Abstract
A gram-negative, budding, catalase negative, oxidase positive and non-motile bacterium (MBLW1T) with a complex endomembrane system has been isolated from a freshwater lake in southeast Queensland, Australia. Phylogeny based on 16S rRNA gene sequence analysis places the strain within the family Planctomycetaceae, related to Zavarzinella formosa (93.3 %), Telmatocola sphagniphila (93.3 %) and Gemmata obscuriglobus (91.9 %). Phenotypic and chemotaxonomic analysis demonstrates considerable differences to the type strains of the related genera. MBLW1T displays modest salt tolerance and grows optimally at pH values of 7.5–8.0 and at temperatures of 32–36 °C. Transmission electron microscopy analysis demonstrates the presence of a complex endomembrane system, however, without the typically condensed nucleoid structure found in related genera. The major fatty acids are 16 : 1 ω5c, 16 : 0 and 18 : 0. Based on discriminatory results from 16S rRNA gene sequence analysis, phenotypic, biochemical and chemotaxonomic analysis, MBLW1T should be considered as a new genus and species, for which the name Tuwongella immobilis gen. nov., sp. nov. is proposed. The type strain is MBLW1T (=CCUG 69661T=DSM 105045T).
Keywords: Planctomycetes, freshwater bacteria, nucleoid, phylogeny, fatty acids
The family Planctomycetaceae belongs to the phylum Planctomycetes within the domain Bacteria, members of which possess distinctive properties such as a complex endomembrane system, budding reproduction and the ability to perform endocytosis-like protein uptake [1, 2]. The family Planctomycetaceae is formed by the described genera Blastopirellula, Gemmata, Gimesia, Pirellula, Planctomicrobium, Planctopirus, Rhodopirellula, Rubinisphaera, Schlesneria, Telmatocola, Thermogutta and Zavarzinella [3–11]. Previously included in the family were the genera of Isosphaera, Singulisphaera, Aquisphaera and Paludisphaera, but these genera have recently been re-classified under the family Isosphaeraceae [12]. Here, we describe an isolate from a freshwater lake that is closely related to members of the genera Gemmata, Telmatocola and Zavarzinella but has a number of distinctive molecular and phenotypic features that distinguish it from the respective type strains G. obscuriglobus, T. sphagniphila and Z. formosa.
Strain MBLW1T was isolated from water samples collected from the freshwater University Lake at the University of Queensland, Brisbane, Australia. Samples were collected, using sterile bottles, from under the water surface at a level no deeper than 30 cm. Water samples were concentrated via filtration through a 0.45 µm membrane filter, with the concentrated particulate material on the filter being resuspended in approximately 3 ml of sterile lake water filtrate. Lake Water (LW) agar plates (15 g agar autoclaved in 250 ml distilled water, upon cooling 25 µg ml−1 filter-sterilised ampicillin added and the volume made up to 1 l with the sterile filtrate that resulted above) were spread with 100 µl of the water concentrate. Plates were then incubated in the light at room temperature in a CO2-enriched atmosphere generated in an anaerobic jar containing an Alka-seltzer dissolved in a beaker of water [13]. After 38 days, colonies were subcultured to purity and maintained on M1 agar [14], in the dark, at 28 °C, aerobically. During the following cultivation on LW agar plates, the incubation time decreased to approximately 5 days at 28 °C. MBLW1T was subsequently cultivated aerobically in the dark on M1 agar plates at 28 °C. Colonies of MBLW1T were smooth, with an entire edge, translucent pink in colour, up to 4 mm in diameter and had a large, flat zone of growth with a slightly raised centre, giving them a ‘fried egg’ appearance.
