFI) ratio of cells stained for pSTAT1 and cells stained using isotype control antibodies. (f) Mock- and SVV.EGFP-66−infected BSC-1 cells were lysed at 72 h p.i. and probed with antibodies specific for STAT1 and β-actin by Western blotting. The arrowheads indicate bands of the appropriate size corresponding to STAT1 (two bands, STAT1α=91 kDa and STAT1β=84 kDa). (g) The signal intensity of STAT1 protein was quantified and normalized to the corresponding β-actin protein abundance (STAT1/β-actin ratio). (h–i) Flow cytometric analysis of GFP fluorescence intensity of BSC-1 cells transiently transfected to express STAT1-GFP and mock- and SVV-infected (48 h p.i.). The frequency of STAT1-GFP-expressing cells (h) and the MFI of STAT1-GFP (i) are shown. (j) Transiently transfected BSC-1 cells expressing STAT1-GFP (green) were mock- or wild-type SVV (SVV.wt)-infected (48 h p.i.), incubated with medium or 1000 U ml−1 IFNγ for 20 min and stained for pSTAT1 (red) and SVV (orange). Nuclei were counterstained with DAPI (blue). Representative confocal microscopy images from two independent experiments are shown. Magnification: 400×. (d, e, g, h) Data represent average values ± sem from three (d, e, h) and four (g) independent experiments. *P<0.05, **P<0.01, ***P<0.001 by unpaired Student’s t-test.
Vaccinia virus and HCMV antagonize IFNγ-induced STAT1 phosphorylation by inducing the expression of cellular and viral tyrosine phosphatases in infected cells [24, 25]. To determine whether SVV-mediated inhibition also involves phosphatases, mock- and SVV.EGFP-66−infected BSC-1 cells (72 h p.i.; 8 : 1 ratio of uninfected to SVV-infected cells) were treated with the phosphatase inhibitors PhosSTOP (Roche) or 1 mM sodium orthovanadate (Na3SOV4; Sigma) [24, 25], and then stimulated with IFNγ and finally analysed for pSTAT1 by flow cytometry. Similar to our previous experiments (Fig. 2a, b), SVV.EGFP-66 infection reduced the frequency and median fluorescence intensity (MFI) of pSTAT1-positive cells upon IFNγ stimulation (Fig. 2e). While phosphatase inhibition increased the frequency and MFI of pSTAT1-positive cells after IFNγ stimulation, treatment with phosphatase inhibitors did not affect SVV.EGFP-66−mediated inhibition of STAT1 phosphorylation (Fig. 2e).
To determine whether SVV affects the protein levels of STAT1 in infected cells, lysates from mock- and SVV.EGFP-66–infected BSC-1 cells (72 h p.i.; 8 : 1 ratio of uninfected to SVV-infected cells) were probed for STAT1 (rabbit anti-STAT1; Aviva Systems Biology) by Western blotting. SVV infection significantly reduced STAT1 protein levels, by 44±14 % (average±sd) compared to mock-infected cells (Fig. 2f, g). Next, we investigated whether the observed SVV-mediated STAT1 downregulation and reduction in IFNγ-induced pSTAT1 are directly linked or are separate processes. For this, BSC-1 cells were transiently transfected with a plasmid expressing STAT1-GFP [26], infected with wild-type SVV (SVV.wt; 8 : 1 ratio of uninfected to SVV-infected cells; 48 h post-transfection) and at 48 h p.i. analysed by flow cytometry. Alternatively, transfected and SVV-infected cells were stimulated with IFNγ (1000 U ml−1) for 20 min, fixed and stained using mouse anti-pY701-STAT1 (clone ST1P-11A5; Thermo Fisher Scientific) and rabbit anti-SVV nucleocapsid serum [22] as primary antibodies, and Alexa Fluor 555 (AF555)-conjugated goat anti-mouse IgG2a and AF633-conjugated goat anti-rabbit Ig as secondary antibodies. Flow cytometric analysis of GFP expression revealed that SVV infection did not reduce the frequency of STAT1-GFP-expressing cells or the STAT1-GFP protein levels in infected cells, suggesting that SVV infection affects the expression of endogenous STAT1 at the transcriptional level (Fig. 2h, i). Confocal microscopy demonstrated that IFNγ stimulation induced phosphorylation and nuclear translocation of endogenous STAT1 and STAT1-GFP in uninfected cells (Fig. 2j). By contrast, only very low amounts of endogenous STAT1 and ectopically expressed STAT1-GFP were phosphorylated in response to the IFNγ stimulation of SVV-infected cells (Fig. 2j). Thus, SVV infection reduces STAT1 protein levels and blocks IFNγ-induced STAT1 phosphorylation via distinct mechanisms.
