Abstract
Diffusion imaging coupled with tractography algorithms allows researchers to image human white matter fiber bundles in-vivo. These bundles are three-dimensional structures with shapes that change over time during the course of development as well as in pathologic states. While most studies on white matter variability focus on analysis of tissue properties estimated from the diffusion data, e.g. fractional anisotropy, the shape variability of white matter fiber bundle is much less explored. In this paper, we present a set of tools for shape analysis of white matter fiber bundles, namely: (1) a concise geometric model of bundle shapes; (2) a method for bundle registration between subjects; (3) a method for deformation estimation. Our framework is useful for analysis of shape variability in white matter fiber bundles.
We demonstrate our framework by applying our methods on two datasets: one consisting of data for 6 normal adults and another consisting of data for 38 normal children of age 11 days to 8.5 years. We suggest a robust and reproducible method to measure changes in the shape of white matter fiber bundles. We demonstrate how this method can be used to create a model to assess age-dependent changes in the shape of specific fiber bundles. We derive such models for an ensemble of white matter fiber bundles on our pediatric dataset and show that our results agree with normative human head and brain growth data. Creating these models for a large pediatric longitudinal dataset may improve understanding of both normal development and pathologic states and propose novel parameters for the examination of the pediatric brain.
Keywords: White Matter, Shape Analysis, Development
1. Introduction
Advances in diffusion magnetic resonance imaging (dMRI) and tractography methods now allow in-vivo high resolution imaging of human white matter (WM) and extraction of the brain connectome – a large collection of WM fibers. As increasing numbers of dMRI datasets become publicly available, much of the recent work in the neuroscientific community revolves around analysis of specific WM bundles – subsets of fibers – with regards to population comparison, lateralization and neurological disorders. Most studies of WM variability focus on analysis of tissue properties estimated by different values extracted from the diffusion data, such as Fractional Anisotropy. The shape variability of white matter, however, is much less studied. The paucity of investigation into white matter shape is due to several factors. It is in part due to the fact that the shape of fiber bundles is irregular and complex, and in part because there are still ongoing debates in the community regarding the ambiguous nature of tractography methods used to extract these structures.
WM fiber bundles are essentially three-dimensional geometric structures with origination and termination regions, anatomically constrained by adjacent brain structures. Their geometric shape has been confirmed by postmortem brain anatomy studies, e.g. Mori et al., 2017, and atlases have been constructed that determine both the common shape and the common location of different fiber bundles, Wakana et al., 2004. At present, shape morphology of WM fiber bundles over the course of normal development and pathologic states represents an unexplored area of investigation that may yield new insights into normal and pathologic brain development. Specifically, the lack of normative data in the pediatric population has constrained efforts to create normal atlases of pediatric brain development using any imaging modality, including WM imaging.
This paper aims to address this gap and presents a framework of computational geometry tools focused on analyzing the shape variability of WM fiber bundles. We present: (1) a concise and novel geometric model for fiber bundle shapes, (2) spatial correspondence mapping between regions of interest on homologous fiber bundles, (3) registration of fiber bundles across subjects in bundle space, and (4) bundle deformation estimation.
To demonstrate the abilities of our framework, we apply our methods to a unique cross-sectional dataset of 38 healthy children aged 0 to 8 years and show how it can be used to model the shape change of WM fiber bundles among subjects of different age. Applying the proposed methods to a large longitudinal pediatric dataset, may provide a novel set of parameters for monitoring pediatric brain development.
1.1. Background
1.1.1. Geometric model of WM fiber bundles
As imaging resolution advances and the extracted connectomes become more and more dense, a need for concise representation of these fiber sets is pressing. In recent years, there have been several efforts to address this need. A method using hierarchical clustering to compress fiber sets by combining analysis in the image space with analysis in fiber space was presented by Guevara et al., 2011. Garyfallidis et al., 2012, presented QuickBundles – a method that reduces fiber clusters to a single centroid streamline. Colby et al., 2012, presented a centralized streamline model for a fiber bundle by assuming that they are tube-like structures with cross-sectional uniformity. Corouge et al., 2004, proposed a clustering scheme that uses the similarity of adjacent curves to group sets of curves to bundles. In this representation, fiber bundles are modeled by a shape prototype swept along a space trajectory. Gori et al., 2016, proposed a weighted prototype scheme for fiber bundles in which several ‘prototype’ fibers are chosen among the streamlines to represent groups of similar streamlines. Alexandroni et al., 2016, compared hierarchical clustering methods with different distance metrics to the Coresets approach (Agarwal et al., 2005), and showed the superiority of the latter in fiber set reduction. The underlying goal of all these methods is data compression – they aim to eliminate redundancies and find the set of representative fibers of a bundle. This reduction is lossy – in the sense that the fine-detailed geometric shape information is lost in the final representation.
In contrast to those methods, Durrleman et al., 2011, proposed using the framework of currents as a metric for analysis of white matter fiber bundles. In this representation, a fiber is represented as a path integral over a vector field which keeps the geometric information of the bundle. The compression ratio that this representation achieves varies with the parameters chosen which control the approximation error. They measure the compression ratio by the number of ‘momenta’ – 3D vectors (represented by 6 floating point numbers – three for location and three for direction) and report 85% compression ratio on average. This is not a fixed-length representation as the number of momenta varies with the size of the dataset and the complexity of the geometry of the fiber bundle. Their work achieves impressive results for registration and variability analysis.
Some efforts are also aimed at characterizing the shape of fiber bundles: Batchelor et al., 2006, used fundamental geometric invariants such as curvature and torsion to estimate an ‘average’ shape for several human fiber bundles. Similarly to Durrleman et al., 2011, our geometric model achieves data compression without compromising the completeness of geometry information. It creates a concise representation of the shape of the fiber bundles, allowing shape analysis at a fine scale as well as bundle registration and deformation estimation. Furthermore, our model achieves fixed-length representation, regardless of the number of subjects in the dataset, or the number of fibers in the analyzed bundles. Our geometric model for a fiber bundle consists of a fixed number, X, of 3D Seed Points (three floating point numbers per Seed Point), where X is determined by the user according to how fine the analysis should be. Since the length and complexity of WM fiber bundles varies greatly within subjects as well as across populations, the ability to adjust the scope of the analysis is essential and useful for different investigations. In the experiments we show in this paper, we chose X=340. The number of nodes in fiber bundles varies, but is usually in the several thousands – thus our representation constitutes a compression ratio of well over 90% on average. The fact that our representation is of fixed length renders it useful for many different types of group analyses and algorithms.
1.1.2. Registration
Most available methods for group analysis of white matter still rely on whole-brain image-based registration, such as Smith et al., 2006, and Ashburner and Friston, 2000. These methods find the optimal diffeomorphic registration by minimizing cost-functions defined on voxels. In dMRI processing, scalar diffusivity measures, such as Fractional Anisotropy (FA) values serve as the basis for registration, e.g. in Groeschel et al., 2014.
The whole-brain registration approach, however, has several challenges when the data analyzed is of brains with ‘non-typical’ gray-level appearance, such as a presence of a mass lesion in the form of a tumor, vascular malformation, abscess, hemorrhage, or brain deformation that might be associated with hydrocephalus. In these cases, since this form of registration relies on the full brain volume for alignment, the abnormal area may cause the registration to fail. Low intensity or poor contrast may also lead to poor results. Additionally, these methods rely on the assumption that the shape variability of white matter fiber bundles is reflected in the variability of diffusivity measures. Most diffusivity measures, however, rely on the diffusion tensor model, which possesses a single major orientation. As a result, this model cannot adequately describe white matter regions that contain two or more differently oriented fiber bundles within the same voxel (crossing, diverging, or kissing fibers) leading to incorrect estimations of diffusion measures and fiber directions Pierpaoli et al., 2001, Alexander et al., 2002, Descoteaux et al., 2009. Thus image-based registration doesn’t necessarily correctly align the neural pathways.
Sotiras et al., 2013, provides an excellent review of methods used for registration in the medical imaging domain. In this work the authors conclude that registration becomes more robust even under the existence of large deformations once landmarks are established. In dMRI imaging such landmarks are the white matter fiber bundles. Indeed, recently there have been much research in this direction – registration in the space of bundles (the anatomical landmarks), rather than in image space. In these methods, the fiber bundles extracted from the dMRI data are treated as stand-alone shapes, and the goal for the registration is to align these shapes across subjects, independently of the tractography algorithms used for their estimation. In Garyfallidis et al., 2015, the authors presented the Streamline-based Linear Registration framework that registers homologous bundles by minimizing a cost function based on local and global geometric properties of the bundles, without using any geometric model for the bundles themselves. They define a distance metric between two bundles that accounts for all the points on the fibers (streamlines) to drive the registration. O’Donnell et al., 2012, proposed to directly register whole tractograms without the indirect step of whole-volume image registration by minimizing a loss function computed only from a random subset of streamline coordinates. Olivetti et al., 2016, find an optimal correspondence between streamlines of one tractogram to streamlines in another in a similar manner to the graph matching problem. Siless et al., 2017, proposed a way to cluster tractography streamlines based on their neighboring anatomical structures, rather than their coordinates, achieving a registration free analysis method across subjects. Zvitia et al., 2010, use Gaussian Mixture Models to represent fiber sets, which are registered by maximizing their correlation ratio. Fishbaugh et al., 2014, proposed a framework based on currents to model and register fiber bundles. The work presented in this paper falls within this category of bundle-space registration.
