Abstract
The Staphylococcus aureus type VII protein secretion system (T7SS) plays important roles in virulence and intra-species competition. Here we show that the T7SS in strain RN6390 is activated by supplementing the growth medium with haemoglobin, and its cofactor haemin (haem B). Transcript analysis and secretion assays suggest that activation by haemin occurs at a transcriptional and a post-translational level. Loss of T7 secretion activity by deletion of essC results in upregulation of genes required for iron acquisition. Taken together these findings suggest that the T7SS plays a role in iron homeostasis in at least some S. aureus strains.
Keywords: Staphylococcus aureus, protein secretion, iron homeostasis, RNA-sequencing
Introduction
Bacteria produce a number of different secretion machineries to transport proteins across their cell envelopes [1]. Secreted proteins play essential roles in environmental adaptation and in pathogenic bacteria are frequently linked with the ability to cause disease. The type VII protein secretion system (T7SS) was discovered almost 15 years ago in pathogenic Mycobacteria. This system, also termed ESX-1, was shown to secrete two small proteinaceous T-cell antigens and to be essential for virulence [2–4].
Mycobacteria produce up to five different T7SSs [5, 6]. In addition to ESX-1, ESX-5 also plays a key role in host interaction during pathogenesis [7]. Of the other ESX systems in Mycobacteria, ESX-3 is the best studied and is critical for siderophore-mediated acquisition of iron [8–10]. Consistent with their diverse roles in the physiology and virulence of Mycobacteria, the ESX systems are differentially regulated. For example, expression of ESX-1 is under indirect transcriptional control of the PhoPR two-component system [11] that appears to respond to low pH, conditions that are found in phagolysosomes [12]. ESX-5 expression is induced in response to phosphate starvation [13] whilst ESX-3 expression is de-repressed when cells are starved of iron or zinc [14, 15].
T7SSs are also found in other bacteria, in particular from the Gram-positive low G+C phylum Firmicutes [16]. Similarity between the T7SS found in Firmicutes and the well-studied mycobacterial ESX T7SSs is limited, with the systems sharing only two common types of components. This has resulted in the T7SS of Firmicutes such as Staphylococcus aureus being designated Ess or T7b to distinguish them from the better-characterized mycobacterial T7a systems [17]. One of the components shared between the two systems is a membrane-bound ATPase of the FtsK/SpoIIIE family termed EccC (T7a) or EssC (T7b). This component forms a hexameric assembly that probably acts as the motor protein and potentially also the translocation channel of the T7SS [18, 19]. The second common component is at least one small protein of the WXG100 family, EsxA, which is secreted by the T7SS. In Mycobacteria, EsxA homologues are secreted as heterodimers with EsxB partner proteins (e.g. [20, 21]) whereas in Firmicutes EsxA is secreted as a homodimer [22].
The T7SS of S. aureus is encoded by the ess locus. In addition to EsxA and EssC, four further proteins encoded by the locus – EsaA, EssA, EssB and EsaB – are essential components of the secretion machinery [23–25]. The ess locus is under complex transcriptional control by the alternative sigma factor σB and expression is also repressed by the two-component SaeSR system [26, 27]. Experiments using mouse models of infection have indicated that the Ess system is required for virulence, in particular for the persistence of abscesses in the liver and kidney [23–25, 28]. It is also required for colonization and for intraspecies competition [25, 29]. The secretion system appears to be highly expressed in mammalian hosts [30], and in at least one strain is transcriptionally activated by pulmonary surfactant [31]. However, in laboratory growth media, although the secretion system components are produced, the machinery is poorly active and the levels of secreted substrates are relatively low [25, 29, 32].
In this study we have attempted to identify factors that activate secretion by the T7SS in vitro. We show that addition of haemin (haem B) enhances T7 secretion in at least two different S. aureus strains. Moreover, we also show that in the absence of a functional T7SS, the laboratory strain RN6390 upregulates numerous genes involved in iron acquisition. Together our findings point to a potential role of the T7SS in S. aureus iron homeostasis.
Methods
Bacterial strains and growth conditions for secretion assays
The S. aureus strains used in this study are listed in Table 1. S. aureus strains were grown overnight at 37 °C with shaking in tryptic soy broth (TSB). To test S. aureus growth with various media additives, strains were subcultured in either TSB or RPMI (Sigma) as indicated, supplemented with the corresponding additives. Additives were made fresh and sterilized by filtration, and were dissolved in distilled water except for the haemin and other protoporhyrins, which were dissolved in DMSO, and haemoglobins, which were dissolved in 0.1 M NaOH. For secretion assays, the indicated strains were grown to an OD600 of 2 and fractionated to give whole cell lysates and supernatant fractions as described previously [25]. Chelation of divalent cations from TSB was achieved after a 30 min treatment with 5 % Chelex-100 (BioRad). Chelex-treated TSB was then supplemented with 25 µM ZnCl2, 25 µM MnCl2, 1 mM MgCl2 or 100 µM CaCl2 [33].
Table 1. S. aureus strains used in this study.
MRSA, methicillin-resistant S. aureus
| Strain | Relevant genotype or description | Source or reference |
|---|---|---|
| RN6390 | NCTC8325 derivative, rbsU, tcaR, cured of φ11, φ12, φ13 | [54] |
| COL | Healthcare acquired MRSA (HA-MRSA) | [55, 56] |
| USA300 | Community-acquired MRSA (CA-MRSA) | [57] |
| 10.1252.X | ST398-like isolate. Livestock-associated | Roslin Institute, Edinburgh, UK |
| MRSA252 | HA-MRSA, representative of Epidemic MRSA-16 | [58] |
| HO 5096 0412 | HA-MRSA, representative of Epidemic MRSA-15 | [59] |
Preparation of a polyclonal EsxB antibody
The EsxB (UniProt accession Q99WT7) coding sequence was PCR-amplified from a synthetic gene (codon optimized for Escherichia coli K12; Genscript) using the forward primer 5′-GCGCGTCGACAATGGGCGGCTATAAAGG C-3′ and the reverse primer 5′-GCGCCTCGAGTTACGGGTTCACGCGATCCAGGC-3′, and cloned into the SalI/XhoI site of a modified pET27b vector (Novagen). The plasmid produces an N-terminal His6-tagged protein with a TEV (tobacco etch virus) protease cleavage site. The protein was expressed and purified as described previously [34], except that in the final size exclusion chromatography step an HR 30/100 GL Superdex75 column (column volume=24 ml; GE Healthcare) equilibrated with 20 mM Tris pH 7.8 and 100 mM NaCl was used. Two milligrams of purified EsxB (retaining a Gly–Ala–Ser–Thr sequence at the N terminus after the cleavage step) was utilized as antigen to immunize rabbits for polyclonal antibody production in a standard three-injection protocol (Seqlab).
