Abstract
In mammals, susceptibility to prion infection is primarily modulated by the host’s cellular prion protein (PrPC) sequence. In the sheep scrapie model, a graded scale of susceptibility has been established both in vivo and in vitro based on PrPC amino acids 136, 154 and 171, leading to global breeding programmes to reduce the prevalence of scrapie in sheep. Chronic wasting disease (CWD) resistance in cervids is often characterized as decreased prevalence and/or protracted disease progression in individuals with specific alleles; at present, no PrPC allele conferring absolute resistance in cervids has been identified. To model the susceptibility of various naturally occurring and hypothetical cervid PrPC alleles in vitro, we compared the amplification rates and amyloid extension efficiencies of eight distinct CWD isolates in recombinant cervid PrPC substrates using real-time quaking-induced conversion. We hypothesized that the in vitro conversion characteristics of these isolates in cervid substrates would correlate to in vivo susceptibility – permitting susceptibility prediction for the rare alleles found in nature. We also predicted that hypothetical alleles with multiple resistance-associated codons would be more resistant to in vitro conversion than natural alleles with a single resistant codon. Our studies demonstrate that in vitro conversion metrics align with in vivo susceptibility, and that alleles with multiple amino acid substitutions, each influencing resistance independently, do not necessarily contribute additively to conversion resistance. Importantly, we found that the naturally occurring whitetail deer QGAK substrate exhibited the slowest amplification rate among those evaluated, suggesting that further investigation of this allele and its resistance in vivo is warranted.
Keywords: Prion, deer, elk, chronic wasting disease, susceptibility, RT-QuIC
Introduction
Chronic wasting disease (CWD) is an efficiently transmitted spongiform encephalopathy of cervids (e.g. deer, elk and moose), and is the only known prion disease affecting both farmed and free-ranging, non-domestic animals [1]. The disease is naturally occurring in white-tailed deer (Odocoileus virginianus), mule deer (Odocoileus hemionus), North American elk (Cervus elaphus nelsoni), moose (Alces alces) and reindeer (Rangifer tarandus), and may be experimentally transmitted to a range of experimental species [2–5]. CWD has now been reported in 24 US states, two Canadian provinces, the Republic of Korea and, most recently, Norway [6, 7].
Within individual cervid species, evidence of CWD resistance – which manifests as decreased prevalence and/or delayed progression of disease – has been reported [8–15]. The resistance mechanisms are incompletely understood, but are thought to depend primarily on the host’s cellular prion protein (PrPC) amino acid sequence, encoded by the PRNP gene [16]. In white-tailed deer, much emphasis has been placed on variations at PrPC position 96 (glycine, G or serine, S), as it is one of the most common polymorphisms found in nature and has repeatedly been linked to susceptibility. The resistant 96S allele has a frequency of roughly 20 % in white-tailed deer populations of North America, a frequency which varies somewhat in both farmed and free-ranging populations. Other codon variations with significantly lower allelic frequency have been identified that have not been systematically evaluated for resistance, including codons 95 (histidine, H, versus wild-type glutamine, Q), 116 (glycine versus wild-type alanine, A) and 226 (lysine, K, versus wild-type glutamine) [11]. Because of their low frequencies, the manifestations of CWD in these alleles (notably in their homozygous state) have not been well studied. In elk, the expression of leucine (L) at codon 132 appears to confer CWD resistance when compared to the more prevalent 132M allele [15, 17–19], while in mule deer, lower CWD prevalence has been reported in animals expressing a phenylalanine (F) at codon 225, versus the wild-type 225S [20, 21]. It is possible that other, as yet undiscovered, alleles (including loss of function mutations) may affect resistance in deer and elk. Resistance in other cervid species has not been well-characterized to date, although studies in a limited number of fallow deer and reindeer have shown that variations from the wild-type PRNP allele likely affect susceptibility [22–25].
The pathogenesis of CWD, like other prion diseases, involves conversion of the host’s cellular prion protein to the abnormal protease-resistant form (PrPres) following exposure to an infectious dose of PrPres and subsequent local and systemic amplification and dissemination [26]. In situ conversion, amplification and accumulation of PrPres in the central nervous system contributes to the fundamental neuropathology associated with progressive and ultimately fatal proteinopathies. Modelling this conversion in vitro, via serial protein misfolding cyclic amplification (sPMCA), for example, has proven useful for understanding the conversion process at the biochemical level and ascertaining non-native host susceptibility, as well as for amplification-based detection of low levels of PrPres in clinical samples [27–30]. Unlike sPMCA, which requires a PrPC conversion substrate derived from whole-brain homogenates of transgenic or wild-type hosts, the real-time quaking-induced conversion assay (RT-QuIC) [13, 31, 32] makes use of recombinant prion protein (rPrPC). This allows investigators to rapidly isolate or synthesize PRNP sequences and express them in a variety of production systems for use as a conversion substrate [33], while at the same time limiting inter-substrate variability to the recombinant proteins’ primary structure.
In the present study, we compared the amplification abilities of eight distinct CWD isolates from a range of cervid hosts, PRNP backgrounds and geographical areas in an array of naturally occurring and hypothetical cervid recombinant PrPC substrates. We hypothesized that resistance in vivo would correlate with two metrics of the in vitro conversion process: (1) amplification rate, measured as the inverse time required for a replicate to cross a predefined threshold, and (2) amyloid extension efficiency, calculated from the slope of the amplification curve during exponential amyloidogenesis. We also hypothesized that when multiple resistance-associated codon substitutions were combined within a hypothetical PRNP allele, there would be an additive effect on slower amplification rates and decreased amyloid extension efficiencies. Our findings lend support to the prospect of using cell-free in vitro conversion assays for estimating host susceptibility to prion diseases, and potentially other protein misfolding disorders as well.
Results
Estimating in vivo susceptibility
In vivo field studies have consistently demonstrated the 95Q/96G/116A/226Q (QGAQ) allele to be the most susceptible cervid allele. Based on several previously reported large datasets, the odds ratios of infection in various cervid species and alleles were compared to the QGAQ standard. Successively lower odds ratios of infection were seen in QSAQ (0.61), QGGQ (0.21), QGAK (0.21), HGAQ (0.18) and 225F (0.055) alleles. In elk, the 132M allele is considered to be more susceptible than its 132L counterpart, and indeed the odds ratio of infection in this allele was 0.25 compared to 132M. Although no data are available to compare susceptibility between specific deer and elk alleles, field studies across data analysis units in endemic areas demonstrate that elk, as a species, have a relative risk of infection of 0.24 compared to sympatric whitetail and mule deer (Tables 1 and 2).
