Abstract
Control of host epigenetics is becoming evident as a mechanism by which symbionts and pathogens survive. Anaplasma phagocytophilum, an obligate intracellular bacterium, downregulates multiple host defense genes where histone deacetylase 1 (HDAC1) binds and histone 3 is deacetylated at their promoters, including the NADPH oxidase component, CYBB. How HDAC1 is targeted to defense gene promoters is unknown. Ankyrin A (AnkA), an A. phagocytophilum T4SS effector, enters the granulocyte nucleus, binds stretches of AT-rich DNA and alters transcription of antimicrobial defense genes, including downregulation of CYBB. Here we found AnkA binds to a predicted matrix attachment region in the proximal CYBB promoter. Using the CYBB promoter as a model of cis-gene silencing, we interrogated the mechanism of AnkA-mediated CYBB repression. The N-terminus of AnkA was critical for nuclear localization, the central ANK repeats and C-terminus were important for DNA binding, and most promoter activity localized to the central ANK repeats. Furthermore, a direct interaction between AnkA and HDAC1 was detected at the CYBB promoter, and was critical for AnkA-mediated CYBB repression. This novel microbial manipulation of host chromatin and gene expression provides important evidence of the direct effects that prokaryotic nuclear effectors can exert over host transcription and function.
Introduction
Global transcriptome analyses of mammalian cells in response to intracellular bacteria have led to the identification of major pathways affected during infection (Bryant et al., 2004). Targeting of individual cellular pathways by single effectors to alter global gene expression is very inefficient for microbes with limited genetic and metabolic resources. Eukaryotic cellular function is governed largely by transcriptional programs that are well-coordinated and tightly regulated, as evident with the reprogramming of differentiated cells toward a pluripotent state with Oct3/4, Sox2, Klf4 and c-Myc (Takahashi et al., 2006, Takahashi et al., 2007). The overarching mechanism of control over transcriptional and functional programs in eukaryotes occurs via the epigenome, including chromatin and associated histone and non-histone elements that regulate transcription both in “cis” from individual genes and in “trans” by modulating and coordinating activity over entire chromosomal domains. Chromatin modification by bacterial nucleomodulins is becoming more evident as a mechanism of host manipulation to allow for pathogen survival (Bierne et al., 2012).
The obligate intracellular bacterial pathogen Anaplasma phagocytophilum survives and propagates within neutrophils, abundant, short-lived, and aggressive phagocytes whose main function is microbial killing. With infection, A. phagocytophilum abrogates key neutrophil antimicrobial functions including oxidative burst, apoptosis, margination, emigration, and phagocytosis while activating proinflammatory responses including degranulation and cytokine/chemokine production (Akkoyunlu et al., 2000, Kim et al., 2000, Mott et al., 2002, Scaife et al., 2003, Choi et al., 2004, Garyu et al., 2005). Changes in these neutrophil functions are in part attributed to A. phagocytophilum-modulated host cell transcription (Borjesson et al., 2005, de la Fuente et al., 2005, Pedra et al., 2005, Lee et al., 2008). Upon A. phagocytophilum infection, downregulation of many host defense genes occurs along with histone deacetylase 1 (HDAC1) binding and histone H3 (H3) deacetylation at their promoters (Garcia-Garcia et al., 2009a). HDAC1 activity and expression are increased with infection and are critical for A. phagocytophilum survival (Garcia-Garcia et al., 2009a). Yet, how HDAC1 is targeted to defense gene promoters with infection is not clear.
One gene target of HDAC1 with A. phagocytophilum infection is CYBB, which encodes the gp91phox component (NOX2) of the NADPH oxidase. Downregulation of CYBB results in decreased superoxide anion production by the NADPH oxidase, a key mechanism of pathogen killing for neutrophils. Ankyrin A (AnkA), an A. phagocytophilum type IV secretion system (T4SS) effector localizes to the host nucleus and binds to the CYBB promoter, along with other defense genes, to decrease transcription. The mechanism of AnkA repression has yet to be determined. We previously showed AnkA binds DNA in a sequence-independent manner, but to regions that are rich in AT nucleotides. AT-richness is a common feature of unstable regions that become base unpaired (BURs) under negative superhelical stress. BURs are found within matrix attachment regions (MARs), specialized DNA structures that serve as attachment sites for nuclear matrix proteins such as lamins, scaffold attachment factor-1, and the special AT binding protein-1 (SATB1) that organize nuclear chromatin for tissue-specific gene expression, chromatin accessibility, and long-range chromatin modifications (Wang et al., 2010). Here we show that AnkA binds known and predicted MARs, and interrogate domains in AnkA that control nuclear localization and DNA binding. Further, we show that AnkA mimics eukaryotic MAR-binding proteins to anchor to DNA, recruits HDAC1 to its binding site in cis to the CYBB promoter, and decreases its transcription, which ultimately provides enhanced microbial survival.
Results
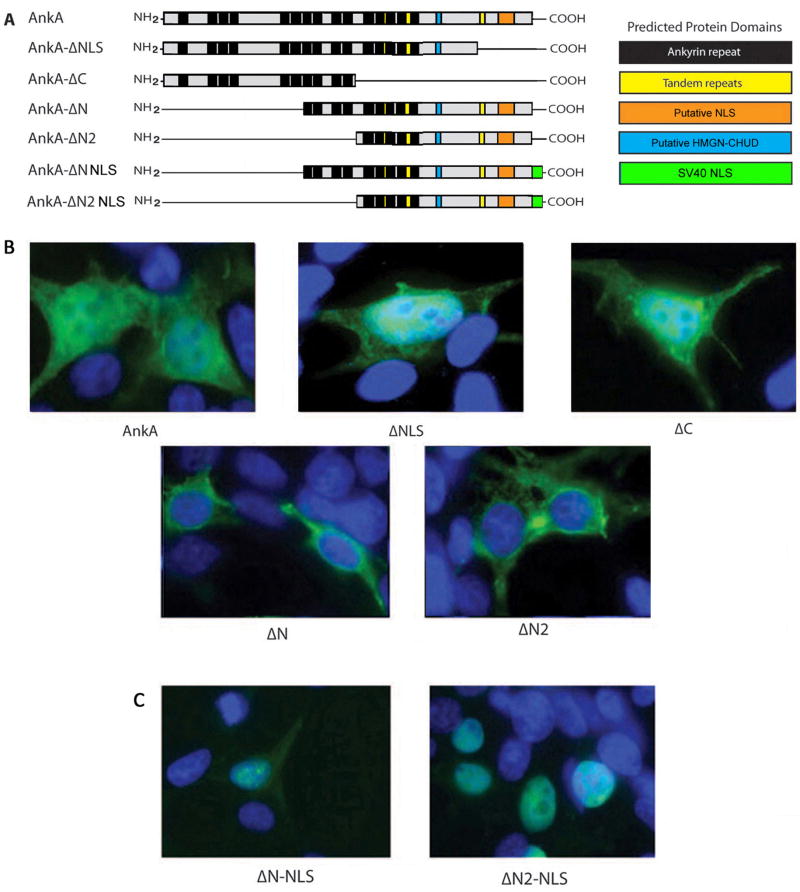
Nuclear localization of AnkA is determined by ANK repeats 1–6
Decreased transcription from the CYBB promoter requires that AnkA enter the nucleus. Suppression of gene transcription by AnkA in cis at CYBB requires the coordinated events of nuclear translocation, DNA binding, and suppressor activity. To investigate this, we constructed AnkA 5’ and 3’ deletion mutants designed to interrogate the roles of predicted eukaryotic domains and motifs, including multiple (8 to 15) ankyrin (ANK) repeats (33-residue motifs that can mediate protein-protein interactions), a putative mammalian nuclear localization sequence (NLS), and a putative high mobility group N protein chromosomal unfolding domain (HMGN-CHUD) (Figure 1A); none have established functions in AnkA. When transfected into HEK293T cells, we found that AnkA-ΔN and AnkA-ΔN2 truncations localized exclusively to the cytoplasm (Figure 1B; Figure S1), whereas AnkA-ΔC and -ΔNLS truncations localized to the nucleus. These results contradicted the prediction of an NLS in the C-terminus and suggested that the NLS was in the N-terminal region of AnkA, probably within the first 4–6 ANK repeats. Eukaryotic ANK repeats can act as nuclear localization signals via RanGDP binding at 2 consecutive ANK domains where the 13th residues in each are hydrophobic (Lu et al., 2014). An alignment of AnkA protein sequences among 31selected from A. phagocytophilum consistently identified 15 ANK repeats. Conserved hydrophobic residues at the 13th positions in ANK repeat regions 1, 2, 3, 4, 7, 8, 9, 11, 12, and 15 were identified in all 31 AnkA proteins (Fig. S2), despite overall amino acid sequence identity that ranged from 68% to 100% for the 31 AnkAs examined.
