Abstract
Biochemically modified proteins have attracted significant attention due to their widespread applications as biomaterials. For instance, chemically modified gelatin derivatives have been widely explored to develop hydrogels for tissue engineering and regenerative medicine applications. Among the reported methods, modification of gelatin with methacrylic anhydride (MA) stands out as a convenient and efficient strategy to introduce functional groups and form hydrogels via photopolymerization. Combining light-activation of modified gelatin with soft lithography has enabled the materialization of microfabricated hydrogels. So far, this gelatin derivative has been referred to in the literature as gelatin methacrylate, gelatin methacrylamide, or gelatin methacryloyl, with the same abbreviation of GelMA. Considering the complex composition of gelatin and the presence of different functional groups on the amino acid residues, both hydroxyl groups and amine groups can possibly react with methacrylic anhydride during functionalization of the protein. This can also apply to the modification of other proteins, such as recombinant human tropoelastin to form MA-modified tropoelastin (MeTro). Here, we employed analytical methods to quantitatively determine the amounts of methacrylate and methacrylamide groups in MA-modified gelatin and tropoelastin to better understand the reaction mechanism. By combining two chemical assays with instrumental techniques, such as proton nuclear magnetic resonance (1H NMR) and liquid chromatography tandem-mass spectrometry (LC-MS/MS), our results indicated that while amine groups had higher reactivity than hydroxyl groups and resulted in a majority of methacrylamide groups, modification of proteins by MA could lead to the formation of both methacrylamide and methacrylate groups. It is therefore suggested that the standard terms for GelMA and MeTro should be defined as gelatin methacryloyl and methacryloyl-substituted tropoelastin, respectively, to remain consistent with the widespread abbreviations used in literature.
Keywords: Protein modification, Gelatin, Tropoelastin, GelMA, Structural analysis
1. Introduction
Biomaterials based on proteins or peptides have been widely studied in the biomedical sciences, partly due to their highly tunable chemical compositions, suitable physical properties, and presentation of bioactive motifs [1]. Specifically, hydrogels based on proteins/peptides are suitable candidates to develop biomimetic matrices that resemble the native extracellular matrix (ECM) for supporting cellular attachment, proliferation, organization, and ultimately tissue regeneration [2,3].
As the most abundant structural protein in ECM, collagen contains various cell-binding motifs (such as the Arg-Gly-Asp, or RGD sequence) and matrix metalloproteinase (MMP)-degradable sequences. Gelatin is a mixture of polypeptides obtained from the partial hydrolysis of collagen. Gelatin is biocompatible and retains the functionality of many of the bioactive peptide sequences found in intact collagen. As a result, gelatin has been widely explored to design and fabricate hydrogels for different tissue engineering and regenerative medicine applications [4,5].
Gelatin is soluble inwater at around 40 °C or above, yet can form a physically crosslinked hydrogel at room temperature by partially restoring the characteristic triple helical structures of the parent collagen. However, thermally reversible gelatin hydrogels have some limitations regarding in vivo applications, including poor mechanical properties and low transition temperatures [6,7]. Consequently, various chemical crosslinking methods have been developed to generate mechanically strong and stable gelatin based hydrogels [8,9]. Initially, glutaraldehyde [8] and diisocyanate [9] were used as crosslinkers to form stable gelatin based hydrogel network. However, the cytotoxicity of these crosslinkers limited their use as cell-laden matrices for tissue engineering applications [10]. To address these limitations, Van Den Bulcke et al. generated photocrosslinkable gelatin derivatives by reacting gelatin with methacrylic anhydride (MA) [7]. The resulting material was termed as gelatin methacrylamide (GelMA).
Since then, GelMA has gained significant attention in the biomaterials community and has become a widely used biomaterial [4,5,11–18]. GelMA hydrogels possess an attractive combination of properties. For example, they are suitable for both two-dimensional (2D) cell seeding and three-dimensional (3D) cell encapsulation [19,20], compatible with various microfabrication techniques [12,15,21], tunable in regards to their physical properties [20,22], cost efficient, and easy to synthesize.
Despite its widespread applications, functional groups in GelMA molecules have been inadequately studied. As a consequence, GelMA is ambiguously described as gelatin methacrylamide [7,23–27], methacrylated gelatin [11,28–31], and gelatin methacrylate [19–22,32–34] (Fig. 1). This confusing nomenclature can apply to all similarly functionalized protein-based biomaterials, such as MA-modified recombinant human tropoelastin, which is a full-length protein and has been developed as a highly elastic biomaterial for cardiovascular tissue engineering in our group and termed as methacrylated tropoelastin (MeTro) [35–37]. We tested the developed method for structural analysis on GelMA and MeTro because we were among the earliest groups to focus on the biomedical applications of GelMA, and because we first reported the synthesis of MeTro.
Fig. 1. Number of academic publications that are related to MA-modified gelatin in each year.
Along with the ever-increasing popularity of this biomaterial, several nomenclatures, including gelatin methacryloyl, gelatin methacrylamide, gelatin methacrylate, or methacrylated gelatin, have been used in parallel by different research groups. Data are based on a survey on www.scopus.com by searching these terms as appeared in the title, keywords, and/or abstract.