Western blotting and protein quantification
Samples were mixed with LDS loading buffer (NuPAGE LDS sample buffer) and boiled for 10 min prior to separation on bis-Tris gels. Western blotting was performed according to standard protocols with the following antibody dilutions: α-EsxA [25] 1 : 2500, α-EsxB 1 : 1000, α-EsxC [25] 1 : 2000, α-TrxA [35] 1 : 25 000. Horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) were used as per the manufacturer’s instructions. For protein quantification, western blots of at least three biological replicates were developed with chemiluminescence (Clarity Western ECL Blotting Substrate; BioRad) and light-emitting bands were visualized with a CCCD camera (GeneGNOME XRQ; Syngene). Densitometry analysis was performed using ImageJ [36].
RNA isolation and qPCR
For the RNA-seq analysis, three biological repeats of the indicated S. aureus strains were grown aerobically in TSB up to an OD600 of 1 at which point mRNA was prepared (in three technical replicates). Total mRNA was extracted, reverse transcribed and sequenced as described previously [37]. The sequence reads from each individual dataset were mapped to the S. aureus NCTC 8325 genome using EDGE-pro [38], and quantification of transcript abundance and calculation of differential gene expression were performed using DEseq2 [39]. DEseq2 uses the Negative Binomial distribution as a model to compute P-values, and we regarded P>0.05 as the probability of observing a transcript's expression levels in different conditions by chance. Reads were aligned using the Tophat aligner [40] and to acquire a single transcriptome for each strain, the three assemblies produced by cufflinks were merged and the abundances of each sample were assembled using cuffquant. Differential expression was analysed using edgeR [41]. Genes were considered to be differentially expressed when log fold change was >2 or <−2 and the q value <0.05. The RNA-seq data from this study were submitted to the European Nucleotide Archive with accession number ERP009279 and in Array express under accession number E-ERAD-362.
To isolate mRNA for RT-PCR, three biological repeats of the indicated S. aureus strains were grown aerobically in TSB in the presence or absence of 1 µM haemin up to an OD600 of 1, at which point mRNA was prepared. Total mRNA was extracted using the SV Total RNA Isolation Kit (Promega) with modifications as described by Kneuper et al. [25]. Briefly, cell samples were stabilized in 5 % phenol/95 % ethanol on ice for at least 30 min and then centrifuged at 2770 g for 10 min. Cells were then resuspended in 100 µl of TE buffer containing 500 µg lysostaphin ml−1 and 50 µg lysozyme ml−1 and incubated at 37 °C for 30 min. Subsequently, the manufacturer’s instructions were followed and the isolated RNA was subjected to a second DNase treatment using a DNA-free kit (Ambion). RNA was stored at −80 °C until use. To determine transcript levels, 500 ng of cDNA template was used with the following primer pairs: esxA (5′-TGGCAATGATTAAGATGAGTCC-3′ and 5′-TCTTGTTCTTGAACGGCATC-3′ [25]), esxC (5′-AAGCATGCTGAAGAGATTGC-3′ and 5′-TCTTCACCCAACATTTCAAGC-3′) and 16S rRNA (5′-GTGCACATCTT GACGGTACCTA-3′ and 5′-CCACTGGTGTTCCTCCATATC-3′ [25]). Quantitative PCR was performed using a thermal cycler. Three technical replicates were prepared for each culture condition, using 2* Quantifast SYBR Green PCR master mix (Qiagen) according to the manufacturer’s instructions. Standard curves were generated from serial 10-fold dilutions of genomic DNA. Amplification results were analysed with MxPro QPCR software (Stratagene) to give the levels of mRNA normalized to the level of 16S rRNA amplification in each sample. Results were further analysed in Microsoft Excel to calculate relative expression levels.
Construction of an esxA-yfp transcriptional/translational fusion
The yfp gene was amplified without its start codon using Yfpfuse1 (5′-GGAACTACTAGATCTTCAAAAGGC-3′) and Xfpfuse2 (5′-CAAATAAGAATTCTGAGCGCCGG-3′) and cloned as a BglII/EcoRI fragment into pRMC2 [42], generating pRMC2-yfp. An approximately 500 bp region covering the esxA promoter, ribosome binding site and start codon was amplified using primers esxprom1 (5′-GAATGGTACCGATTGTTGTTAAGATC-3′) and esxprom2 (5′-TTAGATCTTGCCATAACTAGAAACC-3′) with RN6390 chromosomal DNA as template, and cloned as a KpnI/BglII fragment into pRMC2-yfp to give plasmid pPesxA-yfp.
Results and Discussion
T7SS in strain RN6390 is stimulated by supplementation with calcium ions, haemoglobin and haemin
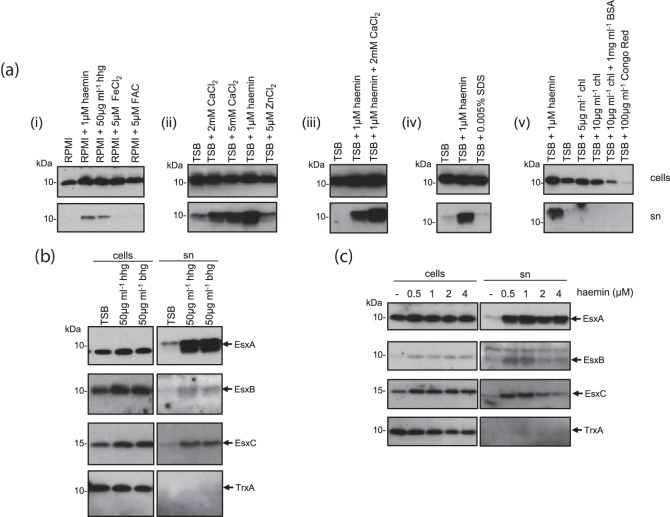
Protein secretion systems are frequently activated in a post-translational manner; for example, type III secretion is activated by addition of the amphipathic dye Congo Red, or by calcium deprivation [43, 44] and the type VI secretion system is activated by protein phosphorylation [45]. We therefore sought to determine whether we could activate secretion by the T7SS in our standard laboratory strain of S. aureus, RN6390, by making empirical additions to the growth media. As shown in Fig. 1(a), panel (i), some secretion of the T7 core component, EsxA, could be detected when the strain was grown in either RPMI or TSB growth media. In general we noted that more EsxA was detected in the supernatant after growth in TSB than in RPMI (Fig. 1a).
Fig. 1.