2. Methods
Diffusion MRI allows imaging the neuroanatomy of white matter structures in-vivo. When comparing the anatomy and function of these structures across populations, it is important to establish spatial correspondence between different sets of tractography data. Such correspondence maps drive the registration (i.e. non-rigid spatial alignment) between the different fiber bundles, from which the shape deformation can be estimated. In this section, we present the full framework for shape analysis of white matter bundles. This framework consists of methods for: (a) creating a geometric model of a WM fiber bundle (section 2.2); (b) fine-scale correspondence mapping between bundles of different subjects (section 2.3); (c) free-form registration for bundle alignment (section 2.4); and (d) shape deformation estimation (section 2.5). Each step is built upon the previous steps.
2.1. Fiber bundle representation and pre-processing
Following tractography processing as elaborated in section 3, the data for each bundle is represented as a fiber bundle – a set of N fibers , each fiber consisting of Mi points in . Each fiber is then resampled to L points, where L is chosen by the user. For longer fiber bundles with elaborate geometry a higher L should be chosen (e.g. L=100), while for shorter bundles a smaller L suffices. In this work we chose L=60. Following the resampling, fibers are flipped such that all [x1, y1, z1] points are on one end of the bundle and all [xL, yL, zL] points are at another end of the bundle.
Following the resampling, we calculate the Centerline – an ‘average’ fiber that follows the curvature of the bundle. The Centerline is a curve consisting of L points where each point is an average of all the points with the same index on all the fibers, calculated as follows: .
2.2. Geometric model of a fiber bundle
Homologous fiber bundles of different subjects may have different shape both in the macro-level and in fine-level details: they may differ in the number of fibers, average length of the fibers, cross-sectional areas along the bundle, the distribution of the fibers within a cross-section etc. We suggest a model that represents the variable geometry of such bundles in a way that is both concise and allows for correspondence mapping between regions of interest on bundles from different subjects. The fundamental observation that guides our model is that the geometry of a bundle has two components – along the fibers and across the fibers. Along the fibers length, the bundle has many fibers aligned in roughly the same direction. At every point along this main direction, a cross-section contains nodes of all the fibers at that point. Our framework models these two components.
Our representation for a fiber bundle is a set of Seed Points . This is a fixed-length representation, since all bundles are represented using the same number of Seed Points. Moreover, the representation is consistent, such that Seed Point of bundle A and Seed Point on bundle B are located in corresponding regions on their respective bundles.
Alg. 1 outlines the process for calculating the representation. As the model essentially creates a division of a bundle into regions, we refer to this process as fiber bundle parcellation. The input to the parcellation algorithm is the fiber bundle, with its fibers resampled and flipped as previously explained. The output is a set of C Seed Points that capture the geometry of the fiber bundle. This is achieved by the following steps:
the Centerline that follows the mean curvature of the fiber bundle is calculated (as explained in section 2.1).
K uniformly spaced (according to geodesic distance) locations along the Centerline are chosen (K is a user-chosen parameter according to the desired parcellation resolution).
the local Centerline direction is calculated at each location.
two points above and below ci along , namely and are found. These points are chosen such that and , effectively creating non-overlapping ranges along the centerline.
two planes perpendicular to the Centerline direction are defined using the local centerline direction as normal and the points and respectively.
the area between these two planes defines a ‘slab’. An example of a ‘slab’ can be seen in figure 2(b). All the nodes on the fibers that are contained within such ‘slab’ create a point-cloud shaped roughly as a short cylinder with an elliptical cross-section.
all nodes in the slab are projected to a cross-sectional plane in the middle of the cylinder to get a 2D ellipsoid. The axes of this ellipsoid are calculated.
the elliptical slab is then partitioned into S equal-angle slices.
The radius of each slice is determined by the node farthest from the corresponding centerline point for that slice.
P Seed Points are placed on each slice, equally spaced, along a radial line passing in the middle of the slice. This creates a total of S × P Seed Points per cross-section.
Figure 2. Seed Points placement procedure.
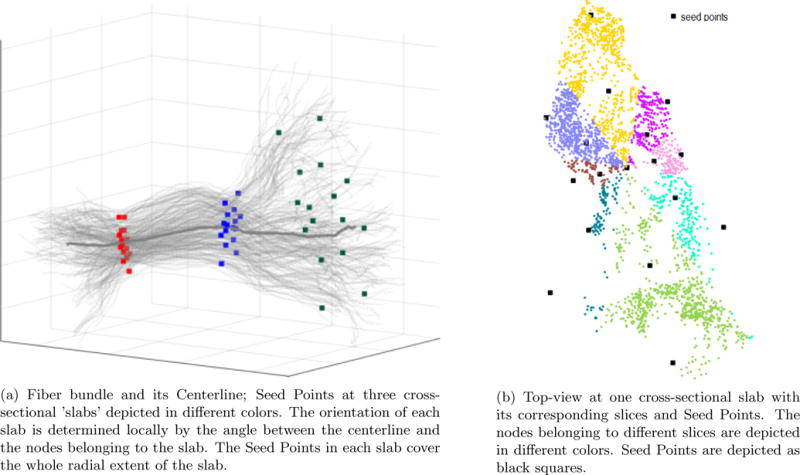
The calculated model captures the nature of the fiber bundle shape along the fibers and across the fibers. Left – fiber bundle (thin gray curves) with centerline (thick gray curve) and Seed Points at three cross-sections. Right – top-view on one cross-sectional ‘slab’.
The model is controlled by four parameters: L – the number of nodes per fiber (defining the distance between nodes on the fiber), K – number of cross-sections per bundle (defining the number of ‘slabs’ per fiber bundle, K<L), S – number of slices per cross-section (defining the number of angular slices each ‘slab’ is cut into) and P – number of Seed Points per slice (defining the number of clusters in each slice). These can be set by the user based on the required correspondence mapping accuracy. A different set of parameters results in a different parcellation scheme, with the resulting cluster locations visually consistent across subjects. Thus for datasets produced by higher resolution white matter imaging methods, or when finer detailed correspondence mapping is required the user may choose to use finer resolution for the shape analysis – requiring more Seed Points. When a coarser analysis is sufficient the user may choose parameters that yield less Seed Points in the model. In this experiment we chose the following parameters: K=20 (number of cross-sections), S=8 (number of slices per cross-section) and P=2 (number of Seed Points per slice). This choice of parameters yields 8 × 2 = 16 SeedPoints per cross-section, resulting in a total (20 × 16) + 20 = 340 Seed Points, thus 340 clusters. For a finer or coarser analysis this number can be set by the user.
An overview of the parcellation process for the left thalamic radiation (LTR) fiber bundle is shown in figure 1. The bundle itself is shown on the left. The middle figure shows the locations of the Seed Points, each Seed Point is a centroid of a corresponding cluster. Note that the Seed Points capture the spatial ‘spread’ of the bundle at each location along the Centerline. The figure on the right shows the result of the parcellation. Points on the fibers are grouped together in clusters based on their spatial location and local geometry using K-nearest-neighbors algorithm. Each cluster is shown in a different color.
Figure 1. Parcellation overview.

(left) Left Thalamic Radiation (LTR) fiber bundle; (middle) Seed Points representation models the geometry of the bundle. Each node on the bundle is assigned to one of the SeedPoints using a nearest-neighbor approach; (right) parcellated LTR bundle – each color represents a different cluster. Cluster location is visually consistent across subjects in a given dataset.
In the illustration in figure 2a we show the fibers (in light gray) with three cross-sectional ‘slabs’, each with s=8 slices and P=2 Seed Points. Figure 2b shows an example of one such ‘slab’ (viewed from above), the nodes belonging to the slab and the assigned Seed Points. This slab was partitioned to S=8 slices of angle 45° each. This partitioning was chosen empirically as an optimal partitioning to capture the fine geometry variations in the dataset while keeping the number of slices as small as possible. The nodes (points on the fibers) belonging to each slice are shown in different color. The radial extent of each slice determines the placement of Seed Points (black squares on the figure) in that slice. The central Seed Point is placed at the centroid of all the nodes belonging to the slab.
Algorithm 1.
Fiber Bundle Parcellation.
| 1: | fibers ← ResampleAndFlipFibers(fibers) |
| 2: | Centerline ← ExtractCenterline(fibers) |
| 3: | for node in [0, 1, …, K] on Centerline do |
| 4: | NodeDir ← CalculateDirection(node, Centerline) |
| 5: | if reference then |
| 6: | NodeDirRef ← NodeDir |
| 7: | p ← GetPointsAround(node) |
| 8: | V ← EigenVectors(p) |
| 9: | V ← Normalize(V) |
| 10: | else |
| 11: | RotMat ← CalcRotationMatrix(NodeDir, NodeDirRef) |
| 12: | V ← RotateVectors(V, RotMat) |
| 13: | end if |
| 14: | SeedPoints ← FindSeeds(V) |
| 15: | end for |
2.3. Fine-scale correspondence mapping between fiber bundles of different subjects
Given a group of fiber bundles from a dataset of subjects, we are interested in finding the spatial correspondence map between these bundles, i.e. a mapping function that indicates the location of a chosen region of interest (ROI) on bundles from different subjects. Such correspondence map can be used for registration and shape deformation estimation (as we do in the rest of this paper). It can also be used for shape analysis or fine-grained tissue properties analysis (similar to the profile analysis in Yeatman et al., 2012, only in a much more fine-grained manner, as the correspondences established relate much smaller regions of interest, rather than cross-sections of the bundle).
