Abstract
The phosphoenopyruvate:carbohydrate phosphotransferase system (PTS) enables Vibrio cholerae – and other bacteria – to recognize and transport exogenous carbon sources for energy, including the six-carbon sugar alcohol, mannitol. The mannitol-specific PTS transporter is encoded by mtlA and its expression is expected to be regulated by the putative repressor encoded by the mtlR gene. Here, we show that mtlR overexpression inhibits V. cholerae growth in medium supplied with mannitol as the sole carbon source and represses MtlA-mediated biofilm formation. We demonstrate that when V. cholerae is grown in non-mannitol medium, knocking out mtlR leads to both increased MtlA protein and mtlA mRNA levels, with these increases being especially pronounced in non-glucose sugars. We propose that in non-mannitol, non-glucose growth conditions, MtlR is a major regulator of mtlA transcription. Surprisingly, with regard to mtlR expression, transcript and protein levels are highest in mannitol medium, conditions where mtlA expression should not be repressed. We further show that MtlR levels increase during growth of the bacteria and linger in cells switched from mannitol to non-mannitol medium. Our data suggests an expression paradigm for mtlA where MtlR acts as a transcriptional repressor responsible for calibrating MtlA levels during environmental transitions.
Keywords: mannitol, Vibrio cholerae, phosphotransferase system
Introduction
As a facultative pathogen, Vibrio cholerae thrives in both marine ecosystems and the gastrointestinal tract of a human host [1–3]. V. cholerae colonization of these disparate climates requires molecular mechanisms to reconcile fluctuations in temperature, pH and carbon source availability [4–8]. One method of adaptation is the recognition and utilization of various carbohydrates for energy, which necessitates the phosphoenolpyruvate (PEP):phosphotransferase system (PTS) [9–12]. Highly conserved across bacteria, the PTS responds to environment-specific stimuli through a phosphorylation cascade [13, 14]. Enzyme I (EI) first transfers a phosphoryl group from PEP to histidine protein (HPr). HPr proceeds to phosphorylate one of several EIIA proteins. Whereas cytosolic EI and HPr are common, core PTS components, sugar transport is facilitated by carbohydrate-specific EII proteins [13]. In V. cholerae, nine EIIA proteins have been identified and are each specific to different carbohydrates [10, 15]. The EIIA protein assembles, with EIIB and EIIC subunits, into a larger permease complex (EII) that catalyses the final step of the PTS: concomitant transport and phosphorylation of the corresponding, extracellular carbon source [13].
For most bacteria, including V. cholerae, transport of exogenous d-mannitol requires the PTS. d-mannitol is a six-carbon sugar alcohol produced by marine algae and is the most abundant hexitol found in nature [16–19]. The mannitol-specific PTS transporter (EIIMtl) is encoded by genes at the mannitol (mtl) locus of both Gram-negative and Gram-positive bacteria. This locus also contains mtlD, encoding a dehydrogenase responsible for oxidizing newly acquired and phosphorylated mannitol (mannitol-1-phophate) into fructose-6-phosphate, the successive metabolic intermediate [20]. In the case of Escherichia coli and other proteobacteria, the mtl locus also encodes mtlR, expressing a regulator of the mtl genes [21]. mtlR is also expressed from the mtl locus of firmicutes, but in the case of Bacillus subtilius, the gene encoding the regulator is located 15 kb downstream from the rest of the mtl genes [22].
The mtl genes of V. cholerae span 3.9 kb and are organized as mtlADR. mtlA encodes the EIIMtl PTS transporter, which shares high sequence homology to the extensively studied E. coli MtlA [23]. In both species, the EIICMtl, EIIBMtl and EIIAMtl subunits are all encoded by the mtlA gene and are fused to each other, in that order [20, 23]. MtlA activity is required for mannitol metabolism in V. cholerae; bacteria lacking functional MtlA are unable to survive in media supplemented with mannitol as the sole carbon source [23, 24]. The persistence of V. cholerae in marine ecosystems is also bolstered by MtlA activity; the EIIB subunit of MtlA is capable of inducing biofilm formation, potentially providing the bacteria a fitness advantage in its aquatic reservoir [3, 25].
Regulation of mtlA in V. cholerae is complex and involves the 3′,5′-cyclic adenosine monophosphate (cAMP) receptor protein (CRP). This regulator is activated by cAMP to directly bind five distinct sites found in the mtlA promoter and is necessary for activation of mtlA [26]. Recent studies further demonstrate that mtlA expression in V. cholerae is negatively regulated by MtlS, a cis-acting small regulatory RNA (sRNA) [27, 28]. One of the least characterized regulators of mtlA in V. cholerae, however, is MtlR.
Based on genomic annotation, the V. cholerae mtlR gene codes for a 195-amino acid protein (22 kD) that shares sequence homology (63 %) to the E. coli homologue [23, 29]. MtlR activity was first studied in the E. coli system, where the protein demonstrated the ability to repress the uptake of exogenous mannitol [29]. In line with this, knocking down mtlR expression in V. cholerae allows the bacteria to achieve exponential growth more quickly when transitioned from rich medium to a mannitol-only medium; this activity appears to be independent of MtlS [28, 30]. It has been hypothesized that MtlR-mediated repression involves the protein directly binding to DNA operators in the mtlA promoter, but interactions between MtlR and DNA have yet to be observed [29, 31]. The solved structure of MtlR from Vibrio parahemeolyticus, furthermore, reveals an absence of established DNA-binding motifs and electrostatic properties unlikely to accommodate direct DNA binding [31]. Thus, the mechanistic details explaining how MtlR affects mannitol metabolism in proteobacteria remain largely unknown.