Table 1. Allelic prevalence of CWD in various cervid species.
A meta-analysis of previously published data, focusing on publications with unbiased sampling methodology, was compiled to determine the prevalence of CWD among various white-tail deer (WTD), mule deer (MD) and elk alleles. The odds ratios of allelic susceptibility are provided by species, including 95 % confidence intervals and probability values.
| Species | QGAQ | QSAQ | HGAQ | QGGQ | QGAK | 225S (QGAQ) | 225F | 132M | 132L | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CWD+ | Total | CWD+ | Total | CWD+ | Total | CWD+ | Total | CWD+ | Total | CWD+ | Total | CWD+ | Total | CWD+ | Total | CWD+ | Total | ||
| WTD | 395 | 469 | 170 | 260 | 5 | 9 | 0 | 0 | 2 | 4 | 13 | ||||||||
| 529 | 749 | 52 | 131 | 0 | 6 | 0 | 0 | 3 | 4 | 9 | |||||||||
| 53 | 293 | 6 | 128 | 0 | 5 | 1 | 17 | 0 | 11 | 47 | |||||||||
| 79 | 132 | 40 | 95 | 1 | 3 | 14 | 36 | 0 | 0 | 14 | |||||||||
| 103 | 122 | 16 | 26 | 0 | 0 | 0 | 0 | 1 | 2 | 51 | |||||||||
| MD | 39 | 339 | 1 | 75 | 20 | ||||||||||||||
| 579 | 2852 | 1 | 112 | 21 | |||||||||||||||
| Elk | 84 | 212 | 4 | 28 | 12 | ||||||||||||||
| Total | 1159 | 1765 | 284 | 640 | 6 | 23 | 15 | 53 | 6 | 21 | 618 | 3191 | 2 | 187 | 84 | 212 | 4 | 28 | |
| Prevalence | 0.66 | 0.42 | 0.26 | 0.28 | 0.29 | 0.194 | 0.011 | 0.40 | 0.14 | ||||||||||
| Odds ratio (95 % CI, P-value) | 1 | 0.61 (0.35–0.50, P<0.0001) | 0.18 (0.07–0.47, P=0.0004) | 0.21 (0.11–0.38, P<0.0001) | 0.21 (0.08–0.54, P=0.001) | 1 | 0.055 (0.014–0.22, P<0.0001) | 1 | 0.25 (0.085–0.76, P=0.014) | ||||||||||
Table 2. Prevalence of CWD among elk and deer in CWD-endemic areas of Colorado in 2005–2007 and 2010.
Test results are presented from CWD-positive data analysis units where both species are present. [48–50].
| WTD and MD | Elk | |||
|---|---|---|---|---|
| CWD+ | Total tested | CWD+ | Total tested | |
| 243 | 8623 | 70 | 10 140 | |
| Prevalence | 0.028 | 0.0069 | ||
| Odds Ratio (95 % CI, P-value | 1 | 0.24 (0.18-0.31, P<0.0001) | ||
The limited number of fallow deer and reindeer naturally or experimentally exposed to CWD makes in vivo susceptibility estimates in these species difficult. The currently available data seem to show, however, that fallow deer and reindeer expressing the 138N, 129S/138S/169M (SSM), and 225Y alleles are not as susceptible as animals with the QGAQ allele [22–25].
Amplification rates vary among naturally occurring cervid and non-cervid substrates
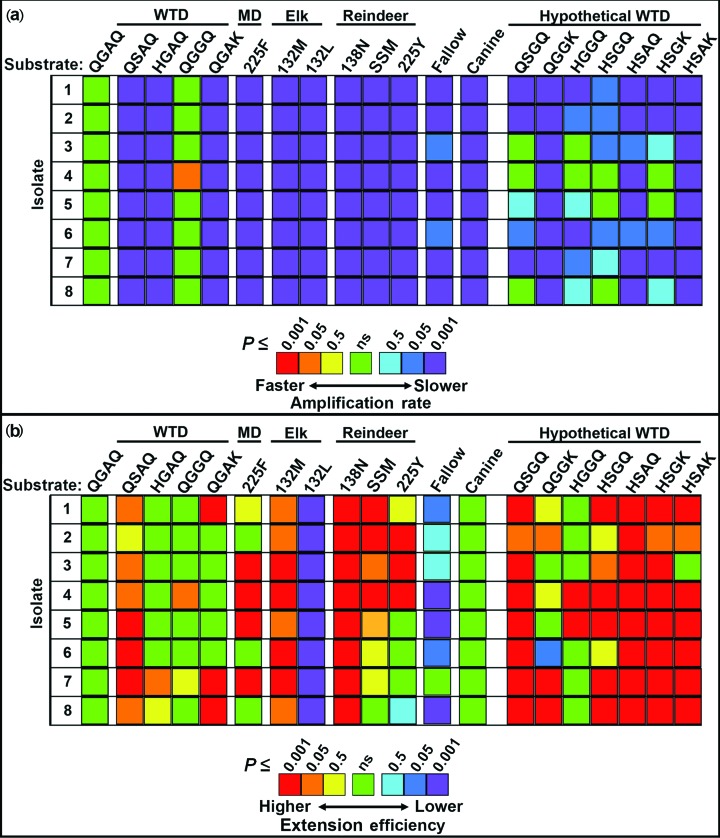
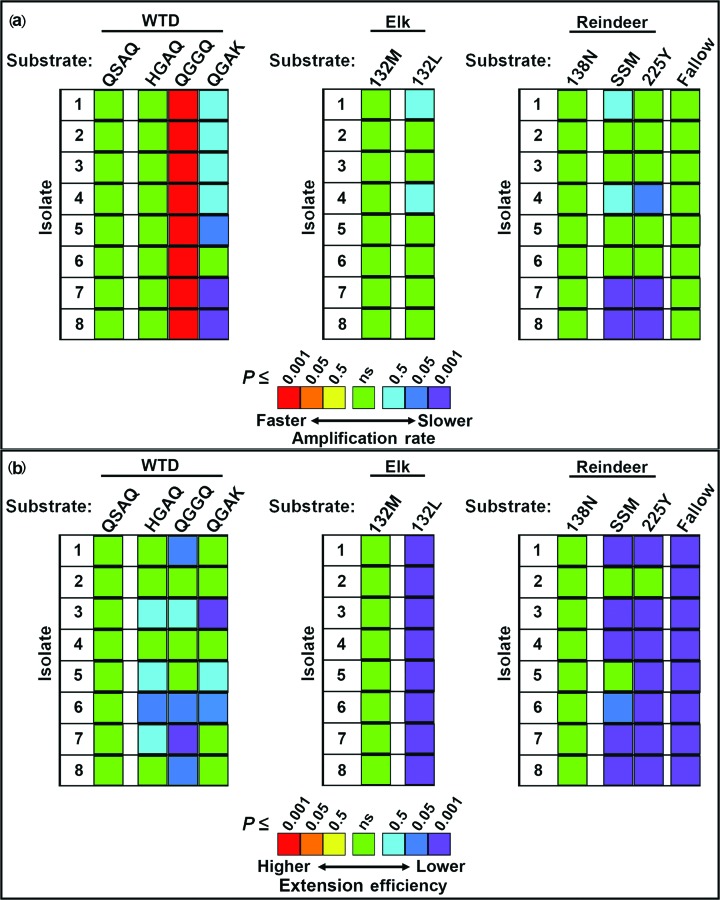
Within each isolate, significant differences were observed in the amplification rates of the naturally occurring cervid PrPC substrates (Figs 1a, b, 2a and 3a, and Table S1, available in the online Supplementary Material). The rates were compared to those of the wild-type QGAQ substrate, which showed the highest amplification rates in some, but not all, isolates. For white-tailed deer isolates 4, the QGGQ substrate was found to have a significantly higher amplification rate than the QGAQ. A species-specific substrate (e.g. QSAQ in white-tailed deer, 132M in elk and 138N in reindeer) was also used as a reference point for rare alleles (Fig. 3a). Allelic substrates associated with resistance, including QSAQ, HGAQ and QGAK, were found to have significantly lower rates of amplification than QGAQ and QGGQ when seeded with any of the isolates examined. Elk-derived 132M and 132L substrates had significantly lower rates of amplification than wild-type QGAQ, and although the 132L substrate in most cases had a lower rate than its 132M counterpart, these differences were not statistically significant. Within reindeer substrates, the 138N, SSM and 225Y substrates had significantly lower amplification rates than the wild-type QGAQ protein, although they rarely differed significantly from one another. Among cervid substrates, the lowest rates of amplification across isolates were observed in the QGAK substrate of white-tailed deer and the mule deer 225F substrate. The canine substrate produced the lowest rates of amplification overall; in most substrates, its rates were not significantly different from those of either QGAK or 225F.