To create the correspondence map for a group of given fiber bundles, we first need to define a reference bundle. A reference can either be a bundle generated by an atlas, or one of the bundles within a given dataset. In this work we arbitrarily chose a fiber bundle of one subject within each dataset as a reference. The parcellation of the reference bundle informs the parcellation of other bundles in the set, such that the ROIs on all fiber bundles will spatially map to the location of the ROIs on the reference. A synthetic reference can also be generated. It is important to note, however, that since the reference bundle serves as a template to which the rest of the bundles are registered, this bundle must have ‘sufficient’ structure to generate non-null Seed Points model. In this context ‘sufficient’ means that each cross-section should have at least S ∗ P number of points. Within this minor constraint, the choice of the reference does not influence the results. Furthermore, this constraint is rarely not satisfied since the number of fibers generated by tractography algorithms is on the order of several hundreds to a few thousands. For the adult dataset, we randomly chose one of the subjects to serve as reference. For the pediatric dataset, since we were interested in the shape change of fiber bundles among the subjects of different ages, we chose the youngest subject (11 days old) to serve as a reference.
The mapping process between a reference bundle and a target bundle is explained in Alg. 1, executing the ‘else’ statements in lines 11-12. The difference between the reference parcellation and the parcellation of the rest of the bundles in a dataset is in the processing of the cross-sectional ‘slabs’: the axes of the ellipse model for the slab (the ellipse axes that were calculated on the reference bundle) are multiplied by a rotation matrix (RotMat, line 11) formed between the local centerline direction on the moving bundle and the corresponding local centerline direction on the reference bundle. This matrix is calculated by finding a rotation needed to align the two vectors. This operation essentially aligns the axes of the cross sectional ellipse on the moving bundle to its corresponding ellipse on the reference bundle. Following this alignment the algorithm proceeds as before, generating Seed Points. Since all fiber bundles are parcellated similarly, mapping between ROIs is a simple index query operation – given an ROI with index j on the reference bundle, its corresponding ROI on other bundles in the dataset has the same index j.
2.4. Fiber bundle registration
One of the major contributions of this work is a method for registration of fiber bundles from different subjects in bundle space. This is enabled by modeling the shape of a fiber bundle by a set of Seed Points, as explained in section 2.2. The Seed Points calculated during the parcellation process constitute the correspondence mapping between the different bundles, as explained in section 2.3. Once such a correspondence is established, we choose a subset of these points as Control points to drive the registration. One of the bundles in the dataset is arbitrarily chosen as reference, as discussed in section 2.3. Reference bundle is considered as fixed bundle and all other bundles in the set are considered as moving bundles and will be registered to the fixed bundle.
Considering the geometric structure of a fiber bundle, the most significant deformation sources are elongation/shortening along the fibers length and radial expansion/reduction. To account for both these major deformation sources, we propose a two-step registration process whereby the first step consists of registration along the fibers length and the second step consists of the radial registration. Section 2.4.1 elaborates on the exact process of free-form deformation. An overview of the registration process is as follows:
all bundles are preprocessed by (a) resampling all fibers to have equal length; (b) flipping fibers if necessary so that all fibers have the same origin and the same destination.1; (c) all bundles are zero-centered.
following the preprocessing, a Centerline for each bundle is calculated as explained in section 2.1.
a rigid transformation is calculated between to Centerline of the moving bundle and the fixed bundle. This step consists of finding a rotation matrix, R, and translation vector, T, as explained in Besl and McKay, 1992.2
the rotation and translation found in the previous step are applied to all fibers in the moving bundle. This results in a very rough alignment between the moving and fixed bundles.
all bundles are parcellated as described in section 2.2 (for the reference) and 2.3 (for the moving bundles).
coarse registration along the fibers length is applied by choosing Centerline Seed Points as Control Points for the free-form registration.
finer ‘wireframe’ registration is applied by choosing peripheral Seed Points as Control Points for the free-form registration.
post-processing – all fibers are lightly smoothed using a 3-point moving average approach and resampled to keep a fixed distance between points. This step is necessary to avoid spurious fiber curves after the registration.
Figure 3 illustrates the control points for the two-step registration. The gray control points are taken from the centerline of the bundle and black control points are on the outer perimeter of the bundle.
Figure 3. Seed Points used in two-step registration.
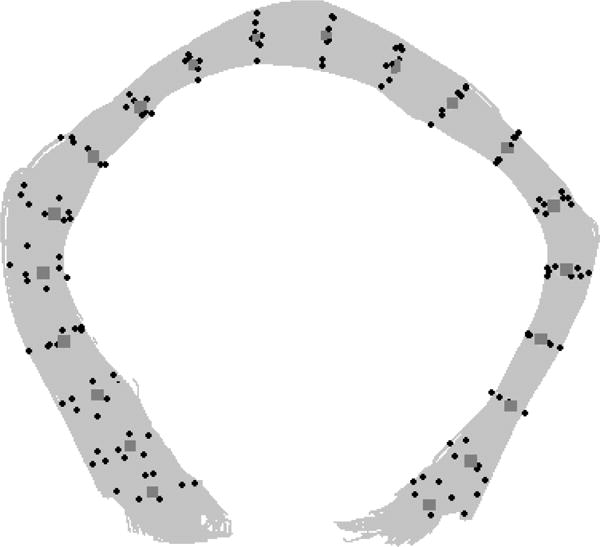
Example of control points used for the two-step registration on callosum forceps major fiber bundle. Control points for the initial coarse registration along the fibers length step are in gray (diamond shaped), control points used for the fine-registration step are in black (dot shaped).
2.4.1. Free-form deformation
Free-form deformation (FFD) by Sederberg et al. is a method for deforming the shape of an object by transforming the three-dimensional space in which the object is embedded. In this work, B-spline FFD is considered. A 3D grid of regularly spaced b-spline control points is constructed to encapsulate a moving bundle. A set of corresponding points on the reference bundle and on the moving bundle is chosen from our previously found Seed Points. Using quasi-newton optimization we find the deformation of the grid that minimizes the euclidean distance between the correspondence points. The 3D grid is refined during this process to allow better accuracy by doubling the number of points in each dimension. The result of this step is a four-dimensional deformation tensor: for each control point, it contains the displacement (scalar value) and the direction of the displacement (3D vector). Subsequently, we calculate the displacement direction and magnitude for each point of the moving bundle fibers by interpolating between the closest control points. This results in a registration of the moving bundle to the reference bundle.
2.4.2. Registration accuracy quantification
To quantify registration accuracy we use the Intersection over Union (IoU) measure, also known as Jaccard similarity coefficient (or Jaccard index) Jaccard, 1901. IoU is a normalized measure of overlap and is defined as the size of the intersection divided by the size of the union of two sample sets: . The index is 1 when there is a perfect overlap and 0 when there is no overlap. We use this index to quantitatively verify registration accuracy and compare our results to whole-brain image-based registration methods – see section 2.4.3. Since our data is three-dimensional, |A ∩ B| is derived by counting the number of jointly occupied voxels in a 3D grid of a bounding box containing both A and B, while |A ∪ B| is derived by counting the number of singly and doubly occupied voxels in that bounding box, that is voxels occupied by nodes from either A or B or both. To calculate IoU between two bundles A and B, we simply divide between the two counts. We use this measure to compare our registration results to nonlinear whole-brain registration transformation applied on specific fiber bundles (see section 2.4.3).
2.4.3. Comparison with nonlinear whole-brain registration (WBR)
To validate our novel fiber-bundle model-based registration (we refer to our method as MBR hereafter), we compare its accuracy with the commonly-used whole-brain image-based nonlinear registration based on FSL [4] (we refer to this method as WBR hereafter). We randomly generated 35 subject pairs (with no duplicates) from dataset 2 (see section 3.2), with the first subject in the pair being the fixed subject and the second subject in the pair being the moving subject. For each subject we examined an ensemble of eight white matter fiber bundles that were identified using AFQ (see section 3.2.3 for details), namely callosum forceps major and minor, left and right corticospinal tracts, left and right inferior fronto-occipital fasciculi and left and right inferior longitudinal fasciculi. The goal of this experiment was to measure the normalized overlap between the registered fiber bundles (IoU, similarly to [23]), and to compare it between the two methods.
For WBR, we aligned the whole-brain FA maps of the moving subject to the fixed subject in each subject-pair using FMRIB Non-linear Image Registration Tool (FNIRT) [4]. First, FMRIB Linear Image Registration Tool [31], [32] was used to compute the initial affine transformation of the two FA maps, which was then fed to FNIRT to compute the nonlinear transformation. The resulting nonlinear transformation was applied to the individual fiber bundles of the moving subject using applywarp (part of FMRIB Software Library) to obtain the moved fiber bundles. The specific parameters used for WBR can be found in Supplementary Material section 2.
For MBR, we directly registered the fiber bundles of the moving subject to the homologous fiber bundles of the fixed subject. The results of this analysis are shown in section 4.1.