T7 secretion in strain RN6390 is stimulated by haemoglobin, haemin and millimolar concentrations of CaCl2. RN6390 was subcultured into either RPMI or TSB media, supplemented with the indicated additives, and grown aerobically until an OD600 of 2 was reached. Samples were fractionated to give cells and supernatant (sn), and supernatant proteins were precipitated using TCA. For each gel, 4 µl of a cell culture adjusted to an OD600 of 1 and 12 µl of culture supernatant were loaded. Final concentrations of additives are as indicated. hhg, Human haemoglobin; bhg, bovine haemoglobin; FAC, ferric ammonium citrate; chl, cholesterol; BSA, bovine serum albumin. (a) Western blots were probed with anti-EsxA antisera. (b and c) Western blots were probed with anti-EsxA, anti-EsxB, anti-EsxC or anti-TrxA (cytoplasmic control) antisera.
Haemoglobin and haemin stimulate T7 secretion
Both TSB and RPMI lack an exogenously added iron source, and RPMI is considered to be iron-limited [46]. We therefore first tested the effect of exogenous iron sources on EsxA secretion. It can be seen that haemoglobin had a strikingly positive effect on EsxA levels in the culture supernatant for RN6390 grown both in RPMI and in TSB media (Fig. 1a, panel i; Fig. 1b). Fig. 1(b) confirms that secreted levels of the T7 substrate proteins EsxB and EsxC [23, 24] were also similarly enhanced in the presence of haemoglobin. We tested whether other iron sources could also stimulate T7 secretion. Fig. 1(a) (panel i) shows that neither ferric ammonium citrate (FAC) nor ferrous chloride stimulated EsxA secretion in RPMI, indicating that it was not a general effect of increased iron availability. The mycobacterial ESX-3 T7SS is transcriptionally regulated by both iron and zinc [14, 15]. However, supplementation of the growth medium with 5 µM zinc had no detectable effect on EsxA secretion (Fig. 1a, panel ii).
We next tested whether the iron-containing cofactor component of haemoglobin, haemin (haem B), could also enhance EsxA secretion. Fig. 1(a) shows that supplementation of both RPMI (panel i) and TSB media (Fig. 1a, panels ii–v) with 1 µM haemin resulted in a marked increase in EsxA secretion. We confirmed that haemin had a similar stimulatory effect on secretion of the T7 substrates EsxB and EsxC (Fig. 1c). Fig. 1(c) also shows that EsxA, EsxB and EsxC secretion were enhanced to similar levels in the presence of 0.5 and 1 µM haemin but at higher haemin concentrations secretion was reduced. We conclude that haemoglobin and its cofactor, haemin, can positively regulate T7 secretion.
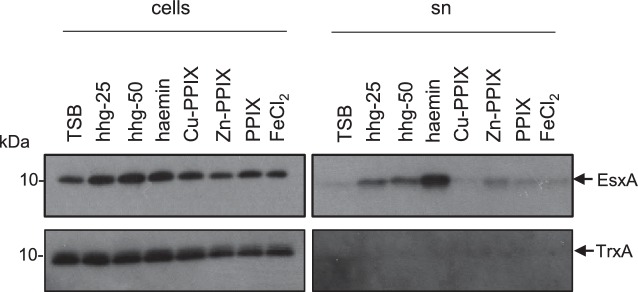
To determine whether the stimulation of T7 secretion was specific to the Fe-loaded form of PPIX, we investigated whether the empty (iron-free) protoporphyrin IX (PPIX) or zinc/copper-loaded PPIX could also stimulate EsxA secretion. Fig. 2 shows that only haemin, the Fe-loaded form of PPIX, enhanced secretion of EsxA.
Fig. 2.
Stimulation of EsxA secretion is specific to the Fe-loaded PPIX. RN6390 was subcultured into TSB medium supplemented with the indicated additives, and grown aerobically until an OD600 of 2 was reached. Samples were fractionated to give cells and supernatant (sn), and supernatant proteins were precipitated using TCA. For each gel, 4 µl of a cell culture adjusted to an OD600 of 1 and 12 µl of culture supernatant were loaded. Human haemoglobin (hhg) was added at a final concentration of either 25 or 50 µg FeCl2 ml−1 at 5 µM and all other supplements at 2 µM. PPIX, protoporphyrin IX. Western blots were probed with anti-EsxA or anti-TrxA (cytoplasmic control) antisera.
Ca2+ also stimulates T7 secretion
Next we tested whether calcium ions could also regulate secretion. Ca2+ is found at millimolar concentrations in mammalian blood and is also highly abundant in pulmonary surfactant [47]. Fig. 1(a) (panel ii) indicates that CaCl2 supplementation of TSB medium, at both 2 and 5 mM, increased the level of EsxA in the supernatant. Inclusion of 2 mM CaCl2 alongside 1 µM haemin appeared to have additive effects over either supplement alone (Fig. 1a, panels ii and iii). SDS, which has been shown to enhance essC mRNA levels [31], did not stimulate EsxA secretion (Fig. 1a, panel iv). Finally, we tested whether either Congo Red or cholesterol, both of which stimulate protein translocation by the type III secretion system [43, 48], could increase EsxA secretion. However, Fig. 1(a) (panel v) indicates that they did not enhance secretion of EsxA and, moreover, Congo Red appeared to downregulate EsxA production.
T7 secretion is not induced by oxidative stress
In addition to acting as an iron source, at high concentrations haemin induces oxidative damage [49, 50]. To determine whether the haemin-induced hypersecretion of EsxA might be an oxidative stress response, we determined the effect of other oxidative stress agents on EsxA secretion. Fig. S1(a) (available with the online version of this article) shows that in the presence of exogenous hydrogen peroxide there was potentially a small increase in EsxA level in the supernatant. However, in the presence of either diamide or methylviologen (paraquat) there was no stimulation of EsxA secretion and indeed the cellular level of EsxA appeared to be lower than the untreated sample (Fig. S1b, c). Co-supplementation of cultures with 1 µM haemin alongside diamide or methylviologen again resulted in haemin-dependent stimulation of EsxA secretion. We conclude that it is unlikely that the haemin induction of EsxA secretion in strain RN6390 is due solely to oxidative stress.
Haemin-induced hyper-secretion of EsxA is strain-dependent
Recent genomic analysis has revealed that there is genetic diversity at the ess locus across S. aureus strains. The ess loci were shown to fall into one of four different groupings, each of which is associated with a specific sequence variant of EssC, and with specific suites of candidate substrate proteins [37]. We therefore undertook experiments to determine whether the haemin-induced stimulation of EsxA secretion was conserved across these groupings. Strain COL is in the same EssC grouping as RN6390 (essC1) – both strains belong to the CC8 clonal complex, but are different sequence types (ST8 and ST250, respectively). COL has been noted previously to have a higher level of in vitro T7SS activity than RN6390 [25, 29]. Fig. 3 shows that EsxA secretion by COL is indeed higher than that of RN6390, and is comparable to the levels seen when RN6390 is grown with 1 µM haemin. Interestingly, haemin addition to cultures of COL grown in TSB had a negligible effect on the level of EsxA secretion.
Fig. 3.