Figure 1. NLS activity in AnkA resides within the first 4 ANK repeat regions.
(A) Schematic of AnkA truncated mutants encoded by the pCMV-tag2b and pT7-FLAG-MAT-Tag-2 vectors and predicted locations of protein domains. (B) Merged confocal immunofluorescence microscopy of AnkA truncations at 24 h post-transfection in HEK293T. (C) Merged confocal immunofluorescence microscopy at 24 h post-transfection in HEK293T with AnkA ΔN and AnkA ΔN2 truncations engineered with the SV40 large T-antigen NLS to re-establish nuclear localization.
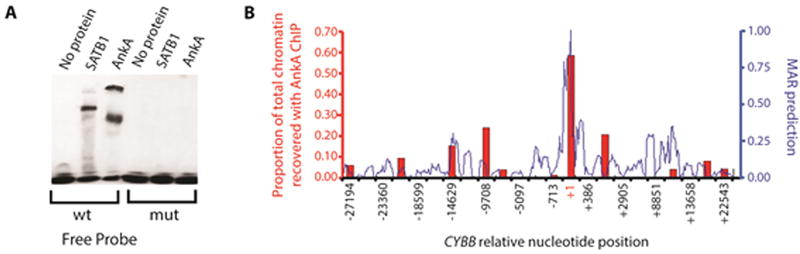
AnkA binds a predicted MAR to decrease CYBB transcription
Once within the granulocyte nucleus, AnkA accumulates to surround heterochromatin and bind AT-rich DNA, a common characteristic of MARs (Caturegli et al., 2000, Lin et al., 2007, Garcia-Garcia et al., 2009b). To determine if AnkA can bind to MARs we used electrophoretic mobility shift assays (EMSAs) to investigate a known SATB1-binding BUR/MAR within the immunoglobulin heavy chain gene promoter. AnkA and SATB1 bound the native MAR, but not when specific AT nucleotides were mutated to GC nucleotides (Figure 2A). Multiple bands appear in the AnkA lane as a result of partial degradation during the purification process (Figure S3).
Figure 2. AnkA binds known and predicted MARs.
(A) EMSA with rAnkA and rSATB1 incubated with the matrix attachment region (MAR) of IGH (wt = 5’-TCTTTAATTTCTAATATATTTAGAAttc-3’) known to bind SATB1, and with a mutated IGH MAR (mut = 5’-TCTTTAATTTCTACTGCTTTAGAAttc-3’). The underscored nucleotides indicate the areas where AT changes were made. Because AnkA is degraded during purification (Figure S1), several bands are present in EMSA. (B) MARs within the CYBB promoter were predicted with MARFinder MAR-Wiz 1.5 (blue line) using default values; a high likelihood was assigned with a value greater than the 0.6 cut-off threshold. AnkA binding along the entire CYBB promoter and downstream positions was assessed by chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) (red bars).
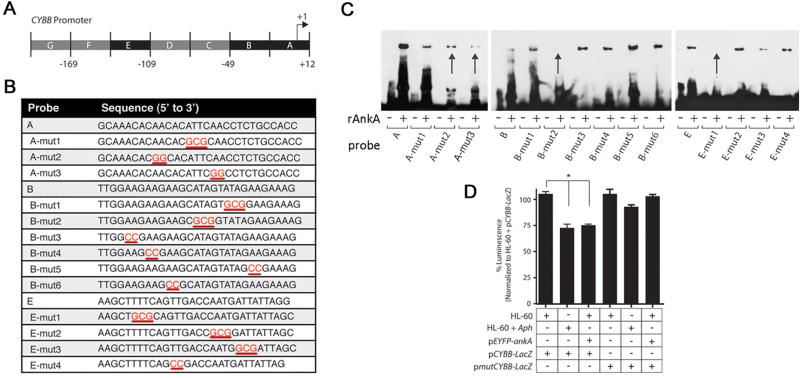
Since SATB1 also binds and represses transcription from the proximal CYBB promoter (Hawkins et al., 2001, Fujii et al., 2003), we used MARFinder MAR-Wiz 1.5 to identify MARs within the CYBB promoter region (Singh et al., 1997). Chromatin immunoprecipitation (ChIP) identified AnkA binding within a predicted MAR located at the proximal CYBB promoter (Figure 2B). AnkA was previously shown to bind within the +12 to −48 and −109 to −138 regions of the CYBB promoter which overlap the predicted MAR (Figure 3A) and to downregulate transcription (Garcia-Garcia et al., 2009b). To interrogate the importance of specific AT nucleotides for AnkA binding at the CYBB promoter, probes containing GC nucleotide substitutions for AT nucleotides were used for EMSA (Figure 3B and 3C). AT to GC nucleotide changes at four positions in the CYBB promoter decreased AnkA binding (indicated by arrows in Figure 3C). To confirm the functional importance of AnkA binding AT nucleotides at these positions, a CYBB promoter containing all 4 mutations (A-mut2, A-mut3, B-mut2, E-mut1) that decreased AnkA binding via EMSA cloned into the pBlueTOPO vector (pmutCYBB-LacZ) and co-transfected into HL-60 cells with the pEYFP-ankA vector. While AnkA significantly decreased expression from the wild-type CYBB promoter, expression from the mutated CYBB promoter did not differ from controls (Figure 3D).
Figure 3. AnkA binds to AT-rich regions in the CYBB promoter that are critical for transcriptional repression.
(A) Schematic of proximal CYBB promoter and regions where AnkA binds (shown in black boxes) (Garcia-Garcia et al., 2009b). (B) Double stranded wild-type and mutated probes used for EMSAs. The mutated bases are shown in red and are underlined. (C) EMSA with rAnkA and mutated CYBB probes. Arrows indicate probes with decreased AnkA binding. (D) Reporter β-galactosidase activity in uninfected HL-60 and A. phagocytophilum-infected HL-60 cells transfected with the indicated plasmids (n=3). *P<0.001 (Student’s t-test, α=0.05). Data are represented as mean ± SEM.