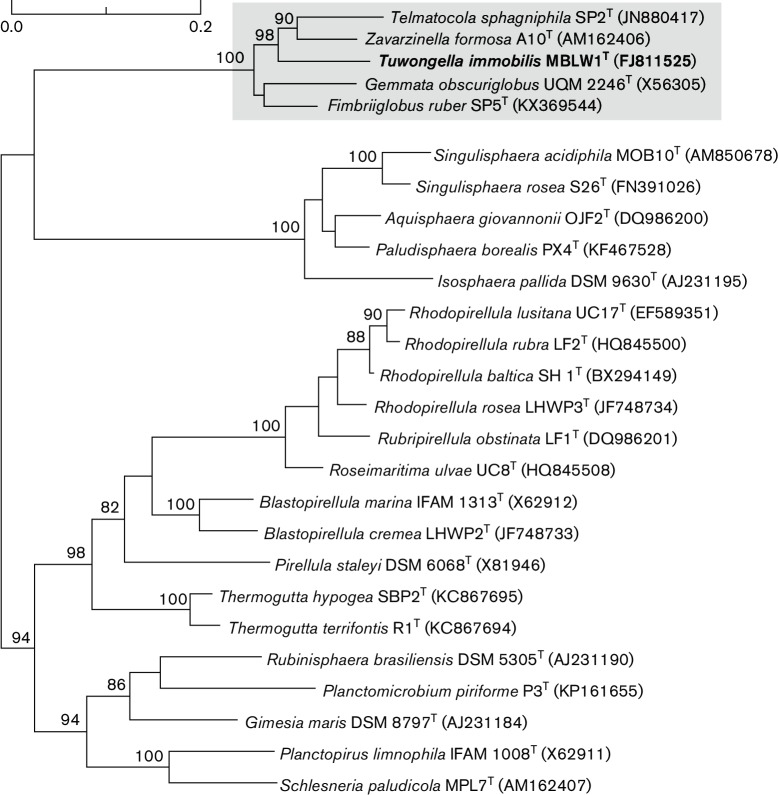
16S rRNA gene sequence of MBLW1T was determined by PCR and sequencing. The phylogenetic analyses of MBLW1T were based on alignments of 16S rRNA sequences from 25 Planctomycetes species. The 16S rRNA gene sequences were aligned using silva Incremental Aligner [15] and the hyper-variable regions were removed from the aligned sequences using the Lane mask [16]. The phylogeny was performed in RaxML version 8.0.26 using the maximum likelihood method with 50 bootstraps and the GTRGAMMAI nucleotide substitution model [17].
The strain MBLW1T shared 91.9 % sequence similarity with G. obscuriglobus which makes it a new sister genus to Gemmata. Pairwise sequence similarity measures among cultivated species showed that MBLW1T was most similar to both Z. formosa and T. sphagniphila, with a sequence similarity of 93.5 and 93.3 % respectively. The maximum likelihood phylogeny confirmed that MBLW1T clusters with G. obscuriglobus, Z. formosa and T. sphagniphila with 100 % bootstrap support, although their internal diversification pattern could not be resolved with significant support solely by 16S rRNA gene phylogeny (Fig. 1). The DNA G+C content of MBLW1T was determined to be 57 mol%, compared to 67 mol% in G. obscuriglobus and 59 mol% in both T. sphagniphila and Z. formosa. The DNA G+C content information for strain MBLW1T was calculated from the genome sequence (Mahajan, Yee, Fuerst, Andersson, unpublished data).
Fig. 1.
Phylogenetic placement of MBWL1T within the order of Planctomycetales. The phylogeny was inferred from a 16S rRNA gene alignment using the maximum likelihood method. Numbers on the nodes refer to bootstrap support values. Only bootstrap support values higher than 75 % are shown. Nucleotide sequence accession numbers are presented next to the species names.
The 16S rRNA gene sequence of strain MBLW1T possess all Planctomycetes signature nucleotides [18], except for the nucleotide in position 983 : 1, which is missing. The MBLW1T 16S rRNA also possess all Gemmata group signature nucleotides [19], except at positions 668 : 738 (U : A instead of the signature C : G), 680 : 710 (G : U instead of the signature C : G/U : A) and 1420 : 1480 (G : C instead of the signature A : U) (Escherichia coli numbering) [20]. Interestingly, in distinction to Z. formosa, G. obscuriglobus and all other Gemmata clade sequences, the MBLW1T16S rDNA sequence lacks the 10-base indel between E. coli positions 998 and 999, which is otherwise characteristic of the Gemmata group [21], including Z. formosa [5] and have an extended helix, Helix 10, compared to other members of the Gemmata group.
