Abstract
Carbapenem-resistance among enterobacteriaceae has become a global health concern. The objective of this study was to understand NDM producing enterobacteriaceae and their genetic basis of resistance, spreading in neonatal intensive care unit. Carbapenem resistant NDM producing enterobacteriaceae isolates were recovered from rectal swab and blood sample of infants admitted in NICU. These were determined by using Carba-NP test. All isolates were identified using BD PhoenixTM−100 and MICs were determined by broth microdilution method. The blaNDM and associated resistant markers were checked by PCR followed by sequencing. Moreover, ERIC-PCR and genetic environment of blaNDM gene were also performed for the analysis of clonal relationship and genetic surrounding of the strains. We characterized 44 isolates with blaNDM variants in Escherichia coli (45.5%), Klebsiella pneumoniae (40.9%), Citrobacter freundii (4.5%), Citrobacter braakii (2.3%), Klebsiella oxytoca (2.3%), Enterobacter cloacae (2.3%), Enterobacter aerogenes (2.2%) from NICU, showing resistance against all antibiotics except colistin and polymixin B. ISAba125 and bleomycin gene were found surrounding all blaNDM variants, besides class I integron on plasmid. (ERIC)-PCR data revealed non-clonal relatedness among most of the isolates. The transfer of resistant markers was confirmed by conjugation experiment. The PCR-based replicon typing was carried out using DNA of transconjugants. These isolates carried NDM-1 (20.45%), NDM-4 (36.36%), NDM-5 (38.64%), NDM-7 (4.55%), along with OXA, CMY, and SHV variants on conjugative plasmid of IncFIA, IncFIC, IncF, IncK, IncFIB, IncB/O, IncHI1, IncP, IncY, IncFIIA, IncI1, and IncN types. An increased number of carbapenem-resistant NDM producing enterobacteriaceae isolates recovered from NICU which is alarming signal for health workers and policy makers. Hence, it is utmost important to think about infection control measures.
Keywords: NDM, carbapenemase, Hospital, NICU, ERIC-PCR, antibiotic resistance, enterobacteriaceae
Introduction
Emergence of New-Delhi Metallo-β-lactamase (NDM) producers is a matter of concern. The spread of MBL-producing enterobacteriaceae has increased from 2008 onward with the discovery of an ST14 Klebsiella pneumoniae with a new MBL gene, blaNDM−1, from a 59-years old Swedish patient who received healthcare in New Delhi, India (Yong et al., 2009). Indian subcontinent are the most endemic region for the spread of NDM-type MBLs and prevalence rates of NDM-producing enterobacteriaceae were found in range of 5–18.5% in Indian and Pakistan hospitals (Perry et al., 2011; Bharadwaj et al., 2012). In other regions (except the Balkan and Middle East countries), NDM-type MBLs are described mostly as periodic occurrences (Dortet et al., 2014). Carbapenem-resistant microorganisms have become an alarming phenomenon in children (Logan, 2012). A recently published study in USA reported that the frequency of carbapenem resistance increased from 0% in 1999–2000 to 0.47% in 2010–2011 among Enterobacteriaceae isolates in children (Logan et al., 2015). To date, 19 variants of NDM-type carbapenemases (NDM-1 to NDM-19) have been identified (http://www.lahey.org/Studies/other.asp#table1). These variants were identified in expanded species of Gram-negative bacteria and were found to have variation either by multiple residues at different positions or by replacing single amino acid. Recently, an NDM-4, NDM-5, and NDM-7 producing Enterobacter aerogenes from NICU of Indian hospital were reported by our group (Ahmad et al., 2017a). The most widespread variants were found in Indian sub-continent, are NDM-1, NDM-4, NDM-5, NDM-6, and NDM-7 (Khan et al., 2017). Whereas, several types of carbapenemases, such as KPC, IMP, OXA-48, VIM, and New Delhi metallo-β-lactamase (NDM), have been identified globally (Pitout et al., 2015; Logan and Weinstein, 2017).
NDM producing bacteria are resistant to almost all antibiotics, except polymyxins (Kumarasamy et al., 2010). But, the hope of colistin and polymyxins as treatment option has become limited after the discovery of MCR-1 gene in human and animals (Liu et al., 2016). The indiscriminate nature of the gene encoding NDM-1 has made major problem in neonatal intensive care units (NICU). In NICU, high consumption of antimicrobial agents, numerous indwelling devices, and staff rotativity, may further complicate the problem (Zaidi et al., 2005).
In enterobacteriaceae, blaNDM−1 is generally located on conjugative plasmids, ranging from 50 to 200 kb in size and belongs to several incompatibility groups, such as IncL/M, IncHI1, IncFIIs, IncF, or untypable, enabling transfer, and rapid dissemination of multidrug resistance (Poirel et al., 2011).
Our study was designed to evaluate retrospectively the spread of NDM producing Enterobacteriaceae and their genetic basis in neonatal intensive care unit of one of the north Indian tertiary care hospital.