For the present study, we sought to further investigate the activity and regulation of MtlR to better understand the collective modulation of mtlA expression in V. cholerae. We investigated the effect of MtlR activity on the physiology of V. cholerae exposed to exogenous mannitol. Additionally, we considered the expression profile of mtlR in the presence and absence of mannitol. Our results suggest that in certain environments, MtlR represses the transcription of mtlA and, therefore, the ability of V. cholerae to respond to extracellular mannitol. We also establish that the expression of mtlR is unexpectedly increased in the presence of mannitol, where it accumulates throughout growth. We suggest a model in which MtlR may serve to fine-tune MtlA levels when V. cholerae transition between areas where mannitol concentrations fluctuate.
Methods
Strain and plasmid construction
All strains and plasmids used in this study are listed in Table S1 (available in the online version of this article). The V. cholerae strains used in this work were El Tor strain N16961 ∆tcpA and derivatives (Table S1). The tcpA mutant is highly attenuated for virulence and was used for safety purposes [32]. Throughout this text, ‘wild-type’ refers to the N16961 ∆tcpA strain. Plasmids with oriR6K were propagated in E. coli DH5αλpir; all other plasmids were propagated in E. coli DH5α or TOP10 strains. Oligonucleotide primers and labelled probes used in this study are listed in Table S2 and were acquired from Integrated DNA Technologies.
Plasmids for generating chromosomal mutations in V. cholerae were constructed in the allelic exchange vector pCVD442 [33]. The mtlR–HA mutant was constructed by combining, using Gibson Assembly [34], two 500 bp gBlock fragments (Integrated DNA Technologies) that introduced the sequence encoding the HA tag at the C-terminus of mtlR. The resulting DNA was PCR-amplified with F1 and R2 primers and cloned into the multiple cloning site of pCVD442 using the SphI and SacI restriction sites. All pCVD442-derived constructs were conjugated into V. cholerae N16961 ∆tcpA from E. coli SM10λpir as described previously [35]. After one passage in Luria–Bertani (LB) broth with streptomycin, sucrose-resistant colonies were selected and subsequently screened for the desired mutation by PCR and sequencing.
Plasmids harbouring mtlR under the control of the Ptrc promoter were constructed by PCR amplifying mtlR from V. cholerae N16961 ∆tcpA using oligonucleotides containing SacI and XbaI restriction sites. PCR products and vector pTrc99A were digested with SacI and XbaI restriction endonucleases. Digestion products were ligated using T4 DNA ligase to produce plasmids containing Ptrc-mtlR alleles.
Bacterial growth
All strains were grown under shaking conditions at 250 r.p.m. at 37 °C in LB medium or M9 minimal medium supplemented with 0.1 % trace metals (5 % MgSO4, 0.5 % MnCl2•4H2O, 0.5 % FeCl3, 0.4 % nitriloacetic acid) and 0.4 % carbon source. Throughout the text, ‘glucose medium’, ‘mannitol medium’, etc., refer to M9 minimal medium supplemented with trace metals and 0.4 % of the respective carbon source. When necessary, cultures were supplemented with 1 mM IPTG to induce the expression of genes inserted into the pTrc99A plasmid under the control of the Ptrc promoter. Antibiotics were used at the following concentrations: streptomycin, 100 µg ml−1; carbenicillin, 50–100 µg ml−1.
Growth curves
Growth of each strain was determined by measuring OD600 using a Bio-Tek microplate reader. Colonies were collected from LB agar plates and grown in LB medium for 2–3 h at 37 °C, 250 r.p.m. Cells were then pelleted and washed with 1× M9 salts, and the OD600 was adjusted to 0.01 (~107 c.f.u. ml−1) in 200 µl of supplemented M9 medium with 0.4 % of the appropriate carbon source. Bacteria were grown with orbital shaking at 37 °C for 12–18 h in the microplate reader at 37 °C. All growth experiments were performed in at least triplicate.
Biofilm assay
Biofilm formation by V. cholerae was quantified as previously described, with minor modifications [25]. Strains were first grown overnight on LB agar plates at 37 °C; colonies were then used to inoculate 2 ml LB cultures, which were grown at 37 °C, 250 r.p.m. After 6–8 h, the cultures were used to inoculate 300 µl of LB medium in borosilicate tubes. After incubation for 17 to 24 h at 22 °C, each planktonic cell suspension was collected and its OD655 was measured using a Bio-Tek microplate reader. To quantify the surface-attached cells, the tubes were rinsed with PBS, followed by the addition of 300 µl of PBS and a small volume of 1 mm diameter glass beads (Biospec,). Through vortexing, the attached cells were dispersed and the OD655 of the resulting cell suspension was measured. The sum of the OD655 values measured for planktonic and surface-associated cell suspensions are reported as the total growth of the cells. Statistical analysis was performed using GraphPad Prism 6 software.
RNA analysis
V. cholerae were cultured to an OD600 ~0.3 and total RNA was extracted using the DirectZol RNA MiniPrep Kit (Zymo) following the manufacturer’s suggested protocol. DNA was further removed from all samples using the TURBO DNA-free kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. To analyse the nature of the mRNA transcripts from the mtl locus, reverse transcription-PCR (RT-PCR) was used. RT of total RNA (100 ng) was performed with reverse primers (2 pmol) specific to mtlADR using Superscript III (Thermo Fisher Scientific) according to the manufacturer’s instructions. The cDNA generated or N16961 genomic DNA was used as a template for PCR with reverse and forward primers specific to monocistronic transcripts or mtlAD, mtlDR or mtlADR polycistronic transcripts. PCR reactions were performed with an initial denaturation step of 5 min at 95 °C, followed by 26 cycles of 30 s at 95 °C, 30 s at 55 °C and 4 m at 72 °C. Control reactions of RNA samples not treated with reverse transcriptase were performed for each run; no product band was observed in any of the no-RT controls.