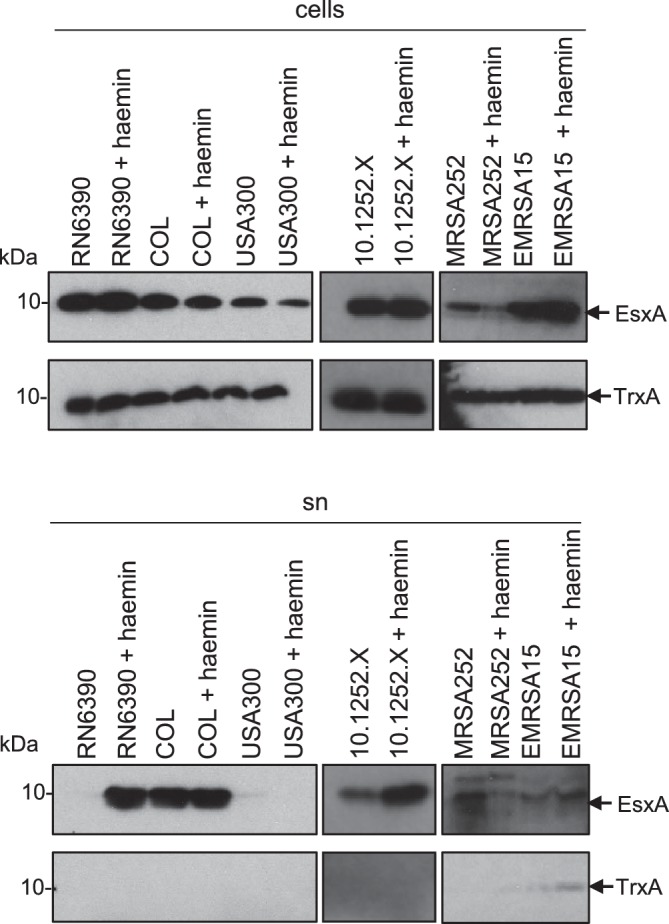
Haemin-induced stimulation of EsxA secretion in S. aureus is strain-specific. The indicated S. aureus strains were subcultured into TSB medium, or TSB medium supplemented with 4 µM haemin, as indicated, and grown aerobically until an OD600 of 2 was reached. Samples were fractionated to give cells and supernatant (sn), and supernatant proteins were precipitated using TCA. For each gel, 4 µl of a cell culture adjusted to an OD600 of 1 and 12 µl of culture supernatant were loaded. Western blots were probed with anti-EsxA or anti-TrxA (cytoplasmic control) antisera.
We next examined the effect of haemin supplementation on an essC2-variant strain, S. aureus 10.1252.X. It can be seen (Fig. 3) that 1 µM haemin also had a positive effect on EsxA secretion in this strain. By contrast, when strain MRSA252 (an essC3 variant) was cultured with haemin, secretion of EsxA was reduced (and there also seemed to be less EsxA associated with the cellular fraction), suggesting a potential repression of ess expression in this strain (Fig. 3). Finally when we examined the essC4 strain variant, EMRSA15, there appeared to be a slight increase of EsxA levels in the supernatant in the presence of haemin, although we noted that there was some cell lysis in this strain as low levels of the cytoplasmic marker protein, TrxA, were also detected in the supernatant fraction. We conclude that the effect of haemin on EsxA secretion is strain-specific but that it clearly enhances secretion in two of the strains we examined.
Haemin has transcriptional and post-translational effects on the T7SS in RN6390
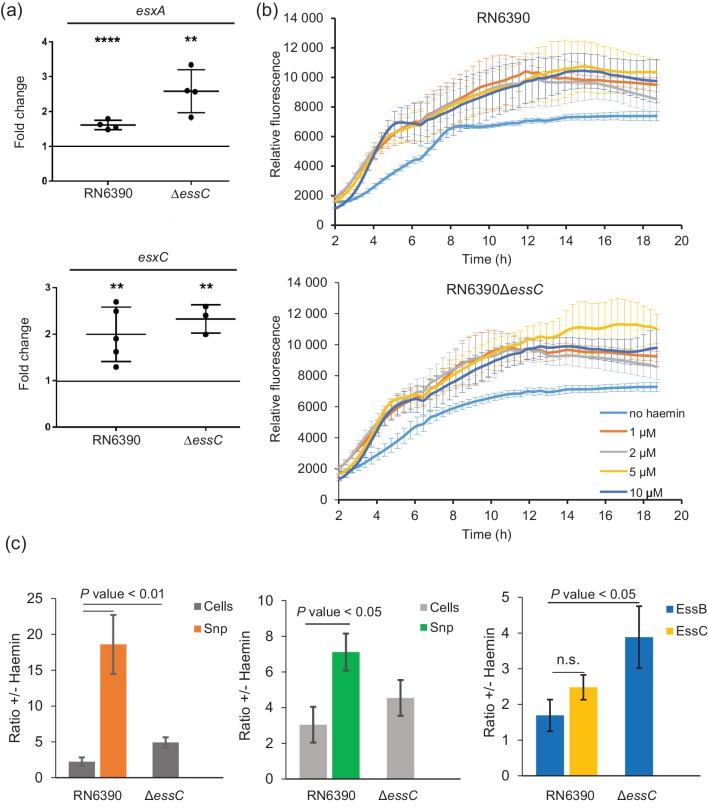
We next addressed whether haemin supplementation increased the level of EsxA in the supernatant due to transcriptional upregulation of the ess gene cluster. To this end, we isolated mRNA from RN6390 and the isogenic essC strain that had been cultured in TSB medium in the presence or absence of 1 µM haemin and used this to prepare cDNA. Since esxA is transcribed separately from the other 11 genes at the ess locus in RN6390, we undertook RT-qPCR with oligonucleotides designed to separately amplify esxA and esxC, normalizing against 16S rRNA as an endogenous control. Fig. 4(a) shows that there is a very small, but statistically significant, effect of haemin on both esxA and esxC transcription in the wild type RN6390 strain (1.5–2-fold). A similar small effect (2–3-fold) was also seen on transcription of these genes in the presence of haemin when the T7SS was inactivated by deletion of essC.
Fig. 4.