Materials and methods
Collection of bacterial strains and hospital setting
A total of 750 Enterobacteriaceae clinical isolates were screened from blood and rectal swab of 1,140 neonates admitted in neonatal intensive care unit (NICU) of Jawaharlal Nehru Medical College and Hospital (JNMCH), Aligarh Muslim University, Aligarh, India, during the period, December 2015 to January 2017 in which 308 isolates were found to be carbapenem resistant. It is a tertiary care hospital of 1,300 bed capacity, in which 90 beds were allotted for pediatric patients and 35 beds for the NICU. Patients enrolled in the study were those who enrolled in the active surveillance system (NICU stay 48 h and weekly surveillance swabs taken at least once). Neonates admitted to the ward before December 2015 and/or discharged after January 2017, were excluded.
Ethical approval
A formal consent from the institutional ethical committee was taken and clearance was obtained from the institute's ethics committee. Participants/guardians had provided written, informed consent to participate in the study. We have a specific format to get the consents of patients/ parents of minors. These formats were made according to the Institutional ethics committee's guidelines. These forms are confidential and cannot be disclosed as per the guide lines. Institutional ethical committee has already approved. The name of committee/board is “Institutional Ethical Committee of Interdisciplinary Biotechnology Unit [Biot/307/01.06.13],” Aligarh Muslim University, Aligarh, India.
Antimicrobial susceptibility, metallo-β-lactamase (MBL), and MICs testing
Antimicrobial susceptibility was determined by the standard disc diffusion method using Mueller Hinton agar plate as per the Clinical and Laboratory Standards Institute guidelines (CLSI, 2016). More than 05 colonies were picked from MH agar plate for antimicrobial susceptibility testing and MBL detection. Detection of metallo-β-lactamase activity was performed, using two imepenem discs (10 μg), one containing 10 μl of 0.1M anhydrous Ethylene Diamine Tetra-Acetic Acid (EDTA). The discs were placed 25 mm distance (center to center) on Mueller-Hinton agar plates (Ahmad et al., 2017b). Minimum Inhibitory Concentrations (MICs) for antimicrobial agents were determined using broth micro dilution method, according to the guidelines of the CLSI.
Carba NP test for detection of carbapenemase
Carba NP test is a biochemical method used for the detection of carbapenemase activity in enterobacteriaceae isolates, performed as described earlier (Nordmann et al., 2012).
Isolate identification
The species level identification of isolates were performed by using BD PhoenixTM−100 automated microbiology system using panel NMIC/ID-55 (Gram negative susceptibility card) and further validated by 16s rRNA sequencing using primer as described previously (Shemesh et al., 2012).
Polymerase chain reaction (PCR) amplification and sequence analysis
PCR (Applied Biosystems model-9902 Verity thermo cycler) amplification was performed using primers as described previously (Poirel et al., 2011; Ali et al., 2014) for blaNDM and other resistant marker (blaVIM, blaOXA−1, blaOXA−9, blaCMY, blaTEM, blaSHV, and blaKPC). Amplicons of NDM were purified from the gel using gel extraction kit (Thermo Fisher Scientific), following manufacturers' protocol and then sequenced for DNA sequencing at Sci Genom Labs Private Ltd, Cochin, India. The nucleotide and deduced protein sequences were analyzed with software available at the National Centre for Biotechnology Information Website (www.ncbi.nlm.nih.gov).
Molecular characterization of plasmid
Plasmid DNA extraction and molecular size of multiple plasmids were identified by Kieser method (Kieser, 1984). Plasmid incompatibility group was determined by a PCR-based replicon typing (PBRT) method. Plasmid DNA was amplified by five multiplex and three simplex PCRs using 18 pair of primers as reported previously (Carattoli et al., 2005) that are recognized as Inc. replicon types: FIA, FIB, FIC, HI1, HI2, I1-Ic, L/M, N, P, W, T, A/C, K, B/O, X, Y, F, and FIIA.
Conjugation experiment
The transfer of resistant markers (blaNDM, blaCMY, blaOXA, and blaSHV) was determined by conjugation, using an azide-resistant E. coli J53 strain as the recipient and isolates as donor (Walsh et al., 2011). Transconjugants were screened on Luria-Bertani agar supplemented with ceftazidime (10 μg ml−1) (Sigma-Aldrich) and sodium azide (100 μg ml−1) (HiMedia Laboratories, India). The PCR amplification confirmed the transconjugants having resistant markers.
Genetic environment analysis
It was performed to identify the genes present at upstream and downstream of blaNDM variants as described previously (Poirel et al., 2011).
Integron analysis
The transconjugants of all the isolates, with blaNDM, were subjected to undergo integron analysis, using PCR amplification of 3′/5′ conserved segment along with Int1 and Sul1 as reported earlier (Dortet et al., 2012).
Molecular genotyping of isolates
The clonally relatedness between NDM producing isolates were investigated by enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) using the primers ERIC-Forward (5′ATGT AAGCTCCTGGGGATTAAC-3′) and ERIC-Reverse (5′AAGTAAGGACTGGGGTGAGCG-3′), was performed as described earlier (Versalovic et al., 1991). Bio-Red Gel Doc system was used to scan gel image and analyzed the bands by PyElph version 1.4 Software to generate a dendrogram by the un weighted pair group method using arithmetic averages (UPGMA) clustering (PyElph) (Pavel and Vasile, 2012).