qRT-PCR was used to analyse relative expression levels using gene-specific primers, a Stratagene MX3005P System, and the Brilliant II SYBR Green qRT-PCR Master Mix Kit (Agilent). The reactions were set up in 96-well optical reaction plates and contained 1× Brilliant SYBR Green qPCR Master Mix, 30 nM ROX reference dye, each primer at 100 nM, 100 ng RNA and 1 µl RT/RNase block enzyme mixture in a 25 µl reaction. The following conditions were used for cDNA synthesis and PCR: 30 m at 50 °C, 10 m at 95 °C, and 40 cycles of 30 s at 95 °C and 1 m at 60 °C (Agilent). MxPro QPCR software (v. 4.10) was used to determine the Ct for each reaction. Relative RNA concentrations were calculated from the Ct values by comparison to standard curves; the relative transcript levels were normalized to an endogenous control (4.5S RNA). No signals were detected in no-template controls and no-RT controls. Statistical analysis was performed using GraphPad Prism 6 software.
Western blot analysis
Bacteria were grown to an OD600 ~0.3 and collected (8000 g, 5 min). For MtlA analysis, lysates were prepared by mixing ~107 cells with SDS-loading buffer and heating at 95 °C for 10 min. For MtlR analysis, cell pellets from 5 to 50 ml cultures were lysed with B-PER Extraction Reagent (Thermo Fisher Scientific) in the presence of DNAse I, following the suggested protocol of the manufacturer; the lysate was then mixed with SDS-loading buffer. The proteins were resolved on 4–20 % TRIS gels (Bio-Rad) and transferred to nitrocellulose membranes. The membranes were treated with polyclonal anti-FLAG (abCam), polyclonal anti-HA (abCam), monoclonal anti-RpoB (abCam), or monoclonal anti-RNAPα (BioLegend) antibodies. Antibodies were detected using secondary antibodies with IR680 and IR800 dyes attached (Licor). As a membrane protein, MtlA has a tendency to oligomerize, even under the denaturing conditions of SDS-PAGE. Signals were visualized and band densities were measured using an Odyssey imager (Licor).
Results
MtlR represses the ability of V. cholerae to use and respond to extracellular mannitol
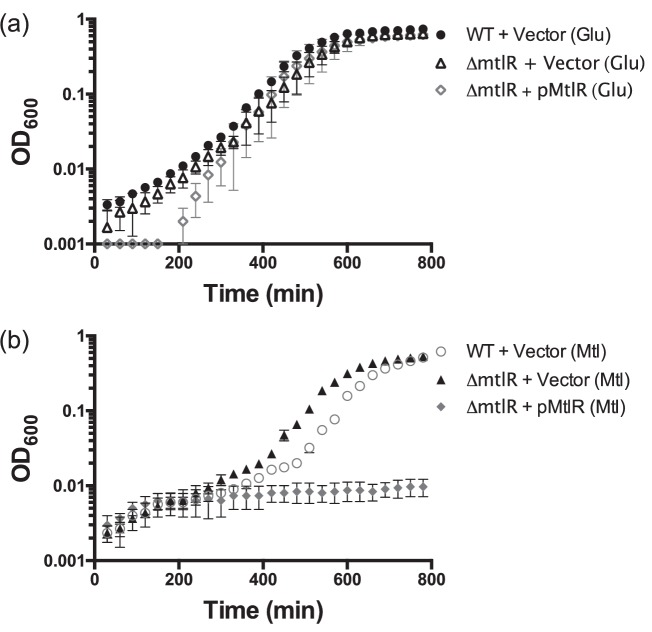
Strains lacking mtlR have a slightly shorter lag phase, compared to the wild-type, when transitioned from LB to mannitol medium [28], conditions that allow one to assess the effect that MtlR has on cells adapting to new conditions that may include mannitol as the primary carbon source. We attempted to complement this phenotype by introducing an mtlR overexpression plasmid into the knockout strain (Fig. S1). This strain had difficulty adapting to mannitol medium (Fig. 1a, b). We hypothesize that the overexpression of mtlR in mannitol prevents the bacteria from producing enough MtlA to take up adequate levels of mannitol to meet their metabolic needs. The effects of overexpressing or deleting mtlR on cell growth appear to be specific to mannitol and not a general effect on carbon uptake or metabolism as all strains grew similarly in glucose medium, although an extended lag phase was observed in the wild-type strain harbouring pMtlR.
Fig. 1.
Overexpression of mtlR negatively impacts growth of V. cholerae on mannitol medium. (a, b) Growth curves of the indicated strains in M9 medium supplemented with the indicated carbon source. Bacteria were first grown to mid-exponential phase in LB medium. Cultures were washed and used to inoculate 200 µl M9 medium with 1 mM IPTG and either glucose (Glu, 0.4 %) or mannitol (Mtl, 0.4 %). Strains were grown at 37 °C with orbital shaking in a 96-well plate. The error bars represent the standard deviation of three biological replicates.
Expression of the B subunit from the EIImtl protein has been shown to activate biofilm formation in V. cholerae. Consistent with prior studies, the mtlR knockout behaves similarly to the wild-type in that both strains increase biofilm formation in response to mannitol (Fig. 2a, b) [25]. Overexpression of mtlR in the knockout strain, however, repressed the ability of the bacteria to induce biofilm formation when mannitol was added to the growth medium, without affecting overall growth (Fig. 2a, b). In summary, both the growth curve and biofilm assay results are consistent with MtlR acting as a negative regulator of mtlA in V. cholerae.
Fig. 2.