Haemin affects transcription of esxA and esxC and secretion of the encoded proteins. (a) S. aureus RN6390 and the isogenic essC deletion strain were grown aerobically in the presence or absence of 1 µM haemin to an OD600 of 1, at which point mRNA from at least three biological replicates was prepared as described in the Methods. Relative transcription levels of the esxA and esxC genes were assayed by RT-qPCR (normalized against the level of 16S rRNA). ****P<0.0001; **P<0.01. (b) S. aureus RN6390 and the isogenic essC deletion strain were cultured in the presence of the indicated concentrations of haemin for 18 h in 96-well plates (200 µl volume) with shaking. Yellow fluorescent protein (YFP) fluorescence of the culture was monitored at 485 nm and was measured in arbitrary units that were normalized to the growth at each time point. Fluorescence was statistically significantly higher in the presence of haemin than in its absence, for both RN6390 and the essC deletion strain (assessed by comparing the relative fluorescence in the absence of haemin with the relative fluorescence measured in the presence of 1, 2, 5 or 10 µM haemin at each time point sampled between 3 and 6 h) (P<0.05 in all cases). (c) Quantification of the levels of T7SS-related proteins in the presence and absence of 1 µM haemin. Cultures of RN6390 and the essC deletion strain grown aerobically in TSB medium in the presence or absence of 1 µM haemin to an OD600 of 2 and separated into cells and supernatant (sn) as described in the Methods. For quantification of protein in the cellular fraction, 5 µl of a cell culture adjusted to an OD600 of 1 was loaded and for the TCA-precipitated supernatant an equivalent of 15 µl of culture supernatant was loaded. Quantification results are expressed as the ratio of the signal obtained in the presence 1 µM haemin to that in the absence of haemin. The results represent the mean±sd of at least three biological replicates. P values from Student's paired t-tests are shown.
To determine whether haemin may also affect translation of the ess genes, we constructed a plasmid-encoded fusion of the esxA promoter and ribosome binding site with yfp and monitored the fluorescence in the same two strains in the presence and absence of exogenous haemin. Fig. 4(b) shows that there is a small (<2-fold) but statistically significant effect of haemin supplementation on YFP fluorescence, consistent with the similar small effect seen on the esxA and esxC transcript levels by RT-qPCR.
To assess whether haemin had a post-translational effect on T7SS activity in RN6390, we quantified the levels of EsxA and EsxC in the cellular and supernatant fractions for strains grown in the presence and absence of haemin. Fig. 4(c) shows that in the presence of haemin there was a 2–3-fold increase in the cellular level of these two proteins, and a comparable increase in the levels of the membrane-bound T7 components EssB and EssC (representative western blots are shown in Fig. S2). In the essC mutant strain, there was also an increase in the cellular level of EssB of approximately 4-fold, and similar increases in the cellular levels of EsxA and EsxC (Fig. 4b). These small but significant increases in cellular levels of protein mirror the similar small increases in transcription of these genes in the presence of haemin. However, analysis of the supernatant fractions shows that in the presence of haemin, there was a 15–20-fold increase in the amount of EsxA in the culture supernatant and a 6–8-fold increase in the supernatant levels of EsxC. These findings are consistent with haemin having a post-translational effect on the secretion activity of the T7SS in strain RN6390. Alternatively, it is possible that haemin affects the levels of extracellular proteases such that the accumulation of EsxA and EsxC in the culture supernatant when haemin is present is an indirect effect due to decreased turnover of these proteins.
Inactivation of RN6390 essC mounts an iron starvation transcriptional response
Taken together, the results so far indicate that haem iron has a striking effect on the secretion activity of the T7SS in S. aureus strain RN6390, potentially implicating the secretion system in iron homeostasis. To investigate this further, we examined differences in the transcriptional profile between wild type reference strain RN6390 and an isogenic essC deletion mutant. Total RNA was prepared from exponentially growing cultures as described in the Methods and RNA-seq was used to investigate gene expression levels. As shown in Table 2 and Fig. S3, a group of 41 genes displayed at least a log 2-fold statistically significant change in expression in an essC mutant, with seven being down-regulated (excluding essC itself) and 34 being up-regulated. Interestingly, 25 of the up-regulated genes have known or implied roles in iron acquisition by S. aureus [51], and these are listed in Table S1. Also included in Table S1 are the fold changes for all of the other genes involved in these iron acquisition pathways but which did not reach our log 2-fold cut-off.
Table 2. Genes differentially regulated (>log 2-fold) in the RN6390 essC deletion mutant, sorted by ascending fold change.
| Locus ID | Gene name | Fold change | Description | Known regulators |
|---|---|---|---|---|
| Downregulated genes | ||||
| SAOUHSC_00262 | essC | −29.4 | T7SS ATPase EssC | |
| SAOUHSC_02290 | – | −7.8 | Unknown, hypothetical protein | |
| SAOUHSC_01942 | splA | −5.4 | Highly specific serine protease specific to S. aureus | Agr (indirect) [60] |
| SAOUHSC_01944 | – | −4.5 | Unknown, hypothetical protein | |
| SAOUHSC_02243 | lukG | −4.5 | Leukocidin-like toxin | |
| SAOUHSC_01941 | splB | −4.3 | Serine protease SplB | Agr [60] |