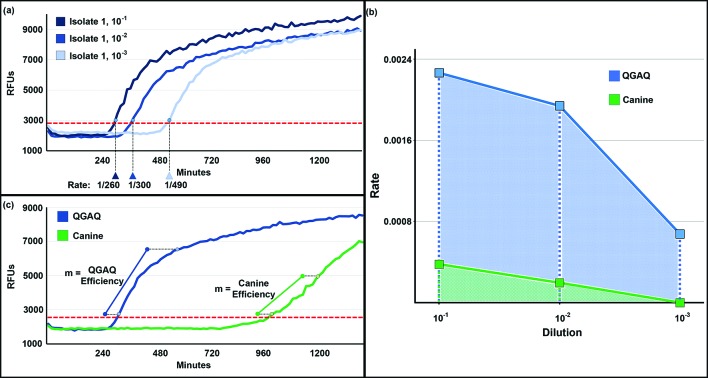
Fig. 1.
Amplification analysis of CWD isolates in cervid substrates. (a) Amplification rates of three dilutions of CWD isolate 1 in 95Q/96G/116A/226Q (QGAQ) substrate. The threshold is indicated by the dashed red line. (b) Plotted amplification rates for three dilutions of isolate 1 in QGAQ and canine substrates. The areas under each of these curves were used to summarize the amplification rate data for each substrate/isolate pair. (c) Amyloid extension efficiencies of the 10−1 dilution of isolate 1 in QGAQ and canine substrates. The threshold is indicated by the dashed red line. RFUs, relative fluorescent units; m, slope of the first four hours of the amplification curves after crossing the threshold.
Fig. 2.
Comparing amplification rates and amyloid extension efficiencies in cervid substrates. The amplification rates (a) and amyloid extension efficiency (b) of eight CWD isolates were compared across naturally occurring and hypothetical cervid alleles. The reference point for amplification rates within each isolate was the 95Q/96G/116A/226Q (QGAQ) allele. Most, but not all, of the substrates had a significant reduction in amplification rate compared to QGAQ. A range of different efficiencies were observed, with many of the reindeer and hypothetical cervid alleles having a significantly higher extension efficiency than QGAQ. Most notably, alleles with apparent in vivo resistance (elk 132L and fallow deer) were found to have a significantly lower efficiency than QGAQ. Where amplification rates were similar, amyloid extension efficiency seemed to help delineate in vivo susceptibilities. Significance is shown as P≤0.5, P≤0.05 and P≤0.001. WTD, white-tailed deer; MD, mule deer; ns, not significant.
Fig. 3.
Comparing the rates and amyloid extension efficiencies of species-specific substrates. With each species, the amplification rates (a) and amyloid extension efficiencies (b) were compared to those of a secondary substrate. In WTD, the secondary substrate that served as a reference point was QSAQ. In elk, 132M was considered as the secondary reference substrate. In reindeer, the 138 N allele served as a secondary reference substrate. Significance is shown as P≤0.5, P≤0.05 and P≤0.001. WTD, white-tailed deer; MD, mule deer; ns, not significant.
Amyloid extension efficiencies vary in naturally occurring cervid and non-cervid substrates
Within each isolate, significant differences were also observed in the amyloid extension efficiencies of the naturally occurring cervid PrPC substrates (Figs 1c, 2b and 3b, and Table S2). The efficiency was compared to that of the wild-type QGAQ substrate – which showed an intermediate extension efficiency in most isolates. Most notably, while the amplification rates of elk 132M and 132L did not significantly differ from one another (as noted above), the amyloid extension efficiency of 132L was significantly reduced compared to that of 132M. Similarly, fallow deer substrate had a markedly reduced extension efficiency compared to the wild-type QGAQ. Separate comparisons were also made between a species-specific substrate in each species (Fig. 3b).
In some cases, substrates that had significantly lower rates of amplification than QGAQ were found to have a significantly higher extension efficiency, and vice versa. For example, both QGAK and 225F were found to have significantly higher amplification efficiencies than the QGAQ protein. Likewise, reindeer substrates 138 N, SSM and 225Y commonly had significantly higher amplification efficiency than QGAQ.