Results
Isolate identification
Of 44 isolates, Escherichia coli (n = 20; 45.5%), K. pneumoniae (n = 18; 40.9%), Citrobacter freundii (n = 2; 4.5%), Citrobacter braakii (n = 1; 2.3%), Klebsiella oxytoca (n = 1; 2.3%), Enterobacter cloacae (n = 1; 2.3%), E. aerogenes (n = 1; 2.2%), were identified.
Antimicrobial susceptibility, metallo-β-lactamase (MBL), and MICs
Of 750 isolates, 44 were found to be New-Delhi Metallo-β-lactamase (NDM) producing enterobacteriaceae strains. All NDM producing strains were found highly resistant antibiotics, including carbapenems (imipenem and meropenem), cephamycin (cefoxitin), extended-spectrum cephalosporins (ceftazidime and cefotaxime), aminoglycoside (gentamicin and amikacin), monobactam (aztreonam), tetracycline (minocycline and tigecycline), fluoroquinolone (ciprofloxacin), except polymyxin and colistin. Metallo-β-lactamase (MBL) activity was present in all 44 NDM producing enterobacteriaceae isolates (Table 1). MICs data revealed high values against all tested antibiotics which were found in the range of 128 ≥ 4,096 μg ml−1 (Supplementary Table S1).
Table 1.
Phenotypic and Genotypic Characterization of (NDM) producing enterobacteriaceae isolates from NICU setting.
| S.No. | Organism name | Isolate Id | Accession no. | NDM variant | Carba NP result | Metallo-β-lactamase | Associated resistance markers* | No. of plasmid/Molecular size in kb* | Plasmid type* | Integron* | Genetic environment of blaNDM | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ISAba125 | bleMBL | |||||||||||
| 1. | Escherichia coli | AK-69 | KX231909 | NDM-7 | Positive | Present | OXA-1, CMY-1 | 38, 6, 4 | FIA, FIC, F, K | Class 1 | Complete | Present |
| 2. | AK-70 | KX231910 | NDM-5 | Positive | Present | OXA-1 | 154, 38, 4 | FIA, FIC, F, K | Class 1 | Truncated | Present | |
| 3. | AK-71 | KX231911 | NDM-5 | Positive | Present | CMY-1 | 66, 38, 6, 4 | FIA, FIB, F, K | Class 1 | Complete | Present | |
| 4. | AK-72 | KX231912 | NDM-5 | Positive | Present | OXA-1 | 154, 66, 38, 6 | FIA, FIC, F, K | Class 1 | Complete | Present | |
| 5. | AK-74 | KX231914 | NDM-5 | Positive | Present | CMY-149 | 66, 38, 6 | I, F, K | Class 1 | Complete | Present | |
| 6. | AK-76 | KX231915 | NDM-5 | Positive | Present | OXA-1 | 154, 38 | FIA, F, K | Class 1 | Complete | Present | |
| 7. | AK-77 | KX231916 | NDM-5 | Positive | Present | OXA-1, CMY-149 | 66, 38, 6, 4 | FIA, FIB, I, B/O, K | Class 1 | Complete | Present | |
| 8. | AK-79 | KX231918 | NDM-5 | Positive | Present | OXA-1, CMY-1 | 38 | FIA, FIB, F, K | Class 1 | Complete | Present | |
| 9. | AK-80 | KX231919 | NDM-5 | Positive | Present | OXA-1 | 38, 2 | FIA, FIB, F, K | Class 1 | Complete | Present | |
| 10. | AK-81 | KX231920 | NDM-5 | Positive | Present | OXA-1, CMY-1 | 38, 6, 4 | I, F, K | Class 1 | Truncated | Present | |
| 11. | AK-83 | KX231922 | NDM-7 | Positive | Present | OXA-1, SHV-1 | 38, 25 | FIA, FIB, F, K | Class 1 | Complete | Present | |
| 12. | AK-86 | KX231925 | NDM-5 | Positive | Present | OXA-1, CMY-1 | 38, 6 | FIA, F, K | Class 1 | Complete | Present | |
| 13. | AK-87 | KX231926 | NDM-5 | Positive | Present | OXA-1 | 38, 6, 4 | FIA, F, K | Class 1 | Complete | Present | |
| 14. | AK-88 | KX231927 | NDM-5 | Positive | Present | OXA-1, OXA-9 | 154, 66 | FIA, F, K | Class 1 | Complete | Present | |
| 15. | AK-90 | KX231929 | NDM-5 | Positive | Present | OXA-1 | 38, 4 | FIA, F, K | ND | Complete | Present | |
| 16. | AK-91 | KX231930 | NDM-5 | Positive | Present | OXA-1 | 154, 66 | FIA, F, I, K | Class 1 | Complete | Present | |
| 17. | AK-105 | KX999132 | NDM-5 | Positive | Present | OXA-1, OXA-9, CMY-1 | 154, 66, 38 | HI1, Y, FIA, FIB, F, K | Class 1 | Truncated | Present | |
| 18. | AK-107 | KX999134 | NDM-4 | Positive | Present | OXA-1, OXA-9, SHV-1 | 66, 38 | I, FIA, FIB, F, FIIA | Class 1 | Complete | Present | |