In this study,we aimed to more accurately describe the chemical modification of protein-based biomaterials by MA and define the correct chemical nomenclature for these biomaterials based on their functional groups. Here, we report the quantification of relative amounts of the different functional groups in GelMA and MeTro prepolymers, namely, methacrylamide groups and methacrylate groups, using a simple microplate assay method. The results are further confirmed by instrumental techniques such as proton nuclear magnetic resonance (1H NMR) and liquid chromatography tandem-mass spectrometry (LC-MS/MS). Our study provides, to our knowledge for the first time, an understanding of the relative reactivity of amine and hydroxyl groups in gelatin and recombinant human tropoelastin with MA, which allows for structurally correct naming of MA-modified proteins, and provides a solid basis for further structural investigations on modified protein/peptide-based biomaterials.
2. Materials and methods
2.1. Materials
Type-A gelatin from porcine skin (300 bloom), methacrylic anhydride (MA, 94%), deuterium oxide (D2O, 99.9% in D), sodium hydroxide (NaOH, 98%), acetohydroxamic acid (98%), iron(III) perchlorate hydrate (crystalline, low chloride), hydroxylamine hydrochloride (>99%), hydrochloric acid (36.5–38.0%, BioReagent), and 3-(trimethylsilyl)-1-propanesulfonic acid sodium salt were purchased from Sigma-Aldrich (St. Louis, MO, USA). Dulbecco’s phosphate buffered saline (DPBS) and Fluoraldehyde OPA reagent were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
2.2. GelMA synthesis
Synthesis of GelMA with different degrees of methacryloyl substitution was performed according to our previously reported procedures [20]. Briefly, 10 g of porcine skin type A gelatin was dissolved in 100 mL DPBS at 50 °C. At a stirring speed of 300 rpm, various amounts of MA (20 mL for ultra-GelMA, 8 mL for high-GelMA, 3 mL for medium-GelMA, or 0.5 mL for low-GelMA) was added to the gelatin solution and the resulting turbid mixture was stirred at 50 °C for 3 h. Under this condition, hydrolysis does typically not take place [7,20]. The mixture was diluted with additional warm DPBS, and dialyzed against an abundance of distilled water using dialysis tubing (12–14 kDa MWCO, Spectrum Lab Inc.) for seven days at 40 °C to remove methacrylic acid and any other impurities. After dialysis, the resulting clear solution was then lyophilized to afford GelMA as a white solid.
2.3. MeTro synthesis
Synthesis of MeTro with different degrees of methacryloyl substitution was performed according to our previously reported procedures with modifications [35,36]. Briefly, tropoelastin was dissolved in DPBS as a 10% (w/v) solution at 4 °C, followed by the addition of various amounts of MA (for the samples used in this study: 20% (v/v) MA for high-MeTro, 15% (v/v) MA for medium-MeTro, or 8% (v/v) MA for low-MeTro). The resulting mixture was stirred at 4 °C for 12 h before the solution was diluted with DPBS, purified by Amicon Ultra-15 Centrifugal Filter Units (10 K MWCO) repeatedly for 6 times at 4 °C and lyophilized.
2.4. Fluoraldehyde assay
GelMA solutions in DPBS and MeTro solutions in DI water were prepared at 0.5 mg/mL. The solutions were mixed with the Fluoraldehyde OPA reagent solution at a 1/1 (v/v) ratio and allowed to react for 60 s to finish the conversion. Fluorescence intensity of the resulting mixtures was monitored in a microplate reader (excitation/emission = 340 nm/455 nm). Gelatin or tropoelastin was used as the standard while pure DPBS or DI water was used as the blank, respectively. Conversion of amine groups (α) was calculated as follows:
2.5. Fe(III)-acetohydroxamic acid complex standard curve
A series of acetohydroxamic acid solutions in DI water with concentrations ranging from 2.5 × 10−4 to 5.0 × 10−3 mol/L was prepared and mixed with Fe(III) solution (0.5 mol/L iron(III) perchloride in 0.5 mol/L hydrochloric acid) at a 1/1 (v/v) ratio. UV–vis absorption spectra of the resulting solutions were recorded (420–700 nm) in microplate (300 μL solution in each well). Absorbance at 500 nm (A500) was plotted against the concentration of acetohydroxamic acid and least square linear fitting was performed to obtain the working curve.
2.6. Sample preparation for UV–vis absorption spectrum measurements
100 μL hydroxylamine hydrochloride solution (0.5 mol/L in DI water) was mixed with 100 μL NaOH solution (1.0 mol/L in DI water), followed by the addition of 200 μL protein solution (for GelMA and gelatin, 50 mg/mL in DPBS; for MeTro and tropoelastin, 12.5 mg/mL in DI water). The resulting mixtures were vortexed for 30 s and allowed to react at room temperature for 10 min. After that, 550 μL hydrochloric acid solution (0.5 mol/L in DI water) was added to acidify the mixture, followed by the addition of 50 μL Fe(III) solution (0.5 mol/L iron(III) perchloride in 0.5 mol/L hydrochloric acid). The resulting mixture was vortexed for 30 s and centrifuged to remove any insoluble solids. UV–vis absorption spectra of the clear solutions were recorded (420–700 nm) in microplate (300 μL solution in each well). The amount of methacrylate groups was calculated using the working curve.
2.7. 1H NMR measurements
GelMA and MeTro samples were dissolved in deuterium oxide (D2O) at a concentration of 30 mg/mL. When necessary, 3-(trimethylsilyl)-1-propanesulfonic acid sodium salt was added as the internal standard in D2O. To avoid precipitation or gel formation of the samples, varied-temperature NMR experiments were performed. For GelMA samples, 1H NMR data were collected at 40 °C, while for MeTro samples the measurements were performed at 4 °C. The data were processed using ACD LABS 12.0 software.