The presence of multiple resistance-associated codons in an artificial cervid substrate does not confer additive effects on amplification rates or amyloid extension efficiency
We hypothesized that substrates encoding multiple resistance-associated amino acids would demonstrate an additive effect on amplification rates and extension efficiencies, with progressively slower conversion rates and/or reduced extension efficiencies as a second, third, or fourth codon was modified. In contrast, we found that both the rates and efficiencies in these artificial cervid substrates were not significantly reduced when compared to their naturally occurring counterparts with a single mutation – in many cases they were modestly higher with respect to efficiency (Fig. 2a, b, and Tables S1 and S2). The exceptions to these findings include conversion in hypothetical QGGK, HSAQ and HSAK substrates – which demonstrated similar rates of amplification compared to QGAK specifically.
Discussion
Among mammalian prion diseases, CWD of cervids has proven to be one of the most problematic to control. While halting the practice of feeding meat and bone meal in 1994 effectively curbed the bovine spongiform encephalopathy (BSE) epidemic, and selective breeding in domestic sheep herds has reduced the prevalence of scrapie by 90 % or more [34–37], an effective management strategy for CWD in farmed and free-ranging cervid herds has proven elusive. In contrast to BSE, CWD is spread quite effectively between cervids through excreta and environmental contamination [6]. Unlike sheep scrapie, no allele or allelic pairing has been found that completely prevents CWD infection in deer and elk species. An important limitation in the search for CWD-resistant cervids is the relative rarity of resistant PRNP alleles. In many cases it is exceedingly uncommon to find these alleles in the homozygous state, somewhat obscuring field studies of the influence of genetics on CWD susceptibility and making their inclusion in challenge studies problematic. The present study sought to examine the misfolding propensities of these rare alleles in vitro, in the hope that our findings might help target specific alleles for further examination in future challenge studies.
We first hypothesized that two metrics of in vitro amplification – rate and efficiency – would parallel in vivo susceptibility across cervid species. Indeed, our findings demonstrate that these two metrics, acting in concert, seem to follow in vivo susceptibility in cervids. The metric that seemed to align most with in vivo susceptibility was the rate of amplification, which likely represents PrPres seed–PrPC substrate compatibility, and the ability of the substrate to form the initial PrPres species. Amyloid extension efficiency may further separate those substrates with statistically similar amplification rates, and likely indicates the capacity of the amyloid product to serve as a seed for amyloid growth, an important step in prion propagation. While the true contributions of these metrics to susceptibility in natural hosts are almost certainly more intricate, our findings suggest that white-tailed deer allele QGGQ, with a limited number of field cases examined, may not be as resistant in nature as it appears – based on rapid amplification rates when seeded with several geographically distinct CWD isolates. Meanwhile, alleles from three different cervid species with similarly low numbers of field cases – 225F in mule deer, QGAK in white-tailed deer and 225Y in reindeer (all found in the Helix 3 domain of PrPC) – demonstrated significantly reduced rates of amplification across multiple CWD isolates and seed dilutions, and warrant further exploration of resistance in the natural host.
Challenge studies in cervid species to estimate susceptibility have so far been limited, and have yet to systematically evaluate alleles other than QSAQ. While field studies under presumably natural conditions have found that CWD-positive 225F mule deer are incredibly uncommon [20, 21], one study involving a small number of orally challenged 225F homozygous mule deer has been reported [8]. These animals were found to have some level of resistance, indicated by prolonged incubation times, although they also presented potential diagnostic challenges based on an unusual neuropathology. Rare challenge studies have also been undertaken in other species – including both fallow deer and reindeer [22–25]. Fallow deer, expressing the 138N/226E allele, appear to be completely resistant to experimental challenge [22–25], while bioassays in a limited number of reindeer have shown that animals that are heterozygous for QGAQ/225Y, QGAQ/138N and QGAQ/SSM alleles may be more resistant than QGAQ homozygous animals. With the discovery of CWD in Norwegian reindeer in 2016 [7], field studies allowing further comparisons of their susceptibility may soon be available following a planned herd reduction over the coming year [38].
We next hypothesized that there would be an additive effect towards delayed amplification rates and/or reduced extension efficiencies in substrates with multiple PrPC mutations associated with natural CWD resistance, based on the additive effects of multiple resistance-associated codons seen in sheep scrapie (e.g. 136V/154R/171Q homozygotes vs 136A/154R/171Q homozygotes vs 136A/154R/171R homozygotes vs heterozygotes of these three alleles) [34, 39–42]. Across CWD isolates, in vitro amplification data in most hypothetical white-tailed deer alleles with multiple mutations showed that this was not the case, with variants commonly demonstrating amplification rates that were comparable to those for QGAQ or naturally occurring alleles with a single mutation. Three proteins (QGGK, HSAQ and HSAK), however, consistently showed significantly reduced rates of amplification, similar to the poorly amplifying QGAK protein. The extension efficiency in these proteins was commonly greater than that of the wild-type QGAQ, a finding that is observed most often in the HSAK substrate. Our examinations with these hypothetical substrates assumed that the various mutations could be found stacked on the same allele (as reported for sheep PRNP variants) – modelling homozygosity in a cervid host. We did not evaluate outcomes from a balanced titration of two white-tailed deer alleles acting together in vitro, however, to model a heterozygous state appropriately (QGGQ × QSAQ, for example). Future experiments examining these permutations may show more clearly the potential for altered susceptibility in cervids that are heterozygous for two different resistance-associated alleles, where the two proteins may act synergistically in vivo to either limit or enhance CWD pathogenesis.
While our experiments were used to ascertain the amplification abilities of various substrates seeded with a specific CWD isolate, we could not examine potential amplification differences between isolates effectively. It would be very tempting to try to draw conclusions from the inter-isolate variability seen among substrates, but a proper approach to answer this question would require well-controlled seeding with a known dose of PrPSc seed across isolates. This may be possible by adjusting seed concentrations to consistent amplification rates in a mutually dissimilar substrate (e.g. truncated Syrian hamster PrPC) or through bioassay titration. Were it possible to overcome this obstacle, it seems plausible that RT-QuIC amplification could be used to examine PrPSc strain typing rigorously, as there is some evidence that several strains may be circulating in cervids [43–45]. This may, in turn, permit a better understanding of the recent source of CWD introductions in North America and Europe.