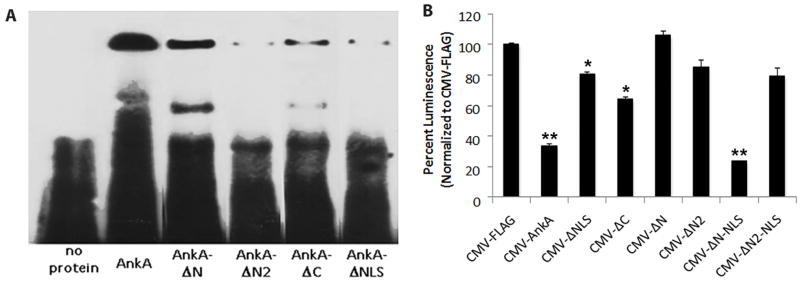
The central ANK repeats of AnkA are critical for binding to the CYBB proximal promoter
To identify the DNA-binding domain(s) within AnkA, the C- and N-terminal truncation mutants were used (Figure 1A). Several recombinant mutants were impaired in binding to the CYBB +12 to −18 promoter probe (Figure 4A). Binding was most dramatically reduced to as low as 5% of wild type AnkA with AnkA-ΔN2 (that lacked 10 ANK repeats); binding was also reduced with AnkA-ΔNLS (ranging from 8 to 23% of AnkA) and AnkA-ΔC (ranging from 23% to 88% of AnkA) (Figure 4A; Supplemental Figure 3). In contrast, binding with AnkA-ΔN was modestly reduced (56%) compared to AnkA. A competitive inhibition EMSA assay using an equimolar quantity of unlabeled CYBB promoter probe was used to confirm specificity of AnkA-CYBB promoter interaction-induced gel shifts (Figure 5). Because AnkA-ΔC localized to the nucleus and bound to DNA, we further examined it for classical DNA-binding motifs; none were identified. As all constructs possessed at least 5 ANK repeats, and binding was most impaired with deletion of ANK repeats 7–10 (AnkA-ΔN2) and to a lesser extent with loss of the flanking ANK repeats (1–6 in AnkA-ΔN and 11–15 in AnkA-ΔC), it is likely that DNA binding is predominantly mediated by the central ANK repeats. The additional reduction in DNA binding observed with AnkA-ΔNLS indicated that the carboxy-terminus of AnkA also contributes to DNA binding. Despite the moderate to significant reduction in DNA binding of AnkA-ΔNLS, it easily translocated into the nucleus and modestly suppressed reporter activity (Figures 4A, 4B, 5). To determine whether the deleted carboxy end of AnkA-ΔNLS possessed other classical DNA binding motifs that could account for these changes, we identified an AT hook-like domain at the C-terminus that was highly conserved among more than 93 AnkA sequences obtained through a NCBI BLAST search of its C-terminal 39 amino acid residues (Supplemental Figure 4). Compared with alignments of known AT hooks in other Alphaproteobacteria, there was strong similarity with non-palindromic type II AT hooks at the core RGR sequence (Figure S4), but lacked other amino acid residues because of its position at the carboxy terminus.
Figure 4. DNA-binding by the N-terminus of AnkA is required to suppress transcription from the CYBB promoter.
(A) Wild-type CYBB probe A (from Figure 3B) EMSA assays with truncated AnkA mutants and labeled proximal CYBB promoter probe. (B) β-galactosidase reporter assay in HL-60 cells co-transfected with pCYBB-LacZ and plasmids expressing the truncated AnkA mutants (n=3); *P<0.05, **P<0.001 vs. FLAG control (Student’s t-test, two-sided α=0.05). Data are represented as mean ± s.d.
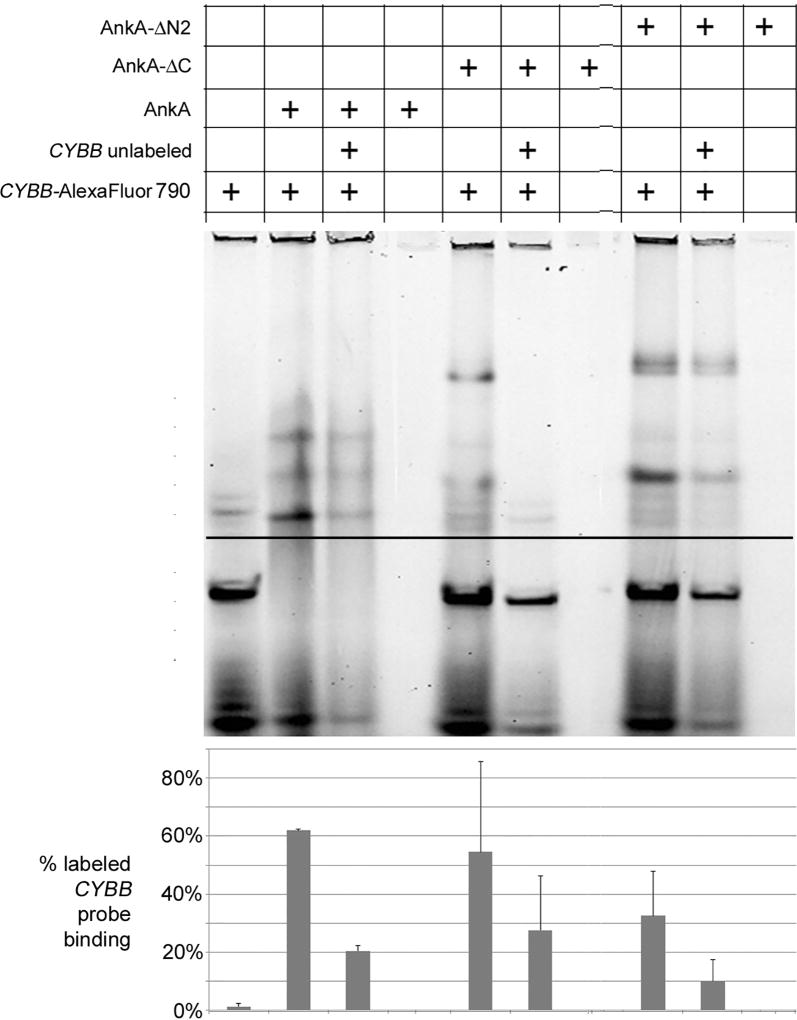
Figure 5. AnkA, AnkA-ΔN2, and AnkA-ΔC binding to the CYBB promoter is specific.
rAnkA, rAnkA-ΔN2, and rAnkA-ΔC binding to the AlexaFluor 790-labeled CYBB promoter probe is antagonized by equimolar unlabeled probe by 67, 69 and 49%, respectively. Recombinant proteins were assayed by EMSA in 4–20% polyacrylamide non-denaturing gels. The pixel density from the LI-COR digital images was analyzed by densitometry using ImageJ 1.48v, comparing pixel density of shifted (above dark line) to total lane density, shown quantified in the graph below the gel image.