The majority of the phenotypic, biochemical and chemotaxonomic analyses were performed with MBLW1T and G. obscuriglobus ACM 2246T. Z. formosa DSM 19928T was only included for TEM analysis. Both T. sphagniphila and Z. formosa require cultivation under acidic conditions and at relatively low temperatures (20–25 °C) [5, 6], while MBLW1T and G. obscuriglobus could be cultivated under comparable conditions on M1 agar plates, at neutral pH and elevated temperature (>30 °C). Furthermore, both T. sphagniphila and Z. formosa grow considerably slower than MBLW1T and G. obscuriglobus.
For analysis by transmission electron microscopy (TEM), MBLW1T and G. obscuriglobus were cultivated for 4 days at 28 °C on M1 agar plates. Z. formosa was cultivated on DSM 1196 agar plates for 30 days at 24 °C. Cells were fixed for 15 min at room temperature in 2 % glutaraldehyde, 1 % paraformaldehyde in 0.01 M phosphate buffer, pH 7.4. The cells were further fixed and stored at 4 °C. Further preparation of the specimen was performed by the electron microscopy unit, Emil, at Karolinska Institutet, Huddinge, Sweden. After fixation, cells were rinsed in 0.01 M phosphate buffer and centrifuged. The pellets were then postfixed in 2 % osmium tetroxide (TAAB) in 0.01 M phosphate buffer, pH 7.4 at 4 °C for 2 h, dehydrated in ethanol, followed by acetone and embedded in LX-112 (Ladd Research). Ultrathin sections (~50–60 nm) were cut by a Leica EM UC 6. Sections were contrasted with uranyl acetate followed by lead citrate and examined in a Hitachi HT 7700 at 80 kV. Digital images were taken by using a Veleta camera (Olympus Soft Imaging Solutions). For negative staining, MBLW1T was grown on M1 agar plates at 28 °C for 2 days, and a suspension of the cells was stained with 1.0 % uranyl acetate in 0.4 % (w/v) sucrose. Cells were examined via a JEOL 1010 transmission electron microscope. For phase contrast microscopy, MBLW1T and G. obscuriglobus were grown for 4 days on M1 agar plates at 28 °C, resuspended in M1 medium and a drop from the cell suspension was placed on a slab of 1 % agarose (w/v) on a slide and allowed to soak in. Cover slips were placed over the agarose and samples viewed using a Zeiss Axioplan2 fluorescence microscope. Motility was checked by phase contrast microscopy of a suspension of cells placed directly onto a slide. For this, a preparation of cells from just-visible colonies growing on an M1 agar plate was used.
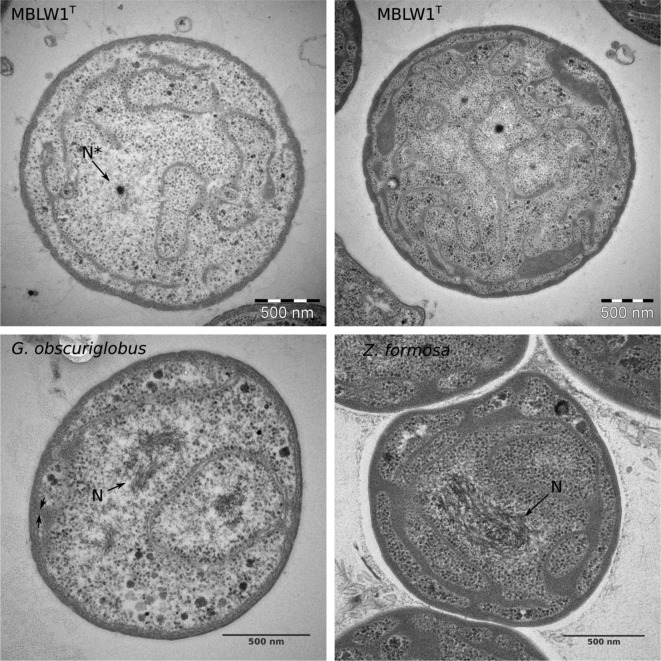
Phase-contrast images of MBLW1T show phase-variable regions within the cells and TEM analysis revealed a complex endomembrane system, similar to G. obscuriglobus and Z. formosa (Fig. 2). Compared to G. obscuriglobus and Z. formosa that both display very distinct nucleoid structures, the nucleoid of MBLW1T is only weakly visible in some TEM micrographs. Crateriform structures, a defining feature within the Planctomycetes phylum [1], were visible over the entire surface of negatively stained MBLW1T cells. Fimbriae could also be observed. Motility of MBLW1T has not been observed, not even in very young cultures and this is consistent with the lack of flagella observed in negatively stained cells (Fig. S1a, available in the online version of this article). This is in contrast to G. obscuriglobus and other Gemmata-like species, like Z. formosa, all of which are motile at some stage of the life cycle [3, 5, 21]. No stalks were apparent in MBLW1T when observed via phase contrast microscopy or TEM of negatively stained cells (Fig. S1b). Mature cells were cocci, with the cell diameter of MBLW1T ranging from 2.2 to 3.1 µm. MBLW1T reproduced by budding. No rosette or chain formation of the cells was observed.