2.8. LC-MS/MS measurements
For tryptic in-solution digestion, 20 μg of either gelatin, GelMA (middle or high), tropoelastin or MeTro (middle or high) dissolved in 100 mM triethylammonium bicarbonate buffer were reduced in 10 mM dithiothreitol at 56 °C for 45 min, then alkylated with 40 mM iodoacetamide at room temperature for 30 min and digested with trypsin (enzyme:protein ratio = 1:20) for 18 h at 37 °C before being extracted into a 60% acetonitrile, and 0.1% formic acid solution. The digested protein was then subjected to reverse phase LC-MS/MS using an Eksigent ekspert 400 nanoLC system coupled to a TripleTOF 5600 + mass spectrometer (SCIEX) in data dependent acquisition mode. The resulting peptide mass spectra were analyzed using the ProteinPilot software against UniProt human or porcine databases including the pig collagen alpha-1(I) sequence published by Bell et al. [38]. The search parameters also included the methacryloyl group as an additional modification. Only peptide sequences identified with a confidence of 99% and above were used to quantify the amount of methacryloyl modifications per amino acid residue using a custom-written R script.
3. Results and discussion
3.1. Determination of amine group conversion
Different functional groups in gelatin and tropoelastin are sensitive to chemical modifications as shown in Fig. 2A. In particular, the reactive functional groups existing in gelatin and tropoelastin are located on the side groups of amino acid residues, including hydroxyl groups (from serine, threonine, hydroxyproline, and hydroxylysine residues), amino groups (from lysine and hydroxylysine residues), and carboxylic acid substitutes (from aspartic acid and glutamic acid residues). It should be noted that reported amino acid compositions of gelatin are influenced by variations in hydrolysis and differences between animal sources of collagen, leading to small alterations in different gelatin products. A typical composition is shown in Fig. 2B [39]. On the other hand, recombinant human tropoelastin has a well-conserved amino acid sequence [40–42]. As a result, the numbers of reactive groups in tropoelastin are more precise (Fig. 2C).
Fig. 2. Scheme of the modification of proteins by MA.
(A) Modifications of proteins and peptides (such as tropoelastin or gelatin) with MA to introduce photocrosslinkable methacryloyl substitutions. Both amine and hydroxyl groups might react with MA in DPBS to form methacrylamide and methacrylate groups, respectively. (B) Typical amino acid residue composition of gelatin [39]. (C) Amino acid composition of recombinant human tropoelastin based on the amino acid residues of the GenBank entry AAC98394 without the signaling peptide (amino acids 1–26) [41]. Amino-containing and hydroxyl-containing amino acid residues are highlighted as blue and red, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
It is worth noting that degree of functionalization of GelMA is of critical importance to determine the physical and biochemical properties of the resulting hydrogels, which has been thoroughly discussed and summarized in our recent review paper on GelMA-based biomaterials [4]. As a result, it is typically necessary to compare and optimize the degree of functionalization toward different specific applications. Conversion of amine groups in biomaterials such as GelMA has conventionally been determined using 1H NMR spectroscopy [20,43]. However, since gelatin is a mixture of polypeptides with complex compositions, it might not be feasible to detect and differentiate the resonance peaks from methacrylamide and methacrylate groups from 1H NMR spectra. Moreover, due to the presence of a mixture of peptides with different lengths/compositions in gelatin and the complex unknown 3D structure of tropoelastin, the use of multi-dimensional NMR techniques in this study would be largely limited.
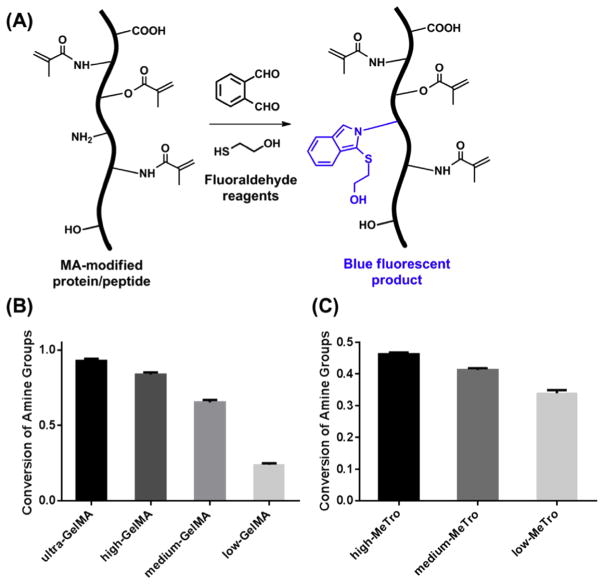
Instead, here we demonstrated that a fluoraldehyde assay [44] allowed for simpler and more accurate determination of the conversion of amine groups. When the modified protein/peptide samples were mixed with the assay solution containing o-phthalaldehyde and 2-mercaptoethanol, all the remaining primary amine groups in the materials were converted into fluorescent species with blue emissions (Fig. 3A). We prepared four distinct GelMA formulations with varying degrees of methacryloyl substitution by adding different amounts of MA to the reaction mixture, while keeping the amount of gelatin unchanged (Table 1) [20]. Depending on the degree of modification, the resulting GelMA formulations were referred to as ultra-GelMA, high-GelMA, medium-GelMA, and low-GelMA, respectively. Using gelatin as the standard in the fluoraldehyde assay protocol, the amount of remaining unreacted primary amine groups in different GelMA samples could be determined by comparing the resulting fluorescent intensity, from which the amount of reacted amine groups and thus the conversion of amine groups could be easily calculated (conversion values: ~93% for ultra-GelMA, ~84% for high-GelMA, ~65% for medium-GelMA, and ~24% for low-GelMA, respectively) (Fig. 3B). Therefore, the conversion of amine groups was positively correlated with the added MA amount, or more precisely, the MA/gelatin feed ratio, at a fixed reaction temperature and reaction time.