Expression silencing of CYBB by AnkA is mediated by the central ANK repeats
Because AnkA-ΔN2 was least able to bind the CYBB promoter, we sought to confirm that it and the other mutants were unable to decrease CYBB transcription. A CYBB – β-galactosidase reporter construct (pCYBB-LacZ) was co-transfected with each of the AnkA truncations. Neither AnkA-ΔN nor -ΔN2 decreased reporter activity, while AnkA-ΔNLS and AnkA-ΔC modestly decreased reporter activity (76% and 64%, respectively). As both DNA binding and nuclear translocation functions localized to the ANK repeat regions in AnkA, to better interrogate and dissociate DNA binding and transcriptional repression, a surrogate NLS, the SV40 large T-antigen NLS, was fused to the carboxy termini of AnkA-ΔN and -ΔN2 constructs to reestablish nuclear translocation (Figure 1C). As anticipated, AnkA reduced expression from the CYBB promoter by approximately 70%, as we observed previously (Garcia-Garcia et al., 2009b), and probably results from transfection efficiencies of about 50% into HL-60 cells. When the SV40 NLS was fused to the AnkA-ΔN truncation, it readily entered the nucleus and expression from the CYBB promoter decreased to levels similar to wild type AnkA. Similarly, the AnkA-ΔN2-SV40 NLS fusion also readily entered the nucleus, but it remained unable to suppress CYBB promoter expression, presumably because it was unable to bind DNA or lacked a distinct domain for recruitment of auxiliary proteins such as HDACs and associated complexes (Figure 4B). These results coupled with the modest reduction in reporter activity with AnkA-ΔC provide evidence that most of the promoter suppressive activity, presumably related to reduced binding and/or auxiliary protein recruitment, resides in ANK repeats 7–10. Since binding is also reduced substantially with loss of flanking ANK repeats and the C-terminal AT hook-like domain, expression silencing best localizes to ANK repeats 7–10.
AnkA interacts with host HDAC1
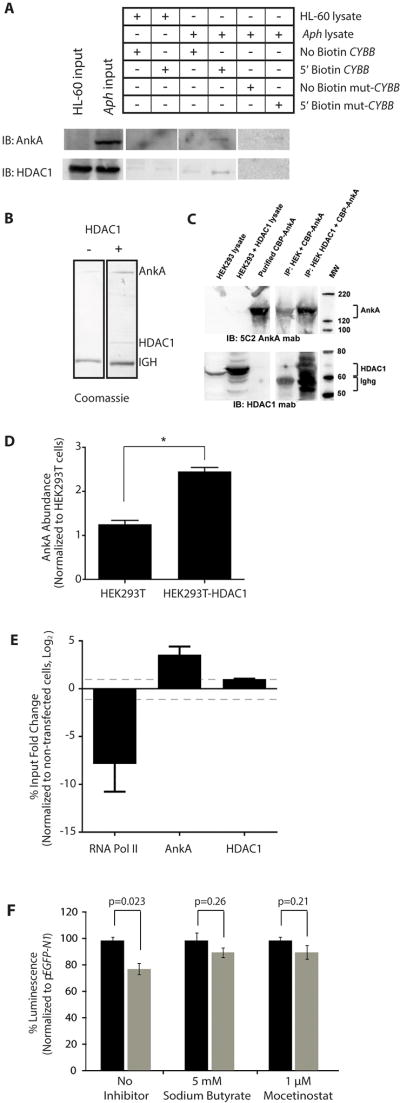
Hypothetically, AnkA binds the CYBB promoter to decrease transcription by recruiting transcriptional repressors, inhibiting transcriptional activator binding, and/or altering chromatin structure. During the course of A. phagocytophilum infection, H3 is deacetylated at the CYBB promoter and HDAC1 binding is increased at defense gene promoters, including CYBB (Garcia-Garcia et al., 2009a). We sought to determine if AnkA plays a direct role in H3 deacetylation. Since recombinant AnkA (rAnkA) lacks HDAC activity (Figure S5), we tested the alternative hypothesis that DNA-bound AnkA recruits histone modifying complexes. Using the proximal 200 bp of the CYBB promoter for DNA pull-down assays, we identified AnkA and HDAC1 co-binding in A. phagocytophilum-infected HL-60 cell lysates (Figure 6A). A direct interaction was implied when binding of both AnkA and HDAC1 was diminished (Figure 6A) using the CYBB promoter containing all four AT nucleotide mutants that abrogate AnkA binding (Figure 3B–C). These results could be interpreted in a way that does not require a direct interaction between AnkA and HDAC1. Therefore, a direct interaction was confirmed using a protein pull down assay where rFLAG-HDAC1 captured from transfected HEK293T cell lysates was used as bait on magnetic beads for purified rAnkA (Figure 6B–D). Recombinant AnkA had limited non-specific binding to the magnetic FLAG beads as bait; however, rAnkA binding greatly increased when rHDAC1 was captured on magnetic beads and used as the bait (Figure 6A–C). Coomassie staining of the eluate from the pull down assay shows abundant AnkA, and minimal contaminating proteins (Figure 6B) demonstrating the direct interaction between AnkA and HDAC1. ChIP further confirmed that HDAC1 binding is increased at CYBB when AnkA is overexpressed in HEK293T cells, while RNA polymerase II binding decreased (Figure 6E), demonstrating that AnkA is sufficient to increase HDAC1 binding at the CYBB promoter and to exclude RNA polymerase from active transcription.
Figure 6. AnkA and HDAC1 directly interact.
(A) DNA pull-down assay with uninfected (HL-60) or A. phagocytophilum-infected HL-60 cell (Aph) lysates and wild-type or mutated CYBB promoters. The first two lanes are controls that demonstrate the presence of AnkA and/or HDAC1 in samples before reaction with the CYBB or mutated probes. The remaining lanes show the results of interactions in the absence of AnkA (HL-60), with AnkA (Aph), with unlabeled promoter or mutated promoter probe (No Biotin CYBB or No Biotin mut-CYBB) that will bind the magnetic beads, and 5’ biotinylated promoter or mutated probe (5’ Biotin CYBB or Biotin mut-CYBB) that will not bind the magnetic beads. (B) Coomassie blue stain and (C) protein immunoblot analysis of eluted proteins from protein pull-down assay with HDAC1-FLAG overexpressed in HEK293T cells mixed with purified rAnkA. The overexpressed HDAC1 (60–65 kDA) readily bound to the FLAG mab matrix and was detected in proximity to large quantities of mouse immunoglobulin gamma heavy chains (Ighg; 55 kDa) dissociated from the matrix in the Laemmli buffer. HDAC1 detection increased simultaneously with a marked increase in AnkA binding compared to cells lacking HDAC1 overexpression. (D) Densitometry analysis of relative AnkA abundance in protein pull-down assay. (E) ChIP analysis of AnkA-, HDAC1- and RNA polymerase II-binding at the proximal CYBB promoter in AnkA-transfected HEK293T cells (n=3). Dashed line indicates 2-fold change in binding. (F) β-galactosidase reporter assay in HL-60 cells co-transfected with pCYBB-LacZ and pEYFP-N1 (black bars) or pEYFP-ankA (gray bars) and treated with 5 mM sodium butyrate HDAC inhibitor or 1µM mocetinostat HDAC1 inhibitor (n=3). Data normalized to empty vector, pEYFP-N1 control. *P<0.001 (Student’s t-test, α=0.05). Data are represented as mean ± SEM.
To determine the importance of HDAC1 for AnkA-mediated repression of CYBB, HDAC activity was pharmacologically inhibited with sodium butyrate or mocetinostat and the CYBB-β-galactosidase reporter assay was used to measure transcription from the CYBB promoter. AnkA significantly decreased transcription of CYBB when HDAC activity was uninhibited, but was unable to represses transcription when HDAC activity was inhibited with 5 mM sodium butyrate, a general HDAC inhibitor, or with 1 µM mocetinostat, an HDAC1-specific inhibitor (Figure 6F). Together these data show that AnkA recruits HDAC1 to the CYBB promoter to decrease transcription.
