Abstract
CD59 protein functions as a negative regulator of the terminal pathway of the complement system by binding to the C8/C9 factors. To date, little is known about the role of CD59 in coronavirus infectious bronchitis virus (IBV) infection. In this study, we discovered that CD59 was downregulated in IBV-infected cells and was associated with IBV virions. This association protected IBV particles from antibody-dependent complement-mediated lysis. IBV titres in the supernatant were significantly increased when CD59 proteins were overexpressed in cells followed by IBV infection, and this observation was further supported by knockdown or cleavage of CD59. Because no considerable change in IBV N protein and viral RNA levels was detected in total cell lysates prepared from the overexpression, knockdown or cleavage of CD59 groups, our data indicated that CD59 was involved in IBV particle release and that IBV had evolved a mechanism to utilize CD59 to evade complement-mediated destruction.
Keywords: Infectious bronchitis virus, complement regulator, CD59, virion lysis
The complement system is one of the key components of innate immunity, which is generally activated by the classical, lectin and alternative pathways [1]. The system plays vital roles in stimulating complement-mediated cell lysis, inflammatory reactions, opsonization, pathogen elimination and also in inducing humoral and cell-mediated immune responses [2, 3]. Activation of the complement system is a proteolytic cascade rigorously regulated by cell surface and soluble factors, such as CD59, complement receptor 1 and decay-accelerating factor [3, 4]. CD59 is considered to be a phosphoinositol-anchored membrane protein that inhibits the formation of the complement terminal membrane attack complex (MAC). To inhibit complement-dependent cytolysis, CD59 can bind to complement components C8 and C9, hence preventing C9 polymerization and insertion into membranes. CD59 expression on tumour cells and virus-infected cells renders these cells resistant to antibody-dependent complement-mediated lysis (ADCML). The complement system is crucial in regard to the immune system's regulation of inflammatory responses and elimination of viruses; nevertheless, some viruses have employed different evasion strategies to combat complement activity [5]. One such strategy is expression of surface proteins that bind to host complement regulators and subsequently block complement activation [6]. Another strategy is the recruitment of host complement regulators into their virions to evade complement attacks [7].
In some enveloped viruses, such as influenza virus, human cytomegalovirus, Ebola virus, human immunodeficiency virus-1 (HIV-1) and herpes simplex virus-1, viral envelopes associate with host complement regulators to escape ADCML [8–12]. Coronavirus infectious bronchitis virus (IBV) is an enveloped, positive-sense, single-stranded RNA virus that belongs to the family Coronaviridae [13], along with severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus [14]. IBV is a highly contagious pathogen of poultry and has caused extensive economic losses to the poultry industry globally [15]. Mannose-binding lectin can inhibit IBV replication in the trachea by activating the complement system [16, 17], but the role of host complement regulators in IBV infection remains unclear. In the present study, we investigated the association of CD59 protein with IBV virions and its potential role in IBV infection, in view of the fact that CD59 is a distinct complement regulator that prevents MAC [C5b-9(n)] formation at the terminal pathway of complement activation [18, 19].