2.5. Shape deformation estimation
The registration process moves all the points on the moving bundle such that the bundle is aligned with the reference bundle. This process does not change the number or the order of nodes on the fibers. Thus, the total deformation-per-node can be calculated by simply taking the L2 norm of the euclidean distance between the location of the node pre- and post-registration. In the developmental dataset, the growth along the fibers length is the major contributor to the total deformation, with the radial growth contributing a smaller component. It is also possible to calculate the deformation components in different orientations from the data. However, in this work we focus on the total absolute deformation, regardless of direction.
3. Data for experiments
We demonstrate the methods above on two very different dMRI datasets: (1) we show how a geometric model is created for left thalamic radiation fiber bundle of six adult normal subjects, and how this allows fine-scale correspondence mapping on these bundles (see section 3.1 for imaging and data acquisition and pre-processing details); (2) we demonstrate the full pipeline starting from the creation of the geometric model and through estimating shape changes on an ensemble of WM fiber bundles: the occipital and frontal radiation of the corpus callosum – the forceps major (C_FMajor) and forceps minor (C_FMinor) respectively, the inferior fronto-occipital fasciculus (IFOF) and the corticospinal tract (CST) – on a cohort of 38 normal pediatric subjects. The first dataset was acquired for research purposes and is thus of high quality and resolution. This small dataset is used primarily to demonstrate the model. The second dataset was initially acquired for clinical purposes to rule out presence of pathological conditions, and is thus of less good quality. Nevertheless, our methods are applicable and work well on both types of data. For the second dataset, we also show how the shape of white matter fiber bundles changes among subjects of different age and suggest an approximated model for shape change of the analyzed bundles during normal development.
3.1. Dataset 1 – normal adult subjects
Magnetic resonance imaging diffusion weighted data of six healthy male adults (age 37-39) were acquired at Stanford’s Center for Cognitive and Neurobiological Imaging using a 3-T General Electric Discovery 750 MRI equipped with a 32-channel head coil (Nova Medical). Data collection procedures were approved by the Stanford University Institutional Review Board. Written consent was collected from each participant. DWI images were measured at 1.5 mm3 uniform spatial resolution with 96 different diffusion directions with b value of 2000 s/mm2 (TE=96.8 ms). The images were corrected for spatial distortions due to B0 field inhomogeneity and motion corrected. Fiber tracking was performed using MRtrix by Tournier et al., 2012.
3.2. Dataset 2 – normal pediatric subjects
3.2.1. Subjects
The subjects chosen for this study were collected from a clinical database of normally developing children with age ranging from 11 days to 8 years. All scans were obtained in the course of normal clinical care based on concerns of the treating physicians and retrospectively reviewed after approval by the Stanford University institutional review board (protocol 28674). Scans were read as normal by the attending pediatric neuroradiologist. Examples of reasons MRI scans were obtained in the normal cohort included: Family history of aneurysm or vascular malformations, sinus disease, peri-orbital dermoid, facial hemangioma, ear infection, and benign strabismus without orbital or intracranial abnormality. The charts of all patients with normal MRIs were interrogated to confirm the absence of any disease states. The total number of subjects was 38, with 21 females and 17 males. The age and gender distributions can be seen in Figure 4.
Figure 4. Age and gender distribution of Dataset 2.
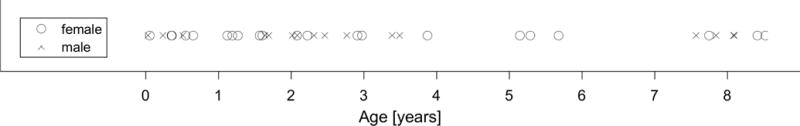
Dataset 2 consisted of 38 clinically normal children, with 21 females and 17 males, of age 11 days old through 8.5 years old.
3.2.2. MRI image acquisition and pre-processing
As part of their clinical evaluation, children were scanned at the Lucile Packard Childrens Hospital at Stanford. Children younger than 3 months of age were scanned using a swaddle-and-feed method; children older than 3 months of age were sedated under general anesthesia; and sedation for children aged 6-8 years was based on individual maturity level and ability to tolerate the MRI exam. High resolution T1-weighted (3D SPGR; TR = 7.75 ms; TE = 3.47 ms; 1 mm isotropic voxels; orientation = axial) and diffusion-weighted images were obtained at 3T (GE MR750 Discovery; GE Healthcare, Waukesha, WI, USA) using an 8-channel head coil.
Diffusion data were acquired with a twice-refocused GRAPPA DT-EPI sequence (TR = 4-6 s depending on slice coverage, acquisition matrix = 128 × 128, acceleration factor = 3, NEX = 3, slice thickness = 3 mm, gap = 0 mm, FOV = 20 cm) using a b-value of 1000 s/mm2 sampling along 25 isotropically distributed diffusion directions. Five T2-weighted (b=0) images were interspersed throughout the acquisition. To reconstruct the data, we used an automated tailored reconstruction software that selected the best GRAPPA and ghost calibration weights; performed 3D rigid-body realignment with importance weighting; and employed phase correction and complex averaging to lower Rician noise and to reduce phase artifacts (see Holdsworth et al., 2012 for details). In addition to the 3D realignment procedure, we quantified the degree of translational head motion in each scan by assessing the magnitude (in voxels) of motion correction required for each volume along the x-y-z- plane. We chose to remove volumes if translational movement exceeded 2 voxels in any direction. No scan was determined to be unusable due to excessive motion as more than 30% of the total number of volumes were maintained in all children. Using a rigid body transformation, diffusion images were registered via the b0 image to the T1-weighted image that had been manually aligned to a canonical AC-PC orientation. For each voxel, a tensor model was fitted using a standard least-square algorithm. Images depicting fractional anisotropy, mean diffusivity, radial diffusivity and axial diffusivity were generated based on the eigenvalue decomposition of the diffusion tensor Pierpaoli and Basser, 1996. Pre-processing was implemented in MATLAB (MathWorks, Natwick, MI) and the analysis tools are publicly available as part of the Vistasoft git repository, VistaLab, 2012.
3.2.3. White matter fiber bundle identification
Automated Fiber Quantification, Yeatman et al., 2012, was used to track and segment cerebral white matter pathways in each subjects native space. In brief, AFQ consists of three main processing steps: i) whole-brain tractography, ii) automatic pathway segmentation, iii) automatic pathway refinement and cleaning. First, a deterministic streamlines tracking algorithm Mori et al., 1999, was used to estimate a whole-brain connectome of fiber bundles. To balance the inclusion of less mature WM regions in very young infants while minimizing the inclusion of GM and partial volume effects, the default tracking parameters were adjusted based on FA values utilized in previous dMRI studies of neonates Rose et al., 2014, de Brune et al., 2011: The tracking algorithm was seeded with a white matter mask defined as all the voxels with FA value above 0.15. Tracking proceeded in all directions and stopped when FA dropped below 0.10 or when the angle between the last path segment and next step direction was greater than 30. Secondly, bundle segmentation was performed using a multiple waypoint ROI procedure in which each fiber from the whole-brain connectome was assigned to a specific fiber group if it passed through two ROIs that defined the trajectory of the fiber group, see Wakana et al., 2007. Because ROIs were defined on a template in MNI space, non-linear transformation, see Friston and Ashburner, 2004, was applied to register these ROIs into each individuals native space. We defined the central portion of the fiber bundle by clipping each streamline to the portion that spans between the two waypoint ROIs and resampling each fiber bundle to 30 equidistant nodes. The final cleaning of the bundle was done by computing the robust mean (3-dimensional Gaussian covariance of the sample points) and removing fibers that were either more than 4 standard deviations above the mean fiber length or that were located more 5 standard deviations away from the core of the fiber bundle. For each child, fiber renderings for the forceps major (C_FMajor), forceps minor (C_FMinor), corticospinal tract (CST), inferior fronto-occipital fasciculus (IFOF) and inferior longitudinal fasciculus (ILF) were visually inspected to ensure that each bundle conformed to anatomical norms prior to any statistical/shape analyses.
4. Experiments and Results
We first validate the proposed novel methods by comparing the results of our registration to nonlinear whole-brain image-based registration based on FA (in section 2.4.3). We then demonstrate the different parts of our framework on the two datasets we elaborated on in section 3 (sections 4.2 – 4.5). Finally, we present a possible application for estimating the shape change of white matter bundles across different ages (section 4.6).
4.1. Comparison with nonlinear whole-brain registration (WBR)
In this experiment we quantitatively assessed the accuracy of our novel fiber bundle model-based registration (MBR) and compared it with the commonly-used whole-brain image-based nonlinear registration based on FSL [4] (WBR). Figure 6 shows the overlap between the moved bundle and the fixed bundle, as quantified by the IoU index, for each method. Visual inspection of both subplots indicates that our MBR method produces a higher overlap than WBR on all fiber bundles considered in this analysis, except for the callosum forceps minor. T-tests confirmed that our overlap values were significantly higher than those of WBR for all bundles except for FMinor (corrected for multiple comparison, see supplementary table 1).
Figure 6. Comparison to FA-based nonlinear Whole-brain based registration.
