Abstract
S-adenosyl-l-methionine (AdoMet) is an essential metabolite, playing a wide variety of metabolic roles. The enzyme that produces AdoMet from l-methionine and ATP (methionine adenosyltransferase, MAT) is thus an attractive target for anti-cancer and antimicrobial agents. It would be very useful to have a system that allows rapid identification of species-specific inhibitors of this essential enzyme. A previously generated E. coli strain, lacking MAT (∆metK) but containing a heterologous AdoMet transporter, was successfully complemented with heterologous metK genes from several bacterial pathogens, as well as with MAT genes from a fungal pathogen and Homo sapiens. The nine tested genes, which vary in both sequence and kinetic properties, all complemented strain MOB1490 well in rich medium. When these strains were grown in glucose minimal medium, growth delays or defects were observed with some specific metK genes, defects that were dramatically reduced if l-methionine was added to the medium.
Keywords: S-adenosyl-L-methionine, MetK, MAT, methionine, Escherichia coli, heterologous complementation
Introduction
S-adenosyl-l-methionine (AdoMet) is an essential metabolite for all known cellular organisms, and is required for a wide range of biochemical processes including everything from methyl transfer reactions to providing adenosyl radicals for ribonucleotide reductase [1, 2]. Methionine adenosyltransferase (MAT; termed MetK in many bacteria) is therefore an attractive target for potential anti-cancer or antimicrobial agents [3]. AdoMet biosynthesis by MAT involves the joining of l-methionine and ATP to produce this critical metabolite [4].
An additional reason for interest in MAT/metK genes is that AdoMet is required for biosynthesis of quorum signalling molecules, including acyl-homoserine lactones (AHLs) [5] and furanosyl borate diesters [6], that control virulence in many Gram-negative bacterial pathogens. There is particular interest in identifying agents that would reduce bacterial pathogenicity by interfering with quorum sensing, but not with growth or viability, to minimize the strong selection for drug resistance [7–10]. Numerous research groups have sought to block quorum sensing, by targeting AHL synthases or the sensor proteins that bind AHLs [10–18], approaches that are promising but have not yet produced compounds with the desired properties and selectivity. Our alternative approach to interference with quorum sensing has been to identify methionine analogues that are substrates for the bacterial MAT/MetK orthologues but yield AdoMet analogues that are selectively non-functional in AHL biosynthesis. This approach takes advantage of the fact that AHL biosynthesis relies upon a unique enzyme-catalysed AdoMet lactonization/cyclization reaction [19]. Accordingly, we have embarked on structural and functional characterization of several AdoMet synthetase orthologues from different bacterial pathogens, and tested their respective abilities to use analogues of l-Met to form AdoMet analogues that are competent for methyl transfer and all other non-AHL-biosynthetic roles, but not the lactonization/cyclization reaction [20, 21].
To facilitate these studies we have developed a screening system that will rapidly detect both inhibitors and alternative substrates for MAT/MetK orthologues from a range of microbial pathogens and, for counter-screening, from humans. We used a ∆metK strain of E. coli, previously developed and generously provided to us [22], that carries an AdoMet transporter from Rickettsia prowazekii. Growth of this strain is completely dependent upon exogenous AdoMet. We found that a surprising range of AdoMet variants, produced in vitro from different methionine analogues, support growth of this strain [23].
We report here that the introduction of MAT/metK genes from a variety of different species, including human, make this metK-deficient E. coli strain independent of exogenous AdoMet. We have also characterized the orthologue-specific growth defects of some of the complemented E. coli strains, seen only during growth in a minimal medium.
Methods
Bacterial strains, plasmids and growth conditions
Escherichia coli K-12 strains JM109, PS2209 and MOB1490 were used in this study. JM109 is endA1 glnV44 thi-1 relA1 gyrA96 recA1 Δ(lac-proAB) e14- [F' traD36 proAB+lacI+ lacZΔM15] hsdR17(rK-mK+) [24]. PS2209 is an essentially wild-type derivative of E. coli W3110, carrying a deletion of lacZ (laboratory stock; originally from B. Wanner). Strain MOB1490, a derivative of BW25113 (lacIq rrnBT14 lacZWJ16 hsdR514 araBADAH33 rhaBADLD78), was kindly provided by Dr David O. Wood [22]. MOB1490 carries ∆metK (metK::kan), plasmid pMW1402 specifying an AdoMet transporter (and AmpR) and pMW1484, a pBAD-derived plasmid carrying rpoB (RifR). Parental MOB1490 requires exogenous AdoMet (the standard concentration used for growth was 32 µM). LB medium or MOPS-based glucose minimal medium (Teknova, Hollister CA) were used for some experiments. MAT/metK genes were cloned into a pUC57 vector, modified to include the synthetic segment illustrated in Fig. 1(b).
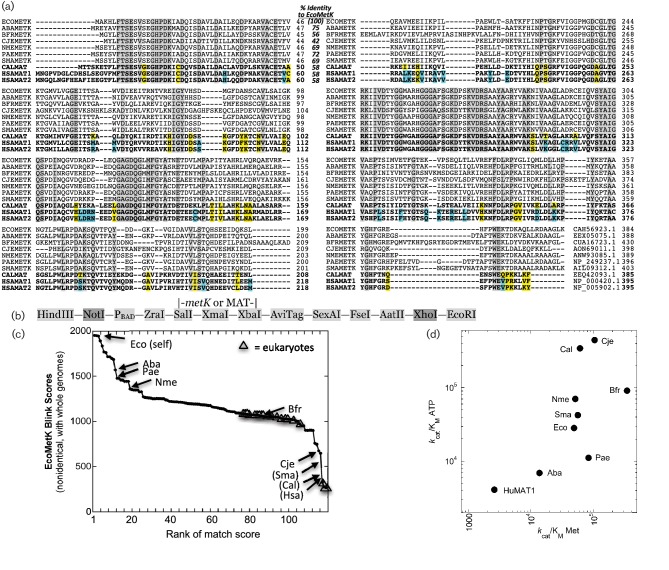
Fig. 1.