MtlR negatively affects mannitol-induced biofilm formation. Quantification of (a) surface-attached cells and (b) total cell growth of the indicated strains. Wild-type V. cholerae or an isogenic ∆mtlR strain harbouring either an empty vector control or pMtlR. Cells were cultured in LB medium supplemented with mannitol (mtl, 0.5 %) or an equivalent volume of water (–). IPTG (1 mM) was also added to induce expression from the pTrc99A vector. Bar graphs represent the means and sds from multiple independent samples. ***P<0.001; ns, not significant, by unpaired two-tailed t-test.
MtlR downregulates mtlA expression at the transcriptional level
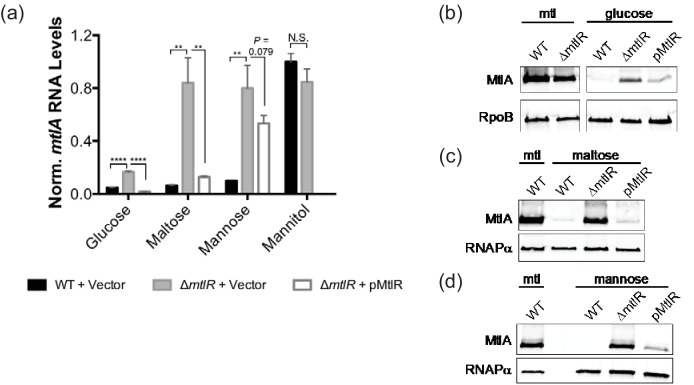
We previously reported conflicting results from qRT-PCR and Northern blot analyses on mtlA mRNA levels in glucose versus mannitol medium [24, 28]. We designed a new probe for mtlA mRNA (Fig. S2) and Northern blotting analysis using this probe entirely agrees with qRT-PCR: mtlA mRNA levels are up-regulated in mannitol medium, compared to glucose medium (Figs 3a and S3). We reasoned that the probes that we used in our previous report were binding to a non-specific target and that the new probes we used here accurately report relative mtlA mRNA levels in V. cholerae.
Fig. 3.
MtlR represses mtlA expression. Wild-type V. cholerae harbouring an empty vector (WT), an isogenic strain lacking mtlR (∆mtlR) and a ∆mtlR strain harbouring pMtlR (pMtlR) were cultured in M9 minimal medium with glucose, mannitol (mtl), maltose or mannose (0.4 % w/v) at 37 °C to an OD600 of 0.3; IPTG was not added to the cell cultures. (a) qRT-PCR was used to measure mtlA mRNA levels, which were first normalized to 4.5S RNA levels, and then to mtlA levels in the wild-type strain grown in mannitol medium, as described in Methods. Analysis of mtlA mRNA in mannitol medium focused on only the wild-type and knockout strains. Bar graphs represent the means and sds from three biological replicates. ** P<0.01; ****P<0.0001; ns, not significant, by unpaired two-tailed t-test. (b–d) MtlA-FLAG protein levels were analysed by Western blot. Cell lysates from an equal number of cells were analysed using anti-FLAG, anti-RpoB and anti-RNAPα antibodies. RpoB and RNAPα served as a loading control. For (b), data is from different regions of the same blot. All data are representative of three independent trials.
To investigate the role of MtlR in mtlA transcription, we repeated the qRT-PCR analysis with an isogenic strain lacking mtlR. In mannitol medium, knockout of mtlR did not lead to higher levels of mtlA transcript or MtlA protein, confirming our expectation that MtlR has no major activity in mannitol growth conditions (Fig. 3a, b). These results are consistent with our growth curve and biofilm data. We therefore focused our efforts on assessing the role of MtlR in non-mannitol growth conditions.
In glucose (conditions where mtlA expression is normally repressed), there was a small but significant increase in mtlA transcript levels when mtlR was knocked out (Fig. 3a). We observed a similar pattern in MtlA protein levels – knocking out mtlR significantly increased MtlA protein levels compared to wild-type, albeit only to 30 % of the amount found in V. cholerae grown in mannitol medium (Fig. 3b). These results suggest that MtlR plays a minor role in regulating mtlA at the transcriptional level when V. cholerae are grown in glucose medium and are consistent with our above analysis that suggested MtlR as a repressor of mtlA. We were able to partially complement the mtlR mutation by introducing mtlR on a plasmid in the mutant strain, indicating that it is the gene product of mtlR that directly impacts MtlA synthesis (Fig. 3a, b). In these complementation experiments, and the ones below, in order to obtain more physiologically relevant levels of MtlR we did not add IPTG to the growth medium (Fig. S1).
In addition to its role in transporting glucose into the cell, EIIAGlc indirectly regulates CRP [13], a known transcriptional activator of mtlA [26]. To avoid the complication of perturbing a major transcription factor known to activate mtlA, we chose to also test the effects of mtlR on mtlA transcription in non-glucose, non-mannitol growth conditions. When the ∆mtlR strain was grown in maltose medium (a non-PTS sugar) and mannose medium (a PTS-sugar), we observed that mtlA transcript levels were highly elevated compared to the wild-type controls (Fig. 3a). Ectopic expression of mtlR was able to partially complement this phenotype (Fig. 3a). Western blot analysis of these same strains grown in maltose or mannose medium further supports a role in which MtlR is a major regulator of mtlA expression in non-mannitol, non-glucose medium (Fig. 3c, d).