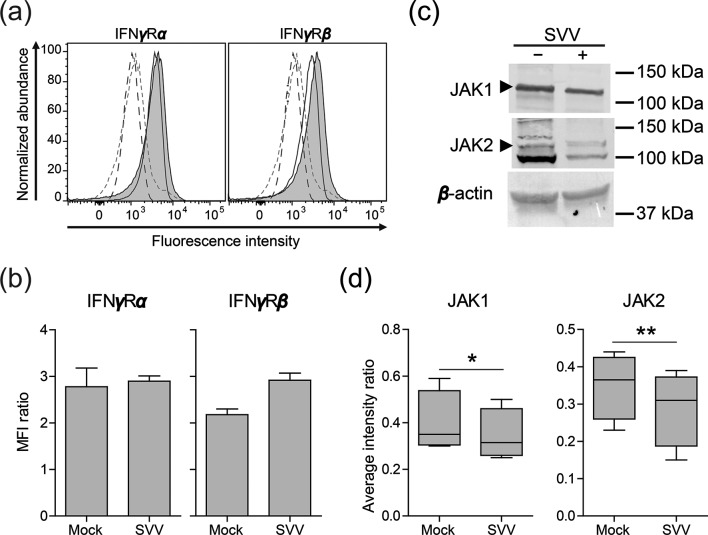
Finally, we analysed whether SVV targets components of the IFNγ signalling pathway upstream of STAT1, i.e. the IFNγ receptor, composed of two homodimers of IFNγRα and IFNγRβ subunits, JAK1 and JAK2 [12]. Expression of the IFNγRα (clone GIR-208; BioLegend) and IFNγRβ chains (clone 2HUB-159; BioLegend) was analysed by flow cytometry on mock- and SVV.EGFP-66−infected BSC-1 cells (72 h p.i.; 8 : 1 ratio of uninfected to SVV-infected cells). However, SVV did not reduce the expression of either of the IFNγ receptor subunits on infected cells (Fig. 3a, b). Next, we investigated JAK1 (rabbit anti-JAK1; clone HR-785; Santa Cruz Biotechnology) and JAK2 (rabbit anti-JAK2; clone C-20; Santa Cruz Biotechnology) protein levels in lysates from mock- and SVV.EGFP-66−infected BSC-1 cells (72 h p.i.) by Western blotting. SVV infection significantly reduced JAK1 protein levels by 14±3.6 % and JAK2 protein levels by 19±10 % (Fig. 3d, e), implying that SVV targets multiple proteins to inhibit IFNγ signalling.
Fig. 3.
SVV infection downregulates JAK1 and JAK 2 protein expression. (a) Histograms showing the expression of the IFNγ receptor alpha (IFNγRα) and beta (IFNγRβ) subunits in mock- and SVV.EGFP-66-infected BSC-1 cells (72 h p.i.) as determined by flow cytometry. Filled (virus-infected) and open (mock-infected) solid lines indicate cells stained with the indicated antibodies. Dashed grey (virus-infected) and black (mock-infected) lines indicate cells stained with isotype control antibodies. (b) The MFI ratio of cells stained for IFNγRα or IFNγRβ, and cells stained using isotype control antibodies, is presented. Data represent average values ± sem from two independent experiments. (c) Mock- and SVV.EGFP-66−infected BSC-1 cells were lysed at 72 h p.i. and probed with antibodies specific for JAK1, JAK2 and β-actin by Western blotting. The arrowheads indicate bands of the appropriate size corresponding to JAK1 (130 kDa) and JAK2 protein (128 kDa). (d) Signal intensities of JAK1 and JAK2 proteins were quantified and normalized to corresponding β-actin protein abundance (e.g. the JAK1/β-actin ratio). Box plots indicate the average intensity ratios from four independent experiments. *P<0.05 and **P<0.01 by paired Student’s t-test.
The extensive co-evolution of herpesviruses with their hosts resulted in a delicate balance between host immunity and viral immune evasion [7, 8] This ensures generally mild disease and lifelong latent infections in the natural host, and facilitates virus dissemination in the population. Compared to those for human herpesviruses, the immune evasion strategies employed by the nonhuman primate alphaherpesvirus SVV remain ill-defined. Recently, it was shown that SVV inhibits type I interferon (IFNα/β) signalling by reducing STAT2 and IRF9 protein levels in infected cells [27]. Furthermore, SVV impaired NFκB signalling by preventing the ubiquitination and degradation of the NFκB inhibitor IκBα [28]. Herein, we have extended these findings by demonstrating that SVV also blocks type II IFN (IFNγ) signalling in infected cells.