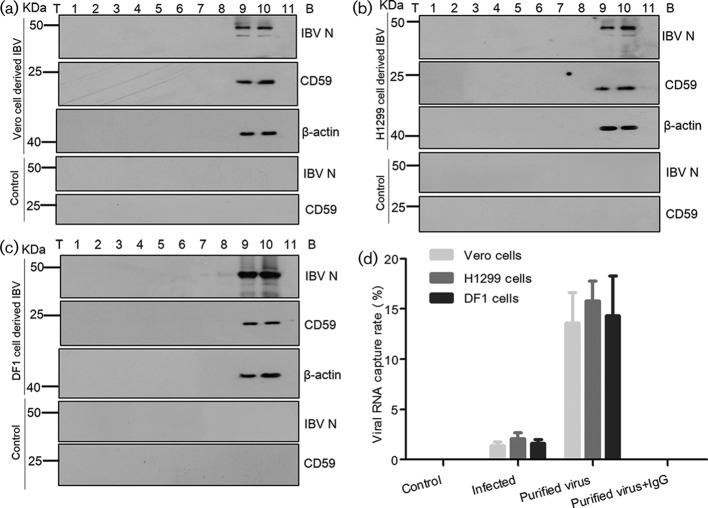
Accordingly, Vero, DF1 and H1299 cells were infected with strain IBV P65 (GenBank accession number DQ001339 [20]) respectively, at a multiplicity of infection (MOI) of 1.0. IBV virions were twice purified from the supernatant of infected cells by sucrose gradient centrifugation as previously described [21]. Aliquots of the fractions from top to bottom were then analysed by immunoblotting to detect viral N proteins, CD59 proteins and beta-actins. Beta-actin was included as a positive control for IBV particles because it was previously reported to be physically associated with IBV [21, 22]. Results showed that IBV N proteins, as well as CD59 proteins and beta-actins, were successfully detected in fractions 9–10 (Fig. 1a–c). Meanwhile, the supernatant from mock-infected cells was fractionated as a control using the same purification protocol, showing that neither CD59 nor IBV N proteins were detected in the supernatant of mock-infected cells (Fig. 1a–c). These data demonstrated that CD59 proteins were likely to be associated with IBV particles.
Fig. 1.
CD59 proteins associated with IBV particles. (a, b and c) IBV virions were twice purified from the supernatant of IBV-infected Vero, H1299 and DF1 cells by 10–50 % sucrose gradient centrifugation. Aliquots of the fractions from top (T) to bottom (B) were analysed by Western blotting for detection of viral N proteins, CD59 and beta-actins. The supernatant from mock-infected cells was set as the control. (d) Immunocapture of IBV virions by anti-CD59 mAbs from the cell-free supernatant of IBV-infected cells and the purified IBV particles (fraction 10 after 10–50 % sucrose gradient centrifugation). Viral genomic RNA capture rates were quantified by sqRT-PCR. The purified viruses captured by mouse immunoglobulin G (IgG) were tested in parallel. All experiments were independently conducted in triplicate.
The potential association of CD59 proteins with IBV virions was further confirmed through virus capture assay, which was performed as previously described [8]. IBV virions were captured by anti-CD59 monoclonal antibodies [mAbs (abcam, UK, catalogue No.: ab79520)] and anti-beta-actin mAbs (Abcam, UK, catalogue No.: ab8226). In the IBV capture assay by anti-CD59 mAbs, an average of 1.4 % (Vero cell-cultured IBV), 2.1 % (H1299 cell-cultured IBV) and 1.6 % (DF1 cell-cultured IBV) of IBV RNAs was captured from an input of 104 50 % tissue culture infective dose (TCID50) IBV in a 100 µl supernatant of the infected cells, as determined by IBV-specific semi-quantitative reverse transcription PCR (sqRT-PCR, Fig. 1d). The use of purified IBV virions significantly enhanced the IBV capture efficiency, resulting in an average of 13.6 % (purified IBV, derived from Vero cells), 15.8 % (purified IBV, derived from H1299 cells) and 14.3 % (purified IBV, derived from DF1 cells) of IBV RNAs being captured from an input of 104 TCID50 purified IBV resuspended in a 100 µl of Tris-NaCl-EDTA (TNE) buffer (Fig. 1d). Meanwhile, the IBV capture assay was carried out using anti-beta-actin mAbs, the results showing that IBV RNAs had been detected in neither the supernatant of infected cells nor the purified IBV samples after capture (data not shown). The rational explanation is that beta-actins probably exist within viral envelopes. Therefore, the results of IBV capture assay confirmed that CD59 proteins were present on the external membranes of the IBV envelopes.