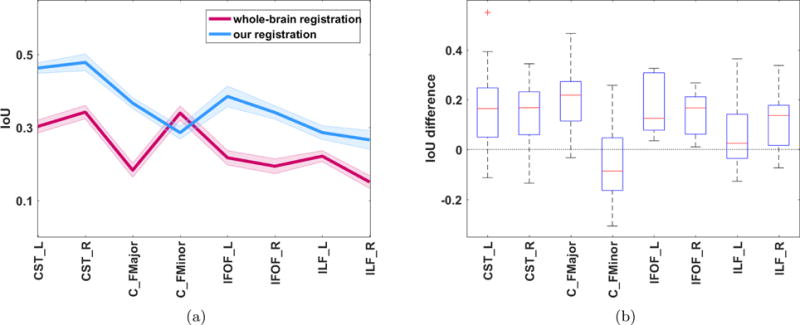
(a) IoU (Intersection over Union) for eight bundles across 35 subject pairs – comparison of WBR and MBR. The plot shows mean values across all 35 pairs are shown in bold lines ± one standard error of the mean. (b) Boxplot of the differences in IoU between MBR and WBR. The central mark (red) represents the median, the edges of the box (blue) are the 25th and 75th percentiles, the whiskers extend to the min and max values. Difference values above 0 indicate that MBR performed better.
4.2. Geometric Model
Figure 7 shows an example of the left inferior fronto-occipital fasciculus of three different subjects, each with its Seed Points. Corresponding Seed Points are plotted with the same color. Visual inspection of the figure allows to qualitatively validate that despite the obvious shape differences between the fiber bundles, both in scale and finer shape details, Seed Points of the same color are located in roughly the same relative location on the bundles, rendering the correspondence mapping correct. The specific values for the four parameters of the geometric model that were used for all experiments in this paper are outlined in section 2.2.
Figure 7. Left inferior fronto-occipital fasciculus of three different subjects with a subset of calculated Seed Points.
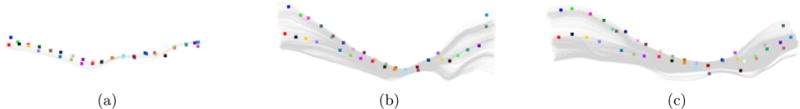
Seed Points corresponding to the same relative location on the different bundles are plotted using the same color. Visual inspection of the Seed Points proves that the correspondence mapping is correct. (a) 2.8 months old; (b) 2 years old; (c) 7.8 years old
4.3. Parcellation
Figure 8 shows an example of a bundle parcellation on the left thalamic radiation fiber bundle of three normal adult subjects from dataset 1. Corresponding regions are plotted in the same color across subjects. For ease of visual inspection, we zoom-in to the central portion of the bundle. Even though the shape of the bundles differs from subject to subject, the correspondence is highly reliable in the majority of the bundle. In the regions of the endpoints, due to their irregular nature, there are some inconsistencies. However, it has become standard practice in white matter variability analysis to use only the central portion of the bundles (see e.g Yeatman et al., 2012) since the diffusion signal towards the endpoints (either on the cortex or in subcortical structures) is much less reliable than the diffusion signal in the central portion of the bundle. Some tractography tools enable ‘clipping’ the extracted fiber bundles to the region between the placed seeds, thus avoiding the endpoints region altogether. We demonstrate the method on clipped fiber bundles from dataset 2 below. Figure 9 shows the resulting parcellation of the left inferior fronto-occipital fasciculus of six different representative subjects from dataset 2. Corresponding ROIs are displayed in the same color. Visual inspection of the images reveals quite satisfactory results – even though the differences between the shapes are significant, regions at corresponding locations share the same color, meaning that they were mapped correctly.
Figure 8. Parcellated Left Thalamic Radiation fiber bundle of three representative subjects from dataset 1.
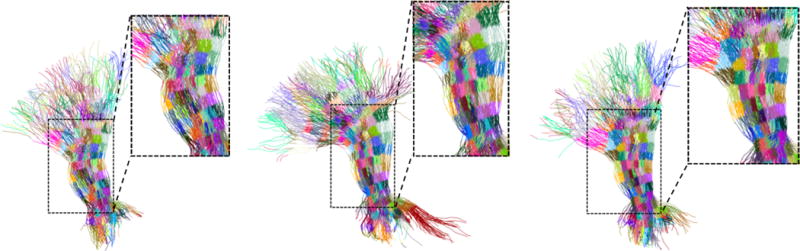
Corresponding regions have the same color across all subjects. In this experiment, no clipping was performed. As a result, it can be seen that while the correspondence mapping at the endpoints has errors, the mapping at the core area of the bundles (shown on the right of each figure) is correct.
Figure 9. Parcellated left inferior fronto-occipital fasciculi with clipped endpoints of 6 different subjects from dataset 2.

Corresponding regions have the same color across all subjects. Due to the clipping, the endpoints of the fiber bundles are not part of the parcellation scheme, yielding a more accurate correspondence.
4.4. Registration
Registration is key tool in medical imaging analysis. It is central to understanding and modeling the shape variability of anatomy structures across subjects. We performed registration between four sets of homologous fiber bundles – callosum forceps major, callosum forceps minor, left IFOF and left corticospinal tract of 38 different subjects. The registration of fiber bundles is evaluated qualitatively by visually inspecting the alignment and quantitatively by comparison to nonlinear whole-brain registration – see section 4.1. Figure 10 shows examples of the registration results we obtain on the callosum forceps major (first row) and left inferior fronto-occipital fasciculus (second row) of an 11 days old infant to a 7.8 years old child. On the left (a) and (d), reference bundles are shown in blue and the “moving” bundle is shown in yellow. The middle figures (b) and (e) show the two bundles after the registration. The right figures (c) and (f) show the “moving” bundle before and after the registration, in purple and yellow colors respectively. As can be seen, after the registration, the “moving” bundle becomes similar in shape and size to the reference bundle.
Figure 10. Registration examples.
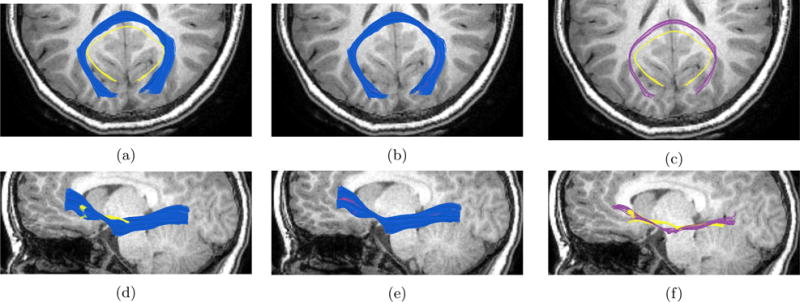
Left image in each row shows the moving bundle (in yellow) and the reference bundle (in blue). Middle image in each line shows the two bundles registered. Right image in each line shows the moving bundle before registration (in yellow) and after registration (in purple). Top row shows an example of callosum forceps major fiber bundle registered between a 8.5 year old female and a 4 months old female. Bottom row shows an example of the left inferior fronto-occipital fasciculus (IFOF) registered between an 8.5 year old female and a 21 days old female. In both rows the background is the T1-image of the reference 8.5 year old child, axial plane on the top row and sagittal plane on the bottom row.
4.5. Deformation mapping
To better understand how the shape of a fiber bundle changes between subjects of different age, we calculate the total deformation for each node on the fibers. Figure 11 shows the left IFOF in four different representative subjects from dataset 2, with the nodes of the bundles color-coded according to the magnitude of deformation they undergo during development, with dark red being the highest deformation (about 15 mm) and dark blue being the lowest deformation (0 mm). In this experiment, the left-IFOF of the youngest subject was used as reference and it can be seen that, as expected, the deformation grows with age.
Figure 11. Shape deformation maps for left inferior fronto-occipital fasciculus color-coded by the amount of deformation in each node.
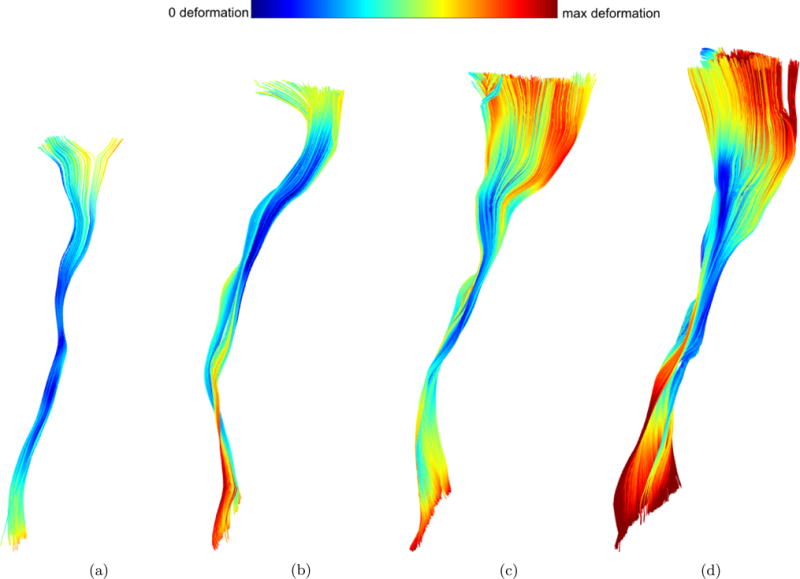
(a) 21 days old; (b) 2.8 months old; (c) 4.4 months old; (d) 20 months old; (e) 7.8 years old. Blue colors indicate low deformation, red colors indicate high deformation as shown in the colorbar on top. Besides the obvious growth in size, there are also subtle changes in shape that are detected and measured.