We demonstrate that SVV infection downregulates STAT1 protein levels and prevents IFNγ-induced STAT1 phosphorylation and nuclear translocation. STAT1 is shared between the IFN-α/β and IFNγ signalling pathways, thereby providing an ideal target for immune evasion [12]. Similarly, previous studies reported that VZV – the human homologue of SVV – reduced STAT1 protein levels in infected human foreskin fibroblasts and brain-derived vascular adventitial fibroblasts [18, 29]. Interestingly, others did not detect SVV- or VZV-induced reduction of STAT1 protein level in rhesus macaque and human lung fibroblasts [27], possibly due to a cell-type specific effect. Although HCMV and vaccinia virus infection block phosphorylation of STAT1 via the induction of cellular or viral tyrosine phosphatase expression [24, 25], we showed that SVV-induced inhibition of IFNγ-induced STAT1 phosphorylation did not involve active dephosphorylation by phosphatases. Our data suggest that SVV infection does not induce degradation of STAT1 protein, suggesting that the virus may reduce STAT1 mRNA expression levels, as was recently demonstrated for VZV [29]. Moreover, we demonstrate that SVV utilizes distinct mechanisms to reduce STAT1 protein levels and block IFNγ-induced phosphorylation of STAT1.
In addition, JAK1 and JAK2 protein levels were decreased in SVV-infected BSC-1 cells. By contrast, VZV infection has been reported to downregulate JAK2, but not JAK1 protein levels [18]. The closely related HSV-1 reduces JAK1 transcript levels via the virion host shutoff factor (vhs; UL41) [13], a gene involved in generalized degradation of cellular mRNA [30 ]. Likewise, the SVV homologue of vhs, ORF17, may be involved in downregulation of JAK1 and/or JAK 2 transcription. JAK1 and JAK2 are tyrosine kinases interact with a wide panel of type I and II cytokine receptors [11, 31]. Consequently, SVV-mediated downregulation of JAK1 and JAK2 may have a broad range of effects on the signalling of various hormones, interleukins and colony-stimulating factors [31].
Robust antiviral immune responses are mounted by NK and T-cells during primary SVV infection, resulting in high amounts of IFNγ in bronchoalveolar lavage and serum samples [32, 33]. The inhibition of IFNγ signalling by SVV may enable the virus to replicate at sufficient levels to establish latency and reactivate later in life. The in vivo relevance of IFNγ signalling blockade during zoster is exemplified by the absence of MHC-II and intercellular adhesion molecule 1 (ICAM-1) in VZV-infected keratinocytes, but not the surrounding uninfected keratinocytes, in the presence of IFNγ-producing infiltrating leukocytes in the skin of human zoster patients [34]. Additionally, we previously reported abundant expression of the ISG, and NK- and T-cell-recruiting chemokine CXCL10 in uninfected, but not SVV-infected neurons and/or infiltrating cells, in the ganglia of monkeys during primary and reactivated SVV infection [21, 35].
In conclusion, SVV evades the antiviral effects of IFNγ by downregulating three essential proteins in the IFNγ signal transduction pathway: STAT1, JAK1 and JAK2. Because these proteins are shared between several signal transduction pathways, SVV is anticipated to affect the signalling of various other cytokines. Given the parallels between the pathogenesis of VZV in humans and SVV in nonhuman primates [36], comparative analysis of immune evasion strategies may identify conserved essential host targets for the development of new intervention strategies.
Funding information
This work was partially supported by National Institutes of Health grant AG032958 (W. J. D. O., R. M. and G. M. G. M. V.).
Acknowledgements
The authors dedicate this work to the memory of our beloved colleague, Don Gilden, MD. The authors thank Paul R. Kinchington (Department of Ophthalmology, University Pittsburgh) for generously providing the VZV.EGFP-66 strain [37] and A. García-Sastre (Departments of Microbiology, Medicine, and Global Health and Emerging Pathogens Institute, Icahn School of Medicine at Mount Sinai) for kindly providing the plasmid encoding STAT1-GFP [26].
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CXCL10, C-X-C motif chemokine 10; EBV, Epstein–Barr virus; HCMV, human cytomegalovirus; p.i., post-infection; HSV-1, herpes simplex virus type 1; ICAM-1, intercellular adhesion molecule 1; IFN, interferon; IFNγR, IFNγ receptor; IRF1, interferon regulatory factor 1; ISG, interferon-stimulated gene; JAK1, Janus tyrosine kinase 1; JAK1, Janus tyrosine kinase 2; MFI, median fluorescence intensity; SVV, simian varicella virus; STAT1, signal transducer and activator of transcription 1; VZV, varicella-zoster virus.