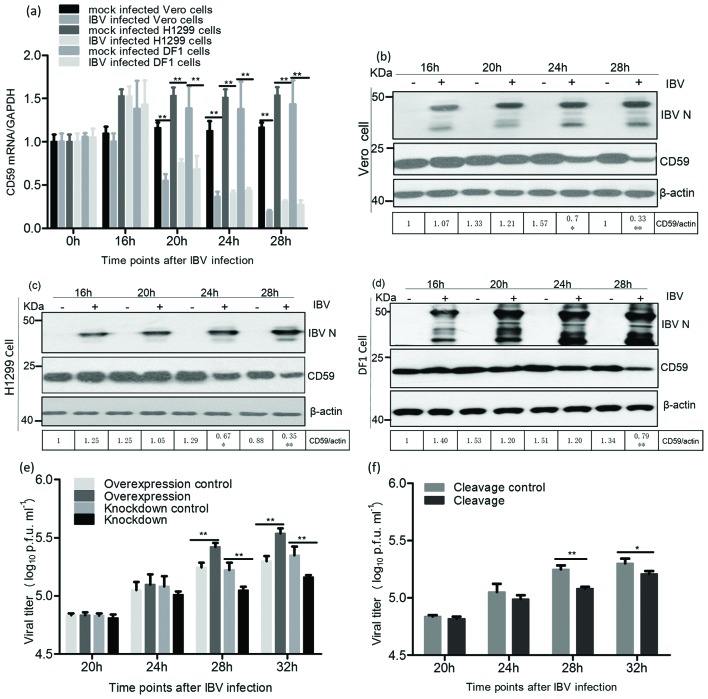
To assess the role of CD59 proteins in the IBV infection process, we investigated CD59 expression at the mRNA level in infected cells, through sqRT-PCR at 0, 16, 20, 24 and 28 h post-infection (pi). The sqRT-PCR results showed that CD59 mRNA levels markedly decreased from 20 h p.i. in the infected Vero, H1299 and DF1 cells (Fig. 2a). Meanwhile, we examined CD59 protein levels on cell membranes during IBV infection; thesewere also markedly reduced on the membranes of infected cells compared to those of mock-infected cells at 28 h p.i. (Fig. 2b–d).
Fig. 2.
CD59 proteins were involved in IBV release. (a) The CD59 mRNA levels in IBV-infected Vero, H1299 and DF1 cells markedly decreased from 20 h p.i. compared with those in mock-infected cells. (b, c and d) CD59 proteins on the membranes of IBV-infected Vero, H1299 and DF1 cells dramatically decreased from 28 h p.i. (e and f) Overexpression of CD59 remarkably increased IBV yields, whereas knockdown of CD59 and cleavage of CD59 significantly reduced IBV titres, in the supernatant of the infected cells from 28 h p.i.
Cells over-/underexpressing CD59 proteins were subsequently infected with IBV, and the effect of CD59 on IBV release was determined at 20, 24, 28 and 32 h p.i. Significantly increased IBV titres in the supernatant of the infected cells were detected from 28 h p.i when CD59 proteins were overexpressed by transfection of the recombinant plasmid pXJ-CD59-flag (Fig. 2e). However, levels of IBV N proteins and viral RNAs in the pXJ-CD59-flag-transfected cells were not markedly higher than those in the empty vector transfected cells (Fig. S1a, b, available in the online Supplementary Material). In this study, the levels of IBV N proteins and viral RNAs were used to reflect IBV replication levels in infected cells. The CD59 mRNAs were knocked down using targeted small interfering RNAs (siRNAs) for further detection of IBV titres in the supernatant of the treated cells after IBV infection. The results showed that viral titres in the supernatant of CD59-knockdown cells markedly decreased from 28 h p.i. compared with those of the control cells using scrambled siRNAs (Fig. 2e). Cleavage of CD59 proteins from cell membranes was conducted by phosphatidylinositol-specific phospholipase C (PI-PLC) treatment assay. The viral titres in the supernatant of CD59 cleavage cells were markedly reduced from 28 h p.i., compared with those of the cells not subjected to PI-PLC treatment (Fig. 2f). Nevertheless, the levels of IBV RNAs and N proteins in cell lysates showed no marked changes between the treatment and control groups in both CD59 knockdown and cleavage experiments (Fig. S1c–f). To rule out the possibility of virus release from the breakdown cells after IBV infection, we determined the viability of mock- and IBV-infected cells in CD59 overexpression, CD59 knockdown and CD59 cleavage experiments using the MTT cell viability assay kit (Beyotime, China). The data showed that there were no marked changes in cell viability between mock- and IBV-infected groups in overexpression, knockdown or cleavage of CD59 experiments (Fig. S2).