4.6. Shape deformation model for fiber bundles
Regular measurements of head circumference are standard of care for children as abnormalities in head growth can allow for early identification of disease states such as hydrocephalus, Barbier et al., 2013. Normal head growth during early development can be estimated by measuring head circumference and can be approximated for the first several years of life by a logarithmic curve, indicating that the growth is very fast during the first two years of life and gradually slowing until about age 5, when the head size is approximately as in adulthood. It is driven by brain growth which pushes the skull out as the skull fuses. The additional growth is smaller as the skull growth thicker in the following years. Similarly, the same trend can be extracted from our bundle deformation measures. Figure 12 shows the growth rate (or deformation) as a function of age for four different bundles extracted from the diffusion data of all 38 subjects in dataset 2: callosum forceps major and minor (a) and (b), left corticospinal tract (c) and left inferior fronto-occipital fasciculi (d). As can be seen in the figures, the deformation increases with age. To find whether this increase is best described with a linear or a logarithmic model, we performed 10-fold cross-validation for the deformation data of each bundle. For each bundle, we plot both the best linear and the best logarithmic model. Logarithmic fitted models are plotted with dashed lines and the linear models are plotted with dotted lines. The equations for the logarithmic and linear models are presented in the caption along with the mean squared error (MSE) for each model. The logarithmic model fits the data better than a linear model for all bundles considered except for the left corticospinal tract, for which the linear model achieves a slightly lower MSE. It is important to note that our pediatric dataset is cross-sectional; therefore, the observed shape deformation cannot solely be associated with age – inter-subject variabilities are also present. Nevertheless, we describe the derivation of the such model primarily to illustrate how our methods can be used to obtain an age-dependent shape deformation model for specific fiber bundles. To generate a model useful for clinical purposes (similarly to the clinically used head circumference measure), a large longitudinal dataset would be required, where several time-points exist per subject. This is further elaborated upon in the discussion section.
Figure 12. Fiber bundle deformation curve data and fitted logarithmic and linear curve for four different pathways.
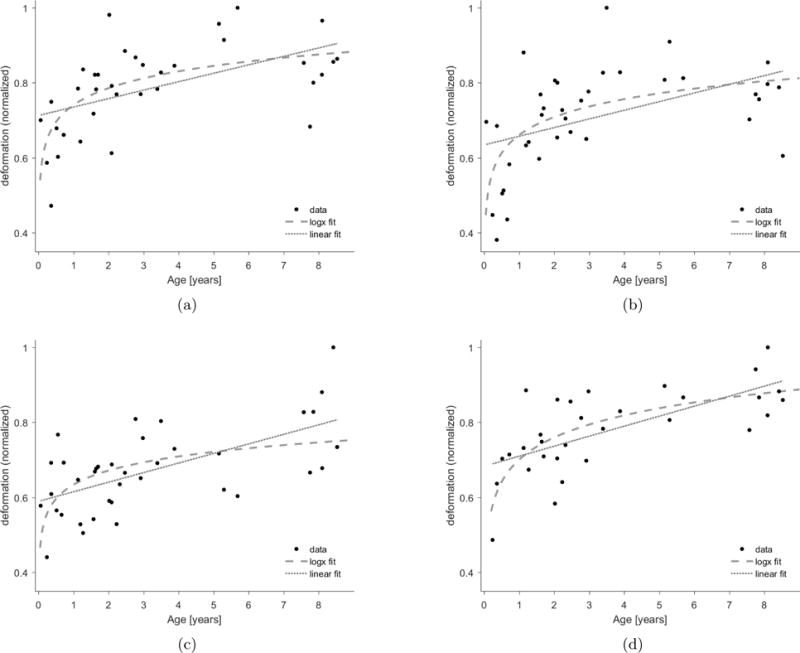
(a) Callosum forceps major; Logarithmic model: y = 0.74 + 0.15 · log(x). (MSE=0.008). Linear model: y = 0.71 + 0.02 · x. (MSE=0.01). (b) callosum forceps minor; Logarithmic model:·y = 0.66 + 0.15 · log(x). (MSE=0.011). Linear model: y = 0.63 + 0.02 · x. (MSE=0.014). (c) left corticospinal; Logarithmic model: y = 0.63 + 0.12 · log(x). (MSE=0.008). Linear model: y = 0.59 + 0.02 · x. (MSE=0.007). (d) left inferior fronto-occipital; Logarithmic· model: y = 0.7 + 0.19 · log(x). (MSE=0.005). Linear model: y = 0.68 + 0.02 · x. (MSE=0.006).
5. Discussion
The shape variability of brain structures extracted from T1-weighted data has been studied and modeled extensively. Regional changes in cortical thickness have been linked to a variety of disorders, including schizophrenia, bipolar disorder, depression, Attention Deficit Hyperactivity Disorder (ADHD) and autism spectrum disorders, see e.g. Rimol et al., 2012, Goldman et al., 2012 and Wang et al., 2016. T1 imaging of Alzheimer’s patients reveals overall volume loss and shape changes in specific brain structures, such as the hippocampus, amygdala, the ventricles and other regions, see e.g. Cuingnet et al., 2011, Ferrarini et al., 2006, as well as assymetry in several brain structures, see Wachinger et al., 2016. Shape descriptors of these structures have been used in machine learning algorithms for disease classification, see e.g. Glozman et al., 2016, Shi et al., 2015. The shape variability of the white matter fiber bundles, however, is much less studied.
Our work aims to close this gap by providing a set of tools for such analysis. The main goal of this work was to generate a simple and flexible framework for shape analysis of fiber bundles. We demonstrate the utility of such framework by generating an approximation to the normative model of white matter shape change across pediatric subjects. We utilize the 3D shape information of the fiber bundle to create a region-to-region map between fiber bundles of different subjects in a given dataset. The geometric model we propose is flexible and adjustable according to the required analysis – finer or coarser Seed Points placement schemes can be defined. It also has the advantage of a fixed-length representation which makes it suitable for applications of group analysis with deep learning architectures, most of which expect a fixed-size input, as well as more traditional machine learning algorithms. By adjusting only four parameters the user controls the resolution of the model and determines the fineness of the analysis.
The methods presented are particularly useful for fiber bundles where the geometry has a major component along the fibers length – e.g. bundles that are relatively long (compared to their cross-sectional ‘footprint’). Fiber bundles that have many branches, such as superior longitudinal fasciculus, or bundles that have very short fibers, such as the uncinate fasciculus will not be a suitable candidate for the geometric model we suggest. The geometry of most other major pathways, however, will be well captured with our geometric model. A limitation of our model is that it does not take into considerations ‘twisting’ of fibers around the central axis, i.e. fiber bundles that have a ‘drill-like’ structure. Based on what is known about fiber bundle organization in the brain, the occurrence of such structures is not very likely: the seminal paper by Bullmore and Sporns, 2012, provides evidence that “many aspects of brain organization can be accounted for by … minimizing the wiring cost involved in anatomically connecting neurons. … governed by laws of conservation of time, space and material…”. The brain has limited volume, bound by the skull size. Thus, fiber bundles must travel between brain areas by the optimal path to minimize the volume they occupy. “Swirling” fibers are unlikely, as they would violate this basic principle.
Our framework establishes correspondences between regions, enabling accurate localization not only for shape variability analysis, but also for comparison of tissue properties. Many studies have confirmed that tissue properties, such as fractional anisotropy, change along the profile of the fiber bundle. Group analysis of FA changes along bundle profiles were found to be associated with a large number of neurologic conditions including Alzheimer’s disease, Parkinson’s disease, age related cognitive decline, Krabbe disease, and delayed neuropsychological effects of treatment of pediatric lymphoid malignancies among others, see Leitner et al., 2015, Feldman et al., 2012a, Feldman et al., 2012b, Poretti et al., 2016, Chan et al., 2016, Schuitema et al., 2013 and Teipel et al., 2012. These analyses were enabled by tools such as AFQ by Yeatman et al., 2012, where the authors show how FA profile comparison can be affected by misalignment of the homologous bundles. They propose a simple procedure to roughly align homologous bundles prior to estimation of the FA profile and show that even this rough alignment significantly reduces confounds caused by crossing fibers, and partial voluming with neighboring structures. Our framework allows mapping corresponding regions on fiber bundles in a much finer scale – thus opening the opportunity of performing a more accurate analyses of localized tissue properties group comparison and detecting differences that might be easily missed in bundle-averaged or even profile-averaged analysis. We plan to explore this direction in future work.
We demonstrated that our methods can be applied to both a high quality research dataset and a clinical dataset, which, by nature of the acquisition protocol, is of lower quality. Our framework extends to a variety of other diffusion models and tractography approaches that output a similar set of fiber bundles with elongated geometry. We showed that our registration method produces a higher overlap for specific bundles compared to whole-brain FA image-based nonlinear registration. The better registration accuracy of MBR as indicated by higher overlap values is due to the local nature of the applied transformation. As mentioned in sections 2.4 and 2.4.1, in our method the Seed Points on the fiber bundles drive the free-form deformation, which allows a very fine local alignment between the fixed and moving bundles. WBR on the other hand creates a deformation field based on voxels intensity values of the 3D volume image – resulting in a less accurate local registration.