Fig. 3. Fluoraldehyde assay to determine conversion of amine groups in GelMA and MeTro.
(A) The reaction between unmodified lysine residues in MA-modified materials and the fluoraldehyde reagents generates blue fluorescent derivatives, which show excitation and emission maxima at wavelengths of 340 nm and 455 nm, respectively. Conversions of amine groups in (B) GelMA and (C) MeTro samples. Gelatin and tropoelastin are used as the controls, respectively. The ultra-GelMA, high-GelMA, medium-GelMA, and low-GelMA refer to samples prepared with 20% (v/v), 8% (v/v), 5% (v/v), and 0.5% (v/v) MA in the reaction, respectively; while high-MeTro, medium-MeTro, and low-MeTro refer to samples prepared with 20% (v/v), 15% (v/v), and 8% (v/v) MA in the reaction, respectively (Table 1). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Summary of molecular parameters of different GelMA and MeTro samples.
| Samples | Conversion of amine groups | Estimated amount of methacrylamide groups (mmol/g) | Estimated amount of methacrylate groups (mmol/g) | Preparation conditions |
|---|---|---|---|---|
| Type-A Ultra-GelMA | 93% | 0.46a | 0.034 | 20% (v/v) MA, 50 °C, 3 h |
| Type-A High-GelMA | 84% | 0.42a | 0.028 | 8% (v/v) MA, 50 °C, 3 h |
| Type-A Medium-GelMA | 65% | 0.32a | 0.022 | 5% (v/v) MA, 50 °C, 3 h |
| Type-A Low-GelMA | 24% | 0.12a | 0.008 | 0.5% (v/v) MA, 50 °C, 3 h |
| High-MeTro | 46% | 0.26 | 0.026 | 20% (v/v) MA, 4 °C, 12 h |
| Medium-MeTro | 41% | 0.23 | 0.019 | 15% (v/v) MA, 4 °C, 12 h |
| Low-MeTro | 31% | 0.18 | 0.007 | 8% (v/v) MA, 4 °C, 12 h |
Estimated data based on the typical amino acid composition of Type-A gelatin and the conversion of amine groups in the corresponding GelMA samples.
We further applied this assay to analyze the conversion of amine groups in MA-modified tropoelastin, MeTro. As tropoelastin is only fully water-soluble at low temperatures (undergoes coacervation at higher temperatures) [41], the reaction with MA was performed under different conditions as compared to the modification of gelatin, i.e., much longer reaction time (12 h) at low temperature (4 °C) [35]. Similarly, we observed that the conversion of amine groups could be tuned by the amount of MA. The determined conversions matched the values measured in our previous publications using 1H NMR [35].
3.2. Quantification of methacrylate groups
In our previous publications on preparation of GelMA [20] and MeTro [35], 1H NMR spectra were used to determine the conversion of amine groups by calculations based on the integration areas of the resonance peak from the amine groups. Quantification of the methacrylate groups was impossible due to the lack of distinguishable resonance peaks of the hydroxyl groups in 1H NMR spectra of the modified peptide or protein samples. Here, we used a Fe(III)-hydroxamic acid-based assay to determine the amount of methacrylate groups in different GelMA samples (Fig. 4). Hydroxamic acid forms a brown-red complex with Fe(III) ions, which can serve as a qualitative test for hydroxamic acid species (Fig. 4A). This class of complexes has an absorption peak centered at around 500 nm(Fig. 4B and C). Formation of the Fe(III)-hydroxamic acid complexes has been used to quantify the reactivity of amine and hydroxyl groups of lysozymes with several different carboxylic acid anhydrides [45], along with other analytic applications such as the quantification of ester group residues in poly(vinyl alcohol) [46]. Acetohydroxamic acid (AHA) was used as the standard to establish the working curve and it is assumed that the complex of acetohydroxamic acid and Fe(III) ions (FeAHA) have similar spectrophotometric properties with that of N-hydroxymethacrylamide and Fe(III) ions (FeHMA) [45]. Iron(III) perchlorate was dissolved in dilute hydrochloric acid to prepare the Fe(III) ion solutions, which were added to the acetohydroxamic acid solutions in large excess to form a 1:1 complex. It has been reported that the apparent extinction coefficient reached its maximum when the molar ratio of Fe(III) and hydroxamic acid is over 20 and will remain independent on the ratio [47]. UV–vis absorption spectra of the series of standard FeAHA solutions were recorded in UV-transparent microplate covering the concentrations of from 1.3 × 10−4 to 2.5 × 10−3 mol/L. Indeed, excellent linearity was achieved when the absorbance was plotted as a function of AHA concentration and analyzed with a linear least-square fit (Fig. 4D).
Fig. 4. Fe(III)-hydroxamic acid assay for quantification of methacrylate groups in GelMA and MeTro prepolymers.