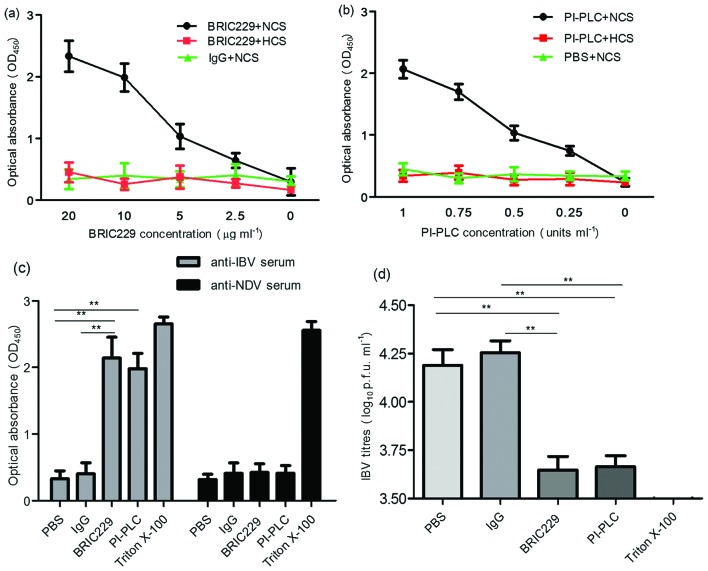
To investigate whether the association of CD59 with IBV particles protects viruses against ADCML, we used CD59-blocking mAbs BRIC229 (IBGRL, Bristol, UK) and PI-PLC to abrogate the function of CD59 on IBV virions, and then analysed the viral lysis of H1299 cell-cultured IBV in the presence or absence of complement activity. Antibody specificity was also investigated in regard to complement-mediated IBV lysis. IBV lysis was quantified by detecting the release of IBV N proteins using antibody sandwich enzyme-linked immunosorbent assay. As shown in Fig. 3a, b, the optical absorbance values of IBV N proteins increased in the BRIC229 mAb groups and PI-PLC treatment groups in a dose-dependent manner. The increased values of optical absorbance of the IBV N proteins were negated when complement activity was inactivated by heating at 56 °C for 30 min (Fig. 3a, b) or when anti-IBV pAbs were replaced with anti-Newcastle disease virus (NDV) pAbs (Fig. 3c). Viral samples from the BRIC229 mAbs treatment assays and PI-PLC cleavage experiments were then subjected to plaque assay to assess infectivity. The results showed that viral samples from both BRIC229 mAbs treatment and PI-PLC cleavage groups had markedly lower viral titres than those from untreated groups (Fig. 3d). These results suggested that abrogation of CD59 function enhanced the ADCML of IBV virions derived from H1299 cells, resulting in a significant reduction in IBV infectivity.
Fig. 3.
Sensitivity of IBV to ADCML. (a and b) Cell-free supernatant from IBV-infected H1299 cells was preincubated with mouse IgG, BRIC229 or PI-PLC at different concentrations, followed by the addition of anti-IBV pAbs plus normal chicken serum (NCS) or heat-inactivated chicken serum (HCS). IBV lysis was quantified by detecting the release of IBV N proteins using antibody sandwich enzyme-linked immunosorbent assay. Dose-dependent analysis of the IBV N proteins released from lysed IBV virions was performed in response to BRIC229 and PI-PLC treatments with anti-IBV pAbs in the presence or absence of the complement components. (c) The effects of antibody specificity on complement-mediated IBV lysis in response to treatment with PBS, mouse IgG, BRIC229, PI-PLC or triton X-100. Chicken-derived complement components in NCS were added in all groups with either anti-IBV pAbs or anti-NDV pAbs. (d) Reduction test for IBV infectivity. A reduction test for IBV infectivity was conducted by inoculating 100 % confluent H1299 cells with viral samples from the groups treated with anti-IBV pAbs in Fig. 3(c). The number of infectious IBV particles retained in the samples of the ADCML experiments was quantified by plaque assay.