Expression of mtlR is induced in mannitol and accumulates throughout growth
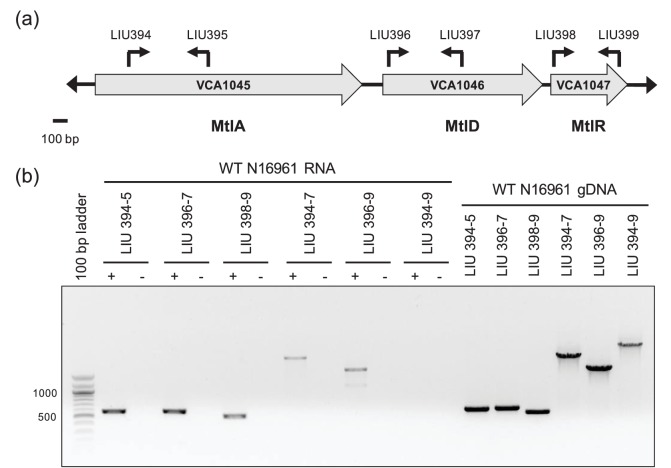
We expected, given the activity of MtlR, that it would be most highly expressed in non-mannitol medium. Instead, we saw that mtlR mRNA levels increase in mannitol (Figs 4 and S4). This may be a result of increased expression of the entire mtl locus during exposure to mannitol. To test this theory, we used RT-PCR to determine the transcriptional organization of the mtl locus when the bacteria were grown in mannitol medium. RT-PCR primers were designed to amplify coding regions spanning two or more mtl genes to probe for the presence of polycistronic transcripts (Fig. 5a). Affirming that all mtl genes were expressed under these growth conditions, the expected RT-PCR products for mtlA (576 bp), mtD (585 bp) and mtlR (515 bp) were detected (Figs 5b and S5). RT-PCR products for mtlAD (2560 bp) and mtlDR (1748 bp) products were also observed, but no product from an mtlADR transcript was obtained, despite multiple attempts (Fig. 5b). These results suggest a transcriptional paradigm in which the mtlD gene can be co-expressed with mtlA and mtlR individually, but the three gene units do not necessarily form a single polycistronic transcript. The existence of multiple promoters within a single operon is a genetic structure that has been previously reported in V. cholerae [36, 37]. Alternatively, the mtlADR transcript may be rapidly processed into smaller transcripts. The primer pair we use for the mtlADR transcript also appears to be less efficient, which may have negatively impacted our ability to observe the mtlADR transcript.
Fig. 4.
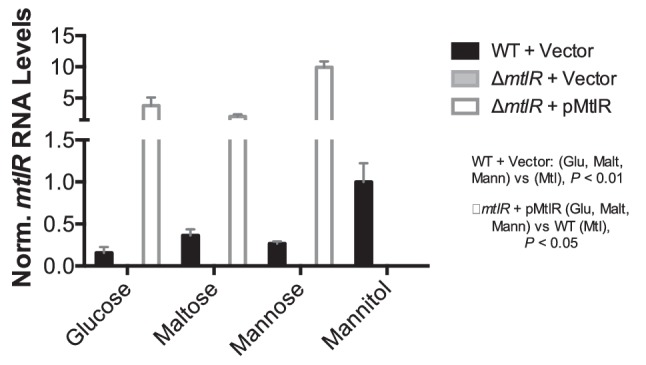
mtlR mRNA levels increase in mannitol medium. The same samples as in Fig. 3 were used to measure mtlR mRNA levels via qRT-PCR. mtlR RNA levels were normalized to 4.5S RNA levels, and then to mtlR levels in the wild-type strain grown in mannitol medium, as described in Methods. Analysis of mtlR mRNA in mannitol medium focused on only the wild-type and knockout strains. Bar graphs represent the means and sds from three biological replicates. mtlR mRNA levels were undetectable, as expected, in the mutant strain. Statistical significance was determined by unpaired two-tailed t-test. All data are representative of three independent trials.
Fig. 5.
mtlAD and mtlDR are co-transcribed. (a) Schematic representation of the 3.9 kb mtl locus in V. cholerae including the mtlA, mltD and mtlR genes, their transcriptional directionality, and the location and names of primers used to determine transcriptional organization. (b) RT-PCR products from total RNA collected from V. cholerae grown in mannitol medium were analysed by gel electrophoresis on an agarose gel. + and − correspond to reactions run in the presence and absence of reverse transcriptase, respectively. PCR was also performed on V. cholerae genomic DNA (gDNA) to provide positive controls. Data are representative of three independent trials.
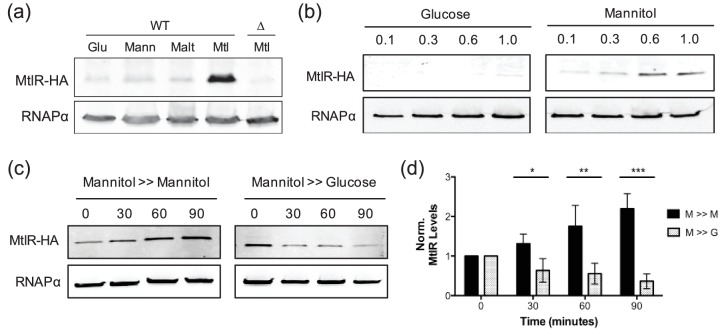
In order to assess MtlR protein levels, we first determined that the N-terminus of mtlR was previously mis-annotated, and that the correct annotation for mtlR encodes a 177-amino acid protein (Figs S6 and S7). Using a strain harbouring an HA-tagged mtlR, we investigated the profile of MtlR in cells cultured to mid-exponential phase in various growth media. Consistent with the mtlR mRNA analysis, we observed the surprising result that MtlR protein levels were markedly elevated in mannitol growth medium; in non-mannitol conditions, the level of MtlR protein was determined to be ~5 % of that found in mannitol medium, as determined by quantification of MtlR band densities (Fig. 6a).
Fig. 6.