References
- 1.Clarkson MJ, Thorpe E, Mccarthy K. A virus disease of captive vervet monkeys (Cercopithecus aethiops) caused by a new herpesvirus. Arch Gesamte Virusforsch. 1967;22:219–234. doi: 10.1007/BF01240517. [DOI] [PubMed] [Google Scholar]
- 2.Gray WL. Simian varicella in Old World monkeys. Comp Med. 2008;58:22–30. [PMC free article] [PubMed] [Google Scholar]
- 3.Messaoudi I, Barron A, Wellish M, Engelmann F, Legasse A, et al. Simian varicella virus infection of rhesus macaques recapitulates essential features of varicella zoster virus infection in humans. PLoS Pathog. 2009;5:e1000657. doi: 10.1371/journal.ppat.1000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahalingam R, Smith D, Wellish M, Wolf W, Dueland AN, et al. Simian varicella virus DNA in dorsal root ganglia. Proc Natl Acad Sci USA. 1991;88:2750–2752. doi: 10.1073/pnas.88.7.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolappaswamy K, Mahalingam R, Traina-Dorge V, Shipley ST, Gilden DH, et al. Disseminated Simian varicella virus infection in an irradiated rhesus macaque (Macaca mulatta) J Virol. 2007;81:411–415. doi: 10.1128/JVI.01825-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahalingam R, Traina-Dorge V, Wellish M, Lorino R, Sanford R, et al. Simian varicella virus reactivation in cynomolgus monkeys. Virology. 2007;368:50–59. doi: 10.1016/j.virol.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 7.Griffin BD, Verweij MC, Wiertz EJHJ. Herpesviruses and immunity: the art of evasion. Vet Microbiol. 2010;143:89–100. doi: 10.1016/j.vetmic.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Vossen M, Westerhout E, Söderberg-Nauclér C, Wiertz E. Viral immune evasion: a masterpiece of evolution. Immunogenetics. 2002;54:527–542. doi: 10.1007/s00251-002-0493-1. [DOI] [PubMed] [Google Scholar]
- 9.Isaacs A, Lindenmann J. Virus interference I. The interferon. Proc R Soc London Ser B, Biol Sci. 1957;147:258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- 10.Vilcek J. Novel interferons. Nat Immunol. 2003;4:8–9. doi: 10.1038/ni0103-8. [DOI] [PubMed] [Google Scholar]
- 11.Bach EA, Aguet M, Schreiber RD. The IFN γ receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 12.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-γ: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 13.Chee AV, Roizman B. Herpes simplex virus 1 gene products occlude the interferon signaling pathway at multiple sites. J Virol. 2004;78:4185–4196. doi: 10.1128/JVI.78.8.4185-4196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisemann J, Mühl-Zürbes P, Steinkasserer A, Kummer M. Infection of mature dendritic cells with herpes simplex virus type 1 interferes with the interferon signaling pathway. Immunobiology. 2007;212:877–886. doi: 10.1016/j.imbio.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Liang L, Roizman B. Expression of gamma interferon-dependent genes is blocked independently by virion host shutoff RNase and by US3 protein kinase. J Virol. 2008;82:4688–4696. doi: 10.1128/JVI.02763-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller DM, Rahill BM, Boss JM, Lairmore MD, Durbin JE, et al. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J Exp Med. 1998;187:675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison TE, Mauser A, Wong A, Ting JP, Kenney SC. Inhibition of IFN-gamma signaling by an Epstein-Barr virus immediate-early protein. Immunity. 2001;15:787–799. doi: 10.1016/S1074-7613(01)00226-6. [DOI] [PubMed] [Google Scholar]
- 18.Abendroth A, Slobedman B, Lee E, Mellins E, Wallace M, et al. Modulation of major histocompatibility class II protein expression by varicella-zoster virus. J Virol. 2000;74:1900–1907. doi: 10.1128/JVI.74.4.1900-1907.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang E, Henriksen MA, Schaefer O, Zakharova N, Darnell JE. Dissociation time from DNA determines transcriptional function in a STAT1 linker mutant. J Biol Chem. 2002;277:13455–13462. doi: 10.1074/jbc.M112038200. [DOI] [PubMed] [Google Scholar]