| SAOUHSC_01938 | splD | −4.3 | Serine protease SplD | Agr [60] |
| SAOUHSC_01121 | hla | −4.1 | α-Haemolysin | Agr [61] |
| Upregulated genes | ||||
| SAOUHSC_02433 | sfaC | 4.1 | Unknown, hypothetical protein | Fur [62] |
| SAOUHSC_02865 | feoA | 4.4 | FeoA domain-containing protein | |
| SAOUHSC_00653 | fhuB | 4.5 | Ferrichrome transport permease FhuB | Fur [63] |
| SAOUHSC_02653 | – | 4.6 | Putative Gcn5-related N-acetyltransferase domain profile | |
| SAOUHSC_02434 | sfaB | 4.6 | Putative siderophore biosynthesis protein | Fur [62] |
| SAOUHSC_00071 | sirC | 4.6 | Involved in staphyloferrin B transport into the cytoplasm | Fur [64] |
| SAOUHSC_00246 | – | 4.7 | Putative transmembrane efflux pump protein | |
| SAOUHSC_01089 | isdG | 4.7 | Haem-degrading monooxygenase IsdG | Fur [65] |
| SAOUHSC_00245 | – | 5.2 | Putative transposase | |
| SAOUHSC_02428 | htsB | 5.4 | Haem transport system permease HtsB | Fur [66] |
| SAOUHSC_01081 | isdA | 5.4 | Iron-regulated haem-iron binding protein | Fur [65] |
| SAOUHSC_02719 | – | 5.5 | ABC transporter ATP-binding protein | |
| SAOUHSC_01082 | isdC | 5.5 | Haem transporter IsdC | Fur [65] |
| SAOUHSC_02654 | trxB2 | 5.5 | Thioredoxin reductase TrxB2 | |
| SAOUHSC_01085 | isdE | 5.6 | Haem-receptor lipoprotein IsdE | Fur [65] |
| SAOUHSC_00130 | isdI | 5.7 | Haem-degrading monooxygenase IsdI | |
| SAOUHSC_01086 | isdF | 6.1 | ABC permease IsdF | Fur [65] |
| SAOUHSC_00131 | – | 6.1 | Putative membrane spanning protein | |
| SAOUHSC_01088 | srtB | 6.2 | Sortase SrtB | Fur [65] |
| SAOUHSC_01084 | isdD | 6.2 | ATP-hydrolysing and haem-binding protein IsdD | Fur [65] |
| SAOUHSC_02432 | – | 6.2 | Unknown, hypothetical protein | |
| SAOUHSC_01087 | – | 6.3 | Iron compound ABC transporter permease | |
| SAOUHSC_02655 | – | 6.3 | Unknown, hypothetical protein | |
| SAOUHSC_02245 | – | 6.5 | Unknown, hypothetical protein | |
| SAOUHSC_02435 | sfaA | 6.7 | Putative transporter | |
| SAOUHSC_02554 | fhuD2 | 6.8 | Ferric hydroxamate receptor 1 FhuD2 | Fur [67] |
| SAOUHSC_00652 | fhuA | 7.0 | Ferrichrome ABC transporter ATP-binding protein FhuA | Fur [63] |
| SAOUHSC_00072 | sirB | 7.4 | Involved in staphyloferrin B transport into the cytoplasm | Fur [64] |
| SAOUHSC_02246 | fhuD1 | 8.0 | Iron compound ABC transporter FhuD1 | Fur [68] |
| SAOUHSC_00747 | sstB | 9.0 | Ferrichrome ABC transporter permease SstB | Fur [69] |
| SAOUHSC_00748 | sstC | 9.1 | Ferrichrome ABC transporter ATP-binding protein SstC | Fur [69] |
| SAOUHSC_02430 | htsA | 10.5 | Haem transport system lipoprotein HtsA | Fur [66] |
| SAOUHSC_00746 | sstA | 10.9 | Ferrichrome ABC transporter permease SstA | Fur [69] |
| SAOUHSC_00074 | sirA | 16.3 | Receptor component of staphyloferrin B | Fur [64] |
It is apparent that many of the genes that encode the Isd machinery, which is involved in haem acquisition, are upregulated. Furthermore, genes for the biosynthesis and uptake of the two S. aureus siderophores, staphyloferrin A and staphyloferrin B, are also upregulated, as are genes encoding the Fhu machinery, which S. aureus uses to import xenosiderophores produced by other bacteria [51]. All of these genes are known to be regulated by the ferric uptake regulatory (Fur) protein (Table 2). These findings indicate that inactivation of the T7SS by deletion of the essC gene prompts S. aureus RN6390 to mount an iron starvation response.
The S. aureus RN6390 and essC strains grow similarly under iron limitation
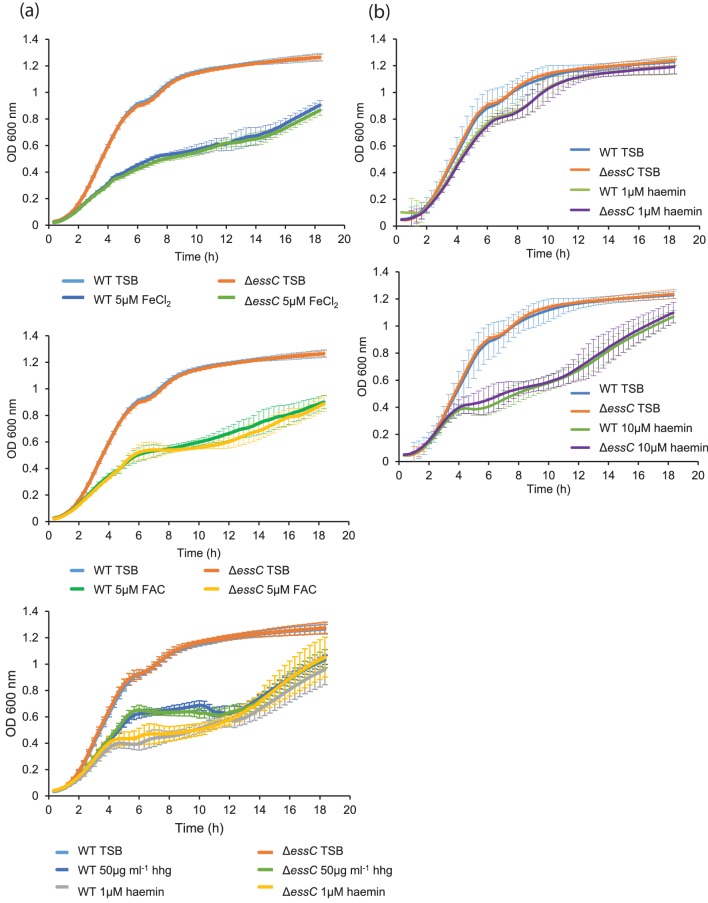
Since RNA-seq analysis suggested that the essC strain was iron-starved, we next investigated the growth behaviour of strain RN6390 and the essC derivative under iron-limited growth conditions. To this end, we used a strong divalent cation chelator to remove iron and other divalent cations from TSB growth medium, and added back the essential cations Zn2+, Mn2+, Mg2+ and Ca2+ [33, 52]. Growth of both the wild type and the ΔessC strains reached a plateau at low optical density, indicating that growth was impaired, presumably because iron had now become a limiting factor (Fig. S4). Next we supplemented the growth medium with 50 μg human haemoglobin ml−1, 5 µM ferrous chloride or ferric ammonium citrate, or 10-fold dilutions of haemin from 10 µM to 1 nM (Figs 5a and S4). Addition of iron sources clearly stimulated growth of both strains, although growth of the essC strain was almost indistinguishable from that of the parental strain regardless of the source of iron (Fig. 5a) or the concentration of haemin added (Fig. S4). We conclude that there is no evidence that the essC strain is impaired in iron acquisition.
Fig. 5.
Effect of iron limitation and haemin on growth of the S. aureus RN6390 and essC mutant strains. (a) S. aureus RN6390 and the essC deletion strain were grown with shaking in either TSB medium or iron-depleted TSB medium (prepared using Chelex-100 to remove all divalent cations followed by re-introduction of Zn2+, Mn2+, Mg2+ and Ca2+ as described in the Methods) that had been supplemented with the indicated iron source. FAC, ferric ammonium citrate; hhg, human haemoglobin. (b) Growth of S. aureus RN6390 and the essC deletion strain in TSB or TSB supplemented with 1 or 10 µM haemin, as indicated. Growth was monitored over 18 h in 96-well plates (200 µl volume). Error bars are ±sd, n=3.
Finally, since haem is toxic to S. aureus at high concentrations (5–10 µM; [49]), we assessed whether the essC mutant was more sensitive to haem toxicity. Haemin was clearly toxic for strain RN6390, since even at 1–2 µM growth was slower than in the absence of haemin (Figs 5b and S5). However, the essC mutant showed similar growth kinetics to the parental strain for all of the haemin concentrations tested, and is therefore not measurably more sensitive to haemin toxicity.