Fig. 2.
Transmission electron micrographs show the complex endomembrane system in MBLW1T, G. obscuriglobus and Z. formosa, a distinct nucleoid (N) in G. obscuriglobus and Z. formosa and diffuse fibrillar nucleoid structures (N*) in one of the two MBWL1T cells.
The growth conditions of MBLW1T were investigated on M1 agar plates and in M1 liquid medium. On M1 agar plates, MBLW1T was able to grow at temperatures of 20 to 40 °C. In M1 liquid medium, growth of MBLW1T was monitored by turbidometry at a wavelength of λ=600 nm and the optimal growth temperature was found between 32 and 36 °C (Fig. S2a) with corresponding generation times of g32 ~11 h and g36 ~6 h, respectively. Growth of MBLW1T under different pH conditions was investigated in M1 liquid media and was performed to distinguish MBLW1T from the acidophilic species Z. formosa and T. sphagniphila, and establish the optimum pH for growth. Variations of the pH between pH 6.0–8.0 were achieved by using a Na2HPO4-NaH2PO4 buffering system, variations from pH 9.0–10.5 were achieved by using a Na2CO3-NaHCO3 buffering system. MBLW1T was grown for 48 h at 32 °C in Nunc Edge 96-well microplates (Thermo Scientific) and the optical density (λ=600 nm) was measured using a Tecan Infinite M200 microplate reader (Tecan). Growth of MBLW1T was observed from pH 6.0–10.5, optimal growth occurred at pH 7.5–8.0 (Fig. S2b). Salt tolerance of MBLW1T and G. obscuriglobus was compared in 24-well M1 agar plates containing increasing NaCl-concentrations [0.0 %–2.0 % (w/v)] after incubation for 5 days at 32 °C. Growth of MBLW1T was observed up to 0.5 % (w/v) NaCl, at concentrations between 0.6 % (w/v) and 0.8 % (w/v), the cells were only growing weakly. Growth of G. obscuriglobus was observed up to 1.5 % (w/v) NaCl but not observed at 2.0 % (w/v) NaCl. Additionally, MBLW1T was grown in M1 liquid media in a 24-well plate format in the presence of increasing NaCl concentrations from 0.0–0.8 % (w/v). After incubation for 5 days at 30 °C, the optical density at λ=600 nm was measured in a UV-spectrophotometer and values of the NaCl-containing samples were compared with the NaCl-free samples to determine the relative growth. Growth of MBLW1T was inhibited by more than 50 % at NaCl concentrations greater than 0.4 % (w/v) (Fig. S2c). At concentrations greater than 0.8 % (w/v) NaCl, essentially no growth of MBLW1T was observed.
For comparing the antibiotic susceptibility of MBLW1T and G. obscuriglobus, cells were grown on M1 agar plates (without CaCO3) for 7 days at 28 °C in the dark using discs containing ampicillin (10 µg), gentamicin (10 µg), kanamycin (30 µg), neomycin (10 µg) and streptomycin (10 µg). MBLW1T and G. obscuriglobus are susceptible to kanamycin and neomycin and resistant to ampicillin and streptomycin. In contrast to G. obscuriglobus, MBLW1T is resistant to gentamicin. Susceptibility of G. obscuriglobus to gentamicin is in accordance with previous results [22]. The comparison of antibiotic susceptibility profiles with Z. formosa and T. sphagniphila is shown in Table 1.
Table 1. Phenotypic characteristics of MBLW1T in comparison to G. obscuriglobus, T. sphagniphila and Z. formosa.