(A) Reactions used for the analysis of methacrylate groups. (i) Methacrylate groups in GelMA or MeTro react with hydroxylamine under basic conditions to generate N-hydroxymethacrylamide in equal molar amount. (ii) N-hydroxymethacrylamide formed a colored complex with Fe(III) ion under acidic conditions. (B) Photographs of an Fe(III) solution before and after the addition of acetohydroxamic acid. The color change indicated the formation of the Fe(III)-acetohydroxamic acid complex. (C) Normalized UV–vis absorption spectrum of the Fe(III)-acetohydroxamic acid complex (FeAHA). The absorption peak was centered at 500 nm. (D) Working curve between absorbance and FeAHA concentration from a series of standard FeAHA solutions. Calculated amounts of methacrylamide and methacrylate groups in (E) GelMA and (F) MeTro samples. Note that the methacrylamide contents in Type-A GelMA samples as shown in (E) were estimated from the typical composition of gelatin shown in Fig. 2B; while the methacrylamide contents in MeTro samples in (F) were calculated from the amino acid composition of tropoelastin shown in Fig. 2C.
To determine the amount of methacrylate groups in GelMA samples, we employed an aminolysis reaction to convert the methacrylate groups to the detectable N-hydroxymethacrylamide compound. In particular, GelMA samples at 50 mg/mL were treated with hydroxylamine solutions at 25 °C for 10 min to generate N-hydroxymethacrylamide. The resulting solution was acidified with hydrochloric acid, followed by the addition of excess Fe(III) ions (see the Experimental Section). Color change upon the addition of Fe(III) ions indicated the formation of the FeHMA complex, which confirmed the existence of methacrylate groups. Concentrations of the FeHMA complex formed in situ were determined from the UV–vis absorption spectra, which could be used to calculate the amounts of methacrylate groups in the GelMA samples (Fig. 4E). For all tested GelMA samples, it was found that methacrylate groups represented below 10% of all methacryloyl substitutions. These results suggested that the amine groups are indeed more reactive than the hydroxyl groups, and the methacrylamide groups are the dominant form in GelMA (Fig. 4E).
When applying this assay to MeTro samples, we found similar results that indicated low amounts of methacrylate groups in MeTro samples (Fig. 4F). From the modification reactions of these two biomaterials, it became evident that the reactivity of hydroxyl groups was lower than that of amine groups.
It is worth mentioning that the quantification of methacrylate groups can also be done by hydrolyzing the methacrylate groups at basic conditions followed by quantifying the amount of released methacrylic acid molecules using high performance liquid chromatography (HPLC) [48]. However, the method developed here, based on a quantitative aminolysis reaction and a colorimetric measurement, could be performed using a microplate reader with satisfying accuracy and reproducibility, which may find wider applications in a biomaterials laboratory.
3.3. Instrumental characterization of GelMA and MeTro samples
To further validate the finding that the major form of methacryloyl groups in GelMA and MeTro was the methacrylamide group, we obtained 1H NMR spectra and LC-MS/MS spectra of some representative samples tested above. 1H NMR spectrum of high-GelMA (Fig. 5A, red curve) showed two additional peaks at chemical shifts of ~5.6 and ~5.8 ppm that were attributed to the methacrylamide groups, which were in agreement with a previous report [49]. Similarly, 1H NMR of high-MeTro showed additional resonance signals at the chemical shift regions of ~5.1 and 5.4 ppm (Fig. 5B, red curve). Neither high-GelMA nor high-MeTro spectra displayed additional resonance peaks that could be attributed to methacrylate groups as observed by others (Fig. S1) [49]. To differentiate the resonance peaks from methacrylamide and methacrylate groups in 1H NMR, we treated the GelMA and MeTro samples with NaOH (0.05 mol/L) at room temperature to hydrolyze the methacrylate groups, which would not affect the methacrylamide groups. The samples after base treatment did not show observable changes in their 1H NMR spectra. Based on these results, we concluded that the peaks shown in the region of 5.5–6.0 ppm were predominately from the methacrylamide groups, confirming that only very small amounts of hydroxyl groups were modified under the typical conditions for GelMA preparation.
Fig. 5. 1H NMR and LC-MS/MS data of selective GelMA and MeTro samples.
(A) 1H NMR spectra of gelatin (black curve) and high-GelMA (red curve) in deuterium oxide (D2O). (B) 1H NMR spectra of tropoelastin (black curve) and high-MeTro (red curve) in deuterium oxide. The asterisk indicates the peaks of 3-(trimethylsilyl)-1-propanesulfonic acid sodium salt as the internal standard, while the double asterisk indicates the residue proton in D2O. (C–D) Degree of methacryloyl substitution on different amino acid residues in (C) GelMA and (D) MeTro samples determined from LC-MS/MS results. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We also applied the LC-MS/MS technique to confirm the distribution of methacryloyl groups on GelMA and MeTro samples among different amino acid residues. Following the optimized procedures, we pretreated the GelMA and MeTro samples to digest them into a library of short oligopeptides, which were briefly separated to record tandem mass spectroscopy. A search algorithm was used to identify the digested peptide sequences and monitor the recovery rates. The unique advantage of this technique is the capability to directly recognize chemically modified amino acid residues in fragmented ions. At good recovery ratios, the results showed that in both GelMA and MeTro samples the methacryloyl groups were attached mostly to the lysine residues (Fig. 5C and D), which was consistent with the results from chemical assays. In addition, amino acid residues with hydroxyl groups were modified at very low degrees (typically <1%) in all the samples.