- 20.Besser D, Bromberg JF, Darnell JE, Hanafusa H. A single amino acid substitution in the v-Eyk intracellular domain results in activation of Stat3 and enhances cellular transformation. Mol Cell Biol. 1999;19:1401–1409. doi: 10.1128/MCB.19.2.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouwendijk WJD, Abendroth A, Traina-Dorge V, Getu S, Steain M, et al. T-cell infiltration correlates with cxcl10 expression in ganglia of cynomolgus macaques with reactivated simian varicella Virus. J Virol. 2013;87:2979–2982. doi: 10.1128/JVI.03181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouwendijk WJ, Mahalingam R, de Swart RL, Haagmans BL, van Amerongen G, et al. T-Cell tropism of simian varicella virus during primary infection. PLoS Pathog. 2013;9:e1003368. doi: 10.1371/journal.ppat.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray WL, Zhou F, Noffke J, Karsten Tischer B, Tischer BK. Cloning the simian varicella virus genome in E. coli as an infectious bacterial artificial chromosome. Arch Virol. 2011;156:739–746. doi: 10.1007/s00705-010-0889-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Najarro P, Traktman P, Lewis JA. Vaccinia virus blocks gamma interferon signal transduction: viral VH1 phosphatase reverses stat1 activation. J Virol. 2001;75:3185–3196. doi: 10.1128/JVI.75.7.3185-3196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baron M, Davignon JL. Inhibition of IFN-γ-induced STAT1 tyrosine phosphorylation by human CMV is mediated by SHP2. J Immunol. 2008;181:5530–5536. doi: 10.4049/jimmunol.181.8.5530. [DOI] [PubMed] [Google Scholar]
- 26.Rose KM, Elliott R, Martínez-Sobrido L, García-Sastre A, Weiss SR. Murine coronavirus delays expression of a subset of interferon-stimulated genes. J Virol. 2010;84:5656–5669. doi: 10.1128/JVI.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verweij MC, Wellish M, Whitmer T, Malouli D, Lapel M, et al. Varicella viruses inhibit interferon-stimulated JAK-STAT signaling through multiple mechanisms. PLoS Pathog. 2015;11:e1004901. doi: 10.1371/journal.ppat.1004901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitmer T, Malouli D, Uebelhoer LS, Defilippis VR, Früh K, et al. The ORF61 Protein encoded by simian varicella virus and varicella-zoster virus inhibits NF-κB signaling by interfering with IκBα degradation. J Virol. 2015;89:8687–8700. doi: 10.1128/JVI.01149-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagel MA, James SF, Traktinskiy I, Wyborny A, Choe A, et al. Inhibition of phosphorylated-STAT1 nuclear translocation and antiviral protein expression in human brain vascular adventitial fibroblasts infected with varicella-zoster virus. J Virol. 2014;88:11634–11637. doi: 10.1128/JVI.01945-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oroskar AA, Read GS. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gadina M, Hilton D, Johnston JA, Morinobu A, Lighvani A, et al. Signaling by type I and II cytokine receptors: ten years after. Curr Opin Immunol. 2001;13:363–373. doi: 10.1016/S0952-7915(00)00228-4. [DOI] [PubMed] [Google Scholar]
- 32.Haberthur K, Meyer C, Arnold N, Engelmann F, Jeske DR, et al. Intrabronchial infection of rhesus macaques with simian varicella virus results in a robust immune response in the lungs. J Virol. 2014;88:12777–12792. doi: 10.1128/JVI.01814-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Traina-Dorge V, Sanford R, James S, Doyle-Meyers LA, de Haro E, et al. Robust pro-inflammatory and lesser anti-inflammatory immune responses during primary simian varicella virus infection and reactivation in rhesus macaques. J Neurovirol. 2014;20:526–530. doi: 10.1007/s13365-014-0274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikkels AF, Sadzot-Delvaux C, Piérard GE. Absence of intercellular adhesion molecule 1 expression in varicella zoster virus–infected keratinocytes during herpes zoster. Am J Dermatopathol. 2004;26:27–32. doi: 10.1097/00000372-200402000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Ouwendijk WJD, Getu S, Mahalingam R, Gilden D, Osterhaus ADME, et al. Characterization of the immune response in ganglia after primary simian varicella virus infection. J Neurovirol. 2016;22:376–388. doi: 10.1007/s13365-015-0408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouwendijk WJD, Verjans G. Pathogenesis of varicelloviruses in primates. J Pathol. 2015;235:298–311. doi: 10.1002/path.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisfeld AJ, Yee MB, Erazo A, Abendroth A, Kinchington PR. Downregulation of class I major histocompatibility complex surface expression by varicella-zoster virus involves open reading frame 66 protein kinase-dependent and -independent mechanisms. J Virol. 2007;81:9034–9049. doi: 10.1128/JVI.00711-07. [DOI] [PMC free article] [PubMed] [Google Scholar]