Concluding remarks
Here we have identified a number of links between the T7SS of S. aureus strain RN6390 and iron. We have shown that haemin affects T7 secretion at a transcriptional level and also significantly enhances the level of secreted substrates. Furthermore, inactivation of the essential T7 secretion gene, essC, resulted in upregulation of a large group of Fur-regulated genes that are required for iron acquisition. In this context, it is interesting to note that the ESX-3 T7SS from both Mycobacterium tuberculosis and Mycobacterium smegmatis plays a key role in iron homeostasis, under iron-replete and iron-sufficient conditions, and secretes at least two substrates involved in siderophore-mediated iron uptake [10, 53]. However, despite these observations the essC mutant strain was not phenotypically iron-starved, nor was it more sensitive to haemin toxicity. Unravelling the links between the S. aureus T7SS and iron homeostasis will require further analysis.
Funding information
This study was supported by the Wellcome Trust (through Investigator Award 10183/Z/15/Z to T. P. and through Clinical PhD studentship support to C. P. H. through grant 104241/z/14/z), the Biotechnology and Biological Sciences Research Council and the Medical Research Council (through grants BB/H007571/1 and MR/M011224/1, respectively).
Acknowledgements
The authors would like to thank Professor Ross Fitzgerald (Roslin Institute/University of Edinburgh) for providing us with the 10.1252.X strain, Dr Francesca Short for her assistance with RNA-seq data analysis, Professor Nicola R. Stanley-Wall for her advice with RNA extraction and Mr Connor Bowen for his assistance in RT-qPCR experiments. Professor Simon Foster (University of Sheffield) is thanked for helpful discussion.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: FAC, ferric ammonium citrate; PPIX, protoporphyrin IX; RPMI, Roswell Park Memorial Institute medium; TSB, tryptic soy broth; T7SS, type VII secretion system; YFP, yellow fluorescent protein.
The RNA-seq data from this study have been submitted to the European Nucleotide Archive with accession number ERP009279 and in Array express under accession number E-ERAD-362.
One supplementary table and five supplementary figures are available with the online version of this article.
Edited by: D. Grainger
References
- 1.Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 2015;13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 2.Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, et al. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci USA. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pym AS, Brodin P, Majlessi L, Brosch R, Demangel C, et al. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat Med. 2003;9:533–539. doi: 10.1038/nm859. [DOI] [PubMed] [Google Scholar]
- 4.Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci USA. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gröschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat Rev Microbiol. 2016;14:677–691. doi: 10.1038/nrmicro.2016.131. [DOI] [PubMed] [Google Scholar]
- 6.Ates LS, Houben EN, Bitter W. Type VII secretion: a highly versatile secretion system. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0011-2015. doi: 10.1128/microbiolspec.VMBF-0011-2015. [DOI] [PubMed] [Google Scholar]
- 7.Abdallah AM, Verboom T, Hannes F, Safi M, Strong M, et al. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol. 2006;62:667–679. doi: 10.1111/j.1365-2958.2006.05409.x. [DOI] [PubMed] [Google Scholar]
- 8.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, et al. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci USA. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serafini A, Boldrin F, Palù G, Manganelli R. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J Bacteriol. 2009;191:6340–6344. doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tufariello JM, Chapman JR, Kerantzas CA, Wong KW, Vilchèze C, et al. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc Natl Acad Sci USA. 2016;113:E348. doi: 10.1073/pnas.1523321113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frigui W, Bottai D, Majlessi L, Monot M, Josselin E, et al. Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog. 2008;4:e33. doi: 10.1371/journal.ppat.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abramovitch RB, Rohde KH, Hsu FF, Russell DG. aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol Microbiol. 2011;80:678–694. doi: 10.1111/j.1365-2958.2011.07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott SR, Tischler AD. Phosphate starvation: a novel signal that triggers ESX-5 secretion in Mycobacterium tuberculosis. Mol Microbiol. 2016;100:510–526. doi: 10.1111/mmi.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, an essential gene in mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, et al. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J Bacteriol. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unnikrishnan M, Constantinidou C, Palmer T, Pallen MJ. The Enigmatic Esx proteins: looking beyond mycobacteria. Trends Microbiol. 2017;25:192–204. doi: 10.1016/j.tim.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, et al. Type VII secretion–mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 18.Rosenberg OS, Dovala D, Li X, Connolly L, Bendebury A, et al. Substrates control multimerization and activation of the multi-domain ATPase motor of type VII secretion. Cell. 2015;161:501–512. doi: 10.1016/j.cell.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zoltner M, Ng WM, Money JJ, Fyfe PK, Kneuper H, et al. EssC: domain structures inform on the elusive translocation channel in the Type VII secretion system. Biochem J. 2016;473:1941–1952. doi: 10.1042/BCJ20160257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renshaw PS, Panagiotidou P, Whelan A, Gordon SV, Hewinson RG, et al. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J Biol Chem. 2002;277:21598–21603. doi: 10.1074/jbc.M201625200. [DOI] [PubMed] [Google Scholar]
- 21.Champion PA, Stanley SA, Champion MM, Brown EJ, Cox JS. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science. 2006;313:1632–1636. doi: 10.1126/science.1131167. [DOI] [PubMed] [Google Scholar]
- 22.Sysoeva TA, Zepeda-Rivera MA, Huppert LA, Burton BM. Dimer recognition and secretion by the ESX secretion system in Bacillus subtilis. Proc Natl Acad Sci USA. 2014;111:7653–7658. doi: 10.1073/pnas.1322200111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burts ML, Williams WA, DeBord K, Missiakas DM. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci USA. 2005;102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burts ML, DeDent AC, Missiakas DM. EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol Microbiol. 2008;69:736–746. doi: 10.1111/j.1365-2958.2008.06324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kneuper H, Cao ZP, Twomey KB, Zoltner M, Jäger F, et al. Heterogeneity in ess transcriptional organization and variable contribution of the Ess/Type VII protein secretion system to virulence across closely related Staphylocccus aureus strains. Mol Microbiol. 2014;93:928–943. doi: 10.1111/mmi.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulthess B, Bloes DA, Berger-Bächi B. Opposing roles of σB and σB-controlled SpoVG in the global regulation of esxA in Staphylococcus aureus. BMC Microbiol. 2012;12:17. doi: 10.1186/1471-2180-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson M, Aly KA, Chen YH, Missiakas D. Secretion of atypical protein substrates by the ESAT-6 secretion system of Staphylococcus aureus. Mol Microbiol. 