Therefore, we observed that the reactivity of primary amine and hydroxyl groups with MA in gelatin and tropoelastin under the typical reaction conditions (10 wt% in DPBS solution) was detectably different. The ratio between methacrylamide and methacrylate groups in GelMA and MeTro samples was more than 9:1. Based on the coexistence of both methacrylamide and methacrylate groups in GelMA and MeTro, here we suggest that the proper nomenclature should be gelatin methacryloyl and methacryloyl-substituted tropoelastin, respectively, to be consistent with the widely used abbreviations (GelMA and MeTro). With this knowledge, we believe that it would be interesting to further study the potentially different biochemical properties of the hydrogels that can be related to the different types of functional groups during the MA modification.
4. Conclusion
In summary, we have developed a quantitative assay method to analyze the composition of functional groups in MA-modified protein derivatives. The amount of methacrylamide groups was calculated based on the lysine residue contents and the conversion of primary amine groups, which could be readily obtained using the fluoraldehyde assay. In addition, the amount of methacrylate groups was determined by the treatment with hydroxylamine to generate hydroxamic acid compounds in an equal molar ratio, which could be further quantified by spectrophotometric measurements via the complexation with Fe(III) ions. We tested the MA-modified biopolymers based on two different protein/peptide systems, i.e., GelMA and MeTro, and revealed the reaction difference between the amine groups and hydroxyl groups. Although both functional groups coexist, the major methacryloyl form is methacrylamide. Based on these results, we advocate that the standard name for these two materials should be gelatin methacryloyl and methacryloyl-substituted tropoelastin, respectively, to match with their widely used abbreviations. Considering the general applicability of the chemical and instrumental assays described here, we believe that these methods can be further extended to other MA-modified protein/peptide materials with complex chemical structures and 3D architectures. This can reveal fundamental structural information of chemically modified natural polymers.
Acknowledgments
This paper is sponsored by the Institute for Soldier Nanotechnology, National Institutes of Health (HL092836, EB02597, AR057837, HL099073), the National Science Foundation (DMR0847287), the Office of Naval Research Young Investigator award, ONR PECASE Award. A.S. acknowledges the Canadian Institutes of Health Research (CIHR) Post-Doctoral Fellowship. Y.S.Z. acknowledges the National Cancer Institute of the National Institutes of Health K99/R00 Pathway to Independence Award (K99CA201603). W.Z. gratefully acknowledges funding from NSFC (31501555), Young Eastern Professorship Award, and STCSM (16391903900). T.J.K. acknowledges funding from the Australian Research Council (FT110100166, DP110103543, ARC Centre in Additive Biomanufacturing). JL acknowledges financial support from Innovative Research Incentives Scheme Veni (#14328) from the Netherlands Organization for Scientific Research (NWO). The authors would also like to thank Dr Daniel Broszczak and Dr Rajesh Gupta, Central Analytical Research Facility at the Institute for Future Environments (Queensland University of Technology) for their technical support in obtaining the MS data. The authors thank Prof. Anthony Weiss from Sydney University for providing us tropoelastin. The authors declare no conflict of interests in this work.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.biomaterials.2017.04.050.
References
- 1.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, et al. Silk-based biomaterials. Biomaterials. 2003;24:401–416. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 2.Jonker AM, Löwik DWPM, van Hest JCM. Peptide- and Protein-Based Hydrogels. Chem Mater. 2012;24:759–773. [Google Scholar]
- 3.Yan C, Pochan DJ. Rheological properties of peptide-based hydrogels for biomedical and other applications. Chem Soc Rev. 2010;39:3528–3540. doi: 10.1039/b919449p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials. 2015;73:254–271. doi: 10.1016/j.biomaterials.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loessner D, Meinert C, Kaemmerer E, Martine LC, Yue K, Levett PA, et al. Functionalization, preparation and use of cell-laden gelatin methacryloyl-based hydrogels as modular tissue culture platforms. Nat Protoc. 2016;11:727–746. doi: 10.1038/nprot.2016.037. [DOI] [PubMed] [Google Scholar]
- 6.Djabourov M, Papon P. Influence of thermal treatments on the structure and stability of gelatin gels. Polymer. 1983;24:537–542. [Google Scholar]
- 7.Van Den Bulcke AI, Bogdanov B, De Rooze N, Schacht EH, Cornelissen M, Berghmans H. Structural and rheological properties of methacrylamide modified gelatin hydrogels. Biomacromolecules. 2000;1:31–38. doi: 10.1021/bm990017d. [DOI] [PubMed] [Google Scholar]
- 8.Jayakrishnan A, Jameela SR. Glutaraldehyde as a fixative in bioprostheses and drug delivery matrices. Biomaterials. 1996;17:471–484. doi: 10.1016/0142-9612(96)82721-9. [DOI] [PubMed] [Google Scholar]