| 19. | AK-109 | KX999136 | NDM-5 | Positive | Present | CMY-149 | 38, 6, 4 | I, F, K | Class 1 | Complete | Present | |
| 20. | AK-116 | KX999143 | NDM-1 | Positive | Present | SHV-2 | 154 | FIA, FIC | Class 1 | Complete | Present | |
| 21. | Klebsiella pneumoniae | AK-66 | KX231906 | NDM-1 | Positive | Present | OXA-1, OXA-9, CMY-1 | 38 | FIIA, FIC. | Class 1 | Complete | Present |
| 22. | AK-78 | KX231917 | NDM-1 | Positive | Present | OXA-1 | 148 | FIIA | Class 1 | Truncated | Present | |
| 23. | AK-85 | KX231924 | NDM-1 | Positive | Present | OXA-9, CMY-145 | 38, 6, 4 | FIA, F, K | Class 1 | Complete | Present | |
| 24. | AK-89 | KX231928 | NDM-1 | Positive | Present | OXA-1 | 38 | FIIA | Class 1 | Complete | Present | |
| 25. | AK-94 | KX999121 | NDM-1 | Positive | Present | CMY-145, SHV-1 | 154, 66, 38 | Y, FIA, K, FIIA | Class 1 | Complete | Present | |
| 26. | AK-97 | KX999124 | NDM-4 | Positive | Present | OXA-1, OXA-9 | 154, 66, 38, 6, 4 | P, FIC, FIA, FIB, F, K | Class 1 | Complete | Present | |
| 27. | AK-98 | KX999125 | NDM-4 | Positive | Present | OXA-1, OXA-9, CMY-1, SHV-1 | 38, 6 | K, FIIA | Class 1 | Truncated | Present | |
| 28. | AK-99 | KX999126 | NDM-4 | Positive | Present | OXA-1, OXA-9, SHV-2 | 38, 6 | K, FIIA | Class 1 | Truncated | Present | |
| 29. | AK-101 | KX999128 | NDM-4 | Positive | Present | OXA-1, OXA-9, CMY-145 | 154, 66, 38, 6, 4 | P, FIC, FIA, FIB, F, K | Class 1 | Complete | Present | |
| 30. | AK-102 | KX999129 | NDM-5 | Positive | Present | OXA-1, OXA-9, CMY-4 | 154, 66, 38, | FIIA | Class 1 | Complete | Present | |
| 31. | AK-103 | KX999130 | NDM-4 | Positive | Present | OXA-1, OXA-9 | 66 | FIC, K | ND | Complete | Present | |
| 32. | AK-104 | KX999131 | NDM-4 | Positive | Present | OXA-1, OXA-9, CMY-4, SHV-1 | 38, 6, 4 | P, FIC, K, FIIA | Class 1 | Complete | Present | |
| 33. | AK-106 | KX999133 | NDM-4 | Positive | Present | OXA-1, OXA-9, SHV-2 | 38, 6, 4 | K | Class 1 | Complete | Present | |
| 34. | AK-110 | KX999137 | NDM-4 | Positive | Present | OXA-1, OXA-9, CMY-145 | 38, 6, 4 | K, FIIA | Class 1 | Truncated | Present | |
| 35. | AK-111 | KX999138 | NDM-4 | Positive | Present | OXA-1, OXA-9 | 38, 6, 4 | K, FIIA | Class 1 | Complete | Present | |
| 36. | AK-112 | KX999139 | NDM-1 | Positive | Present | OXA-1 | 66, 38 | K, FIIA | Class 1 | Truncated | Present | |
| 37. | AK-114 | KX999141 | NDM-4 | Positive | Present | OXA-1, OXA-9, SHV-1 | 66, 38 | K, FIIA | Class 1 | Complete | Present | |
| 38. | AK-115 | KX999142 | NDM-4 | Positive | Present | OXA-1, OXA-9 | 38, 6 | Y, FIA, FIB, F, K, FIIA | Class 1 | Complete | Present | |
| 39. | Citrobacter freundii | AK-82 | KX231921 | NDM-4 | Positive | Present | OXA-9, SHV-1, CMY-149 | 38 | N, F, K | Class 1 | Complete | Present |
| 40. | AK-113 | KX999140 | NDM-1 | Positive | Present | OXA-1, SHV-2, CMY-149 | 66 | FIC, K | Class 1 | Truncated | Present | |
| 41. | Citrobacter braakii | AK-84 | KX231923 | NDM-4 | Positive | Present | OXA-1, CMY-145 | 38 | F | Class 1 | Complete | Present |
| 42. | Klebsiella oxytoca | AK-100 | KX999127 | NDM-4 | Positive | Present | OXA-1, OXA-9 | 154, 66, 38 | I, Y, FIA, F, K, FIIA | Class 1 | Complete | Present |
| 43. | Enterobacter cloacae | AK-108 | KX999135 | NDM-4 | Positive | Present | OXA-1, OXA-9, CMY-149 | 66,38 | FIA, FIB | Class 1 | Truncated | Present |
| 44. | Enterobacter aerogenes | AK-67 | KX231907 | NDM-1 | Positive | Present | OXA-1, SHV-2 | 154, 38, 6, 4 | N, FIIA, FIC, K | Class 1 | Truncated | Present |
These features were also found on transconjugants.
Carbapenemase production
All 44 NDM- producing enterobacteriaceae isolates were found positive for Carba-NP test, indicating the production of a carbapenemase as shown in Table 1.