In summary, evaluating the amplification rates and efficiencies of recombinant PrPC substrates by RT-QuIC could be a useful tool for estimating the susceptibility of rare or newly discovered PRNP alleles, allowing researchers to target specific alleles for downstream evaluation in challenge studies. In the face of an ever-expanding CWD-endemic area, it is increasingly important to characterize the natural susceptibility of these alleles, as well as their geographical distribution and the evolutionary basis for their rarity. Do the QGAK, 225F and 225Y alleles represent recent, random anomalies, or are they more primitive mutations that adversely affect reproductive fitness? Perhaps they are an indication that cervids with these rare alleles were themselves once the target of a primordial prion strain. While some evidence has been presented for distinct strains of CWD, little is known about their geographical distribution or virulence in cervid hosts of diverse PRNP backgrounds. It is possible that, with the appropriate framework, RT-QuIC could allow for the discrimination of known and novel prion strains. Without further research into disease management and prevention, including resistance, the only certainty seems to be that CWD will continue its insidious spread, with further discoveries in new hosts and geographical locations.
Methods
CWD isolates
Recto-anal mucosa-associated lymphoid tissue (RAMALT) biopsies were collected during previous studies from eight CWD-positive white-tailed deer, mule deer or elk. The animals represented various PRNP genotypic backgrounds and geographical locations across North America (Table 3). The tissues were homogenized at ~2 % (weight/volume) in phosphate-buffered saline (PBS) to be used as stock isolates. These stock preparations were then diluted 1 : 10 in RT-QuIC dilution buffer [(PBS with 0.05 % sodium dodecyl sulfate (SDS)] and aliquoted as single-use preparations for each experiment. Single-use aliquots were then frozen at −80 °C until just prior to experimental setup.
Table 3. Detailed information on the species, geographical source and genetic background of the CWD isolates used.
| Isolate | Species | Geographical source | Genotype |
|---|---|---|---|
| 1 | White-tailed deer | Pennsylvania | QGAQ/QGAQ |
| 2 | White-tailed deer | Wisconsin | QGAQ/QGAQ |
| 3 | White-tailed deer | Iowa | QGAQ/QGAQ |
| 4 | White-tailed deer | Iowa | QGAQ/QSAQ |
| 5 | White-tailed deer | Iowa | QSAQ/QSAQ |
| 6 | Elk | Saskatchewan | 132M/L |
| 7 | Elk | Saskatchewan | 132M/M |
| 8 | Mule Deer | Texas | 225 S/S (QGAQ/QGAQ) |
Recombinant cervid PrPC design and development
Recombinant prion protein sequences (rPrPC, residues 25–232) were derived from naturally occurring white-tailed deer, mule deer, elk, fallow deer, reindeer, or canine (25-233) alleles collected in the field; hypothetical alleles were synthesized in vitro (Genewiz, South Plainfield, NJ, USA). The alleles included the wild-type 95Q/96G/116A/226Q (QGAQ) allele commonly found in white-tailed deer, mule deer and reindeer, the putatively resistant 95 h (HGAQ), 96S (QSAQ), 116G (QGGQ) and 226K (QGAK) alleles found in white-tailed deer, as well as several hypothetical stacked permutations of these white-tailed deer alleles (e.g. HSAQ, QGGK, etc.). Both 132M/226E and 132L/226E alleles of North American elk (132M and 132L, respectively) were also evaluated, as well as the 225F/226Q allele of mule deer, the 138N/226E allele of fallow deer, and the 138 N, 129S/138S/169M (SSM) and 225Y alleles of reindeer. A canine PRNP substrate, cloned specifically for these experiments using postmortem tissue collected from a German wirehaired pointer dog, was used as a reference control, as no studies have found canines to be susceptible to CWD or other prion diseases. The canine sequence cloned is a novel one, with a PrPY→H substitution (GenBank accession number MG018983). The frequency of this allele in the canine population is unknown, though there is no reason to assume that it would perform any differently in the assay than previously published canine PrP sequences. A comprehensive summary of the alleles used and their respective codon variances is presented in Fig. 4. Naturally occurring or hypothetical cervid sequences were PCR-amplified from tissue or plasmid preparations using the forward primer 208F (5′-CAC CAA GAA GCG ACC AAA ACC-3′) and the reverse primer 208R (5′-TCA CCC TCT TTG GTA ATA AGC C-3′). Canine sequences were amplified using the forward primer 209F (5′-CAC CAA GAA GCG GCC GAA GCC-3′) and the reverse primer 209R (5′-TCA CCC TCT TTG GTA GTA AGC C-3′). Each forward primer incorporated an overhang to allow for incorporation into the plasmid vector and properly orient the translation reading frame (indicated in bold), while the reverse primers allowed for termination of transcription through a stop codon (underlined). Amplification was achieved using 35 cycles of denaturation at 95 °C for 45 s, annealing at 54 °C for 45 s and extension at 72 °C for 45 s, with a final 5′ extension step. Each product was run on a 1.5 % agarose gel to verify amplicon size, and was then purified and transfected into a pET100d plasmid using a TOPO-TA cloning system according to the manufacturer’s instructions (Thermo Fisher Scientific). Selected colonies were cultured, and their plasmids isolated and sequences confirmed. Plasmids were then transfected into BL21 cells, reisolated and resequenced, and with transfected cultures frozen in 50 % glycerol at −80 °C until expression.
Fig. 4.
Comprehensive summary of substrates used in the experiments. Both naturally occurring and hypothetical cervid alleles were examined. Canine substrate was also evaluated as a reference control, as this species is thought to be resistant to prion infection.
PRNP expression and preparation
Recombinant prion proteins were expressed and purified as previously described [46]. In brief, 5 ml cultures of lysogeny broth with 0.01 % ampicillin (w/v) were inoculated with rPrPC expressing BL21 strain E. coli, grown overnight, and transferred to 0.5 L cultures of lysogeny broth (LB) containing ampicillin (0.01 %) and autoinduction reagents [final concentrations, 0.5 M (NH4)2SO4, 1 M KH2PO4, 0.5 % glycerol, 0.05 % glucose, 0.2 % lactose, 0.001 M MgSO4], before being harvested when an optical density (OD, 600 nm) of ~4 was reached. The cells were lysed with Bug Buster reagent and Lysonase (EMD Biosciences) with DNAse (Sigma-Aldrich) according to the manufacturer’s instructions, and inclusion bodies (IBs) were harvested by centrifugation of the lysate at 15 000 g. IB pellets were washed twice and solubilized overnight in 8M guanidine hydrochloride (GuHCl) in 100 mM NaPO4 and 10 mM Tris pH 8.0, then clarified by centrifugation at 15 000 g for 15 min. Supernatants were added to Super Flow nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) pre-equilibrated with denaturing buffer (6.0M GuHCl, 100 mM NaPO4, 10 mM Tris pH 8.0). Denatured rPrPC and Ni-NTA resin were incubated by rotating at room temperature for 1 h and then added to an XK fast protein liquid chromatography column (GE Healthcare). Refolding was achieved on a column using a linear refolding gradient of denaturing buffer (6M GndCl, 100 mM NaPO4, 10 mM Tris pH 8.0) to refold the buffer (100 mM NaPO4, 10 mM Tris pH 8.0) over 4 h at 1.0 ml min−1. Recombinant PrPC was eluted with a linear gradient of refold buffer to elution buffer (100 mM NaPO4, 10 mM Tris pH 8.0, 1M imidazole pH 5.8) over 45 min at 2.0 ml min−1. Peak UV 280 nm fractions were pooled and dialyzed overnight against two changes of 4.0L of dialysis buffer (20 mM NaPO4 pH 5.8). Recovered rPrPC concentrations were measured using a Nanodrop1000 spectrophotometer (ThermoFisher Scientific) to determine absorbance at 280 nm (A280). The molar concentrations were determined using the equation c=A280/ε, where A280 was the average absorbance across four readings and ε was the calculated extinction coefficient. Proteins were diluted to ~1.00•10−5 M (~0.27 mg ml−1) and stored at 4 °C for up to 45 days.