Discussion
In part, as an agent of an emerging infection with an unusual intracellular niche, but more as a model of how an intracellular microbe manipulates its hosts’ gene expression to improve fitness, it is important to understand how A. phagocytophilum survives and propagates within the neutrophil. What makes it capable of differentially regulating transcription of critical genes such as CYBB or those involved with proinflammatory or anti-apoptosis programs? Due to the large number of genes that are differentially regulated with infection and the relatively limited genomic capacity of the bacterium, we predicted that A. phagocytophilum influences global granulocyte transcription through chromatin/histone remodeling to broadly impact transcriptional programs. AnkA, an A. phagocytophilum effector protein that translocates to the neutrophil nucleus and binds DNA to interact with nuclear proteins, was predicted to play a role in host transcriptional reprogramming since over-expression resulted in host defense gene downregulation (Garcia-Garcia et al., 2009b). Among many genomic targets, AnkA binds to the CYBB promoter in close proximity to regions of decreased histone H3 acetylation (Garcia-Garcia et al., 2009a). Using the CYBB promoter as a model of AnkA-mediated cis transcriptional control we identified a unique mechanism by which a prokaryotic protein alters host transcription. The multiple ankyrin repeats in AnkA were identified to be critical for AnkA nuclear localization and transcriptional regulation, and to a great extent, its binding to CYBB, whereas deletion of both the putative HGMN-CHUD domain and the tandem repeats did not substantially effect nuclear localization, DNA binding or transcriptional activity, although these were not examined in detail.
Ankyrin repeats are commonly occurring motifs among proteins with diverse biological functions, usually mediating protein-protein interactions. In addition to its previous morphologic distribution around heterochromatin in transfected cells, here AnkA was shown to have additional properties of a MAR binding protein. SATB1, a known MAR binding protein that binds to the CYBB promoter and represses transcription early during myeloid differentiation, recruits the histone deacetylase contained in the NURD chromatin remodeling complex to deacetylate histones and repress gene expression (Hawkins et al., 2001, McKinsey et al., 2006). Similar to SATB1, here we showed AnkA recruited a histone deacetylase (HDAC1) to alter histone H3 acetylation and was critical for AnkA-mediated CYBB repression. Precedent for proteins with ANK repeat domains and HDAC recruitment include the eukaryotic protein, RFXANK, which interacts with HDAC4 and 5 through its ANK repeats to repress major histocompatibility complex II gene expression (McKinsey et al., 2006), and ANKRA2, a paralog of RFXANK, which binds HDAC4 and 5 to decrease expression from xenobiotic responsive elements as a complex with the aryl hydrocarbon receptor repressor (Oshima et al., 2007).
How AnkA binds to DNA to recruit HDAC1 is of great interest, as most MAR-binding proteins do so to areas that easily uncoil. There are currently no reports of ankyrin repeat-containing MAR-binding proteins or any ankyrin repeat containing proteins that directly bind DNA. In fact, most examples of ankyrin-containing proteins that localize to DNA, such as GABPα/β, involve heterodimers for which one component binds DNA (GABPα) and the other recruits interacting proteins (GABPβ) (Rosmarin et al., 2004). Of interest here was that decreasing the repertoire of AnkA ankyrin repeats with increasing truncations from the N-terminus was associated with diminished DNA binding, an observation congruent with contemporary understanding of ankyrin proteins as highly stable nanosprings (Itzhaki et al., 2012) for which stability diminishes as the number of ankyrin repeats is reduced (Hagai et al., 2012), as observed for AnkA-ΔN2. Of further interest was the reduced DNA binding observed with the minimal truncation in AnkA-ΔNLS at the carboxy end that deleted a putative bipartite NLS. While nuclear localization was not changed by this deletion, inspection of the sequence identified a potential AT hook-like region at the AnkA C-terminus that was deleted in this mutant. Whether the identified AT hook that has similarities to other known Alphaproteobacteria AT hooks functions in DNA binding will require further experimental validation.
Likewise, ankyrin proteins are not well recognized to serve as nuclear localization signals. Yet even here precedent exists since, the second ankyrin repeat of IκBα is critical for its nuclear import (Sachdev et al., 1998), and the data for AnkA are consistent with those observations. More recent investigations of eukaryotic ANK repeats in proteins that translocate into the nucleus, such as IκBα, GABPβ, mutated p16ink4a, and ASPP2, do so in part via the presence of hydrophobic amino acid residues at the 13th positions in 2 consecutive ANK repeats (Lu et al., 2014). An analysis of multiple AnkA ANK repeats demonstrates that the first four N-terminal ANK repeats possess highly conserved hydrophobic residues in these positions, and the deletion of all 4 of these ANK repeats (AnkA-ΔN) is sufficient to inhibit nuclear localization. Whether this relates specifically to those hydrophobic positions will also need to be determined experimentally.
The specific domains or regions to which HDAC1 is recruited are still not established. Deletion of the 4 N-terminal ANK repeats inhibited nuclear localization but not DNA binding or promoter activity. Deletion of the C-terminus did not change nuclear localization or significantly reduce promoter activity, but moderately abrogated DNA binding. Similarly, the AnkA-ΔC mutant that lacked both the C-terminus and ANK repeats 11–15 entered the nucleus unimpaired, had a modest defect in DNA binding, and a 25% reduction in promoter activity. The most dramatic effect on promoter activity, and perhaps the ability to recruit HDAC1, was found with deletion of the first 10 ANK repeats for which nuclear localization, DNA binding, and promoter activity were all significantly impaired. While nuclear localization could be easily restored by an exogenous NLS in both AnkA-ΔN and AnkA-ΔN2, promoter activity was still significantly abrogated only in AnkA-ΔN2 that lacked the central ANK repeats. Thus, it could be that the central ANK repeats, in part shared with the deletion in AnkA-ΔC, provide most function toward suppression of promoter activity (Figure 7). Thus, this region is a prime candidate to examine for recruitment of HDAC1, and perhaps other proteins with which it complexes to achieve histone deacetylation and cis regulation of gene expression.
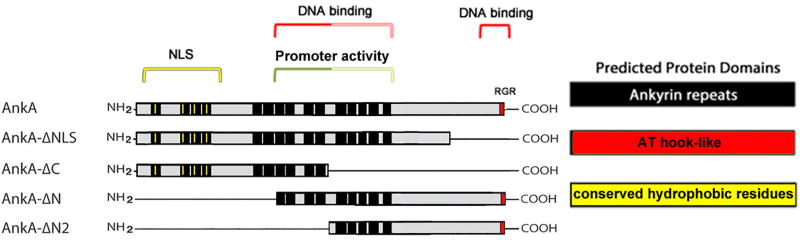
Figure 7. Schematic of AnkA regions for nuclear localization, DNA binding, and promoter activity.
The diagram depicts the specific regions in AnkA that mediate nuclear localization (red lines in first 4 ANK repeats show conserved hydrophobic domains at the 13th residues), regions most associated with DNA binding, specifically localized to the central ANK repeats deleted in AnkA-ΔN2 or to the AT hook-like C-terminal residues (RGR), and regions most strongly associated with promoter activity deleted in AnkA-ΔN2 and less in AnkA-ΔC.
A prokaryotic protein that binds host DNA and recruits chromatin modifying complexes to alter the epigenetic state is fundamentally distinct from bacterial effectors that modify signaling pathways, even in the nucleus, to alter host transcription. Currently, only a few bacterial pathogens are known to translocate effectors to the host cell nucleus and alter chromatin; the mechanism(s) of gene and transcriptional program targeting and chromatin modifications are poorly understood. Shigella flexneri OspF (Arbibe et al., 2007), Listeria monocytogenes LntA (Lebreton et al., 2014), Chlamydia trachomatis NUE (Pennini et al., 2010), and Legionella pneumophila RomA (Rolando et al., 2013) are nuclear-translocated proteins (nucleomodulins) that influence host cell transcription by modifying histone structure.