Detection of antibiotic resistance markers
PCR amplification and sequencing confirmed that all isolates harbored blaNDM of which NDM-1 (9; 20.45%), NDM-4 (16; 36.36%), NDM-5 (17; 38.64%), and NDM-7 (2; 4.55%) were found to be prevalent. Sequences were submitted to NCBI database (Table 1). Further blaCMY was detected in 20 isolates (08; blaCMY−1, 02; blaCMY−4, 05; blaCMY−145, and 05; blaCMY−149) whereas, blaOXA−1 was detected in 37 isolates, and blaOXA−9 was found in 20 isolates. Moreover, 07 blaSHV−1 and 05 blaSHV−2 were also found in this study (Table 1). However, blaTEM, blaVIM, blaIMP, and blaKPC were not detected in any of these isolates. Conjugation experiment, further confirmed the presence of these resistance markers on plasmid in each isolate.
Conjugation
The plasmidic location of resistant markers was determined by conjugation, using an azide-resistant E. coli J53 strain as the recipient [12]. Transconjugants were obtained at the frequencies of 10−3 to 10−5 cells, showing that plasmid from the donors (E. coli, K. pneumoniae, C. freundii, C. braakii, K. oxytoca, E. cloacae, E. aerogenes), were found stable in E. coli J53.
Replicon typing
These studied NDM producing isolates contained detectable plasmid size (154kb, 66kb, 38kb, 6kb, and 4kb) as shown in Table 1. Number of plasmids were found in the isolates, 1(n = 09), 2(n = 14), 3(n = 15), 4(n = 04), 5(n = 02). PBRT method identified 12 of 18 replicons types in our study while, IncHI2, IncL/M, IncW, IncT, IncA/C, and IncX were not detected in this study. IncFIA (n = 24), IncFIC (n = 11), IncF (n = 25), IncK (n = 36), IncFIB (n = 11), IncB/O (n = 01), IncHI1 (n = 01), IncP (n = 03), IncY (n = 04), IncFIIA (n = 16), IncI1 (n = 07), and IncN (n = 02), replicon types were predominant in the present study and IncFIA, IncFIC, IncF, IncK, and IncFIB were found to be the most frequent types in this study.
Integron analysis
The transconjugants of all isolates harbored plasmid carrying class 1 integron, except two isolates (AK-90 and AK-103) which were confirmed by PCR amplification of 5′/3′ CS, IntI, and SulI genes. We further confirmed that no resistant marker was present in the integron cassette as shown by a PCR using amplicon of 5′/3′ CS as template.
Genetic relatedness of the carbapenem resistant NDM producing enterobacteriaceae isolates
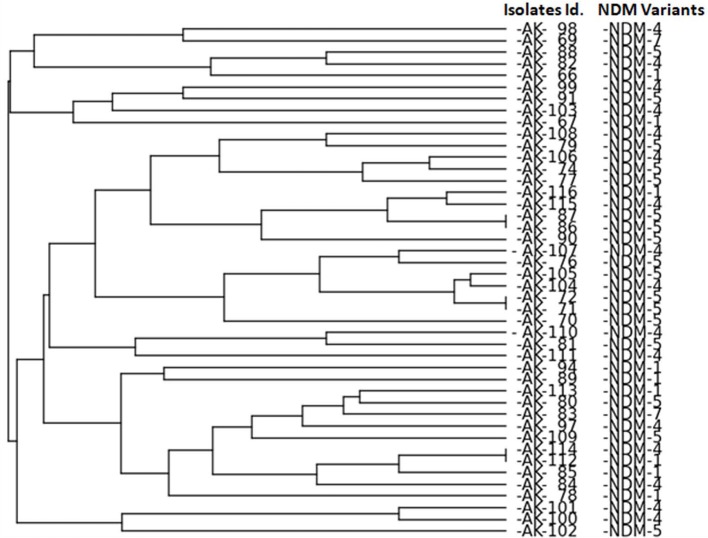
ERIC-PCR analysis revealed no clonal relatedness among isolates except for the isolates of K. pneumoniae (AK-86 with AK-87, AK-71 with AK-72 and AK-112 with AK-114) as shown in Figure 1.
Figure 1.
ERIC PCR analysis of NDM producing isolates. Bio-Red Gel Doc system was used to analyze the bands by PyElph version 1.4 Software generate a dendrogram by the unweighted pair group method using arithmetic averages (UPGMA) clustering. Generated dendogram showing genetic relationship among NDM producing isolates.