2013;90:734–743. doi: 10.1111/mmi.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Hu M, Liu Q, Qin J, Dai Y, et al. Role of the ESAT-6 secretion system in virulence of the emerging community-associated Staphylococcus aureus lineage ST398. Sci Rep. 2016;6:25163. doi: 10.1038/srep25163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol. 2016;2:16183. doi: 10.1038/nmicrobiol.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Windmüller N, Witten A, Block D, Bunk B, Spröer C, et al. Transcriptional adaptations during long-term persistence of Staphylococcus aureus in the airways of a cystic fibrosis patient. Int J Med Microbiol. 2015;305:38–46. doi: 10.1016/j.ijmm.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Ishii K, Adachi T, Yasukawa J, Suzuki Y, Hamamoto H, et al. Induction of virulence gene expression in Staphylococcus aureus by pulmonary surfactant. Infect Immun. 2014;82:1500–1510. doi: 10.1128/IAI.01635-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jäger F, Zoltner M, Kneuper H, Hunter WN, Palmer T. Membrane interactions and self-association of components of the Ess/Type VII secretion system of Staphylococcus aureus. FEBS Lett. 2016;590:349–357. doi: 10.1002/1873-3468.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006;188:8421–8429. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoltner M, Fyfe PK, Palmer T, Hunter WN. Characterization of Staphylococcus aureus EssB, an integral membrane component of the Type VII secretion system: atomic resolution crystal structure of the cytoplasmic segment. Biochem J. 2013;449:469–477. doi: 10.1042/BJ20121209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller M, Donat S, Rakette S, Stehle T, Kouwen TR, et al. Staphylococcal PknB as the first prokaryotic representative of the proline-directed kinases. PLoS One. 2010;5:e9057. doi: 10.1371/journal.pone.0009057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis Nature Meth. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warne B, Harkins CP, Harris SR, Vatsiou A, Stanley-Wall N, et al. The Ess/Type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genomics. 2016;17:222. doi: 10.1186/s12864-016-2426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magoc T, Wood D, Salzberg SL. EDGE-pro: estimated degree of gene expression in prokaryotic genomes. Evol Bioinform Online. 2013;9:127–136. doi: 10.4137/EBO.S11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank KL, del Pozo JL, Patel R. From clinical microbiology to infection pathogenesis: how daring to be different works for Staphylococcus lugdunensis. Clin Microbiol Rev. 2008;21:111–133. doi: 10.1128/CMR.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corrigan RM, Foster TJ. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid. 2009;61:126–129. doi: 10.1016/j.plasmid.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Parsot C, Ménard R, Gounon P, Sansonetti PJ. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol Microbiol. 1995;16:291–300. doi: 10.1111/j.1365-2958.1995.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 44.Heesemann J, Gross U, Schmidt N, Laufs R. Immunochemical analysis of plasmid-encoded proteins released by enteropathogenic Yersinia sp. grown in calcium-deficient media. Infect Immun. 1986;54:561–567. doi: 10.1128/iai.54.2.561-567.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 46.Critchley IA, Basker MJ. Conventional laboratory agar media provide an iron-limited environment for bacterial growth. FEMS Microbiol Lett. 1988;50:35–39. doi: 10.1111/j.1574-6968.1988.tb02907.x. [DOI] [Google Scholar]
- 47.Benson BJ, Williams MC, Sueishi K, Goerke J, Sargeant T. Role of calcium ions the structure and function of pulmonary surfactant. Biochim Biophys Acta. 1984;793:18–27. doi: 10.1016/0005-2760(84)90048-1. [DOI] [PubMed] [Google Scholar]
- 48.Hayward RD, Cain RJ, McGhie EJ, Phillips N, Garner MJ, et al. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol Microbiol. 2005;56:590–603. doi: 10.1111/j.1365-2958.2005.04568.x. [DOI] [PubMed] [Google Scholar]
- 49.Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, et al. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe. 2007;1:109–119. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wakeman CA, Hammer ND, Stauff DL, Attia AS, Anzaldi LL, et al. Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Mol Microbiol. 2012;86:1376–1392. doi: 10.1111/mmi.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol. 2011;65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hurd AF, Garcia-Lara J, Rauter Y, Cartron M, Mohamed R, et al. The iron-regulated surface proteins IsdA, IsdB, and IsdH are not required for heme iron utilization in Staphylococcus aureus. FEMS Microbiol Lett. 2012;329:93–100. doi: 10.1111/j.1574-6968.2012.02502.x. [DOI] [PubMed] [Google Scholar]
- 53.Serafini A, Pisu D, Palù G, Rodriguez GM, Manganelli R. The ESX-3 secretion system is necessary for iron and zinc homeostasis in Mycobacterium tuberculosis. PLoS One. 2013;8:e78351. doi: 10.1371/journal.pone.0078351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, et al. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. Embo J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dyke KG, Jevons MP, Parker MT. Penicillinase production and intrinsic resistance to penicillins in Staphylococcus aures. Lancet. 1966;1:835–838. doi: 10.1016/s0140-6736(66)90182-6. [DOI] [PubMed] [Google Scholar]
- 56.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holden MT, Lindsay JA, Corton C, Quail MA, Cockfield JD, et al. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW) J Bacteriol. 2010;192:888–892. doi: 10.1128/JB.01255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holden MT, Hsu LY, Kurt K, Weinert LA, Mather AE, et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013;23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, et al. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vojtov N, Ross HF, Novick RP. Global repression of exotoxin synthesis by staphylococcal superantigens. Proc Natl Acad Sci USA. 2002;99:10102–10107. doi: 10.1073/pnas.152152499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beasley FC, Vinés ED, Grigg JC, Zheng Q, Liu S, et al. Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol Microbiol. 2009;72:947–963. doi: 10.1111/j.1365-2958.2009.06698.x. [DOI] [PubMed] [Google Scholar]
- 63.Xiong A, Singh VK, Cabrera G, Jayaswal RK. Molecular characterization of the ferric-uptake regulator, fur, from Staphylococcus aureus. Microbiology. 2000;146:659–668. doi: 10.1099/00221287-146-3-659. [DOI] [PubMed] [Google Scholar]
- 64.Heinrichs JH, Gatlin LE, Kunsch C, Choi GH, Hanson MS. Identification and characterization of SirA, an iron-regulated protein from Staphylococcus aureus. J Bacteriol. 1999;181:1436–1443. doi: 10.1128/jb.181.5.1436-1443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 66.Skaar EP, Humayun M, Bae T, Debord KL, Schneewind O. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 67.Horsburgh MJ, Ingham E, Foster SJ. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol. 2001;183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sebulsky MT, Speziali CD, Shilton BH, Edgell DR, Heinrichs DE. FhuD1, a ferric hydroxamate-binding lipoprotein in Staphylococcus aureus: a case of gene duplication and lateral transfer. J Biol Chem. 2004;279:53152–53159. doi: 10.1074/jbc.M409793200. [DOI] [PubMed] [Google Scholar]
- 69.Morrissey JA, Cockayne A, Hill PJ, Williams P. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect Immun. 2000;68:6281–6288. doi: 10.1128/IAI.68.11.6281-6288.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.