The experiments described above indicated that CD59 proteins were associated with IBV particles, potentially protecting IBV from ADCML, and were involved in the IBV release process. Evolution has forced viruses to develop diverse mechanisms to avoid or delay destruction by the complement system [6]. To our knowledge, this article is the first to report that CD59 proteins are associated with IBV virions, although parainfluenza virus 5, mumps virus, vesicular stomatitis virus and NDV have been found to be associated with complement regulators through their envelopes [23–27]. Proteomic analysis of IBV particles has demonstrated that dozens of host proteins are present in the purified IBV virions, but CD59 was not detected [22, 28]. Several factors may have contributed to this discrepancy. First, Kong et al. used bromelain protease to strip proteins from the outside of virus particles, whereas CD59 may have been present on the external surface of IBV virions and it is probable that the CD59 on virus particles was stripped away before proteomic analysis was conducted. Second, IBV virions used in proteomic studies were derived from the allantoic fluid of embryonated eggs, while we used the cell-cultured IBV P65 strain to purify virions. Previous studies were conducted using an in ovo system, which is far less complex proteomically than cell-culture preparations. In addition, the viral purification approaches used in the studies above differ from those used in our work.
The role of CD59 protein was also investigated in IBV-infected cells. CD59 protein levels were markedly reduced on the plasma membranes of IBV-infected cells at 28 h p.i. compared with those of mock-infected cells, implying that IBV may have displaced CD59 from cell membranes when viral particles were budding. The effect of CD59 on IBV release was further determined after CD59 proteins were over-/underexpressed in cells followed by IBV infection. Overexpression of CD59 dramatically elevated viral titres in the supernatant of IBV-infected cells, whereas knockdown of CD59 mRNAs or cleavage of CD59 from cell membranes markedly decreased viral titres. Because no marked changes in the levels of IBV N proteins and viral RNAs were observed in cell lysates prepared from the overexpression, knockdown or cleavage of CD59 groups compared with respective control groups, this suggests that these treatments did not affect IBV replication in cells. Taken together, these results indicate that CD59 is involved in IBV release.
CD59 proteins are mainly located in lipid rafts, and many enveloped viruses are assembled in these subdomains. Knockdown or removal of CD59 can affect the lipid raft structure, resulting in reduction of IBV particle assembly. In fact, the assembly and release processes of the IBV replication cycle were not dramatically affected after lipid raft disruption by cholesterol depletion [29]. Perhaps the distribution of CD59 proteins was decreased in lipid rafts after depleting plasma membrane cholesterol with methyl-β-cyclodextrin or mevastatin in that study, although the level of CD59 proteins on plasma membranes remained stable. In the present study, we found that IBV titres in the supernatant of IBV-infected cells were significantly decreased when CD59 proteins were knocked down or removed from cells followed by IBV infection.ThusCD59 proteins not distributed on lipid rafts may play a vital role in IBV release process using a novel mechanism.
Functional inhibition of CD59 reportedly restores the activities of neutralizing and non-neutralizing antibodies in initiating the ADCML of HIV-1 virions and provirus-activated, latently infected cells [12]. Thus, we abolished CD59 function by its blocker and PI-PLC cleavage, and determined the sensitivity of IBV virions to ADCML. IBV infectivity decreased markedly after the viral particles were treated with CD59 blocker and PI-PLC cleavage, indicating that the sensitivity of IBV virions to ADCML increased after functional inhibition of CD59; and also that CD59 is present on the external surface of IBV envelopes and provides protection against ADCML. This resistance to ADCML possibly explains why some viruses cannot be neutralized by the complement system in body fluids even when viruses elicit high antibody titres. Interfering with complement-mediated destruction by CD59 may help IBV extend its lifespan in vivo and afford it further time for replication and transmission. The direct influences of the association of CD59 with IBV particles on complement evasion in vivo during IBV delivery require further investigation in the clinical setting.