Genetic environment of the blaNDM gene
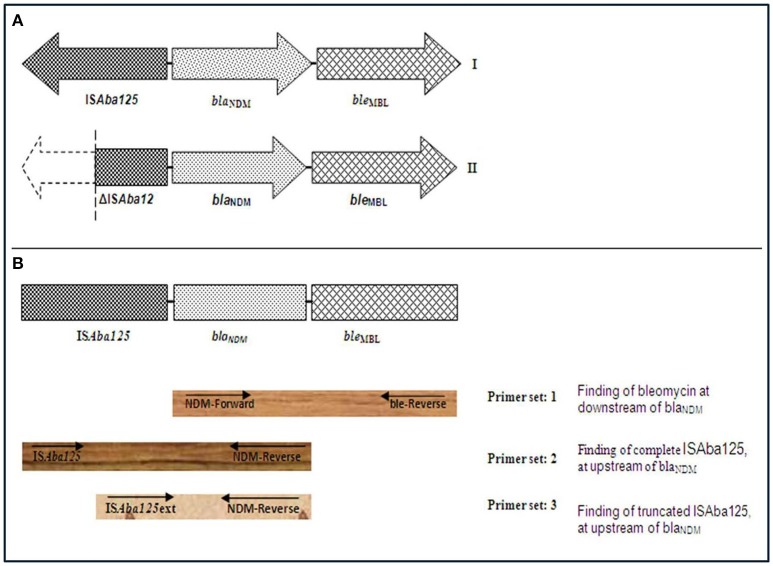
PCR based genetic environment analysis of blaNDM gene was performed and bleMBL was found at downstream of blaNDM variants in all isolates (Figure 2). A complete ISAba125 sequence was found at upstream of blaNDM in one blaNDM−1 (AK-116), one blaNDM−4 (AK-107), 13 blaNDM−5 (AK-71, AK-72, AK-74, AK-76, AK-77, AK-79, AK-80, AK-86, AK-87, AK-88, AK-90, AK-91, and AK-109,) and two NDM-7 producing E. coli (AK-69 and AK-83). Further, complete ISAba125 was amplified in 4 isolates of NDM-1(AK-66, AK-85, AK-89, and AK-94), eight isolates of NDM-4 (AK-97, AK-101, AK-103 AK-104, AK-106, AK-111, AK-114, and AK-115) and one (AK-102) NDM-5 producing K. pneumoniae (Figure 2). A complete ISAba125 was amplified in three isolates of NDM-4 producing C. freundii, C. braakii, and K. oxytoca, respectively (AK-84, AK-82, and AK-100). However, truncated ISAba125 was detected in three isolates of NDM-5 producing E. coli (AK-70, AK-81, and AK-105). Moreover, 2; NDM-1 (AK-78, AK-112), 3; NDM-4 (AK-98, AK-99, and AK-110), producing K. pneumoniae and one NDM-1 (AK-113) producing C. freundii, one NDM-4 (AK-108) producing E. cloacae and one (AK-67) NDM-1 producing E. aerogenes had truncated ISAba125 at upstream of blaNDM (Table 1, Figure 2).
Figure 2.
(A) A schematic representation of genetic elements surrounding blaNDM. (I) In AK-69, AK-71, AK-72, AK-74, AK-76, AK-77, AK-79, AK-80, AK-83, AK-86, AK-87, AK-88, AK-90, AK-91, AK-107, AK-109, AK-116, AK-66, AK-85, AK-89, AK-94, AK-97, AK-101, AK-102, AK-103, AK-104, AK-106, AK-111, AK-114, AK-115, AK-84, AK-82, and AK-100, complete element of ISAba125 at upstream and bleomycin gene at downstream to blaNDM was found. (II) In AK-70, AK-81, AK-105, AK-78, AK-98, AK-99, AK-110, AK-112, AK-113, AK-108, and AK-67, truncated ISAba125 at upstream and bleomycin gene at downstream to blaNDM was found. (B) A schematic representation for PCR-based genetic environment analysis of blaNDM. Arrow indicates the position of primer (use of primers as described in reference Poirel et al., 2011).
Discussion
Emergence of NDM-producing enterobacteriaceae has become a globally serious concern. NDM producers led to limited therapeutic options hence it has become a threat to public health. Epidemiological investigation and surveillance of NDMs are of importance to clinical infection control. This study revealed outbreak of multiple variants of blaNDM (9; blaNDM−1, 16; blaNDM−4, 17; blaNDM−5, and 2; blaNDM−7) in clinically important bacteria (20 E. coli, 18 K. pneumoniae, 02 C. freundii, 01 C. braakii, 01 K. oxytoca, 01 E. cloacae, 01 E. aerogenes), as shown in Figure 3.
Figure 3.
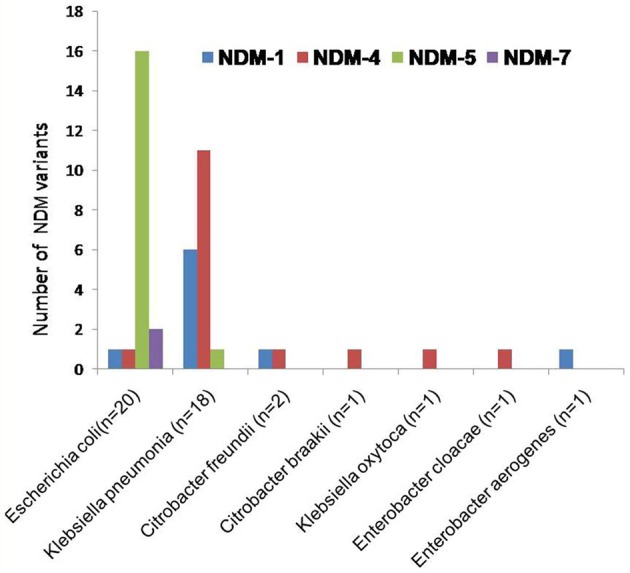
The clustered bar graph presents the number of NDM variants (each is represented by its own bar) distributed among NDM-producing enterobacteriaceae collected from NICU. The horizontal axis represents the NDM-producing enterobacteriaceae while the vertical axis represents the number of NDM variants.