RT-QuIC procedure
Single-use aliquots of CWD-positive tissue homogenates (0.2 %) were thawed and diluted 1 : 10, 1 : 100 and 1 : 1000 in RT-QuIC dilution buffer (0.05 % SDS). Five µl of these 10−1, 10−2 and 10−3 preparations was added to 95 µl of RT-QuIC reaction buffer, consisting of 50 mM NaPO4, 350 mM NaCl, 1.0 mM ethylenediaminetetraacetic acid tetrasodium salt (EDTA), 10 µM thioflavin T (ThT) and 3.70•10−6 M (~0.1 mg ml−1) cervid or non-cervid rPrPC to yield a final volume of 100 µl. For each isolate, the dilution series was repeated in triplicate on a single plate in three separate experiments. Two groups of negative controls, the first consisting of a 5 µl of a ~0.002 % homogenate of CWD-negative RAMALT from a white-tailed deer (sample control) and the second consisting of 5 µl RT-QuIC dilution buffer (untreated control) spiked into 95 µl of RT-QuIC reaction buffer, were included in each experiment. Reactions were prepared in a black 96-well optical-bottom plate (NalgeNunc), and were then sealed and incubated in a BMG Labtech Polarstar fluorimeter at 42°C for 24 h (96 15-minute cycles) with intermittent shaking cycles; specifically, 1 min shakes (700 r.p.m., double orbital pattern) interrupted by 1 min rest periods. ThT fluorescence measurements (450 nm excitation and 480 nm emission) were taken every 15 min, with the gain set at 1200. The relative fluorescence units (RFU) for each triplicate sample were progressively monitored against time with orbital averaging and 20 flashes/well at the 4 mm setting.
In vivo susceptibility estimates
Historical studies and publications evaluating CWD prevalence in various cervid species were systematically evaluated to ascertain the allelic susceptibilities of white-tailed deer, mule deer and elk [9, 12–14, 20, 21, 47–51]. Prevalence among the various naturally occurring cervid alleles was calculated across groups and studies, allowing for the determination of the relative risks of infection within and among species.
Data analysis
Amplification rate
Replicates were considered positive when the relative fluorescence crossed a pre-defined positive threshold, calculated as 10 standard deviations above the mean fluorescence of all sample wells from cycles 2–8. The rate of amplification, e.g. 1/threshold time, for each isolate and dilution (10−1, 10−2 and 10−3) within cervid rPrPC substrates was recorded (Fig. 1a).
For each isolate and substrate, the amplification rates for each isolate dilution were plotted. The area under the curve (AUC) of these rate points was then used to summarize the amplification rates for each substrate and isolate (Fig. 1b). Within each isolate, the mean AUC and standard errors (SEM) were compared across substrates using ANOVA with Tukey’s multiple comparisons test. The substrate amplification rates were only compared within isolates, because a standard seeding dose could not be adequately determined across isolates a priori to allow for inter-isolate comparisons.
Amyloid extension efficiency
For the first dilution of each isolate (e.g. 1 : 10 in RT-QuIC dilution buffer), amplification curves for each replicate crossing the threshold prior to the 20 h mark were used to estimate amyloid extension efficiency. Because some positive replicates crossed the threshold much later during the experiment (while some failed to amplify at all), we sought to restrict our analysis to those replicates that had at least 16 cycles (4 h) of amplification; this constraint enabled us to include >92 % of replicates for the first dilution. The origin for each curve was standardized to the point where it crossed the threshold. For each isolate and substrate, mean relative fluorescence means and SEM for each cycle of amplification were calculated from the nine replicates. A linear regression of each curve was performed, which allowed for the calculation of a mean slope ±standard error (Fig. 1c). Within each isolate, slopes were compared across substrates using ANOVA with Tukey’s multiple comparisons test.
Funding information
This work was supported in part by NIH NCRR K01OD010994 and Merial.
Acknowledgements
The authors would like to thank the many deer and elk farmers who provided tissues for this particular study, including Brad Farmer for providing fallow deer tissue for DNA extraction.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
No animals were used for the purposes of this study. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health, the United States Department of Agriculture or the Canadian Food Inspection Agency. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the US Government.
Supplementary Data
Footnotes
Abbreviations: AUC, area under the curve; ANOVA, analysis of variance; BSE, bovine spongiform encephalopathy; CWD, chronic wasting disease; IB, inclusion body; PRNP, prion protein gene; PrPC, cellular prion protein; PrPres, misfolded prion protein; QGAQ, cervid prion protein with amino acids glutamine (Q), glycine (G), alanine (A) and glycine at PrPC residues 96, 96, 116 and 226, respectively; RAMALT, recto-anal mucosa-associated lymphoid tissue; RFU, relative fluorescence units; RT-QuIC, real time quaking-induced conversion; sPMCA, serial protein misfolding cyclic amplification; SSM, cervid prion protein with amino acids serine (S), serine and methionine (M) at PrPC residues 129, 138 and 169, respectively; ThT, thioflavin T.
Two supplementary tables are available with the online Supplementary Material.
