Among more than 100 different types of chemical marks on RNA species, N6-methyladenosine (m6A) is the most abundant modification in the internal regions of mammalian messenger RNA (mRNA). m6A is installed on mRNA by the Mettl3–Mettl14 methyltransferase complex and could be dynamically erased by demethylases Fto and Alkbh5. Through being recognized by an assortment of selective binding proteins, m6A has been shown to regulate almost every fundamental step of the mRNA life cycle, including mRNA processing, export, translation, and degradation.1 One of the m6A binding proteins, YTHDF2, accelerates decay of m6A-modified mRNA by recruiting the CCR4–NOT deadenylase complex.2,3
The physiological significance of m6A to mark groups of transcripts for coordinated turnover has been emphasized in a growing number of studies of cell fate transitions, including the maternal-to-zygotic transition, immune cell differentiation, and blood progenitor cell formation. A recent study by Yoon et al.4 reported how m6A regulates mammalian brain development, specifically cortical neurogenesis.
Neurogenesis is the process in which neural stem cells (NSCs) produce neurons and begins during embryonic development. In the mouse cortex, radial glia cells (RGCs) function as NSCs and give birth radially to a sequence of layered neurons. This precise and characteristic temporal progression in cell identities necessitates dynamic yet tightly regulated waves of transcriptome change.5 Various factors have been indicated to regulate the timing of neuronal cell differentiation, including sequential expression of transcription factors, cell-cycle regulation, and the microRNA pathway.5 The work by Yoon et al.4 has added m6A to the repertoire of contributing factors.
The authors investigated the expression patterns of molecular players in the m6A pathway and focused on Mettl14, which is strongly expressed in RGCs but not neuronal cells produced later in the lineage. Using the Nestin-Cre;Mettl14f/f conditional knockout (cKO) mouse as a model, the authors were able to study how the deletion of Mettl14 starting on embryonic day 11.5 (E11.5) affects murine nervous system development.
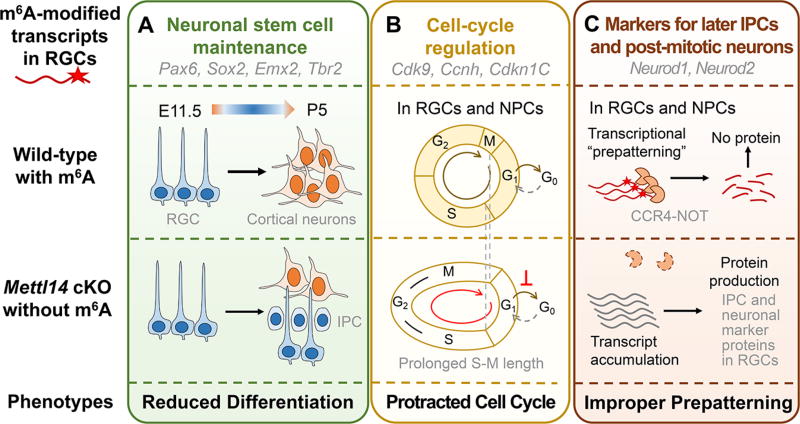
The authors first examined the cortical structures of cKO mice on postnatal day 5 (P5) using immunohistological staining of the markers for different neurogenesis stages combined with ethynyldeoxyuridine (EdU) labeling of cell proliferation. The presence of neurogenic and proliferating RGCs and intermediate progenitor cells (IPCs) in cKO but not wild-type (WT) mice showed that cortical neurogenesis in cKO animals was delayed and continued to postnatal stages (Figure 1A).
Figure 1.
m6A signaling mediates temporal progression of mammalian cortical neurogenesis by regulating the turnover rate of m6A-modified transcripts. Three categories of genes enriched with m6A modification were demonstrated. Abbreviations: RGC, radial glia cells; IPC, intermediate progenitor cells; NPC, neural progenitor cells; E11.5, embryonic day 11.5; P5, postnatal day 5; cKO, conditional knockout.