Sequences relevant to the study. (a) Aligned amino acid sequences for the nine MAT/metK genes used in this study, plus a second human sequence for comparison. Residues fully conserved among this set are shaded grey, yellow shading indicates that all three eukaryotic sequences differ from the bacterial ones, while cyan shading indicates that both human sequences differ from the others, including the eukaryotic (fungal) pathogen. Abbreviations: bacterial MetK proteins from Acinetobacter baumannii (Aba), Bacteroides fragilis (Bfr), Campylobacter jejuni (Cje), Escherichia coli (Eco), Neisseria meningitidis (Nme), Pseudomonas aeruginosa (Pae) and Stenotrophomonas maltophilia (Sma); and eukaryote MAT proteins from Candida albicans (Cal) and Homo sapiens (Hsa). The reference sequence (EcoMetK) is shown on the top line, while the three eukaryote sequences (CalMAT, HsaMAT1 and HsaMAT2) are at the bottom in bold type. Percentage identity to the reference sequence, and accession numbers for each sequence, are indicated. (b) Synthesized sequence inserted into vector pUC57 for the various MAT/metK complementation plasmids, indicating restriction sites, a promoter and location of gene insertion. (c) Sequence diversity of a total of 125 MAT (▲) and metK (⚫) genes, with the nine tested orthologues indicated as rank position of the Blast-link (BLink) match score [51] to EcoMetK. Only non-identical sequences from complete genomes were included, so some match scores were individually determined and added (names in parentheses). (d) Kinetic diversity of the nine enzymes is illustrated by plotting kcat/Km for l-methionine vs that for ATP.
Amino acids and AdoMet
l-Methionine was prepared as a concentrated stock solution in distilled water, sterilized using a 0.22 µm syringe filter and diluted as needed with M63 medium [25] to achieve the final concentrations. Standard AdoMet was obtained as a 32 mM stock solution from New England Biolabs.
Growth behaviour of orthologue-complemented strains of E. coli MOB1490
Each strain was grown overnight after inoculation at various dilutions into LB medium, so that still-growing cultures could be used to start the experiments. Following inoculation at 1 : 2500 into 100 µl of LB or MOPS-glucose minimal medium in 96-well microtitre plates, the plates were incubated at 37 °C in a heated FLUOstar Omega plate reader (BMG Labtech, Ortenberg, Germany), that took hourly readings of optical density at 600 nm. When added, d-arabinose was 2 %, l-Met was 100 µM,and AdoMet was 32 µM (final concentrations).
Purification and kinetic assay of orthologues
Purification of the AdoMet synthetases from A. baumanni (Aba), B. fragilis (Bfr), C. albicans (Cal), S. maltophilia (Sma) and human MAT1A was performed as previously described [20], with some modifications. E. coli BL21 (DE3) cells, containing a pET28b or pET41a vector coding for a polyhistidine-tagged AdoMet synthetase, were grown at 37 °C in LB broth containing 50 µg ml−1 of kanamycin up to an OD600 of 0.6 to 0.8. Cultures were induced by adding 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG), and further incubated for 18–24 h at 16 °C. Cells were harvested by centrifugation and stored at −80 °C. Five grams of cell paste were resuspended in Buffer A (50 mM Tris-HCl pH 8.0, 500 mM KCl, 5 mM β-mercaptoethanol, 10 % glycerol and 25 mM imidazole) and lysed by sonication. The lysate was centrifuged at 11 000 g, and the filtered supernatant was loaded into a 20 ml Ni-IMAC column equilibrated with Buffer A. A linear gradient of 25–400 mM imidazole was used to elute the bound enzyme. The active fractions containing Aba, Sma and human MAT1A were dialysed against Buffer B (50 mM Tris-HCl, pH 8.0, 500 mM KCl, 10 % glycerol and 1.0 mM DTT) and stored at −80 °C. In contrast, fractions containing Bfr and Cal were pooled and dialysed overnight against Buffer C [50 mM Tris-HCl, pH 8.0, 50 mM KCl, 5 % glycerol and 1.0 mM dithiothreitol (DTT)]. The protein samples were then loaded onto a 20 ml Source 30Q anion exchange column. A linear gradient of 50– 500 mM KCl was used to elute each enzyme. The collected samples were then dialysed against Buffer D (50 mM Tris-HCl, pH 8.0, 50 mM KCl, 5 % glycerol and 1.0 mM DTT) and stored at −80 °C.
Measurement of AdoMet synthetase activity and kinetic parameters, with ATP, l-methionine and the alternative substrates, used our previously published coupled enzyme assay protocol [20]. Briefly, the production of NADPH at 340 nm (ε=6.22 mM−1 cm−1) was monitored on a SpectraMax 190 microplate reader (Molecular Devices) at room temperature. The assay buffer contains 20 mM Tris-HCl, pH 7.6, 16 mM MgCl2, 0.1 mM ATP, 0.1 mM l-methionine, 1 mM uridine-5′-diphosphoglucose, 1 mM NADP, 10 µM glucose 1,6-bisphosphate, 0.8 units of glucose-6-phosphate dehydrogenase, 0.8 units of phosphoglucomutase and 0.1 units of uridine-5′-diphosphoglucose pyrophosphorylase.