MtlR protein levels are higher in mannitol medium compared to glucose medium. (a) MtlR-HA protein levels from bacteria (wild-type, WT; isogenic ∆mtlR, ∆) cultured to an OD600 of 0.3 at 37 °C in M9 minimal medium with the indicated carbon source (0.4 %): glucose (Glu), mannose (Mann), maltose (Malt), mannitol (Mtl). (b) Wild-type V. cholerae cells cultured at 37 °C in M9 minimal medium with glucose or mannitol (0.4 %) were harvested at the indicated OD600. (c) Wild-type V. cholerae were cultured at 37 °C in M9 minimal medium with 0.4 % mannitol. At an OD600 of 0.3, cells were collected, re-suspended in an equal volume of M9 minimal media with 0.4 % mannitol or glucose and harvested at 0, 30, 60 and 90 min relative to the medium switch. (d) Quantification of MtlR-HA from Western blot analyses, as in (c), normalizing to RNAPα levels and then to MtlR-HA levels at time point 0. Shown are the means and sds from three independent experiments. *P<0.05; **P<0.01; ***P<0.001 by unpaired two-tailed t-test. For Western blots, whole cell lysates from an equal number of cells were analysed by Western blot with anti-HA and anti-RNAPα antibodies. RNAPα is acting as the loading control. All blots are representative of at least three independent trials.
Our initial analysis of mtlR expression was based on analysing bacteria grown to mid-log phase (OD600 ~0.3). We considered that at different growth phases, the bacteria might exhibit a more expected expression profile, with MtlR levels higher in non-mannitol medium compared to mannitol medium. Thus, cells were grown to varying optical densities in glucose and mannitol medium and subsequently analysed by Western blot (Fig. 6b). Although these data suggest that cellular MtlR levels are highest at elevated optical densities regardless of the carbon source, the levels of MtlR appear to be higher in cells grown in mannitol medium versus glucose medium at any point during growth.
Given the unexpected expression pattern of mtlR, we considered that the bacteria may maintain high levels of the repressor during transitions from mannitol to non-mannitol growth conditions to assist with adaptation. To test this theory, we cultured cells in mannitol medium to an OD600 of 0.3 before switching the growth conditions to either fresh glucose or mannitol medium. V. cholerae cells were harvested at time points subsequent to the media switch and MtlR levels were analysed. Upon moving bacteria from mannitol to glucose medium, we observe a marked decrease to approximately 60 % of initial MtlR levels 30 min after the media switch (Fig. 6c, d). MtlR levels linger around 50 % of initial measurements 60 and even 90 min after the media switch to glucose growth medium (Fig. 6c, d). Similar results were observed when moving the bacteria from mannitol into maltose or mannose medium (Fig. S8). In terms of MtlA, we generally observe a consistent decrease in transporter levels after moving V. cholerae from mannitol to glucose medium (Fig. S8) [28]. That MtlR protein levels remain appreciable up to 1.5 h subsequent to a media switch out of mannitol suggests that, in V. cholerae, MtlR assists with adaptation to new, non-mannitol media by precluding mtlA expression during the early stages of transition.
Discussion
Regulation of mtlA expression in V. cholerae is a process involving a range of different cellular machinery, including the global regulator CRP [26] and the cis-acting small regulatory RNA, MtlS [27, 28]. Taken together, the established framework for mtlA regulation includes both transcriptional and post-transcriptional mechanisms. Here, we characterize a third regulator of mtlA expression: MtlR, a transcriptional regulator of mtlA that plays a role in regulating mannitol transporter levels in non-mannitol growth conditions. Our results are further supported by a study from Wang et al., suggesting that MtlR acts as a transcriptional regulator of mtlA expression in both toxigenic and non-toxigenic El Tor strains of V. cholerae [30]. We note, however, that the role of MtlR does not appear to be equal in all non-mannitol media. In glucose, for example, MtlR appears to play a minor role in regulating mtlA transcription (Fig. 3a). Given the known role of CRP in activating mtlA expression, it could be that in glucose growth conditions, mtlA transcription is largely governed by cAMP levels – low adenylyl cyclase activity results in low mtlA mRNA levels. However, in the case of mannose and maltose, MtlR appears to be the major transcriptional repressor of mtlA (Fig. 3a). Indeed, in these non-glucose, non-mannitol conditions, MtlR may counteract elevated cAMP levels to maintain low mtlA expression when the transporter is not needed.
Our data also suggest MtlR represses mtlA expression with considerable efficacy. MtlR levels in V. cholerae grown to mid-exponential phase in non-mannitol conditions are almost imperceptible in our traditional Western blot assay, but necessary for full repression of mtlA expression (Figs 3 and 6). This being said, MtlR alone is insufficient for full repression of mtlA, as evident by our observations that maximal levels of MtlA are not observed in ∆mtlR bacteria grown under non-mannitol conditions (Fig. 3). It is likely that MtlR acts additively alongside MtlS, an sRNA that is similarly expressed under non-mannitol conditions and also represses mtlA. MtlR- and MtlS-mediated repression of mtlA appear to be independent of each other, moreover, as MtlR exhibits no observable influence on MtlS synthesis [28].
We also noted that our complementation strain was not able to bring mtlA expression back to wild-type levels. This could be because the complementation assays were done in the absence of IPTG, which does allow for ectopic mtlR expression, but at low levels (Fig. S1). Alternatively, there may be some cis-effects with regard to MtlR’s regulation of mtlA. For example, chromosomal mtlR may allow for MtlR to be synthesized and localized to specific regions of the cell that would allow for maximal activity. The mtlR locus may also express additional transcripts that affect mtlA expression, which were not appropriately expressed from the pMtlR construct.