The four strains utilize N-acetylglucosamine, glucose, galactose, maltose and rhamnose (weak growth in MBLW1T and G. obscuriglobus). The four strains do not utilize fructose, mannitol and glycerol. MBLW1T and G. obscuriglobus grow weakly on arabinose (not tested for T. sphagniphila and Z. formosa). The four strains are resistant to ampicillin and streptomycin but susceptible to kanamycin. R, resistant; S, susceptible.
| Characteristic | MBLW1T | G. obscuriglobus | Z. formosa* | T. sphagniphila* |
|---|---|---|---|---|
| Cell shape | Spherical | Spherical to ovoid | Ellipsoidal | Spherical |
| Cell size (µm) | 2.2–3.1 | 1.9–2.4 | 2.5–3.2 x 2.0–2.5 | 1.2–2.0 |
| Rosette formation | − | − | + | + |
| Stalked | − | − | + | + |
| Motile | − | + | + | − |
| Growth pH <5.0 | − | −* | + | + |
| Growth >30 °C | + | + | − | − |
| Major fatty acids | 16 : 1ω5c, 16 : 0, 18 : 0 | 18 : 0, 16 : 1ω5c, 15 : 0 ANTEISO | 18 : 1ω5c, 16 : 1ω5c, 18 : 0 | 16 : 1ω5c, 18 : 0, 18 : 1ω5c |
| DNA G+C content (mol%) | 57 | 67† | 59† | 59 |
| NaCl tolerance | 0.5 % | 1.5 % | 0.6 % | 0.1 % |
| Oxidase test | + | − | + | − |
| Catalase test | − | + | + | + |
| Urease test | + | − | − | − |
| Carbon sources | ||||
| Sucrose | − | + | + | + |
| Xylose | − | − | + | + |
| Antibiotic susceptibility | ||||
| Gentamicin | R | S | S | S |
| Neomycin | S | S | S | R |
The oxidase test based on the cytochrome oxidase catalysed reaction of N,N-dimethyl-p-phenylenediamine oxalate and α-naphthol to indophenol blue [23, 24] was performed using oxidase test discs (Sigma Aldrich). MBLW1T and G. obscuriglobus were grown for 4 days at 28 °C on M1 agar plates and cells were transferred with a sterile cotton tip onto oxidase test discs. The species were deemed oxidase positive if the colour on the disc changed to deep purple blue within 2 min at room temperature. For the catalase test, 4 day-old cultures (M1 agar, 28 °C) of MBLW1T and G. obscuriglobus were transferred with a sterile cotton tip into 1 ml 3 % (v/v) H2O2. Immediate formation of bubbles was indicative for a positive catalase test. For the urease test, MBLW1T and G. obscuriglobus were grown in M1 liquid medium supplemented with 2 % (w/v) urea, 0.1 % (w/v) glucose and 0.002 % (w/v) phenol red for 7 days at 32 °C in 24-well plates under orbital shaking at 100 r.p.m. Acid production (red to yellow) is indicative for a urease negative result, while base production (red to orange) is indicative for a urease positive result. The oxidation-fermentation test was essentially performed as described by Hugh and Leifson [25] to test for fermentative metabolism of the relevant sugars. Test tubes with 4 ml M1 media, supplemented with 0.003 % Bromothymol blue, 0.3 % (w/v) agar and 0.2 % (w/v) glucose and 0.2 % (w/v) N-acetyl glucosamine, respectively, were inoculated with MBLW1T and G. obscuriglobus and incubated for 6 days at 32 °C in the dark under oxidative and fermentative conditions (addition of a layer of mineral oil). In addition to the Hugh-Leifson test, carbohydrate utilization of MBLW1T and G. obscuriglobus was determined on M1 agar plates supplemented with 0.2 % (w/v) of different carbohydrates after cultivation for 6 days at 32 °C.
Carbohydrate utilization profiles of G. obscuriglobus and Z. formosa are similar, with no apparent difference regarding the two main preferred carbohydrates, N-acetylglucosamine and glucose [5]. In the Hugh-Leifson test, both MBLW1T and G. obscuriglobus display fermentative metabolism of N-acetylglucosamine and glucose (Table 1). Hence MBLW1T grows best under aerobic conditions with carbohydrates, but displays similar facultative aerobic characteristics to Schlesneria paludicola [4] and Planctopirus limnophila [8, 26]. Further investigation of carbohydrate utilization showed similar profiles, except that MBLW1T, in contrast to G. obscuriglobus, does not grow on sucrose. Strain MBLW1T displays additional distinct biochemical characteristics compared to G. obscuriglobus. MBLW1T is catalase negative, oxidase positive and urease positive, while G. obscuriglobus is catalase positive, oxidase negative and urease negative. Z. formosa has been reported to be catalase positive, oxidase positive and urease negative [5].