Results
Cloning of metK genes
We cloned the MAT/metK genes from a variety of Gram-negative pathogenic or opportunistic bacteria, including Acinetobacter baumannii (Aba, [26]), Bacteroides fragilis (Bfr, [27]), Campylobacter jejuni (Cje, [28]), Escherichia coli [Eco, [29]], Neisseria meningitidis [Nme, [30]], Pseudomonas aeruginosa (Pae, [31]) and Stenotrophomonas maltophilia (Sma, [32]). In addition, we cloned the human MAT1A gene and the equivalent gene from the fungal pathogen Candida albicans (Cal, [33]). Humans and other mammals produce three forms of MAT, and MAT1A yields two of those forms that differ in being either homodimeric or homotetrameric [34]. The amino acid sequences of these AdoMet synthetases are shown in Fig. 1(a), along with the second human gene for MAT (MAT2A, the catalytic subunit of MAT II) for comparison. These sequences range, in identity to EcoMetK, from 75 % (AbaMetK) down to 42 % (CjeMetK), with the three eukaryote sequences each 58 % identical to EcoMetK. This sequence-level range is illustrated in Fig. 1(c), with these nine sequences annotated among the diverse family of over 40 000 nonidentical MAT/MetK genes in GenBank’s non-redundant protein database. These enzymes also vary kinetically. While there are limitations to comparing the catalytic efficiencies of related enzymes via the kcat/Km ratios for their substrates [35], we have done so here not to assert that one enzyme is a better catalyst than another, but just to test whether their kinetic diversity mirrors their sequence diversity. We determined kcat/Km for each of the orthologues with respect to the natural substrates (l-Met and ATP; Table 1), and the results show that these enzymes do indeed have a range of kinetic properties (Fig. 1d) in addition to their range of sequences. Table 1 also shows that the various MetK/MAT orthologues varied considerably in their ability to utilize three different l-methionine analogues. These nine enzymes thus represent a reasonable test of the flexibility of an E. coli-based system for identifying species-specific MAT inhibitors or substrates.
Table 1. Kinetic constants for MetK/MAT orthologues.
| l-Methionine | MgATP | LMEE | LMME | NAM | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme form | kcat (s−1) | KM (mM) |
kcat/KM
(M−1s−1) |
Relative kcat/KM |
KM (mM) |
kcat/KM
(M−1s−1) |
Relative kcat/KM |
kcat (s−1) | KM (mM) | kcat (s−1) | KM (mM) | kcat (s−1) | KM (mM) |
| E. coli* | 3.42±0.34† | 0.070±0.001 | 4.89×104 | 1.0 | 0.120±0.008 | 2.85×104 | 1.0 | 2.27±0.31 | 0.12±0.01 | 1.20±0.08 | 0.17±0.05 | – | >20 |
| HuMAT1 | 0.70±0.35 | 0.27±0.02 | 2.59×103 | 0.05 | 0.17±0.03 | 4.11×103 | 0.1 | 3.75±0.52 | 1.43±0.33 | – | >10 | – | (Non-substrate) |
| A. baumannii | 2.07±0.03 | 0.193±0.016 | 1.38×104 | 0.3 | 0.300±0.044 | 6.90×103 | 0.2 | 12.9±0.6 | 0.59±0.05 | – | >10 | – | (Non-substrate) |
| P. aeruginosa* | 3.33±0.71 | 0.040±0.005 | 8.33×104 | 1.7 | 0.300±0.020 | 1.11×104 | 0.4 | 0.43±0.09 | 0.65±0.09 | 0.17±0.03 | 0.62±0.09 | 7.0±0.6 | 15.7±0.7 |
| N. meningitidis* | 5.63±0.48 | 0.110±0.011 | 5.12×104 | 1.1 | 0.080±0.001 | 7.04×104 | 2.5 | 8.46±0.06 | 0.18±0.02 | 8.07±0.11 | 0.24±0.01 | 4.3±1.4 | 2.7±0.40 |
| C. jejuni* | 9.35±0.58 | 0.090±0.002 | 1.04×105 | 2.1 | 0.021±0.020 | 4.46×105 | 16 | 7.28±0.15 | 0.53±0.04 | 0.38±0.04 | 0.52±0.06 | – | >20 |
| B. fragilis | 10.3±0.2 | 0.033±0.001 | 3.42×105 | 7.0 | 0.113±0.080 | 9.08×104 | 3.2 | 14.2±0.3 | 0.29±0.02 | – | >10 | – | (Non-substrate) |
| S. maltophilia | 10.7±0.4 | 0.194±0.018 | 5.61×104 | 1.1 | 0.251±0.027 | 4.25×104 | 1.5 | 37.6±4.4 | 0.19±0.02 | – | >10 | – | (Non-substrate) |
| C. albicans | 17.7±0.1 | 0.691±0.070 | 6.09×104 | 1.2 | 0.051±0.090 | 3.46×105 | 12 | 7.89±1.97 | 1.5±0.5 | – | >10 | – | (Non-substrate) |
LMEE, l-methionine ethyl ester; LMME, l-methionine methyl ester; NAM, N-acetyl-l-methionine.
*Eco, Cje, Nme and Pae values are calculated from previously published data [20].
†Error values are all standard error of the mean, from triplicate determinations.
These MAT/metK genes, some PCR-amplified from genomic DNA and others obtained by synthesis, were each cloned into a pUC57 plasmid vector and driven by a PBAD promoter for expression (Fig. 1b). Thus the nine genes have the same vector (so presumably the same copy number), and the same expression sequences which do not include the native regulatory elements even in the case of E. coli metK. Each of these plasmids was transformed into an E. coli strain (MOB1490), in which the chromosomal metK gene is disabled. In every tested organism, metK deletion is lethal; however, some intracellular parasites produce transport proteins that allow them to take up AdoMet produced by their hosts [36, 37]. Strain MOB1490 carries the gene for a rickettsial AdoMet transporter, and can grow essentially normally without metK complementation if sufficient exogenous AdoMet is provided [22].