As a repressor of mtlA, excess MtlR protein can impair V. cholerae growth in mannitol medium. Accordingly, we observed that ectopic expression of mtlR, even in the absence of IPTG, can impede growth of the bacteria when initially switched from LB to mannitol medium (data not shown). We also attempted to grow our mtlR complementation strain in mannitol in the absence of IPTG. Surprisingly, these cells harbouring the pMtlR plasmid successfully grew overnight and were able to reach mid-log phase following back-dilution. However, subsequent Western blot analysis revealed that less mtlR is expressed from pMtlR in V. cholerae grown in mannitol medium compared to non-mannitol medium (Fig. S1c). This decrease in ectopic mtlR expression could allow for enough MtlA to be made such that the cells are able to grow in mannitol medium. We reason that, through overnight growth, the bacteria adapted (potentially through suppressor mutations) to grow robustly in mannitol despite harbouring the pMtlR plasmid.
In wild-type V. cholerae, MtlR demonstrates a highly unusual expression pattern as a repressor, as expression is highest in conditions where it is not expected to be active (mannitol medium). We have yet to ascertain a mechanistic explanation for this expression profile. One possibility could be that MtlR acts as part of a regulatory complex. That is, although MtlR levels are high in mannitol, its putative protein partner(s) may be more active in non-mannitol medium, allowing for mtlA repression. Although MtlR structurally displays no favourable DNA-binding domains, the regulator could be acting in a larger transcriptional complex with other proteins – such as CRP or FruR – to modulate mtlA expression from the mtl locus in bacteria [31]. These potential MtlR protein-binding partners may confer the ability to bind macromolecules such as DNA or RNA, or these MtlR protein contacts could be responsible for activation and repression of MtlR activity. Our lab is currently investigating these potential binding partners.
From a functional and evolutionary standpoint, we have evidence to believe that expressing high levels of the repressor under non-repressive conditions could be advantageous for V. cholerae. The fact that MtlR lingers during a media switch from mannitol to non-mannitol media could mean that V. cholerae rely on high levels of the repressor to be present during transitions in order to accelerate adaptation to different media (Figs 6 and S8). In other words, the high levels of MtlR in mannitol medium allow the bacteria to rapidly shut-off MtlA production when the transporter is no longer needed. Indeed, one expects that in their aquatic reservoirs, V. cholerae would be subject to constant fluctuations, at a microscale level, of available nutrients and signalling molecules [4]. MtlR may be calibrating MtlA levels during the transition to better suit the lower extracellular levels of mannitol.
Expression of mtlA from the mtl locus is a highly regulated biological process in bacteria. The complexity of genetic regulation along the mtl locus is matched by the diverse functionality of MtlA in metabolic and environmental sensing processes. Mannitol is an important carbon source throughout the V. cholerae lifecycle; accordingly, the ability to quickly sense, acquire and process extracellular mannitol for energy remains important. In growth conditions without mannitol, though, the energy expenditure for robust expression of an unnecessary protein must be curbed. It is likely expression of mtlA is never fully shut off, however. V. cholerae may maintain MtlA protein at low levels to act as an environmental sensor for extracellular mannitol and to trigger the formation of a protective biofilm under permitting growth conditions. This complex mtlA expression cycle requires an equally elaborate regulation paradigm, which currently involves CRP, MtlS and MtlR.
Funding information
This research was supported by Pomona College, the Rose Hills Foundation (to N. V.), the Arnold and Mabel Beckman Foundation (to M. G. Z.), a National Science Foundation grant (CBET-1258307 to J. M. L.) and a National Institutes of Health grant (AI-090606 to J. M. L.). No one employed by the funders (other than the authors) played any role in the study or in the preparation of the article or in any decision to publish the article.
Acknowledgements
We thank Marc Kimball and Robert Scheffler for assistance in cloning various constructs used in this work. We thank Daniel Stoebel and Rou-Jia Sung for their comments during the preparation of this manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest associated with this research or the publication of this manuscript.
Supplementary Data
Footnotes
Abbreviations: glu, glucose; malt, maltose; mann, mannose; mtl, mannitol; PTS, phosphotrasferase system.
Eight supplementary figures and two supplementary tables are available with the online version of this article.