In conclusion, we report for the first time that CD59 is a novel protein that enables IBV to evade complement-mediated destruction, and is involved in the IBV release process. Further investigation on the role of complement in IBV pathogenesis can provide valuable insights into virus–host interactions, and may facilitate the discovery of novel targets for preventing IBV-induced diseases.
Funding information
This research was supported by grants from the National Natural Science Foundation of China (31602038, 31672592 and 31602083), International Science & Technology Cooperation Program of China (2014DFA31890) and Elite Youth program of Chinese Academy of Agricultural Sciences, China.
Acknowledgements
We thank Zifu Zhong for critical reading of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: ADCML, antibody-dependent complement-mediated lysis; HIV-1, human immunodeficiency virus-1; IBV, infectious bronchitis virus; MAC, membrane attack complex; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; NDV, Newcastle disease virus; PI-PLC, phosphatidylinositol-specific phospholipase C; siRNA, small interfering RNA; TCID 50, 50% tissue culture infective dose; TNE, Tris-NaCl-EDTA.
Two supplementary figures are available with the online Supplementary Material.
Reference
- 1.Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat Rev Immunol. 2009;9:729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
- 2.Ricklin D, Lambris JD. Complement in immune and inflammatory disorders: pathophysiological mechanisms. J Immunol. 2013;190:3831–3838. doi: 10.4049/jimmunol.1203487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathern DR, Heeger PS. Molecules great and small: the complement system. Clin J Am Soc Nephrol. 2015;10:1636–1650. doi: 10.2215/CJN.06230614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20:34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 6.Favoreel HW, van de Walle GR, Nauwynck HJ, Pensaert MB. Virus complement evasion strategies. J Gen Virol. 2003;84:1–15. doi: 10.1099/vir.0.18709-0. [DOI] [PubMed] [Google Scholar]
- 7.Stoermer KA, Morrison TE. Complement and viral pathogenesis. Virology. 2011;411:362–373. doi: 10.1016/j.virol.2010.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amet T, Ghabril M, Chalasani N, Byrd D, Hu N, et al. CD59 incorporation protects hepatitis C virus against complement-mediated destruction. Hepatology. 2012;55:354–363. doi: 10.1002/hep.24686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu W, Yu Q, Hu N, Byrd D, Amet T, et al. A high-affinity inhibitor of human CD59 enhances complement-mediated virolysis of HIV-1: implications for treatment of HIV-1/AIDS. J Immunol. 2010;184:359–368. doi: 10.4049/jimmunol.0902278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. Cellular proteins in influenza virus particles. PLoS Pathog. 2008;4:e1000085. doi: 10.1371/journal.ppat.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spear GT, Lurain NS, Parker CJ, Ghassemi M, Payne GH, et al. Host cell-derived complement control proteins CD55 and CD59 are incorporated into the virions of two unrelated enveloped viruses. Human T cell leukemia/lymphoma virus type I (HTLV-I) and human cytomegalovirus (HCMV) J Immunol. 1995;155:4376–4381. [PubMed] [Google Scholar]
- 12.Yang K, Lan J, Shepherd N, Hu N, Xing Y, et al. Blockage of CD59 function restores activities of neutralizing and nonneutralizing antibodies in triggering antibody-dependent complement-mediated lysis of HIV-1 virions and provirus-activated latently infected clls. J Virol. 2015;89:9393–9406. doi: 10.1128/JVI.01614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valastro V, Holmes EC, Britton P, Fusaro A, Jackwood MW, et al. S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect Genet Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westerbeck JW, Machamer CE. A coronavirus E protein is present in two distinct pools with different effects on assembly and the secretory pathway. J Virol. 2015;89:9313–9323. doi: 10.1128/JVI.01237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maier HJ, Neuman BW, Bickerton E, Keep SM, Alrashedi H, et al. Extensive coronavirus-induced membrane rearrangements are not a determinant of pathogenicity. Sci Rep. 2016;6:27126. doi: 10.1038/srep27126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juul-Madsen HR, Norup LR, Handberg KJ, Jørgensen PH. Mannan-binding lectin (MBL) serum concentration in relation to propagation of infectious bronchitis virus (IBV) in chickens. Viral Immunol. 2007;20:562–570. doi: 10.1089/vim.2007.0036. [DOI] [PubMed] [Google Scholar]