References
- 1.Williams ES, Young S. Chronic wasting disease of captive mule deer: a spongiform encephalopathy. J Wildl Dis. 1980;16:89–98. doi: 10.7589/0090-3558-16.1.89. [DOI] [PubMed] [Google Scholar]
- 2.Heisey DM, Mickelsen NA, Schneider JR, Johnson CJ, Johnson CJ, et al. Chronic wasting disease (CWD) susceptibility of several North American rodents that are sympatric with cervid CWD epidemics. J Virol. 2010;84:210–215. doi: 10.1128/JVI.00560-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nalls AV, Mcnulty E, Powers J, Seelig DM, Hoover C, et al. Mother to offspring transmission of chronic wasting disease in reeves' muntjac deer. PLoS One. 2013;8:e71844. doi: 10.1371/journal.pone.0071844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh RF, Kincaid AE, Bessen RA, Bartz JC. Interspecies transmission of chronic wasting disease prions to squirrel monkeys (Saimiri sciureus) J Virol. 2005;79:13794–13796. doi: 10.1128/JVI.79.21.13794-13796.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Race B, Meade-White KD, Phillips K, Striebel J, Race R, et al. Chronic wasting disease agents in nonhuman primates. Emerg Infect Dis. 2014;20:833–837. doi: 10.3201/eid2005.130778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haley NJ, Hoover EA. Chronic wasting disease of cervids: current knowledge and future perspectives. Annu Rev Anim Biosci. 2015;3:305–325. doi: 10.1146/annurev-animal-022114-111001. [DOI] [PubMed] [Google Scholar]
- 7.Benestad SL, Mitchell G, Simmons M, Ytrehus B, Vikøren T. First case of chronic wasting disease in Europe in a Norwegian free-ranging reindeer. Vet Res. 2016;47:88. doi: 10.1186/s13567-016-0375-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfe LL, Fox KA, Miller MW. "Atypical" chronic wasting disease in PRNP genotype 225FF mule deer. J Wildl Dis. 2014;50:660–665. doi: 10.7589/2013-10-274. [DOI] [PubMed] [Google Scholar]
- 9.Johnson C, Johnson J, Vanderloo JP, Keane D, Aiken JM, et al. Prion protein polymorphisms in white-tailed deer influence susceptibility to chronic wasting disease. J Gen Virol. 2006;87:2109–2114. doi: 10.1099/vir.0.81615-0. [DOI] [PubMed] [Google Scholar]
- 10.Kelly AC, Mateus-Pinilla NE, Diffendorfer J, Jewell E, Ruiz MO, et al. Prion sequence polymorphisms and chronic wasting disease resistance in Illinois white-tailed deer (Odocoileus virginianus) Prion. 2008;2:28–36. doi: 10.4161/pri.2.1.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson SJ, Samuel MD, O'Rourke KI, Johnson CJ. The role of genetics in chronic wasting disease of North American cervids. Prion. 2012;6:153–162. doi: 10.4161/pri.19640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haley NJ, Siepker C, Hoon-Hanks LL, Mitchell G, Walter WD, et al. Seeded amplification of chronic wasting disease prions in nasal brushings and recto-anal mucosa-associated lymphoid tissues from elk by real-time quaking-induced conversion. J Clin Microbiol. 2016;54:1117–1126. doi: 10.1128/JCM.02700-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haley NJ, Siepker C, Walter WD, Thomsen BV, Greenlee JJ, et al. Antemortem detection of chronic wasting disease prions in nasal brush collections and rectal biopsy specimens from white-tailed deer by real-time quaking-induced conversion. J Clin Microbiol. 2016;54:1108–1116. doi: 10.1128/JCM.02699-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Rourke KI, Spraker TR, Hamburg LK, Besser TE, Brayton KA, et al. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J Gen Virol. 2004;85:1339–1346. doi: 10.1099/vir.0.79785-0. [DOI] [PubMed] [Google Scholar]
- 15.O'Rourke KI, Spraker TR, Zhuang D, Greenlee JJ, Gidlewski TE, et al. Elk with a long incubation prion disease phenotype have a unique PrPd profile. Neuroreport. 2007;18:1935–1938. doi: 10.1097/WNR.0b013e3282f1ca2f. [DOI] [PubMed] [Google Scholar]
- 16.Lloyd SE, Mead S, Collinge J. Genetics of prion diseases. Curr Opin Genet Dev. 2013;23:345–351. doi: 10.1016/j.gde.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monello RJ, Powers JG, Hobbs NT, Spraker TR, O'Rourke KI, et al. Efficacy of antemortem rectal biopsies to diagnose and estimate prevalence of chronic wasting disease in free-ranging cow elk (Cervus elaphus nelsoni) J Wildl Dis. 2013;49:270–278. doi: 10.7589/2011-12-362. [DOI] [PubMed] [Google Scholar]
- 18.O'Rourke KI, Besser TE, Miller MW, Cline TF, Spraker TR, et al. PrP genotypes of captive and free-ranging Rocky Mountain elk (Cervus elaphus nelsoni) with chronic wasting disease. J Gen Virol. 1999;80:2765–2679. doi: 10.1099/0022-1317-80-10-2765. [DOI] [PubMed] [Google Scholar]
- 19.Perucchini M, Griffin K, Miller MW, Goldmann W. PrP genotypes of free-ranging wapiti (Cervus elaphus nelsoni) with chronic wasting disease. J Gen Virol. 2008;89:1324–1328. doi: 10.1099/vir.0.83424-0. [DOI] [PubMed] [Google Scholar]
- 20.Geremia C, Hoeting JA, Wolfe LL, Galloway NL, Antolin MF, et al. Age and repeated biopsy influence antemortem PRPCWD testing in mule deer (Odocoileus hemionus) in Colorado, USA. J Wildl Dis. 2015;51:801–810. doi: 10.7589/2014-12-284. [DOI] [PubMed] [Google Scholar]
- 21.Jewell JE, Conner MM, Wolfe LL, Miller MW, Williams ES. Low frequency of PrP genotype 225SF among free-ranging mule deer (Odocoileus hemionus) with chronic wasting disease. J Gen Virol. 2005;86:2127–2134. doi: 10.1099/vir.0.81077-0. [DOI] [PubMed] [Google Scholar]
- 22.Hamir AN, Greenlee JJ, Nicholson EM, Kunkle RA, Richt JA, et al. Experimental transmission of chronic wasting disease (CWD) from elk and white-tailed deer to fallow deer by intracerebral route: final report. Can J Vet Res. 2011;75:152–156. [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell GB, Sigurdson CJ, O'Rourke KI, Algire J, Harrington NP, et al. Experimental oral transmission of chronic wasting disease to reindeer (Rangifer tarandus tarandus) PLoS One. 2012;7:e39055. doi: 10.1371/journal.pone.0039055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore SJ, Kunkle R, Greenlee MH, Nicholson E, Richt J, et al. Horizontal transmission of chronic wasting disease in reindeer. Emerg Infect Dis. 2016;22:2142–2145. doi: 10.3201/eid2212.160635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhyan JC, Miller MW, Spraker TR, Mccollum M, Nol P, et al. Failure of fallow deer (Dama dama) to develop chronic wasting disease when exposed to a contaminated environment and infected mule deer (Odocoileus hemionus) J Wildl Dis. 2011;47:739–744. doi: 10.7589/0090-3558-47.3.739. [DOI] [PubMed] [Google Scholar]