In E. coli the predominant NDM variant was found to be blaNDM−1, followed by blaNDM−4, blaNDM−5, and blaNDM−7 (Figure 3). Although this is not first description of these NDM variants being produced by E. coli (Zhang et al., 2013; Qin et al., 2016; Zhu et al., 2016; Pál et al., 2017). Moreover, in these strains existence of NDM and its variants, with CMY, OXA, SHV, and VIM variants and other resistant determinants are documented. Of 20 NDM producing E. coli, one NDM-1 isolate (AK-116) was coexisting with blaSHV−2 and one NDM-4 isolate (AK-107) coexisting with blaOXA−1, blaOXA−9, and blaSHV−1. Further, two isolates of NDM-7 (AK-69, AK-83) were associated with blaOXA−1, blaSHV−1, blaCMY−1, and 16 isolates of blaNDM−5 were linked to blaOXA−1, blaOXA−9, blaSHV−1, blaCMY−1, or blaCMY1−49 in different combinations.
The most prevalent NDM variants in K. pneumoniae is blaNDM−4, followed by blaNDM−5 and blaNDM−1 (Figure 3). It has also been shown in earlier studies in Klebsiella pneumonia (Khalifa et al., 2016; Petersen-Morfin et al., 2017). Of 18 NDM producing K. pneumoniae, 6 were NDM-1 isolates, coexisting with blaOXA−1, blaOXA−9, blaSHV−1, blaCMY−1, and blaCMY−145. Further, 11 NDM-4 isolates were found associated with blaOXA−1, blaOXA−9, blaSHV−1, blaSHV−2, blaCMY−1, blaCMY−149, and blaOXA−1, blaOXA−9, blaCMY−4 in association with blaNDM−5.
Citrobacter species are rare opportunistic nosocomial pathogens (Ryan and Ray, 2004). It normally causes urinary tract infections, blood stream infections, intra-abdominal sepsis, brain abscesses, pneumonia, and other neonatal infection (Pepperell et al., 2002) such as meningitis, neonatal sepsis, joint infection, or general bacteremia (Doran, 1999). The principal NDM variant found in C. freundii was blaNDM−1 which was followed by blaNDM−4. It is a first report of NDM-4 producing C. freundii, (AK-82) co-associated with blaOXA−9, blaSHV−1, and blaCMY−149. Further, C. freundii (AK-113) was also found to have blaOXA−1, blaSHV−2, and blaCMY−149 in association with blaNDM−1.
Moreover, for the first time NDM-4 producing C. braakii (AK-84), K. oxytoca (AK-100), and E. cloacae (AK-108) were identified in association with blaOXA−1 and blaCMY−145, blaOXA−1 and blaOXA−9 and, blaOXA−1, blaOXA−9, and blaCMY−149, respectively.
We have also identified NDM-1 producing E. aerogenes co-associated with blaOXA−1 and blaSHV−2 in AK-67. NDM-1 producing C. braakii, in Pakistan (Pesesky et al., 2015), NDM-1 producing K. oxytoca in China (Wang et al., 2017), NDM-1 producing E. cloacae in Turkey (Haciseyitoglu et al., 2017) and Coratia (Petrosillo et al., 2016), have been reported in earlier studies.
The transconjugants were stable and carried all the resistant determinants from donor. Moreover, the presence of class 1 integron in all isolates except AK-90 and AK-103, suggests that the resistant markers can competently exchange among species leading to its spread in the hospital (Martinez-Freijo et al., 1998). Presence of resistance genes on plasmids of varying sizes (4–154 kb) were identified in this study. Previous studies have proved to have these resistance genes on plasmid of size 7–200 kb (Mshana et al., 2009). The replicon typing revealed varying replicon types (IncFIA, IncFIB, IncFIC, IncFIIA, IncF, IncN, IncK, IncB/O, IncHI1, IncY, IncI1, and IncP). In previous studies, blaNDM gene was shown to be associated with plasmid type (IncFIA IncFIB) (Gamal et al., 2016), (IncX3) (Zhang et al., 2016), (IncFIC, IncF, and IncK) (Ahmad et al., 2017a), (IncB/O) (An et al., 2016), (IncHI1, IncN, and IncFIIA) (Sartor et al., 2014), (IncY, IncA/C IncI1) (Kapmaz et al., 2016). Moreover, first time we have identified three NDM-4 producing Klebsiella pnemoniae with incompatibility group IncP in AK-97, AK-101, and AK-104 strains.
Complete ISAba125 sequence was observed at upstream of blaNDM in most of the isolates implies that this factor may play a main role in horizontal gene transfer of the blaNDM among enterobacteriaceae members (Poirel et al., 2011). In all blaNDM variants, bleMBL was found at it downstream. The occurrence of bleMBL, associated with blaNDM gene, suggests that they might have mobilized simultaneously from same progenitor and is thought to protect blaNDM (Dortet et al., 2012). These results suggest that the plasmids encoding for carbapenem resistant NDM variants can easily spread among the enterobacteriaceae isolates. These results are in conformity with previous reports that clarified the horizontal transfer of plasmids encoding for carbapenemases among enterobacteriaceae including K. pneumoniae (Dortet et al., 2014; Jin et al., 2015).