Analyses of fatty acid methyl esters (FAME) in strain MBLW1T and the reference strain G. obscuriglobus were performed with aid from the Identification Service of the DSMZ, Braunschweig, Germany. Prior to FAME analysis, MBLW1T and G. obscuriglobus were cultivated on M1 agar plates for 5 days at 32 °C, harvested, washed in 50 mM Tris, pH 7.5 and dried in a Speedvac at 45 °C.
The most abundant fatty acids in MBLW1T were 16 : 1 ω5c, 16 : 0 and 18 : 0, while the most abundant fatty acids in G. obscuriglobus were 18 : 0, 16 : 1 ω5c and 15 : 0 ANTEISO (Table 2). Three fatty acids (16 : 0 3OH, 14 : 0, 13 : 0 iso 3OH) were found with low abundance in MBLW1T that were not identified in G. obscuriglobus. MBLW1T displays therefore a distinct fatty acid profile compared to G. obscuriglobus. Furthermore, the fatty acid profile of MBLW1T differs from T. sphagniphila and Z. formosa, obtained by phospholipid fatty acid analysis [5, 6], where the major fatty acids are 16 : 1 ω5c, 18 : 1 ω5c and 18 : 0. Fatty acids with relative abundance of less than 1 % are shown in Table S1.
Table 2. Fatty acid composition of MBLW1T in comparison to G. obscuriglobus based on FAME analysis.
Values are percentages of total fatty acids. The major fatty acids of each species are highlighted in bold. Only fatty acids with a relative abundance of more than 1% are shown. A detailed summary of the fatty acid analysis is shown in Table S1.
| Fatty acid | MBLW1T | G. obscuriglobus |
|---|---|---|
| 15 : 0 ANTEISO | nd | 7.9 |
| 16 : 0 | 25.0 | 3.6 |
| 16 : 0 ISO | nd | 1.4 |
| 16 : 0 3OH | 2.3 | nd |
| 16 : 1 ω5c | 59.0 | 27.5 |
| 17 : 0 | 2.2 | 0.3 |
| 17 : 0 ANTEISO | nd | 4.6 |
| 18 : 0 | 6.7 | 49.2 |
| 18 : 1 ω5c | 2.5 | 2.5 |
| 18 : 1 ω7c | nd | 1.6 |
The 16S rRNA gene sequence from MBLW1T has been deposited in the EMBL/NCBI databases under accession number FJ811525.
The major characteristics that distinguish MBLW1T from G. obscuriglobus, T. sphagniphila and Z. formosa are summarized in Table 1. Strain MBLW1T differs from the type strains of related genera in the absence of motility, except for T. sphagniphila, and flagella, in the phylogenetic position based on 16S rRNA, in the absence of a Gemmata-group characteristic 10-base indel in 16S rRNA, several biochemical properties and in the fatty acid composition, including the presence of three fatty acids, C14 : 0, C13 : 0iso3OH and C16 : 03OH. MBLW1T shares with G. obscuriglobus the presence of a complex endomembrane system, spherical cell morphology and colony colour. We therefore propose strain MBLW1T to be classified as the representative of a novel genus and species for which the name Tuwongella immobilis gen. nov., sp. nov. is proposed.
Description of Tuwongella gen. nov.
Tuwongella (Tu.wong.el’la. L. dim. ending –ella; N.L. fem. n. Tuwongella referring to Tu-wong, a region of the Brisbane River, Australia).
Gram-negative cells are spherical, non-motile and reproduce by budding. Crateriform structures are visible over the entire cell surface. Cells do not have stalks and do not form rosettes. Colonies are translucent and pink. Mesophilic and neutrophilic cells are chemoorganotrophic facultative aerobes. The major fatty acids are 16 : 1 ω5 c and 16 : 0. Cells possess a complex endomembrane system characteristic for members of the phylum Planctomycetes. The genus is a member of the phylum Planctomycetes, order Planctomycetales, family Planctomycetaceae. The type species is Tuwongella immobilis.