- 9.Olde Damink LHH, Dijkstra PJ, Van Luyn MJA, Van Wachem PB, Nieuwenhuis P, Feijen J. Crosslinking of dermal sheep collagen using hexamethylene diisocyanate. J Mater Sci Mater Med. 1995;6:429–434. [Google Scholar]
- 10.Speer DP, Chvapil M, Eskelson CD, Ulreich J. Biological effects of residual glutaraldehyde in glutaraldehyde-tanned collagen biomaterials. J Biomed Mater Res. 1980;14:753–764. doi: 10.1002/jbm.820140607. [DOI] [PubMed] [Google Scholar]
- 11.Benton JA, DeForest CA, Vivekanandan V, Anseth KS. Photocrosslinking of gelatin macromers to synthesize porous hydrogels that promote valvular interstitial cell function. Tissue Eng Part A. 2009;15:3221–3230. doi: 10.1089/ten.tea.2008.0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koshy ST, Ferrante TC, Lewin SA, Mooney DJ. Injectable, porous, and cell-responsive gelatin cryogels. Biomaterials. 2014;35:2477–2487. doi: 10.1016/j.biomaterials.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ovsianikov A, Deiwick A, Van Vlierberghe S, Dubruel P, Moeller L, Draeger G, et al. Laser fabrication of three-dimensional CAD scaffolds from photosensitive gelatin for applications in tissue engineering. Biomacromolecules. 2011;12:851–858. doi: 10.1021/bm1015305. [DOI] [PubMed] [Google Scholar]
- 14.Eng G, Lee BW, Parsa H, Chin CD, Schneider J, Linkov G, et al. Assembly of complex cell microenvironments using geometrically docked hydrogel shapes. Proc Natl Acad Sci. 2013;110:4551–4556. doi: 10.1073/pnas.1300569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv Mater. 2014;26:3124–3130. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 16.Jeon O, Wolfson DW, Alsberg E. In-situ formation of growth-factor-loaded coacervate microparticle-embedded hydrogels for directing encapsulated stem cell fate. Adv Mater. 2015;27:2216–2223. doi: 10.1002/adma.201405337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visser J, Melchels FPW, Jeon JE, van Bussel EM, Kimpton LS, Byrne HM, et al. Reinforcement of hydrogels using three-dimensionally printed microfibres. Nat Commun. 2015;6 doi: 10.1038/ncomms7933. http://dx.doi.org/10.1038/ncomms7933. [DOI] [PubMed] [Google Scholar]
- 18.Shirahama H, Lee BH, Tan LP, Cho NJ. Precise tuning of facile one-pot gelatin methacryloyl (GelMA) synthesis. Sci Rep. 2016;6:31036. doi: 10.1038/srep31036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aubin H, Nichol JW, Hutson CB, Bae H, Sieminski AL, Cropek DM, et al. Directed 3D cell alignment and elongation in microengineered hydrogels. Biomaterials. 2010;31:6941–6951. doi: 10.1016/j.biomaterials.2010.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials. 2010;31:5536–5544. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikkhah M, Eshak N, Zorlutuna P, Annabi N, Castello M, Kim K, et al. Directed endothelial cell morphogenesis in micropatterned gelatin methacrylate hydrogels. Biomaterials. 2012;33:9009–9018. doi: 10.1016/j.biomaterials.2012.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YC, Lin RZ, Qi H, Yang Y, Bae H, Melero-Martin JM, et al. Functional human vascular network generated in photocrosslinkable gelatin methacrylate hydrogels. Adv Funct Mater. 2012;22:2027–2039. doi: 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billiet T, Van Gasse B, Gevaert E, Cornelissen M, Martins JC, Dubruel P. Quantitative contrasts in the photopolymerization of acrylamide and methacrylamide-functionalized gelatin hydrogel building blocks. Macromol Biosci. 2013;13:1531–1545. doi: 10.1002/mabi.201300143. [DOI] [PubMed] [Google Scholar]
- 24.Daniele MA, Adams AA, Naciri J, North SH, Ligler FS. Interpenetrating networks based on gelatin methacrylamide and PEG formed using concurrent thiol click chemistries for hydrogel tissue engineering scaffolds. Biomaterials. 2014;35:1845–1856. doi: 10.1016/j.biomaterials.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Kaemmerer E, Melchels FPW, Holzapfel BM, Meckel T, Hutmacher DW, Loessner D. Gelatine methacrylamide-based hydrogels: an alternative three-dimensional cancer cell culture system. Acta Biomater. 2014;10:2551–2562. doi: 10.1016/j.actbio.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 26.Schuurman W, Levett PA, Pot MW, van Weeren PR, Dhert WJA, Hutmacher DW, et al. Gelatin-methacrylamide hydrogels as potential biomaterials for fabrication of tissue-engineered cartilage constructs. Macromol Biosci. 2013;13:551–561. doi: 10.1002/mabi.201200471. [DOI] [PubMed] [Google Scholar]
- 27.Visser J, Gawlitta D, Benders KEM, Toma SMH, Pouran B, van Weeren PR, et al. Endochondral bone formation in gelatin methacrylamide hydrogel with embedded cartilage-derived matrix particles. Biomaterials. 2015;37:174–182. doi: 10.1016/j.biomaterials.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Bertassoni LE, Cardoso JC, Manoharan V, Cristino AL, Bhise NS, Araujo WA, et al. Direct-write bioprinting of cell-laden methacrylated gelatin hydrogels. Biofabrication. 2014;6:024105. doi: 10.1088/1758-5082/6/2/024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen MB, Srigunapalan S, Wheeler AR, Simmons CA. A 3D microfluidic platform incorporating methacrylated gelatin hydrogels to study physiological cardiovascular cell-cell interactions. Lab Chip. 2013;13:2591–2598. doi: 10.1039/c3lc00051f. [DOI] [PubMed] [Google Scholar]