Our registration and deformation estimation tools create a quantifiable measure for shape difference in the space of fiber bundles. This metric may serve as an additional tool for assessing both individual variability (e.g. in brain lateralization and longitudinal studies) and population variability (e.g. for estimating the normative shape variability per fiber bundle in a cohort of subjects. We have demonstrated in this study how to use our framework to generate an atlas of fiber bundle shape change across different ages for individual white matter pathways.
In [47], the authors show that using a cross-sectional data may estimate a wrong longitudinal effect, since in such dataset, the inter-subject differences are not taken into account. The pediatric dataset we are using is cross-sectional and thus the age-dependent changes we presented need further verification using large-scale longitudinal data. Nevertheless, to the best of our knowledge, this work is the first to suggest a method to derive a model for shape deformation growth for white matter fiber bundles. Applying our analysis to longitudinal data would yield normative growth curves for individual fiber bundles – a measurement may possibly have clinical potential. In future work, we therefore plan to use our framework to assess typical and pathologic development. Further analysis may focus on evaluating recovery following neurosurgical procedures such as tumor extraction or hydrocephalus shunt installation. All the code developed for this work is freely available at https://github.com/tanyagl/TractGeometry.
Supplementary Material
Figure 5. Bundle ROI placement for clipping endpoints on four different bundles.
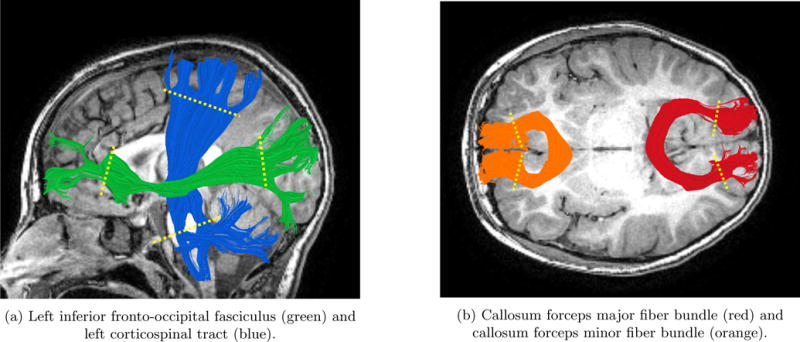
Location of ROIs is indicated by yellow dotted line on each fiber bundle. This procedure was part of the pre-processing prior to the geometry modeling.
Acknowledgments
TG was supported by Stanford Interdisciplinary Graduate Fellowship; FP was supported by NSF IIS 1636893 and NIH ULTTR001108; DWY was supported by NIH/NINDS R25 Grant, Tashia and John Morgridge Endowed Postdoctoral Fellowship, the Child Health Research Institute and the Stanford NIH-NCATS-CTSA (grant no. UL1 TR001085). The authors would like to thank Prof. Brian Wandell and Prof. Heidi Feldman for helpful discussions and suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We denote the origin of fiber i, [x1, y1, z1] of {fi}, as Oi. We denote the termination coordinate of fiber i, [xL, yL, zL] of {fi}, as Ti. A fiber is flipped if ‖O1 − Oi‖ > ‖O1 − Ti‖
Since the Centerlines of the moving and fixed bundles have the same number of nodes, L, we are looking for the optimal rotation matrix (R) and translation vector (T) between two sets of corresponding 3D points. To do this, we first calculate the centroids, CM and CF, for the moving and fixed Centerlines, respectively. We then use Singular Value Decomposition of the covariance matrix of the two centered Centerlines to find the rotation matrix, R, as follows: ; [U,S,V] = SV D(H). The rotation matrix is then simply R = VUT. The translation is T = −R × CM + CF.
Bibliography
- Agarwal P, Har-Peled S, Varadarajan K. Geometric approximation via coresets. Combinatorial and Computational Geometry. 2005;52 [Google Scholar]
- Alexander D, Barker G, Arridge S. Detection and modeling of non-gaussian apparent diffusion coefficient profiles in human brain data. Magnetic Resonance in Medicine. 2002;48:331–340. doi: 10.1002/mrm.10209. [DOI] [PubMed] [Google Scholar]
- Alexandroni G, Zimmerman-Moreno G, Sochen N, Greenspan H. Coresets vs clustering: comparison of methods for redundancy reduction in very large white matter fiber sets. Medical Imaging 2016: Image Processing. 2016 [Google Scholar]
- Andersson J, Jenkinson M, Smith S. Non-linear registration, aka spatial normalization. FMRIB Technical Report TR07JA2. 2010 [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometrythe methods. NeuroImage. 2000;11(6):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Barbier A, Boivin A, Yoon W, Vallerand D, Platt R, Audibert F, Barrington K, Shah P, Nuyt A. New reference curves for head circumference at birth, by gestational age. Pediatrics. 2013;131 doi: 10.1542/peds.2011-3846. [DOI] [PubMed] [Google Scholar]
- Batchelor P, Calamante F, Tournier J, Atkinson D, Hill D, Connelly A. Quantification of the shape of fiber tracts. Magnetic Resonance in Medicine. 2006 Mar;55(4) doi: 10.1002/mrm.20858. [DOI] [PubMed] [Google Scholar]
- Besl P, McKay N. A method for registration of 3d shapes. IEEE Trans on Pattern Analysis and Machine Intelligence. 1992;14(2) [Google Scholar]
- Bullmore E, Sporns O. The economy of brain network organization. Nature Reviews Neuroscience. 2012;13(5) doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- Chan L, Ng K, Yeoh C, Rumpel H, Li H, Tan E. Putaminal diffusivity correlates with disease progression in parkinsons disease: Prospective 6-year study. Medicine. 2016;95(6) doi: 10.1097/MD.0000000000002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby J, Soderberg L, Lebel C, Dinov I, Thompson P, Sowell E. Along-tract statistics allow for enhanced tractography analysis. NeuroImage. 2012;59 doi: 10.1016/j.neuroimage.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corouge I, Gouttard S, Gerig G. Towards a shape model of white matter fiber bundles using diffusion tensor mri. Proc 2004 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2004:1158–67. [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehricy S, Habert MO, Chupin M, Benali H, Colliot O. Automatic classification of patients with alzheimer’s disease from structural mri: A comparison of ten methods using the {ADNI} database. NeuroImage. 2011;56(2):766–781. doi: 10.1016/j.neuroimage.2010.06.013. ISSN 1053-8119. Multivariate Decoding and Brain Reading. [DOI] [PubMed] [Google Scholar]
- de Brune F, van Wezel-Meijler G, Leijser L, van den Berg-Huysmans A, van Steenis A, van Buchem M, van der Grond J. Tractography of developing white matter of the internal capsule and corpus callosum in very preterm infants. European Radiology. 2011;21(3):538–547. doi: 10.1007/s00330-010-1945-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descoteaux M, Deriche R, Knosche T, Anwander A. Deterministic and probabilistic tractography based on complex fibre orientation distributions. IEEE Trans Med Imaging. 2009;28:269–286. doi: 10.1109/TMI.2008.2004424. [DOI] [PubMed] [Google Scholar]
- Durrleman S, Fillard P, Pennec X, Trouv A, Ayache N. Registration, atlas estimation and variability analysis of white matter fiber bundles modeled as currents. NeuroImage. 2011;55(3):1073–1090. doi: 10.1016/j.neuroimage.2010.11.056. [DOI] [PubMed] [Google Scholar]
- Feldman H, Lee E, Loe I, Yeom K, Grill-Spector K, Luna B. White matter microstructure on diffusion tensor imaging is associated with conventional mri findings and cognitive function in adolescents born preterm. Developmental Medicine and Child Neurology. 2012a;54(9):809–14. doi: 10.1111/j.1469-8749.2012.04378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman H, Lee E, Yeatman J, Yeom K. Language and reading skills in school-aged children and adolescents born preterm are associated with white matter properties on diffusion tensor imaging. Developmental Medicine and Child Neurology. 2012b;50(14):3348–62. doi: 10.1016/j.neuropsychologia.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarini L, Palm W, Olofsen H, van Buchem M, Reiber J, Admiraal-Behloul F. Shape differences of the brain ventricles in alzheimer’s disease. NeuroImage. 2006;32(3):1060–1069. doi: 10.1016/j.neuroimage.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Fishbaugh J, Prastawa M, Gerig G, Durrleman S. Geodesic regression of image and shape data for improved modeling of 4d trajectories. 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI) 2014;55:385–388. doi: 10.1109/ISBI.2014.6867889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K, Ashburner J. Generative and recognition models for neuroanatomy. NeuroImage. 2004;23(1):21–24. doi: 10.1016/j.neuroimage.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Garyfallidis E, Brett M, Morgado-Correia M, Williams GB, Nimmo-Smith I. Quickbundles, a method for tractography simplification. Frontiers in Neuroscience. 2012;6 doi: 10.3389/fnins.2012.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garyfallidis E, Ocegueda O, Wassermann D, Descoteaux M. Robust and efficient linear registration of white-matter fascicles in the space of streamlines. NeuroImage. 2015;117:124–140. doi: 10.1016/j.neuroimage.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Glozman T, Solomon J, Pestilli F, Guibas L. Shape-attributes of brain structures as biomarkers for alzheimer’s disease. Journal of Alzheimer’s Disease. 2016;56(1):287–295. doi: 10.3233/JAD-160900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman A, Pezawas L, Doz P, Mattay V, Fischl B, Verchinski B, Chen Q, Weinberger D, Meyer-Lindenberg A. Widespread reductions of cortical thickness in schizophrenia and spectrum disorders and evidence of heritability. Arch Gen Psychiatry. 2012 May;66(5):467–477. doi: 10.1001/archgenpsychiatry.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori P, Colliot O, Marrakchi-Kacem L, Worbe Y, De F, Chavez M, Poupon C, Hartmann A, Ayache N, Durrleman S. Parsimonious approximation of streamline trajectories in white matter fiber bundles. IEEE transactions on medical imaging. 2016 Dec; doi: 10.1109/TMI.2016.2591080. [DOI] [PubMed] [Google Scholar]