Description of Tuwongella immobilis sp. nov.
Tuwongella immobilis (im.mo'bi.lis. L. fem. adj. immobilis non-motile) referring to a non-motile member of the family Planctomycetaceae.
Displays the properties given in the genus description with following traits. Spherical cells have a diameter of 2.2–3.1 µm. Growth occurs at pH 6.0–10.5 (optimum at pH 7.5–8.0) and at temperatures of 20–40 °C (optimum at 32–36 °C). Growth is inhibited by more than 50 % at NaCl concentrations greater than 0.4 % (w/v). DNA G+C content is 57 mol%. Oxidase positive, catalase negative and urease positive. Carbon sources are N-acetylglucoseamine, arabinose, galactose, glucose, maltose and rhamnose. Does not utilize sucrose, fructose, mannitol, xylose and glycerol. Fermentative metabolism of N-acetylglucoseamine and glucose. Resistant to ampicillin, gentamicin and streptomycin. Sensitive to kanamycin and neomycin.
The type strain MBLW1T (=CCUG 69661 T=DSM 105045T) was isolated from the University Lake, on the St. Lucia campus of The University of Queensland, Brisbane, Queensland, Australia.
Funding information
This project has been funded by grants to S. G. E. A. from the Swedish Research Council (349-2007-8732, 621-2014-4460) and the Knut and Alice Wallenberg Foundation (2011.0148, 2012.0075), and to J. A. F. from the Australian Research Council.
Acknowledgements
We thank Richard I. Webb from the University of Queensland, Brisbane, Australia for technical assistance and Paul Memmott, Ray Kerkhove and Alex Bond, of the Aboriginal Environments Research Centre, for advice on indigenous Australian place names of the Brisbane region.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: FAME, fatty acid methyl ester; LW, lake water; TAAB, osmium tetroxide; TEM, transmission electron microscopy.
The 16S rRNA gene sequence of Tuwongella immobilis has been deposited in the NCBI/EBI databases under accession number FJ811525.
One supplementary table and two supplementary figures are available with the online version of this article.
References
- 1.Fuerst JA. The planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology. 1995;141:1493–1506. doi: 10.1099/13500872-141-7-1493. [DOI] [PubMed] [Google Scholar]
- 2.Lonhienne TG, Sagulenko E, Webb RI, Lee KC, Franke J, et al. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA. 2010;107:12883–12888. doi: 10.1073/pnas.1001085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franzmann PD, Skerman VB. Gemmata obscuriglobus, a new genus and species of the budding bacteria. Antonie van Leeuwenhoek. 1984;50:261–268. doi: 10.1007/BF02342136. [DOI] [PubMed] [Google Scholar]
- 4.Kulichevskaya IS, Ivanova AO, Belova SE, Baulina OI, Bodelier PL, et al. Schlesneria paludicola gen. nov., sp. nov., the first acidophilic member of the order Planctomycetales, from sphagnum-dominated boreal wetlands. Int J Syst Evol Microbiol. 2007;57:2680–2687. doi: 10.1099/ijs.0.65157-0. [DOI] [PubMed] [Google Scholar]
- 5.Kulichevskaya IS, Baulina OI, Bodelier PL, Rijpstra WI, Damsté JS, et al. Zavarzinella formosa gen. nov., sp. nov., a novel stalked, Gemmata-like planctomycete from a Siberian peat bog. Int J Syst Evol Microbiol. 2009;59:357–364. doi: 10.1099/ijs.0.002378-0. [DOI] [PubMed] [Google Scholar]
- 6.Kulichevskaya IS, Serkebaeva YM, Kim Y, Rijpstra WI, Damsté JS, et al. Telmatocola sphagniphila gen. nov., sp. nov., a novel dendriform planctomycete from northern wetlands. Front Microbiol. 2012;3:1–9. doi: 10.3389/fmicb.2012.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulichevskaya IS, Ivanova AA, Detkova EN, Rijpstra WI, Sinninghe Damsté JS, et al. Planctomicrobium piriforme gen. nov., sp. nov., a stalked planctomycete from a littoral wetland of a boreal lake. Int J Syst Evol Microbiol. 2015;65:1659–1665. doi: 10.1099/ijs.0.000154. [DOI] [PubMed] [Google Scholar]