- 30.Hosseini V, Ahadian S, Ostrovidov S, Camci-Unal G, Chen S, Kaji H, et al. Engineered contractile skeletal muscle tissue on a microgrooved methacrylated gelatin substrate. Tissue Eng Part A. 2012;18:2453–2465. doi: 10.1089/ten.tea.2012.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin RZ, Chen YC, Moreno-Luna R, Khademhosseini A, Melero-Martin JM. Transdermal regulation of vascular network bioengineering using a photo-polymerizable methacrylated gelatin hydrogel. Biomaterials. 2013;34:6785–6796. doi: 10.1016/j.biomaterials.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutson CB, Nichol JW, Aubin H, Bae H, Yamanlar S, Al-Haque S, et al. Synthesis and characterization of tunable poly(ethylene glycol): gelatin methacrylate composite hydrogels. Tissue Eng Part A. 2011;17:1713–1723. doi: 10.1089/ten.tea.2010.0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan G, Zhou L, Ning C, Tan Y, Ni G, Liao J, et al. Biomimetically-mineralized composite coatings on titanium functionalized with gelatin methacrylate hydrogels. Appl Surf Sci. 2013;279:293–299. [Google Scholar]
- 34.Topkaya SN. Gelatin methacrylate (GelMA) mediated electrochemical DNA biosensor for DNA hybridization. Biosens Bioelectron. 2015;64:456–461. doi: 10.1016/j.bios.2014.09.060. [DOI] [PubMed] [Google Scholar]
- 35.Annabi N, Mithieux SM, Zorlutuna P, Camci-Unal G, Weiss AS, Khademhosseini A. Engineered cell-laden human protein-based elastomer. Biomaterials. 2013;34:5496–5505. doi: 10.1016/j.biomaterials.2013.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Annabi N, Tsang K, Mithieux SM, Nikkhah M, Ameri A, Khademhosseini A, et al. Highly elastic micropatterned hydrogel for engineering functional cardiac tissue. Adv Funct Mater. 2013;23:4950–4959. doi: 10.1002/adfm.201300570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Annabi N, Selimovic S, Acevedo Cox JP, Ribas J, Bakooshli MA, Heintze D, et al. Hydrogel-coated microfluidic channels for cardiomyocyte culture. Lab Chip. 2013;13:3569–3577. doi: 10.1039/c3lc50252j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell MP, Neff TB, Polarek JW, Seeley TW. Animal collagens and gelatins. Priority number: US19990439058 19991112 espacenet patent search. 2001 [Google Scholar]
- 39.Stevens PV. Food Aust. 1992;44:320–324. [Google Scholar]
- 40.Vrhovski B, Weiss AS. Biochemistry of tropoelastin. Eur J Biochem. 1998;258:1–18. doi: 10.1046/j.1432-1327.1998.2580001.x. [DOI] [PubMed] [Google Scholar]
- 41.Baldock C, Oberhauser AF, Ma L, Lammie D, Siegler V, Mithieux SM, et al. Shape of tropoelastin, the highly extensible protein that controls human tissue elasticity. Proc Natl Acad Sci. 2011;108:4322–4327. doi: 10.1073/pnas.1014280108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Getie M, Schmelzer CEH, Weiss AS, Neubert RHH. Complementary mass spectrometric techniques to achieve complete sequence coverage of recombinant human tropoelastin. Rapid Commun Mass Spectrom. 2005;19:2989–2993. doi: 10.1002/rcm.2164. [DOI] [PubMed] [Google Scholar]
- 43.Annabi N, Mithieux SM, Zorlutuna P, Camci-Unal G, Weiss AS, Khademhosseini A. Engineered cell-laden human protein-based elastomer. Biomaterials. 2013;34:5496–5505. doi: 10.1016/j.biomaterials.2013.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munoz Z, Shih H, Lin CC. Gelatin hydrogels formed by orthogonal thiolnorbornene photochemistry for cell encapsulation. Biomater Sci. 2014;2:1063–1072. doi: 10.1039/c4bm00070f. [DOI] [PubMed] [Google Scholar]
- 45.Bernad A, Nieto MA, Vioque A, Palaciáan E. Modification of the amino and hydroxyl groups of lysozyme with carboxylic acid anhydrides: a comparative study. Biochimica Biophysica Acta (BBA) - Protein Struct Mol Enzym. 1986;873:350–355. [Google Scholar]
- 46.Andermann G, Zimmermann G, Schilling E. Application of iron(III)-hydroxamic acid complexes in the spectrophotometric determination of poly(vinyl alcohol) in pharmaceutical preparations. Analyst. 1980;105:575–580. doi: 10.1039/an9800500575. [DOI] [PubMed] [Google Scholar]
- 47.Monzyk B, Crumbliss AL. Mechanism of ligand substitution on high-spin iron(III) by hydroxamic acid chelators. Thermodynamic and kinetic studies on the formation and dissociation of a series of monohydroxamatoiron(III) complexes. J Am Chem Soc. 1979;101:6203–6213. [Google Scholar]
- 48.Abbadessa A, Mouser VHM, Blokzijl MM, Gawlitta D, Dhert WJA, Hennink WE, et al. A synthetic thermosensitive hydrogel for cartilage bioprinting and its biofunctionalization with polysaccharides. Biomacromolecules. 2016;17:2137–2147. doi: 10.1021/acs.biomac.6b00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoch E, Schuh C, Hirth T, Tovar GEM, Borchers K. Stiff gelatin hydrogels can be photo-chemically synthesized from low viscous gelatin solutions using molecularly functionalized gelatin with a high degree of methacrylation. J Mater Sci - Mater Med. 2012;23:2607–2617. doi: 10.1007/s10856-012-4731-2. [DOI] [PubMed] [Google Scholar]