A. phagocytophilum AnkA is the first for which a mechanism that directly involves DNA binding and cis-recruitment of histone modifying enzymes to chromatin at an individual gene. The influence AnkA exerts over CYBB and neutrophil respiratory burst provides the pathogen a significant fitness advantage, as expression from CYBB under the control of distinct constitutive promoter reverses pathogen growth (Carlyon et al., 2002). While CYBB expression occurs dominantly during neutrophil maturation, AnkA is bound to host DNA genome-wide (Park et al., 2004) and histone deacetylation or methylation is frequently observed at promoters of host defense genes in A. phagocytophilum-infected cells (Garcia-Garcia et al., 2009a, Garcia-Garcia et al., 2009b), suggesting a much broader and robust effect on transcription and cellular functions. Lastly, microbial survival depends on upregulation of HDAC expression (Garcia-Garcia et al., 2009b). Considering these observations and the need for economy in reduced-genome intracellular prokaryotes, more investigation is needed to determine if AnkA and/or similar effectors in other prokaryotes behave similarly, but also act in “trans” to modify the epigenome and chromosomal territory architecture as a broad regulatory mechanism over global transcriptional and functional programs (Sinclair et al., 2014). Prokaryote-induced host epigenetic alteration is an emerging topic that will lead to a better understanding of both microbes and hosts. How small genome intracellular prokaryotes broadly manipulate eukaryotic cell functions via transcriptional regulation is of significant interest to explain microbial fitness, and pathogenesis, as well as reprogrammed cellular functions.
Experimental Procedures
Cell Lines and Cell Culture
Promyeloblast HL-60 cells (ATCC CCL-240) and the Webster strain of A. phagocytophilum were maintained in HL-60 cells as previously described (Garcia-Garcia et al., 2009b). HEK293T cells were cultured in DMEM media supplemented with 10% FBS and Glutamax (Life Technologies, Carlsbad, CA) in a humidified incubator at 37°C with 5% CO2.
Expression of AnkA and SATB1 in E. coli and mammalian cells
Full-length and truncated ankA coding sequences were amplified via PCR (Table S1) using Webster strain genomic DNA and cloned into pT7-FLAG-MAT-Tag-2 (Sigma-Aldrich, St. Louis, MO) or pCAL-n-EK (Agilent Technologies, Santa Clara CA) expression vectors for expression in E. coli. Recombinant protein was purified using a HisTrap HP column (GE Healthcare, Buckinghamshire, United Kingdom) or calmodulin sepharose (GE Healthcare Bio-Sciences, Pittsburg, PA) following the manufacturer's recommendations. Plasmid pGEX2T-SATB1, donated by Terumi Kohwi-Shigematsu, Lawrence Berkeley National Laboratory, Berkeley, CA was used to overexpress SATB1 in E. coli, purified as described previously(Dickinson et al., 1997). The ankA coding sequences were also cloned the pCMV-Tag2b vector for expression in HL-60 and HEK293T cells (Table S2). pEYFP-ankA plasmid was constructed as described previously(Garcia-Garcia et al., 2009b). Plasmids were transfected into HL-60 cells using a Lonza 96-well Nucleofector, solution SF, program EN-138 (Lonza, Basel, Switzerland) or into HEK293T cells using Xfect transfection reagent (Clontech Laboratories, Mountain View, CA).
Electrophoretic Mobility Shift Assay
Purified full-length or truncated rAnkAs and rSATB1 were assayed by EMSA for their ability to bind to the wild-type or mutated immunoglobulin heavy chain MAR or specific CYBB DNA probes that included a putative MAR. Oligonucleotides were biotinylated by the supplier (Figure 1A and 2B) (Integrated DNA Technologies, Coralville, IA). EMSA reactions were performed as previously described using 20 fmoles of CYBB probe and 25 ng/µL protein (Garcia-Garcia et al., 2009b). To prove interaction specificity, an N-terminal amine-modified CYBB probe was fluorescently labeled with AlexaFluor 790 (Life Technologies) and 1280 to 5000 fmoles was competed against equimolar unlabeled probe for binding to 20 µM protein, separated in TBE Criterion (Bio-Rad Laboratories, Hercules, CA) 4–20% polyacrylamide gels for in-gel infrared imaging (LI-COR Odyssey, LI-COR Biosciences, Lincoln, NE). The pixel density from scanned blots or LI-COR digital images was analyzed by densitometry using ImageJ 1.48v, comparing pixel density of shifted (Figure 5 above dark line) to total lane density.
Reporter Assay
Plasmid pCYBB-LacZ, kindly donated by Erol Fikrig, Yale University, New Haven, CT was used to express the lacZ gene under the control of the −209 to +12 region of the CYBB promoter (Thomas et al., 2005). A mutant CYBB promoter (pmutCYBB-LacZ) was synthesized in pIDT Blue vector (Integrated DNA Technologies, Coralville, IA) to contain mutations A-mut2, A-mut3, B-mut2 and E-mut1 (Figure 2B). The mutated promoter was amplified by PCR and cloned into the pBlueTOPO vector (Life Technologies, Coralville, IA) (Table S3). HL-60 cells were transfected as described above with indicated plasmids. For HDAC inhibition reporter assays, 18 h post transfection cells were treated for 6 h with 5 mM sodium butyrate (Sigma-Aldrich) and 1 µM mocetinostat. β-galactosidase activity was measured using the Galacto-Star System (Life Technologies, Coralville, IA). Luminescence was read every 2 min for 2 h. For analysis, relative light units between 54 and 64 min were averaged.
Immunofluorescence
pCMV-Tag2b-ankA or ankA truncation plasmids were transfected into HEK293T cells. 24 h post-transfection, cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton-X in PBS. Rinsed cells were incubated with anti-FLAG antibody (Sigma-Aldrich, St. Louis, MO) for 1 h, washed 4 times with 0.2% Tween-20 in PBS, and incubated with AlexaFluor488 goat anti-mouse secondary antibody (Life Technologies, Coralville, IA) for 1 h. Nuclei were stained with 1:5000 dilution of Hoechst 33342, trihydrochloride, trihydrate (Life Technologies, Coralville, IA) and washed again 4 times with 0.2% Tween-20 in PBS. For confocal microscopy, cells were viewed using a Zeiss LSM 510 Meta Confocal Microscope (University of Maryland, Baltimore Confocal Core Facility).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was completed using the ChIP-IT Express Chromatin Immunoprecipitation Kit (Active Motif, Carlsbad, CA) or as previously reported (Garcia-Garcia et al., 2009b). DNA was immunoprecipitated with the AnkA monoclonal antibody 5C2 (prepared against a recombinant AnkA devoid of most ANK repeats in the N-terminus), RNA polymerase II and HDAC1 antibodies (Active Motif, Carlsbad, CA). ChIP using goat anti-mouse IgG was also included as negative control in experiments. Immunoprecipitation of the proximal CYBB promoter was analyzed via qPCR using USB Veriquest SYBR Green (Affymetrix, Santa Clara, CA) or regions along the X-chromosome using iQSYBR Green Supermix (Bio-Rad, Hercules, CA) (Tables S3 and S4). Percent of input chromatin was calculated and normalized to ChIP with non-transfected HEK293T cells or uninfected HL-60 cells.
HDAC activity assay
rAnkA was tested for HDAC activity using the fluorescent HDAC assay kit (Active Motif, Carlsbad, CA) following the manufacturer's instructions. 5 µg rAnkA was diluted in assay buffer and activity expressed as pmol of fluorescent product formed after incubation at 37°C for 1 h. Fluorescence intensity was measured using a Perkin Elmer Victor II plate reader with an excitation wavelength at 355 nm and emission at 460 nm. HeLa cell extract was used as a positive control.