- Groeschel S, Tournier JD, Northam G, Baldeweg T, Wyatt J, Vollmer B, Connelly A. Identification and interpretation of microstructural abnormalities in motor pathways in adolescents born preterm. NeuroImage. 2014;87:209–219. doi: 10.1016/j.neuroimage.2013.10.034. [DOI] [PubMed] [Google Scholar]
- Guevara P, Poupon C, Rivire D, Cointepas Y, Descoteaux M, Thirion B, Mangin J. Robust clustering of massive tractography datasets. NeuroImage. 2011;54(3):1975–1993. doi: 10.1016/j.neuroimage.2010.10.028. [DOI] [PubMed] [Google Scholar]
- Holdsworth S, Aksoy M, Newbould R, Yeom K, Van A, Ooi M, S S. Diffusion tensor imaging (dti) with retrospective motion correction for large-scale pediatric imaging. Journal of Magnetic Resonance Imaging. 2012;36(4):961–971. doi: 10.1002/jmri.23710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccard P. tude comparative de la distribution florale dans une portion des alpes et des jura. Bulletin de la Socit Vaudoise des Sciences Naturelles. 1901 [Google Scholar]
- Jenkinson M, Smith S. Global optimization method for robust affine registration of brain images. Medical Image Analysis. 2001;5(2) doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady J, Smith S. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2) doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Leitner Y, Travis K, Ben-Shachar M, Yeom K, Feldman H. Tract profiles of the cerebellar white matter pathways in children and adolescents. Cerebellum. 2015;14(6):613–23. doi: 10.1007/s12311-015-0652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Crain B, Chacko V, van Zijl P. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annals of Neurology. 1999;45(2):265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Mori S, Kageyama Y, Hou Z, Aggarwal M, Patel J, Brown T, Miller M, Wu D, Troncoso J. Elucidation of white matter tracts of the human amygdala by detailed comparison between high-resolution postmortem magnetic resonance imaging and histology. Frontiers in Neuroanatomy. 2017;11 doi: 10.3389/fnana.2017.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell L, Wells R, Golby A, Westin C. Unbiased groupwise registration of white matter tractography. Medical image computing and computer-assisted intervention: MICCAI … International Conference on Medical Image Computing and Computer-Assisted Intervention. 2012 doi: 10.1007/978-3-642-33454-2_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivetti E, Sharmin N, Avesani P. Alignment of tractograms as graph matching. Frontiers in Neuroscience. 2016 doi: 10.3389/fnins.2016.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpaoli C, Basser P. Toward a quantitative assessment of diffusion anisotropy. Magnetic Resonance in Medicine. 1996;36(6):893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C, Barnett A, Pajevic S, Chen R, Penix L, Virta A, Basser P. Water diffusion changes in wallerian degeneration and their dependence on white matter architecture. Neuroimage. 2001;13:1174–1185. doi: 10.1006/nimg.2001.0765. [DOI] [PubMed] [Google Scholar]
- Poretti A, Meoded A, Fatemi A. Diffusion tensor imaging: A biomarker of outcome in krabbe’s disease. Journal of Neuroscience Research. 2016;94(11):1108–1115. doi: 10.1002/jnr.23769. [DOI] [PubMed] [Google Scholar]
- Rimol LM, Nesvg R, Jr, D JH, Bergmann Rjan, Fennema-Notestine C, Hartberg CB, Haukvik UK, Lange E, Pung CJ, Server A, Melle I, Andreassen OA, Agartz I, Dale AM. Cortical volume, surface area, and thickness in schizophrenia and bipolar disorder. Biological Psychiatry. 2012 Mar;71(6):552–560. doi: 10.1016/j.biopsych.2011.11.026. [DOI] [PubMed] [Google Scholar]
- Rose J, Vassar R, Cahill-Rowley K, Guzman X, Stevenson D, Barnea-Goraly N. Brain microstructural development at near-term age in very-low-birth-weight preterm infants: An atlas-based diffusion imaging study. Neuroimage. 2014;86:244–256. doi: 10.1016/j.neuroimage.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuitema I, Deprez S, Hecke WV, Daams M, Uyttebroeck A, Sunaert S, Barkhof F, van Dulmenden Broeder E, van der Pal E, van den Bos C, Veerman A, de Sonneville L. Accelerated aging, decreased white matter integrity, and associated neuropsychological dysfunction 25 years after pediatric lymphoid malignancies. Journal of Clinical Oncology. 2013;31(27) doi: 10.1200/JCO.2012.46.7050. [DOI] [PubMed] [Google Scholar]
- Sederberg J, Thomas W, Scott RP. Free-form deformation of solid geometric models. SIGGRAPH Computer Graphics. 1986;20 [Google Scholar]
- Shi J, Stonnington C, Thompson P, Chen K, Gutman B, Reschke C, Baxter L, Reiman E, Caselli R, Wang Y. Studying ventricular abnormalities in mild cognitive impairment with hyperbolic ricci flow and tensor-based morphometry. NeuroImage. 2015;104(0):1–20. doi: 10.1016/j.neuroimage.2014.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siless V, Davidow J, Nielsen J, Fan Q, Hedden T, Hollinshead M, Bustamante C, Drews M, Dijk KV, Sheridan M, Buckner R, Fischl B, Somerville L, Yendiki A. Registration-free analysis of diffusion mri tractography data across subjects through the human lifespan. ISMRM. 2017 doi: 10.1016/j.neuroimage.2020.116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Hinkle J, Joshi S, Fletcher PT. Hierarchical geodesic models in diffeomorphisms. International Journal of Computer Vision. 2016 Mar;117 [Google Scholar]
- Smith S, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols T, Mackay C, Watkins K, Ciccarelli O, Cader M, Matthews P, Behrens T. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. 2006;31(4):1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Sotiras A, Davatzikos C, Paragios N. Deformable medical image registration: A survey. IEEE Transactions on Medical Imaging. 2013;32(7):1153–1190. doi: 10.1109/TMI.2013.2265603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teipel S, Wegrzyn M, Meindl T, Frisoni G, Bokde A, Fellgiebel A, Filippi M, Hampel H, Klppel S, Hauenstein K, Ewers M. Anatomical mri and dti in the diagnosis of alzheimer’s disease: a european multicenter study. Journal of Alzheimer’s Disease. 2012;31 doi: 10.3233/JAD-2012-112118. [DOI] [PubMed] [Google Scholar]
- Tournier D, Calamante F, Connelly A. Mrtrix: Diffusion tractography in crossing fiber regions. International Journal of Imaging Systems and Technology. 2012;22(1):53–66. [Google Scholar]
- VistaLab. mrdiffusion. 2012 http://github.com/vistalab/vistasoft/mrDiffusion.
- Wachinger C, Salat D, Weiner M, Reuter M. Whole-brain analysis reveals increased neuroanatomical asymmetries in dementia for hippocampus and amygdala. Brain. 2016 Oct;139(12):32533266. doi: 10.1093/brain/aww243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakana S, Jiang H, Nagae-Poetscher L, van Zijl P, Mori S. Fiber tractbased atlas of human white matter anatomy. Radiology. 2004;230(1):77–87. doi: 10.1148/radiol.2301021640. [DOI] [PubMed] [Google Scholar]
- Wakana S, Caprihan A, Panzenboeck M, Fallon J, Perry M, Gollub R, Hua K, Zhang J, Jiang H, Dubey P, et al. Reproducibility of quantitative tractography methods applied to cerebral white matter. NeuroImage. 2007;36(3):630–644. doi: 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wang K, Qu H, Zhou J, Li Q, Deng Z, Du X, Lv F, Ren G, Guo J, Qiu J, Xie P. Disorganized cortical thickness covariance network in major depressive disorder implicated by aberrant hubs in large-scale networks. Nature Scientific Reports. 2016 Jun;6 doi: 10.1038/srep27964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatman J, Dougherty R, Myall N, Wandell B, Feldman H. Tract profiles of white matter properties: Automating fiber-tract quantification. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0049790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvitia O, Mayer A, Shadmi R, Miron S, Greenspan HK. Co-registration of white matter tractographies by adaptive-mean-shift and gaussian mixture modeling. In IEEE Transactions on Medical Imaging. 2010;29 doi: 10.1109/TMI.2009.2029097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