Growth of E. coli MOB1490 with heterologous metK genes
Each of the nine complementation plasmids rescued the growth of the metK knock-out E. coli strain in rich medium (LB broth) in the absence of exogenous AdoMet, demonstrating that these heterologous genes all produce functional MAT enzymes in E. coli (Fig. 2a). The group that developed strain MOB1490 had used it to show that the metK gene from Rickettsia prowazekii also complements in LB medium [22]. LB is a complex, undefined medium that does not contain AdoMet, but has amino acids, peptides, sugars, vitamins and a variety of other components [38, 39]. The log-phase growth rates were similar, with ~1 h doubling times for each of the E. coli strains complemented with bacterial orthologues. For comparison, this is the same growth rate observed when the uncomplemented strain MOB1490 is provided with sufficient exogenous AdoMet in LB medium (Fig. 2 of [23]). For the strains complemented with MAT/metK orthologues from the eukaryotes C. albicans and H. sapiens, there was a longer lag time compared to the bacterial metK-complemented strains. However, once that lag time ended, the log-phase growth rate was only slightly lower for the eukaryotic-complemented strains than those observed with the bacterial MetK orthologues. Similarly, the maximum cell densities achieved were similar for each of these complemented strains, except for the human MAT1-complemented strain, which ceased growing at a final cell density roughly 20 % lower than that of the other strains (Fig. 2a). So, when grown in LB medium, this E. coli strain can be rescued with a fairly wide range of MAT/MetK orthologues possessing quite different kinetic properties.
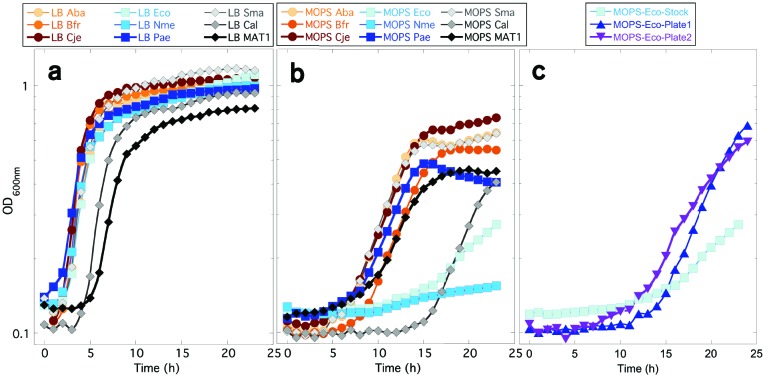
Fig. 2.
Ability of various MAT/metK genes to complement the AdoMet dependence of ∆metK E. coli. (a) Growth curves in LB broth. The nine strains are all in the same background (E. coli MOB1490), and all with a pUC57-derived plasmid carrying the sequence shown in Fig. 1(b) and a gene for each of the nine AdoMet synthetases shown in Fig. 1(a). (b) Growth curves as in (a), but in MOPS minimal glucose medium. (c) Growth curves as in (b), but showing only the EcoMetK strain, where the overnight culture was inoculated with a colony from an LB agar plate (‘stock’) or from a minimal glucose plate (‘plate’).
Because some of the compounds we seek to study are l-Met analogues [23], it was necessary also to examine cell growth in defined minimal media to avoid background chemical contamination. When grown in MOPS glucose minimal medium [40], the various complemented strains behaved in more diverse manners (Fig. 2b). Under these conditions the growth curve for the human MAT1-complemented strain was now found to be similar to those of most of the other strains. Each of these complemented strains required at least 5 h before growth commenced. However, a lag nearly three times that long was shown by the strain containing CalMAT, and the strain with NmeMetK never appeared to initiate exponential growth (Fig. 2b). Surprisingly, the strain containing EcoMetK, which is the native orthologue, showed a roughly 12 h lag. Furthermore, once the EcoMetK-complemented strain began growth, the doubling time was about fourfold longer than most of the other strains (10 h vs ~2.5 h for all others except the NmeMetK strain). For comparison, when the uncomplemented strain MOB1490 is provided with 8 µM exogenous AdoMet in MOPS glucose medium, the doubling time is just under 4 h (Fig. 2 of [23]).
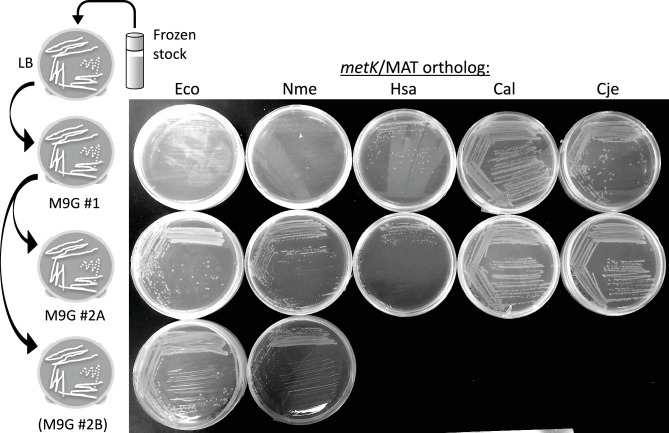
We observed that the lag, and slow growth, associated with the EcoMetK strain were only seen when colonies taken from LB agar plates were used to inoculate the MOPS-glucose overnight cultures, before our growth experiments. This lag likely reflects selection of some mutation in the host E. coli strain. If the strain is first plated onto glucose minimal agar, and then those colonies are used to inoculate overnight cultures, growth was substantially improved (Fig. 2c). Specifically, when colonies are re-streaked from LB agar onto minimal glucose agar, the initial plate shows poor growth for the two poorly growing strains in Fig. 2(b) (EcoMetK and NmeMetK, top row of Fig. 3), while the other strains grow well. However, when colonies were re-streaked from the initial minimal glucose agar onto new glucose plates, growth was robust for those strains (bottom rows of Fig. 3).