DNA Pull Down Assay
DNA pull down assays were performed using biotinylated 5’ wild-type or mutated −209 to +12 CYBB promoter probes. The mutated probe contained all four mutations found to decrease AnkA binding via EMSA (Figure 2B and C). Probes used for DNA pull down were synthesized using primers in Table S3 and the mutated probes were amplified from the pmutCYBB-LacZ plasmid. Uninfected and A. phagocytophilum-infected HL-60 cells were lysed with lysis buffer (20 mM HEPES pH7.5 with KOH, 150 mM KCl, 0.5 mM DTT, 10% glycerol) for 15 min on ice. NP-40 was added to a concentration of 0.5%. Cells were sonicated 3 times for 10 sec with a Branson Sonifier 250 (Branson Ultrasonics, Danbury, CT) at output 4 and centrifuged at 13,000 rpm for 10 min. Lysates were pre-cleared for 1 h at 4°C with 1 µg/mL salmon sperm DNA and 25 µL streptavidin paramagnetic beads (Promega, Madison, WI) in DNA pull-down buffer (20 mM Tris, 15% glycerol, 150 mM KCl, 1 mM EDTA, 0.5 mg/mL BSA, 0.05% NP-40). Approximately 500 µg of pre-cleared lysates were then incubated with 500 ng biotinylated CYBB probe, 1 µg/mL salmon sperm DNA, and 25 µL fresh streptavidin paramagnetic beads (Promega, Madison, WI) for 1 h at 4°C. As controls, lysates were also incubated with non-biotinylated probes. Proteins bound to the CYBB promoter were eluted in SDS-PAGE loading buffer and protein immunoblot analysis was performed to detect AnkA and HDAC1.
Protein Pull Down Assay
HEK293T cells were transfected with pFLAG-HDAC1 plasmid (Addgene plasmid 13820 (Emiliani et al., 1998)) using Xfect transfection reagent (Clontech Laboratories, Mountain View, CA). 48 h post transfection, cells were lysed as described in the DNA pull down assay procedure above. HDAC1-FLAG was incubated with mouse monoclonal IgG1 anti-FLAG M2 affinity gel (Sigma-Aldrich, St. Louis, MO) overnight at 4°C and washed with 1X Tris-buffered saline with 1 M NaCl and 5% Tween-20. rCBP-AnkA was purified as described above. Approximately 5 µg of rCBP-AnkA was incubated with HDAC1-FLAG beads overnight at 4°C and washed with 1X PBS with 0.02% Tween-20. rCBP-AnkA was also incubated with Anti-FLAG M2 affinity gel incubated with non-transfected HEK293T cellular lysate as a negative control. Proteins were eluted with Laemmli sample buffer at 95°C for 5 min. Samples were analyzed by SDS-PAGE with Coomassie blue stain and protein immunoblots using anti-mouse IgG-HRP (Cell Signaling Technology, Danvers MA). Images were obtained with a Kodak Image Station 4000R and band intensities measured using ImageJ (http://rsb.info.nih.gov/ij/). The AnkA band intensities were normalized to immunoglobulin heavy chain band intensity and the average for the negative control was set to one.
Bioinformatics
Full-length AnkA was examined for DNA binding motifs using GYM 2.0 (for helix-turn-helix motifs; http://users.cis.fiu.edu/~giri/bioinf/GYM2/prog.html), 2ZIP (for leucine zippers; http://2zip.molgen.mpg.de/index.html), and DP-Bind (for evolutionarily-conserved binding sequences; http://lcg.rit.albany.edu/dp-bind/). To assess the potential for an AT hook at the carboxy terminus of AnkA, previously identified AT hook regions among 48 Alphaproteobacteria were downloaded from the Simple Modular Architecture Research Tool (SMART; http://smart.embl.de/smart/do_annotation.pl?DOMAIN=SM00384; Table S6) and aligned using CLUSTALX. Similarly, the C-terminal 39 amino acid residues of A. phagocytophilum Webster strain AnkA were used as template in a BLAST (Altschul et al., 1990) search of the NCBI Non-redundant protein sequences database to obtain those most similar (Table S7). The CLUSTALX (Thompson et al., 1997) aligned sequences were used as a template for sequence logo creation (Crooks et al., 2004) and examination of similar linear features. To assess the diversity of hydrophobic residues in the 13th positions of the ANK repeats of AnkA, 31 AnkA protein sequences representing diverse strains from human and animals, and from several continents were obtained (Table S5), aligned by CLUSTALX, and ANK repeats were characterized by screening the consensus sequence using the Eukaryotic Linear Motif resource for Functional Sites in Proteins (ELM; http://elm.eu.org/).
Supplementary Material
Acknowledgments
This work was supported by grant R01 AI044102 to JSD. We thank Dennis J. Grab, The Johns Hopkins University School of Medicine, for critical discussion and reading the manuscript. Erol Fikrig, Yale University, kindly provided the pCYBB-LacZreporter plasmid. Eric Verdin, Gladstone Institute for Virology and Immunology, University of California, San Francisco, provided the HDAC1-FLAG plasmid. Terumi Kohwi-Shigematsu, Lawrence Berkeley National Laboratory, assisted with MAR prediction and provided the pGEX2T-SATB1 plasmid, as well as critical discussion.
Footnotes
Author Contributions
K.E.R.-B. designed and did most of the experiments and prepared the manuscript. J.C.G.-G. demonstrated AnkA binding to MARs. S.H.S. tested AnkA for intrinsic histone deacetylase activity. J.S.D. helped analyze data and interpret results and supervised all studies. All authors contributed to discussions and manuscript preparation.