- 26.Johnson RT. Prion diseases. Lancet Neurol. 2005;4:635–642. doi: 10.1016/S1474-4422(05)70192-7. [DOI] [PubMed] [Google Scholar]
- 27.Kurt TD, Perrott MR, Wilusz CJ, Wilusz J, Supattapone S, et al. Efficient in vitro amplification of chronic wasting disease PrPRES. J Virol. 2007;81:9605–9608. doi: 10.1128/JVI.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 29.Haley NJ, Mathiason CK, Carver S, Telling GC, Zabel MD, et al. Sensitivity of protein misfolding cyclic amplification versus immunohistochemistry in ante-mortem detection of chronic wasting disease. J Gen Virol. 2012;93:1141–1150. doi: 10.1099/vir.0.039073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saá P, Castilla J, Soto C. Ultra-efficient replication of infectious prions by automated protein misfolding cyclic amplification. J Biol Chem. 2006;281:35245–35252. doi: 10.1074/jbc.M603964200. [DOI] [PubMed] [Google Scholar]
- 31.Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, et al. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods. 2007;4:645–650. doi: 10.1038/nmeth1066. [DOI] [PubMed] [Google Scholar]
- 32.Orrú CD, Favole A, Corona C, Mazza M, Manca M, et al. Detection and discrimination of classical and atypical L-type bovine spongiform encephalopathy by real-time quaking-induced conversion. J Clin Microbiol. 2015;53:1115–1120. doi: 10.1128/JCM.02906-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davenport KA, Henderson DM, Bian J, Telling GC, Mathiason CK, et al. Insights into chronic wasting disease and bovine spongiform encephalopathy species barriers by use of real-time conversion. J Virol. 2015;89:9524–9531. doi: 10.1128/JVI.01439-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnold M, Ortiz-Pelaez A. The evolution of the prevalence of classical scrapie in sheep in Great Britain using surveillance data between 2005 and 2012. Prev Vet Med. 2014;117:242–250. doi: 10.1016/j.prevetmed.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 35.Hagenaars TJ, Melchior MB, Bossers A, Davidse A, Engel B, et al. Scrapie prevalence in sheep of susceptible genotype is declining in a population subject to breeding for resistance. BMC Vet Res. 2010;6:25. doi: 10.1186/1746-6148-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutton D. USDA-APHIS Scrapie Program Update and Scrapie Surveillance Projects. Providence, RI: 2015. (editor). One Hundred and Nineteenth Annual Meeting of the United States Animal Health Association. [Google Scholar]
- 37.Bradley R, Wilesmith JW. Epidemiology and control of bovine spongiform encephalopathy (BSE) Br Med Bull. 1993;49:932–959. doi: 10.1093/oxfordjournals.bmb.a072654. [DOI] [PubMed] [Google Scholar]
- 38.Stokstad E. Norway plans to exterminate a large reindeer herd to stop a fatal infectious brain disease. Science. 2017 doi: 10.1126/science.aal0996. [DOI] [Google Scholar]
- 39.Baylis M, Goldmann W. The genetics of scrapie in sheep and goats. Curr Mol Med. 2004;4:385–396. doi: 10.2174/1566524043360672. [DOI] [PubMed] [Google Scholar]
- 40.Francois D, Elsen JM, Barillet F, Lajous D, Eychenne F, et al. Breeding Programmes for Improving the Quality and Safety of Products New Traits, Tools, Rules and Organization [Internet] Zaragoza: CIHEAM; 2003. Breeding sheep for scrapie resistance; pp. 29–35.http://om.iamm.fr/om/pdf/a55/03600060.pdf [Google Scholar]
- 41.Thorne L, Holder T, Ramsay A, Edwards J, Taema MM, et al. In vitro amplification of ovine prions from scrapie-infected sheep from Great Britain reveals distinct patterns of propagation. BMC Vet Res. 2012;8:223. doi: 10.1186/1746-6148-8-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bucalossi C, Cosseddu G, D'Agostino C, di Bari MA, Chiappini B, et al. Assessment of the genetic susceptibility of sheep to scrapie by protein misfolding cyclic amplification and comparison with experimental scrapie transmission studies. J Virol. 2011;85:8386–8392. doi: 10.1128/JVI.00241-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Angers RC, Kang HE, Napier D, Browning S, Seward T, et al. Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science. 2010;328:1154–1158. doi: 10.1126/science.1187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raymond GJ, Raymond LD, Meade-White KD, Hughson AG, Favara C, et al. Transmission and adaptation of chronic wasting disease to hamsters and transgenic mice: evidence for strains. J Virol. 2007;81:4305–4314. doi: 10.1128/JVI.02474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrott MR, Sigurdson CJ, Mason GL, Hoover EA. Evidence for distinct chronic wasting disease (CWD) strains in experimental CWD in ferrets. J Gen Virol. 2012;93:212–221. doi: 10.1099/vir.0.035006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilham JM, Orrú CD, Bessen RA, Atarashi R, Sano K, et al. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010;6:e1001217. doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson GA, Nakada SM, Bollinger TK, Pybus MJ, Merrill EH, et al. Polymorphisms at the PRNP gene influence susceptibility to chronic wasting disease in two species of deer (Odocoileus Spp.) in western Canada. J Toxicol Environ Health A. 2009;72:1025–1029. doi: 10.1080/15287390903084264. [DOI] [PubMed] [Google Scholar]
- 48.2010 CWD Prevalence for Deer by DAU http://cpw.state.co.us/Documents/Hunting/BigGame/CWD/PDF/TestResults/CWDDeer2010.pdf
- 49.2010 CWD Prevalence for Elk by DAU http://cpw.state.co.us/Documents/Hunting/BigGame/CWD/PDF/TestResults/CWDElk2010.pdf
- 50.Miller M. Chronic wasting disease in Colorado 2005−2007. 2008. http://hermes.cde.state.co.us/drupal/islandora/object/co%3A3458/datastream/OBJ/download/Chronic_wasting_disease_in_Colorado__2005-2007.pdf
- 51.Keane DP, Barr DJ, Bochsler PN, Hall SM, Gidlewski T, et al. Chronic wasting disease in a Wisconsin white-tailed deer farm. J Vet Diagn Invest. 2008;20:698–703. doi: 10.1177/104063870802000534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.