Fig. 3.
Growth of various complemented strains on minimal glucose agar plates. The experiment is shown schematically at the left, with cells taken from a frozen stock (LB broth +20 % glycerol, stored at −80 °C), streaked onto LB agar (not shown in the photograph) and then isolated colonies re-streaked onto minimal glucose agar (top row). Colonies from these glucose plates were then re-streaked onto fresh minimal glucose plates (bottom two rows). All plates were incubated for 24 h at 37 °C and then refrigerated until photographed.
Effect of induction and supplementation on the growth of metK-complemented strains
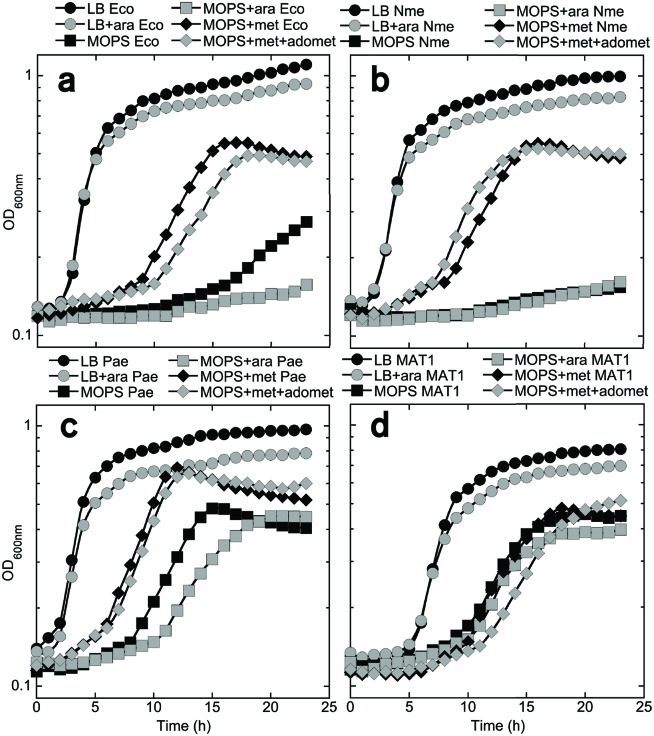
While each of the MAT/MetK orthologues supported E. coli growth in glucose minimal medium to some extent, to be able to test the poorly growing strains with methionine analogues it was necessary to understand why these strains had orthologue-specific growth defects in minimal medium. Accordingly, we next explored the unexpectedly poor growth of the EcoMetK and NmeMetK strains in the glucose minimal medium, and examined the PaeMetK and human MAT1 strains in parallel for comparison. First, to determine whether cell growth is limited by the expression levels of the different MAT orthologues, despite their genes being carried on multi-copy plasmids, the cells were regrown in the presence of arabinose. This compound induces MAT/metK gene expression, as the cloned genes are under control of the PBAD promoter (Fig. 1b). When grown in LB medium (Fig. 4, circles), arabinose slightly reduced growth in all four of the metK-complemented strains, suggesting possible mild toxicity due to higher-level expression. In unsupplemented MOPS glucose medium (Fig. 4, squares), the growth defect for EcoMetK and NmeMetK is again evident, and arabinose induction either had no effect (NmeMetK; panel B) or reduced growth (EcoMetK; panel A). This suggests that the growth defect with these strains is not due to underexpression of MetK. For those strains without growth defects (panels C and D), arabinose has either no effect (HuMAT1; panel D) or, as in LB, a slight negative effect on growth (PaeMetK; panel C).
Fig. 4.
Effects of various supplements on the growth of complemented E. coli strains. For growth of E. coli strain MOB1490, carrying different metK/MAT genes, was monitored in rich LB medium ±2 % arabinose (circles), MOPS minimal glucose medium ±arabinose (squares) or MOPS medium supplemented with l-Met ±AdoMet (diamonds). Arabinose addition tests the effects of elevated expression of the cloned MAT/metK gene, which is under the control of PBAD. 32 mM AdoMet tests for insufficient activity of the cloned MAT/metK gene, while 100 mM l-Met tests for excessive activity of those genes (explained in Fig. 4) (a) E. coli-complemented (EcoMetK), (b) N. meningitidis-complemented (NmeMetK), (c) P. aeruginosa-complemented (PaeMetK) and (d) human MAT1-complemented MOB1490 strains.