- 17.Kjærup RM, Dalgaard TS, Norup LR, Hamzic E, Sørensen P, et al. Characterization of cellular and humoral immune responses after IBV infection in chicken lines differing in MBL serum concentration. Viral Immunol. 2014;27:529–542. doi: 10.1089/vim.2014.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, Hu W, Qin X. The role of complement in the mechanism of action of rituximab for B-cell lymphoma: implications for therapy. Oncologist. 2008;13:954–966. doi: 10.1634/theoncologist.2008-0089. [DOI] [PubMed] [Google Scholar]
- 19.Sjöberg AP, Trouw LA, Blom AM. Complement activation and inhibition: a delicate balance. Trends Immunol. 2009;30:83–90. doi: 10.1016/j.it.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Wei YQ, Guo HC, Dong H, Wang HM, Xu J, et al. Development and characterization of a recombinant infectious bronchitis virus expressing the ectodomain region of S1 gene of H120 strain. Appl Microbiol Biotechnol. 2014;98:1727–1735. doi: 10.1007/s00253-013-5352-5. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Fang S, Xiao H, Chen B, Tam JP, et al. Interaction of the coronavirus infectious bronchitis virus membrane protein with beta-actin and its implication in virion assembly and budding. PLoS One. 2009;4:e4908. doi: 10.1371/journal.pone.0004908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong Q, Xue C, Ren X, Zhang C, Li L, et al. Proteomic analysis of purified coronavirus infectious bronchitis virus particles. Proteome Sci. 2010;8:29. doi: 10.1186/1477-5956-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biswas M, Johnson JB, Kumar SR, Parks GD, Elankumarana S, et al. Incorporation of host complement regulatory proteins into Newcastle disease virus enhances complement evasion. J Virol. 2012;86:12708–12716. doi: 10.1128/JVI.00886-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rangaswamy US, Cotter CR, Cheng X, Jin H, Chen Z. CD55 is a key complement regulatory protein that counteracts complement-mediated inactivation of Newcastle Disease Virus. J Gen Virol. 2016;97:1765–1770. doi: 10.1099/jgv.0.000498. [DOI] [PubMed] [Google Scholar]
- 25.Yu K, Deng S, Wang H, Zhang Y, Chen X, et al. Small interfering RNA expression inhibits avian infectious bronchitis virus replication and inflammatory response. Antivir Ther. 2016;21:469–479. doi: 10.3851/IMP3027. [DOI] [PubMed] [Google Scholar]
- 26.Moerdyk-Schauwecker M, Hwang SI, Grdzelishvili VZ. Cellular proteins associated with the interior and exterior of vesicular stomatitis virus virions. PLoS One. 2014;9:e104688. doi: 10.1371/journal.pone.0104688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson JB, Lyles DS, Alexander-Miller MA, Parks GD. Virion-associated complement regulator CD55 is more potent than CD46 in mediating resistance of mumps virus and vesicular stomatitis virus to neutralization. J Virol. 2012;86:9929–9940. doi: 10.1128/JVI.01154-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dent SD, Xia D, Wastling JM, Neuman BW, Britton P, et al. The proteome of the infectious bronchitis virus Beau-R virion. J Gen Virol. 2015;96:3499–3506. doi: 10.1099/jgv.0.000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo H, Huang M, Yuan Q, Wei Y, Gao Y, et al. The important role of lipid raft-mediated attachment in the infection of cultured cells by coronavirus infectious bronchitis virus beaudette strain. PLoS One. 2017;12:e0170123. doi: 10.1371/journal.pone.0170123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.