The authors then investigated whether there were defects in RGCs in embryonic cKO mice. Taking advantage of the periodic movement of the cell nucleus during the cell cycle and the staining of cell-cycle markers, the authors showed that the cell-cycle progression was delayed in embryonic RGCs in vivo, with a prolonged S to M phase transition and a delayed cell-cycle exit. They further confirmed the cell-cycle deficits in cortical neural progenitor cells (NPCs) cultured from E13.5 cortex. cKO NPCs transfected with a dual-reporter system for cell-cycle phase tracking showed protracted S–G2–M phases and an unaltered G1 phase length (Figure 1B).
The importance of the m6A pathway in regulating RGC differentiation and cell-cycle progression was supported by knockdown of Mettl3, which led to consistent phenotypes with Mettl14 cKO both in vitro and in vivo, as shown by the authors. To investigate the mechanisms underlying m6A-mediated regulation of cortical neurogenesis, the authors mapped m6A-modified transcripts in both cortical NPCs and mouse embryonic forebrain at the stage enriched for NSCs, using anti-m6A antibody pull down followed by high-throughput sequencing (m6A-seq). m6A-modified transcripts were found to be enriched with transcription factors and genes related to cell-cycle and neuronal differentiation. By profiling the lifetime of mRNAs in WT and cKO cortical NPCs, the authors demonstrated that m6A-modified transcripts exhibit a turnover rate significantly faster than those of non-m6A-modified transcripts in a Mettl14-dependent manner.
It is of interest that the authors found that markers for IPC and neurons were readily m6A-modified in progenitor cells, and their mRNA levels—and the resulting protein levels—were increased in cKO NPCs. They further showed that this increase was not due to upregulated transcription by measuring nascent mRNA levels, supporting a model of reduced mRNA decay in cKO cortical NPCs. Additionally, in vivo CCR4–NOT complex knockdown resulted in phenotypes similar to Mettl14 cKO. The authors thus proposed that in NPCs, neuronal lineage genes are already transcribed yet actively suppressed at the posttranscriptional level by the m6A-mediated decay, to transcriptionally “prepattern” NPCs for further differentiation (Figure 1C).
Finally, the authors extended the study to human neurogenesis using human NPCs and a forebrain organoid model derived from human-induced pluripotent stem cells, where METTL14 KD led to a conserved delay in cell-cycle progression. They performed m6A-seq of human forebrain organoids and fetal brain, both at a developing stage similar to that of the mouse embryonic forebrain examined. Comparison of m6A landscapes in mice and humans revealed that conserved m6A-modified transcripts were enriched for neurogenesis and oncogenic processes, while human- unique ones were enriched for mental disorders.
In summary, this work by Yoon et al.4 revealed a critical and conserved role of m6A methylation in mammalian cortical neurogenesis in vivo. Besides supporting the function of m6A to promote decay of a group of existing key transcripts for timely cell fate transition as exemplified in previous studies, the authors proposed “prepatterning” as an additional mechanism for temporal specification of cell identity enabled by m6A-mediated decay.
Future studies could focus on how the specificity and extent of mRNA decay are regulated for various groups of m6A-modified transcripts present in these cells, and how genetic and other epigenetic factors for neurogenesis interact with m6A signaling. An equally interesting question is how the role of m6A in regulating translation would contribute to physiological processes and human diseases. Considering the variety of m6A reader-mediated functions in mRNA metabolism, more substantial and diversified roles of m6A could be anticipated in the spatiotemporal regulation of gene expression.
Acknowledgments
The authors thank Mr. Allen C. Zhu for editing and suggestions.
Funding
C.H. is an investigator of the Howard Hughes Medical Institute and is supported by the National Institutes of Health (Grants GM071440 and HG008935).
Footnotes
The authors declare the following competing financial interest(s): C.H. is a scientific founder of Accent Therapeutics, Inc.
References
- 1.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, Kim S, Wang X, Doré LC, Jin P, Regot S, Zhuang X, Canzar S, He C, Ming GL, Song H. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171:877–889. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohwi M, Doe CQ. Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 2013;14:823–838. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]