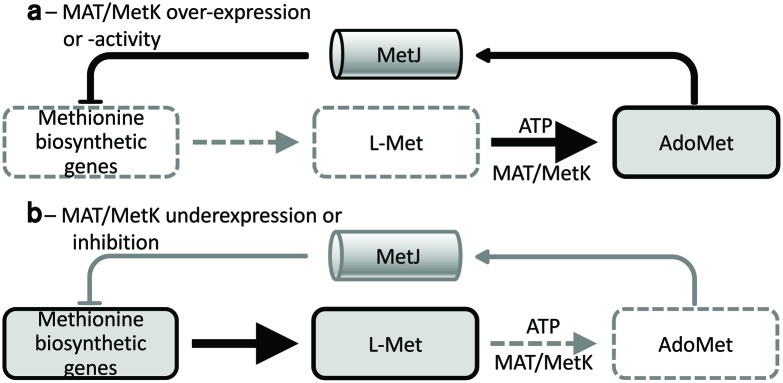
Next, we tested whether the poor growth of the EcoMetK and NmeMetK strains in glucose minimal medium seen in Fig. 2(b) could be reversed by the addition of l-Met with or without exogenous AdoMet. In these experiments the l-Met was included because AdoMet is a co-repressor for MetJ (Fig. 5), which represses methionine biosynthesis [41]. Under normal circumstances the cell would never contain AdoMet in the absence of l-Met, but strain MOB1490 in minimal medium supplemented with AdoMet results in starvation for l-Met [42]. Unlike our minimal medium conditions, LB medium contains ~6 mM l-Met [39]. As shown in Fig. 4 (panels A and B, diamonds), the growth defect with the EcoMetK- and NmeMetK-complemented strains was reversed by the addition of 0.1 mM l-Met, whether or not AdoMet was also added. l-Met also had lesser stimulatory effects on the PaeMetK strain (panel C), and had no effect on growth of the MAT1 strain (panel D). These results suggest that the observed growth defects with some strains are due to starvation of l-Met. One possible explanation is that the EcoMetK and NmeMetK enzymes might accumulate to higher levels, and/or have higher activity, than the other enzyme forms (at least in the E. coli environment). This could lead to over-accumulation of AdoMet and strong MetJ-dependent repression of methionine biosynthetic genes (Fig. 5a). If this model is correct, then partial inhibitors of EcoMetK or NmeMetK would actually promote growth of those strains in minimal glucose medium, and could be screened for in that manner. Alternatively, putting the EcoMetK and NmeMetK genes onto lower-copy plasmid vectors might also eliminate the growth limitation.
Fig. 5.
Effects of altered MAT/MetK activity. (a) Excess activity leads to high AdoMet levels, which in E. coli repress methionine biosynthesis via MetJ. In minimal medium, this would result in starvation for l-Met. (b) Insufficient activity leads to AdoMet levels too low to support growth, despite active methionine biosynthesis.
Discussion
We found that a wide range of MAT/metK genes, with as little as 42 % identity to the native E. coli metK, were capable of complementing a ∆metK defect in E. coli. This extends the work of the group that originally developed strain MOB1490 [22], who had demonstrated complementation by metK genes from Rickettsia species (66 % identical to the E. coli orthologue). The ability of the human MAT1a gene to function in E. coli is particularly significant, as it will allow counter-screening by growth for compounds that are inhibitors for bacterial but not human MAT/MetK enzymes. We have not yet tested the human MAT II, in part because – unlike MAT I/III – it is a heterodimer with a catalytic and a regulatory subunit [34]. However we have included its sequence in Fig. 1(a), which reveals that 17 % of residues (66/395; yellow and cyan shading) are distinct between the set of bacterial MetKs and the two human genes for MAT catalytic subunits. High-resolution structures of MetKs have previously been determined for the enzyme from E. coli [43] and the human MAT1A [44], as well as our structure for the C. jejuni MetK [20]. The campylobacterial orthologue is a dimer in solution, unlike the tetrameric structures of the other family members. A comparison of the E. coli and C. jejuni enzyme structures [20] has identified critical differences that support the possibility that inhibitors specific for bacterial MetKs can be found. We note that growth-based screening using E. coli dependent on HsaMAT1 might reveal small molecules stimulatory for that enzyme, which could ultimately be useful in treating some forms of human MATI/III deficiency, known as Mudd’s Disease [45, 46].
These results support the use of a library of E. coli MOB1490 strains, each carrying a distinct MetK/MAT orthologue, in growth-based screening for orthologue-specific inhibitors (or, in the case of the human orthologue, stimulatory molecules). The fact that such a wide range of orthologues function well enough in E. coli to support growth indicates that neither post-translational modifications nor protein–protein interactions are key to the proper functioning of any of these enzymes – at least to the level of providing sufficient levels of AdoMet to support growth. This is despite the existence of evidence in support of each phenomenon – for example, EcoMetK activity is modulated by lysine acetylation [47] while the HsaMAT1 enzyme interacts in human cells with the oncogene product PDRG1 [48]. This functional flexibility of AdoMet synthetases is striking, but consistent with other studies in which a range of orthologues are used to complement a specific defect (e.g. [49, 50]).
The results of these complementation studies clearly indicate that a wide variety of MAT/MetK orthologues can function sufficiently well to support growth of E. coli, even in the more metabolically demanding conditions imposed by minimal medium.
Funding information
This work was supported by an NIH grant (AI098702) to REV and RMB.
Acknowledgements
The authors thank Dr David Wood (Univ. of S. Alabama) for strain MOB1490, and Drs Jyl Matson and Akira Takashima (Univ. of Toledo) for use of plate readers.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This work includes no research involving human or animal subjects.
Footnotes
Abbreviations: Aba, Acinetobacter baumannii; AdoMet, S-adenosyl-L-methionine; AHL, acylhomoserine lactone; Bfr, Bacteroides fragilis; Cal, Candida albicans; Cje, Campylobacter jejuni; Eco, Escherichia coli; Hsa, Homo sapiens; L-Met, L-methionine; MAT, methionine adenosyltransferase; MetK/metK, protein/gene name for methionine adenosyltransferase in most bacteria; Nme, Neisseria meningitidis; Pae, Pseudomonas aeruginosa; Sma, Stenotrophomonas maltophilia; XxxMAT, the MAT1A ortholog from a given eukaryote; XxxMetK, the MetK ortholog from a given bacterium.