Bibliography and References Cited
- Akkoyunlu M, Fikrig E. Gamma interferon dominates the murine cytokine response to the agent of human granulocytic ehrlichiosis and helps to control the degree of early rickettsemia. Infect. Immun. 2000;68:1827–1833. doi: 10.1128/iai.68.4.1827-1833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–- 410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, Muchardt C, et al. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 2007;8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- Bierne H, Cossart P. When bacteria target the nucleus: the emerging family of nucleomodulins. Cell. Microbiol. 2012;14:622–633. doi: 10.1111/j.1462-5822.2012.01758.x. [DOI] [PubMed] [Google Scholar]
- Borjesson DL, Kobayashi SD, Whitney AR, Voyich JM, Argue CM, Deleo FR. Insights into pathogen immune evasion mechanisms: Anaplasma phagocytophilum fails to induce an apoptosis differentiation program in human neutrophils. J. Immunol. 2005;174:6364–6372. doi: 10.4049/jimmunol.174.10.6364. [DOI] [PubMed] [Google Scholar]
- Bryant PA, Venter D, Robins-Browne R, Curtis N. Chips with everything: DNA microarrays in infectious diseases. The Lancet infectious diseases. 2004;4:100–111. doi: 10.1016/S1473-3099(04)00930-2. [DOI] [PubMed] [Google Scholar]
- Carlyon JA, Chan WT, Galan J, Roos D, Fikrig E. Repression of rac2 mRNA expression by Anaplasma phagocytophila is essential to the inhibition of superoxide production and bacterial proliferation. J. Immunol. 2002;169:7009–7018. doi: 10.4049/jimmunol.169.12.7009. [DOI] [PubMed] [Google Scholar]
- Caturegli P, Asanovich KM, Walls JJ, Bakken JS, Madigan JE, Popov VL, Dumler JS. ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect. Immun. 2000;68:5277–5283. doi: 10.1128/iai.68.9.5277-5283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KS, Grab DJ, Dumler JS. Anaplasma phagocytophilum infection induces protracted neutrophil degranulation. Infect Immun. 2004;72:3680–3683. doi: 10.1128/IAI.72.6.3680-3683.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente J, Ayoubi P, Blouin EF, Almazan C, Naranjo V, Kocan KM. Gene expression profiling of human promyelocytic cells in response to infection with Anaplasma phagocytophilum. Cell. Microbiol. 2005;7:549–559. doi: 10.1111/j.1462-5822.2004.00485.x. [DOI] [PubMed] [Google Scholar]
- Dickinson LA, Dickinson CD, Kohwi-Shigematsu T. An atypical homeodomain in SATB1 promotes specific recognition of the key structural element in a matrix attachment region. J. Biol. Chem. 1997;272:11463–11470. doi: 10.1074/jbc.272.17.11463. [DOI] [PubMed] [Google Scholar]
- Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y, Kumatori A, Nakamura M. SATB1 makes a complex with p300 and represses gp91(phox) promoter activity. Microbiology and immunology. 2003;47:803–811. doi: 10.1111/j.1348-0421.2003.tb03438.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia JC, Barat NC, Trembley SJ, Dumler JS. Epigenetic silencing of host cell defense genes enhances intracellular survival of the rickettsial pathogen Anaplasma phagocytophilum. PLoS pathogens. 2009a;5:e1000488. doi: 10.1371/journal.ppat.1000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia JC, Rennoll-Bankert KE, Pelly S, Milstone AM, Dumler JS. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect. Immun. 2009b;77:2385–2391. doi: 10.1128/IAI.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garyu JW, Choi KS, Grab DJ, Dumler JS. Defective phagocytosis in Anaplasma phagocytophilum-infected neutrophils. Infect Immun. 2005;73:1187–1190. doi: 10.1128/IAI.73.2.1187-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagai T, Azia A, Trizac E, Levy Y. Modulation of folding kinetics of repeat proteins: interplay between intra- and interdomain interactions. Biophys. J. 2012;103:1555–1565. doi: 10.1016/j.bpj.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins SM, Kohwi-Shigematsu T, Skalnik DG. The matrix attachment region-binding protein SATB1 interacts with multiple elements within the gp91phox promoter and is down-regulated during myeloid differentiation. J. Biol. Chem. 2001;276:44472–44480. doi: 10.1074/jbc.M104193200. [DOI] [PubMed] [Google Scholar]
- Itzhaki LS, Lowe AR. From artificial antibodies to nanosprings: the biophysical properties of repeat proteins. Advances in experimental medicine and biology. 2012;747:153–166. doi: 10.1007/978-1-4614-3229-6_10. [DOI] [PubMed] [Google Scholar]
- Kim HY, Rikihisa Y. Expression of interleukin-1beta, tumor necrosis factor alpha, and interleukin-6 in human peripheral blood leukocytes exposed to human granulocytic ehrlichiosis agent or recombinant major surface protein P44. Infect Immun. 2000;68:3394–3402. doi: 10.1128/iai.68.6.3394-3402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton A, Job V, Ragon M, Le Monnier A, Dessen A, Cossart P, Bierne H. Structural basis for the inhibition of the chromatin repressor BAHD1 by the bacterial nucleomodulin LntA. MBio. 2014;5:e00775-13. doi: 10.1128/mBio.00775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Kioi M, Han J, Puri RK, Goodman JL. Anaplasma phagocytophilum-induced gene expression in both human neutrophils and HL-60 cells. Genomics. 2008;92:144–151. doi: 10.1016/j.ygeno.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol. 2007;9:2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x. [DOI] [PubMed] [Google Scholar]
- Lu M, Zak J, Chen S, Sanchez-Pulido L, Severson DT, Endicott J, et al. A code for RanGDP binding in ankyrin repeats defines a nuclear import pathway. Cell. 2014;157:1130–1145. doi: 10.1016/j.cell.2014.05.006. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Kuwahara K, Bezprozvannaya S, Olson EN. Class II histone deacetylases confer signal responsiveness to the ankyrin-repeat proteins ANKRA2 and RFXANK. Mol. Biol. Cell. 2006;17:438–447. doi: 10.1091/mbc.E05-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott J, Rikihisa Y, Tsunawaki S. Effects of Anaplasma phagocytophila on NADPH oxidase components in human neutrophils and HL-60 cells. Infect. Immun. 2002;70:1359–1366. doi: 10.1128/IAI.70.3.1359-1366.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima M, Mimura J, Yamamoto M, Fujii-Kuriyama Y. Molecular mechanism of transcriptional repression of AhR repressor involving ANKRA2, HDAC4, and HDAC5. Biochemical and biophysical research communications. 2007;364:276–282. doi: 10.1016/j.bbrc.2007.09.131. [DOI] [PubMed] [Google Scholar]
- Park J, Kim KJ, Choi KS, Grab DJ, Dumler JS. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell. Microbiol. 2004;6:743–751. doi: 10.1111/j.1462-5822.2004.00400.x. [DOI] [PubMed] [Google Scholar]
- Pedra JH, Sukumaran B, Carlyon JA, Berliner N, Fikrig E. Modulation of NB4 promyelocytic leukemic cell machinery by Anaplasma phagocytophilum. Genomics. 2005;86:365–377. doi: 10.1016/j.ygeno.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Pennini ME, Perrinet S, Dautry-Varsat A, Subtil A. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog. 2010;6:e1000995. doi: 10.1371/journal.ppat.1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando M, Sanulli S, Rusniok C, Gomez-Valero L, Bertholet C, Sahr T, et al. Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell host & microbe. 2013;13:395–405. doi: 10.1016/j.chom.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Rosmarin AG, Resendes KK, Yang Z, McMillan JN, Fleming SL. GA-binding protein transcription factor: a review of GABP as an integrator of intracellular signaling and protein-protein interactions. Blood Cells Mol. Dis. 2004;32:143–154. doi: 10.1016/j.bcmd.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Sachdev S, Hoffmann A, Hannink M. Nuclear localization of IkappaB alpha is mediated by the second ankyrin repeat: the IkappaB alpha ankyrin repeats define a novel class of cis-acting nuclear import sequences. Mol. Cell. Biol. 1998;18:2524–2534. doi: 10.1128/mcb.18.5.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaife H, Woldehiwet Z, Hart CA, Edwards SW. Anaplasma phagocytophilum reduces neutrophil apoptosis in vivo. Infect. Immun. 2003;71:1995–2001. doi: 10.1128/IAI.71.4.1995-2001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair SH, Rennoll-Bankert KE, Dumler JS. Effector bottleneck: Microbial reprogramming of parasitized host cell transcription by epigenetic remodeling of chromatin structure. Front. Genet. 2014;5:274. doi: 10.3389/fgene.2014.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh GB, Kramer JA, Krawetz SA. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 1997;25:1419–1425. doi: 10.1093/nar/25.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thomas V, Samanta S, Wu C, Berliner N, Fikrig E. Anaplasma phagocytophilum modulates gp91phox gene expression through altered interferon regulatory factor 1 and PU.1 levels and binding of CCAAT displacement protein. Infect. Immun. 2005;73:208–218. doi: 10.1128/IAI.73.1.208-218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TY, Han ZM, Chai YR, Zhang JH. A mini review of MAR-binding proteins. Mol. Biol. Rep. 2010;37:3553–3560. doi: 10.1007/s11033-010-0003-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.