Edited by: P. Langford and M. Whiteley
References
- 1.Ali M, Lopez AL, You YA, Kim YE, Sah B, et al. The global burden of cholera. Bull World Health Organ. 2012;90:209–218. doi: 10.2471/BLT.11.093427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colwell RR. Infectious disease and environment: cholera as a paradigm for waterborne disease. Int Microbiol. 2004;7:285–289. [PubMed] [Google Scholar]
- 3.Reidl J, Klose KE. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev. 2002;26:125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 4.Lutz C, Erken M, Noorian P, Sun S, McDougald D. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front Microbiol. 2013;4:375. doi: 10.3389/fmicb.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merrell DS, Hava DL, Camilli A. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol. 2002;43:1471–1491. doi: 10.1046/j.1365-2958.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- 6.Moorthy S, Watnick PI. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol Microbiol. 2004;52:573–587. doi: 10.1111/j.1365-2958.2004.04000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, et al. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe. 2007;2:264–277. doi: 10.1016/j.chom.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Q, Dziejman M, Mekalanos JJ. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci USA. 2003;100:1286–1291. doi: 10.1073/pnas.0337479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deutscher J, Aké FM, Derkaoui M, Zébré AC, Cao TN, et al. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev. 2014;78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houot L, Chang S, Pickering BS, Absalon C, Watnick PI. The phosphoenolpyruvate phosphotransferase system regulates Vibrio cholerae biofilm formation through multiple independent pathways. J Bacteriol. 2010;192:3055–3067. doi: 10.1128/JB.00213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houot L, Watnick PI. A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm. J Bacteriol. 2008;190:311–320. doi: 10.1128/JB.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moorthy S, Watnick PI. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol Microbiol. 2005;57:1623–1635. doi: 10.1111/j.1365-2958.2005.04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houot L, Chang S, Absalon C, Watnick PI. Vibrio cholerae phosphoenolpyruvate phosphotransferase system control of carbohydrate transport, biofilm formation, and colonization of the germfree mouse intestine. Infect Immun. 2010;78:1482–1494. doi: 10.1128/IAI.01356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groisillier A, Shao Z, Michel G, Goulitquer S, Bonin P, et al. Mannitol metabolism in brown algae involves a new phosphatase family. J Exp Bot. 2014;65:559–570. doi: 10.1093/jxb/ert405. [DOI] [PubMed] [Google Scholar]
- 17.Iwamoto K, Shiraiwa Y. Salt-regulated mannitol metabolism in algae. Mar Biotechnol. 2005;7:407–415. doi: 10.1007/s10126-005-0029-4. [DOI] [PubMed] [Google Scholar]
- 18.Oddo E, Saiano F, Alonzo G, Bellini E. An investigation of the seasonal pattern of mannitol content in deciduous and evergreen species of the Oleaceae growing in northern Sicily. Ann Bot. 2002;90:239–243. doi: 10.1093/aob/mcf177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rousvoal S, Groisillier A, Dittami SM, Michel G, Boyen C, et al. Mannitol-1-phosphate dehydrogenase activity in Ectocarpus siliculosus, a key role for mannitol synthesis in brown algae. Planta. 2011;233:261–273. doi: 10.1007/s00425-010-1295-6. [DOI] [PubMed] [Google Scholar]
- 20.Rambhatla P, Kumar S, Floyd JT, Varela MF. Molecular cloning and characterization of mannitol-1-phosphate dehydrogenase from Vibrio cholerae. J Microbiol Biotechnol. 2011;21:914–920. doi: 10.4014/jmb.1104.04020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joyet P, Derkaoui M, Bouraoui H, Deutscher J. PTS-mediated regulation of the transcription activator MtlR from different species: surprising differences despite strong sequence conservation. J Mol Microbiol Biotechnol. 2015;25:94–105. doi: 10.1159/000369619. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe S, Hamano M, Kakeshita H, Bunai K, Tojo S, et al. Mannitol-1-phosphate dehydrogenase (MtlD) is required for mannitol and glucitol assimilation in Bacillus subtilis: possible cooperation of mtl and gut operons. J Bacteriol. 2003;185:4816–4824. doi: 10.1128/JB.185.16.4816-4824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Smith KP, Floyd JL, Varela MF. Cloning and molecular analysis of a mannitol operon of phosphoenolpyruvate-dependent phosphotransferase (PTS) type from Vibrio cholerae O395. Arch Microbiol. 2011;193:201–208. doi: 10.1007/s00203-010-0663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu JM, Livny J, Lawrence MS, Kimball MD, Waldor MK, et al. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res. 2009;37:e46. doi: 10.1093/nar/gkp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ymele-Leki P, Houot L, Watnick PI. Mannitol and the mannitol-specific enzyme IIB subunit activate Vibrio cholerae biofilm formation. Appl Environ Microbiol. 2013;79:4675–4683. doi: 10.1128/AEM.01184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou YY, Zhang HZ, Liang WL, Zhang LJ, Zhu J, et al. Plasticity of regulation of mannitol phosphotransferase system operon by CRP-cAMP complex in Vibrio cholerae. Biomed Environ Sci. 2013;26:831–840. doi: 10.3967/bes2013.006. [DOI] [PubMed] [Google Scholar]
- 27.Chang H, Replogle JM, Vather N, Tsao-Wu M, Mistry R, et al. A cis-regulatory antisense RNA represses translation in Vibrio cholerae through extensive complementarity and proximity to the target locus. RNA Biol. 2015;12:136–148. doi: 10.1080/15476286.2015.1017203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mustachio LM, Aksit S, Mistry RH, Scheffler R, Yamada A, et al. The Vibrio cholerae mannitol transporter is regulated posttranscriptionally by the MtlS small regulatory RNA. J Bacteriol. 2012;194:598–606. doi: 10.1128/JB.06153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Figge RM, Ramseier TM, Saier MH. The mannitol repressor (MtlR) of Escherichia coli. J Bacteriol. 1994;176:840–847. doi: 10.1128/jb.176.3.840-847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang HY, Yan MY, Zhao YW, Kan B. [Transcriptional repressor gene - - mtlR of mannitol PTS operon in Vibrio cholerae] Wei Sheng Wu Xue Bao. 2007;47:522–525. [PubMed] [Google Scholar]
- 31.Tan K, Clancy S, Borovilos M, Zhou M, Hörer S, et al. The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria. J Biol Chem. 2009;284:36670–36679. doi: 10.1074/jbc.M109.062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, et al. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 35.Lee SH, Angelichio MJ, Mekalanos JJ, Camilli A. Nucleotide sequence and spatiotemporal expression of the Vibrio cholerae vieSAB genes during infection. J Bacteriol. 1998;180:2298–2305. doi: 10.1128/jb.180.9.2298-2305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papenfort K, Förstner KU, Cong JP, Sharma CM, Bassler BL. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc Natl Acad Sci USA. 2015;112:E766. doi: 10.1073/pnas.1500203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D, Manos J, Ma X, Belas R, Karaolis DK. Transcriptional analysis and operon structure of the tagA-orf2-orf3-mop-tagD region on the Vibrio pathogenicity island in epidemic V. cholerae. FEMS Microbiol Lett. 2004;235:199–207. doi: 10.1016/j.femsle.2004.04.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.