Edited by: C. Dahl and I. Martin-Verstraete
References
- 1.Cheng X, Blumenthal RM. S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. Singapore: World Scientific; 1999. (editors) [Google Scholar]
- 2.Fontecave M, Atta M, Mulliez E. S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Taylor JC, Bock CW, Takusagawa F, Markham GD. Discovery of novel types of inhibitors of S-adenosylmethionine synthesis by virtual screening. J Med Chem. 2009;52:5967–5973. doi: 10.1021/jm9006142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pajares MA, Markham GD. Methionine adenosyltransferase (S-adenosylmethionine synthetase) Adv Enzymol Relat Areas Mol Biol. 2011;78:449–521. doi: 10.1002/9781118105771.ch11. [DOI] [PubMed] [Google Scholar]
- 5.Churchill ME, Chen L. Structural basis of acyl-homoserine lactone-dependent signaling. Chem Rev. 2011;111:68–85. doi: 10.1021/cr1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galloway WR, Hodgkinson JT, Bowden SD, Welch M, Spring DR. Quorum sensing in gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem Rev. 2011;111:28–67. doi: 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]
- 7.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Defoirdt T, Boon N, Bossier P. Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 2010;6:e1000989. doi: 10.1371/journal.ppat.1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalia VC. Quorum sensing inhibitors: an overview. Biotechnol Adv. 2013;31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Hodgkinson JT, Galloway WR, Wright M, Mati IK, Nicholson RL, et al. Design, synthesis and biological evaluation of non-natural modulators of quorum sensing in Pseudomonas aeruginosa. Org Biomol Chem. 2012;10:6032–6044. doi: 10.1039/c2ob25198a. [DOI] [PubMed] [Google Scholar]
- 11.Bjarnsholt T, van Gennip M, Jakobsen TH, Christensen LD, Jensen PØ, et al. In vitro screens for quorum sensing inhibitors and in vivo confirmation of their effect. Nat Protoc. 2010;5:282–293. doi: 10.1038/nprot.2009.205. [DOI] [PubMed] [Google Scholar]
- 12.Christensen QH, Grove TL, Booker SJ, Greenberg EP. A high-throughput screen for quorum-sensing inhibitors that target acyl-homoserine lactone synthases. Proc Natl Acad Sci USA. 2013;110:13815–13820. doi: 10.1073/pnas.1313098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung J, Goo E, Yu S, Choi O, Lee J, et al. Small-molecule inhibitor binding to an N-acyl-homoserine lactone synthase. Proc Natl Acad Sci USA. 2011;108:12089–12094. doi: 10.1073/pnas.1103165108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, et al. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci USA. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng WL, Perez L, Cong J, Semmelhack MF, Bassler BL. Broad spectrum pro-quorum-sensing molecules as inhibitors of virulence in vibrios. PLoS Pathog. 2012;8:e1002767. doi: 10.1371/journal.ppat.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Annapoorani A, Umamageswaran V, Parameswari R, Pandian SK, Ravi AV. Computational discovery of putative quorum sensing inhibitors against LasR and RhlR receptor proteins of Pseudomonas aeruginosa. J Comput Aided Mol Des. 2012;26:1067–1077. doi: 10.1007/s10822-012-9599-1. [DOI] [PubMed] [Google Scholar]
- 17.Ahumedo M, Drosos JC, Vivas-Reyes R. Application of molecular docking and ONIOM methods for the description of interactions between anti-quorum sensing active (AHL) analogues and the Pseudomonas aeruginosa LasR binding site. Mol Biosyst. 2014;10:1162–1171. doi: 10.1039/c3mb70181f. [DOI] [PubMed] [Google Scholar]
- 18.Kai K, Fujii H, Ikenaka R, Akagawa M, Hayashi H. An acyl-SAM analog as an affinity ligand for identifying quorum sensing signal synthases. Chem Commun. 2014;50:8586–8589. doi: 10.1039/C4CC03094J. [DOI] [PubMed] [Google Scholar]
- 19.Raychaudhuri A, Jerga A, Tipton PA. Chemical mechanism and substrate specificity of RhlI, an acylhomoserine lactone synthase from Pseudomonas aeruginosa. Biochemistry. 2005;44:2974–2981. doi: 10.1021/bi048005m. [DOI] [PubMed] [Google Scholar]
- 20.Zano SP, Bhansali P, Luniwal A, Viola RE. Alternative substrates selective for S-adenosylmethionine synthetases from pathogenic bacteria. Arch Biochem Biophys. 2013;536:64–71. doi: 10.1016/j.abb.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wijayasinghe YS, Blumenthal RM, Viola RE. Producing proficient methyl donors from alternative substrates of S-adenosylmethionine synthetase. Biochemistry. 2014;53:1521–1526. doi: 10.1021/bi401556p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Driskell LO, Tucker AM, Winkler HH, Wood DO. Rickettsial metK-encoded methionine adenosyltransferase expression in an Escherichia coli metK deletion strain. J Bacteriol. 2005;187:5719–5722. doi: 10.1128/JB.187.16.5719-5722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao M, Wijayasinghe YS, Bhansali P, Viola RE, Blumenthal RM. A surprising range of modified-methionyl S-adenosylmethionine analogues support bacterial growth. Microbiology. 2015;161:674–682. doi: 10.1099/mic.0.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 25.Pardee AB, Jacob F, Monod J. The genetic control and cytoplasmic expression of “Inducibility” in the synthesis of β-galactosidase by E. coli. J Mol Biol. 1959;1:165–178. doi: 10.1016/S0022-2836(59)80045-0. [DOI] [Google Scholar]