- 8.Scheuner C, Tindall BJ, Lu M, Nolan M, Lapidus A, et al. Complete genome sequence of Planctomyces brasiliensis type strain (DSM 5305T), phylogenomic analysis and reclassification of Planctomycetes including the descriptions of Gimesia gen. nov., Planctopirus gen. nov. and Rubinisphaera gen. nov. and emended descriptions of the order Planctomycetales and the family Planctomycetaceae. Stand Genomic Sci. 2014;9:10. doi: 10.1186/1944-3277-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlesner H, Rensmann C, Tindall BJ, Gade D, Rabus R, et al. Taxonomic heterogeneity within the Planctomycetales as derived by DNA-DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int J Syst Evol Microbiol. 2004;54:1567–1580. doi: 10.1099/ijs.0.63113-0. [DOI] [PubMed] [Google Scholar]
- 10.Schlesner H, Hirsch P. Rejection of the genus name Pirella for pear-shaped budding bacteria and proposal to create the genus Pirellula gen. nov. Int J Syst Bacteriol. 1987;37:441. doi: 10.1099/00207713-37-4-441. [DOI] [Google Scholar]
- 11.Slobodkina GB, Kovaleva OL, Miroshnichenko ML, Slobodkin AI, Kolganova TV, et al. Thermogutta terrifontis gen. nov., sp. nov. and Thermogutta hypogea sp. nov., thermophilic anaerobic representatives of the phylum Planctomycetes. Int J Syst Evol Microbiol. 2015;65:760–765. doi: 10.1099/ijs.0.000009. [DOI] [PubMed] [Google Scholar]
- 12.Kulichevskaya IS, Ivanova AA, Suzina NE, Rijpstra WI, Sinninghe Damsté JS, et al. Paludisphaera borealis gen. nov., sp. nov., a hydrolytic planctomycete from northern wetlands, and the proposal of Isosphaeraceae fam. nov. Int J Syst Evol Microbiol. 2015;66:837–844. doi: 10.1099/ijsem.0.000799. [DOI] [PubMed] [Google Scholar]
- 13.Giovannoni SJ, Godchaux W, Schabtach E, Castenholz RW. Cell wall and lipid composition of Isosphaera pallida, a budding eubacterium from hot springs. J Bacteriol. 1987;169:2702–2707. doi: 10.1128/jb.169.6.2702-2707.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlesner H. The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst Appl Microbiol. 1994;17:135–145. doi: 10.1016/S0723-2020(11)80042-1. [DOI] [Google Scholar]
- 15.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane DJ. 16S/23S rRNA Sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. Chichester, UK: John Wiley and Sons; 1991. pp. 115–175. (editors) [Google Scholar]
- 17.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuerst JA, Gwilliam HG, Lindsay M, Lichanska A, Belcher C, et al. Isolation and molecular identification of planctomycete bacteria from postlarvae of the giant tiger prawn, Penaeus monodon. Appl Environ Microbiol. 1997;63:254–262. doi: 10.1128/aem.63.1.254-262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brosius J, Dull TJ, Sleeter DD, Noller HF. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Jenkins C, Webb RI, Fuerst JA. Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl Environ Microbiol. 2002;68:417–422. doi: 10.1128/AEM.68.1.417-422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cayrou C, Raoult D, Drancourt M. Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J Antimicrob Chemother. 2010;65:2119–2122. doi: 10.1093/jac/dkq290. [DOI] [PubMed] [Google Scholar]
- 23.Gaby WL, Hadley C. Practical laboratory test for the identification of Pseudomonas aeruginosa. J Bacteriol. 1957;74:356–358. doi: 10.1128/jb.74.3.356-358.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon J, Mcleod JW. The practical application of the direct oxidase reaction in bacteriology. J Pathol Bacteriol. 1928;31:185–190. doi: 10.1002/path.1700310206. [DOI] [Google Scholar]
- 25.Hugh R, Leifson E. The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various gram negative bacteria. J Bacteriol. 1953;66:24–26. doi: 10.1128/jb.66.1.24-26.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirsch P, Müller M. Planctomyces limnophilus sp. nov., a stalked and budding bacterium from freshwater. Syst Appl Microbiol. 1985;6:276–280. doi: 10.1016/S0723-2020(85)80031-X. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.