- 26.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents. 2013;41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest. 2014;124:4166–4172. doi: 10.1172/JCI72334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stahl M, Butcher J, Stintzi A. Nutrient acquisition and metabolism by Campylobacter jejuni. Front Cell Infect Microbiol. 2012;2:5. doi: 10.3389/fcimb.2012.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robins-Browne RM, Holt KE, Ingle DJ, Hocking DM, Yang J, et al. Are Escherichia coli pathotypes still relevant in the era of whole-genome sequencing? Front Cell Infect Microbiol. 2016;6:141. doi: 10.3389/fcimb.2016.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens DS, Greenwood B, Brandtzaeg P. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet. 2007;369:2196–2210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- 31.Lorenz A, Pawar V, Häussler S, Weiss S. Insights into host-pathogen interactions from state-of-the-art animal models of respiratory Pseudomonas aeruginosa infections. FEBS Lett. 2016;590:3941–3959. doi: 10.1002/1873-3468.12454. [DOI] [PubMed] [Google Scholar]
- 32.Chang YT, Lin CY, Chen YH, Hsueh PR. Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front Microbiol. 2015;6:893. doi: 10.3389/fmicb.2015.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sellam A, Whiteway M. Recent advances on Candida albicans biology and virulence. F1000Res. 2016;5:2582. doi: 10.12688/f1000research.9617.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shafqat N, Muniz JR, Pilka ES, Papagrigoriou E, von Delft F, et al. Insight into S-adenosylmethionine biosynthesis from the crystal structures of the human methionine adenosyltransferase catalytic and regulatory subunits. Biochem J. 2013;452:27–36. doi: 10.1042/BJ20121580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenthal R, Danson MJ, Hough DW. Catalytic efficiency and kcat/KM: a useful comparator? Trends Biotechnol. 2007;25:247–249. doi: 10.1016/j.tibtech.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Leal O, Moncada C, Clarkson AB, Merali S. Pneumocystis S-adenosylmethionine transport: a potential drug target. Am J Respir Cell Mol Biol. 2011;45:1142–1146. doi: 10.1165/rcmb.2011-0009OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tucker AM, Winkler HH, Driskell LO, Wood DO. S-adenosylmethionine transport in Rickettsia prowazekii. J Bacteriol. 2003;185:3031–3035. doi: 10.1128/JB.185.10.3031-3035.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baev MV, Baev D, Radek AJ, Campbell JW. Growth of Escherichia coli MG1655 on LB medium: determining metabolic strategy with transcriptional microarrays. Appl Microbiol Biotechnol. 2006;71:323–328. doi: 10.1007/s00253-006-0392-8. [DOI] [PubMed] [Google Scholar]
- 39.Sezonov G, Joseleau-Petit D, D'Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shoeman R, Coleman T, Redfield B, Greene RC, Smith AA, et al. Regulation of methionine synthesis in Escherichia coli: effect of metJ gene product and S-adenosylmethionine on the in vitro expression of the metB, metL and metJ genes. Biochem Biophys Res Commun. 1985;133:731–739. doi: 10.1016/0006-291X(85)90965-9. [DOI] [PubMed] [Google Scholar]
- 42.El-Hajj ZW, Reyes-Lamothe R, Newman EB. Cell division, one-carbon metabolism and methionine synthesis in a metK-deficient Escherichia coli mutant, and a role for MmuM. Microbiology. 2013;159:2036–2048. doi: 10.1099/mic.0.069682-0. [DOI] [PubMed] [Google Scholar]
- 43.Komoto J, Yamada T, Takata Y, Markham GD, Takusagawa F. Crystal structure of the S-adenosylmethionine synthetase ternary complex: a novel catalytic mechanism of S-adenosylmethionine synthesis from ATP and Met. Biochemistry. 2004;43:1821–1831. doi: 10.1021/bi035611t. [DOI] [PubMed] [Google Scholar]
- 44.Murray B, Antonyuk SV, Marina A, Lu SC, Mato JM, et al. Crystallography captures catalytic steps in human methionine adenosyltransferase enzymes. Proc Natl Acad Sci USA. 2016;113:2104–2109. doi: 10.1073/pnas.1510959113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barić I, Staufner C, Augoustides-Savvopoulou P, Chien YH, Dobbelaere D, et al. Consensus recommendations for the diagnosis, treatment and follow-up of inherited methylation disorders. J Inherit Metab Dis. 2017;40:5–20. doi: 10.1007/s10545-016-9972-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chien YH, Abdenur JE, Baronio F, Bannick AA, Corrales F, et al. Mudd's disease (MAT I/III deficiency): a survey of data for MAT1A homozygotes and compound heterozygotes. Orphanet J Rare Dis. 2015;10:99. doi: 10.1186/s13023-015-0321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun M, Guo H, Lu G, Gu J, Wang X, et al. Lysine acetylation regulates the activity of Escherichia coli S-adenosylmethionine synthase. Acta Biochim Biophys Sin. 2016;48:723–731. doi: 10.1093/abbs/gmw066. [DOI] [PubMed] [Google Scholar]
- 48.Pérez C, Pérez-Zúñiga FJ, Garrido F, Reytor E, Portillo F, et al. The oncogene PDRG1 Is an interaction target of methionine adenosyltransferases. PLoS One. 2016;11:e0161672. doi: 10.1371/journal.pone.0161672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kachroo AH, Laurent JM, Yellman CM, Meyer AG, Wilke CO, et al. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science. 2015;348:921–925. doi: 10.1126/science.aaa0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein JA, Dave BM, Raphenya AR, Mcarthur AG, Knodler LA. Functional relatedness in the Inv/Mxi-Spa type III secretion system family. Mol Microbiol. 2017;103:973–991. doi: 10.1111/mmi.13602. [DOI] [PubMed] [Google Scholar]
- 51.NCBI RC Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013;41:D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]