Conclusions
Carbapenem resistance among enterobacteriaceae has been considered as one of the most significant menaces to the global healthcare, and the prevalence of NDM variants in enterobacteriaceae has further increased the threat. Therefore, the early detection of the blaNDM possessing enterobacteriaceae isolates with any decreased sensitivity to the carbapenems is crucial for the choice of the most appropriate antibiotic therapy and the application of efficient infection control measures. The emergence of such resistance patterns may be reduced by the restricted implementation of antibiotics, especially for carbapenems and cephalosporins. Moreover, a strong infection control management in the hospital is necessary to check such infection.
Author contributions
NA: performed experiments, wrote draft manuscript; SK: performed experiments; SA: provided samples, and interpreted clinical data; AK: designed study and checked draft manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling Editor declared a past co-authorship with one of the authors AK.
Acknowledgments
The authors are also highly grateful to technical support of Pediatrics department and NICU of J. N. Medical College and Hospital, Aligarh, India, for providing samples. This work was supported by DBT (India) grants; BT/PR8281/BID/7/448/2013.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00407/full#supplementary-material
References
- Ahmad N., Ali S. M., Khan A. U. (2017a). Detection of New Delhi metallo-β-lactamase variants NDM-4, NDM-5, and NDM-7 in Enterobacter aerogenes isolated from a neonatal intensive care unit of a North India Hospital: a first report. Microb. Drug Resist. [Epub ahead of print]. 10.1089/mdr.2017.0038 [DOI] [PubMed] [Google Scholar]
- Ahmad N., Ali S. M., Khan A. U. (2017b). First reported New Delhi metallo-β-lactamase-1 producing Cedecea lapagei. Int. J. Antimicrob. Agents 49, 118–119. 10.1016/j.ijantimicag.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Ali S. Z., Ali S. M., Khan A. U. (2014). Prevalence of IncI1-Iγ and IncFIA-FIB type plasmids in extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains isolated from the NICU of a North Indian Hospital. Microbiology 160, 1153–1161. 10.1099/mic.0.075762-0 [DOI] [PubMed] [Google Scholar]
- An J., Guo L., Zhou L., Ma Y., Luo Y., Tao C., et al. (2016). NDM-producing Enterobacteriaceae in a Chinese hospital, 2014–2015: identification of NDM-producing Citrobacter werkmanii and acquisition of blaNDM-1 carrying plasmid in vivo in a clinical Escherichia coli isolate. J. Med. Microbial. 65, 1253–1259. 10.1099/jmm.0.000357 [DOI] [PubMed] [Google Scholar]
- Bharadwaj R., Joshi S., Dohe V., Gaikwad V., Kulkarni G., Shouche Y. (2012). Prevalence of New Delhi metallo-β-lactamase (NDM-1)-positive bacteria in a tertiary care centre in Pune, India. Int. J. Antimicrob. Agents 39, 265–266. 10.1016/j.ijantimicag.2011.09.027 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Six Informational Supplement M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Doran T. I. (1999). The role of Citrobacter in clinical disease of children: review. Clin. Infect. Dis. 28, 384–394. 10.1086/515106 [DOI] [PubMed] [Google Scholar]
- Dortet L., Nordmann P., Poirel L. (2012). Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1693–1697. 10.1128/AAC.05583-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet L., Poirel L., Nordmann P. (2014). Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed. Res. Int. 2014:249856. 10.1155/2014/249856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamal D., Fernández-Martínez M., El-Defrawy I., Ocampo-Sosa A. A., Martínez-Martínez L. (2016). First identification of NDM-5 associated with OXA-181 in Escherichia coli from Egypt. Emer. Microb. Infect. 5:e30. 10.1038/emi.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haciseyitoglu D., Dokutan A., Abulaila A., Erdem F., Cag Y., Ozer S., et al. (2017). The first Enterobacter cloacae co-producing, N. D. M., and OXA-48 carbapenemases and interhospital spread of OXA-48 and NDM-producing Klebsiella pneumoniae in Turkey. Clin. Lab. 63, 1213–1222. 10.7754/Clin.Lab.2017.170120 [DOI] [PubMed] [Google Scholar]
- Jin Y., Shao C., Li J., Fan H., Bai Y., Wang Y. (2015). Outbreak of multidrug resistant NDM-1-producing Klebsiella pneumoniae from a neonatal unit in Shandong Province, China. PLoS ONE 10:e0119571. 10.1371/journal.pone.0119571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapmaz M., Erdem F., Abulaila A., Yeniaras E., Oncul O., Aktas Z. (2016). First detection of NDM-1 with CTX-M-9, TEM, SHV and rmtC in Escherichia coli ST471 carrying IncI2, A/C and Y plasmids from clinical isolates in Turkey. J. Glob. Antimicrob. Resist. 7, 152–153. 10.1016/j.jgar.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Khalifa H. O., Soliman A. M., Ahmed A. M., Shimamoto T., Shimamoto T. (2016). NDM-4 and NDM-5 producing Klebsiella pneumoniae coinfection in a 6-month-old infant. Antimicrob. Agents Chemother. 60, 4416–4417. 10.1128/AAC.00479-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. U., Maryam L., Zarrilli R. (2017). Structure, genetics and Worldwide spread of New Delhi Metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol. 17:101. 10.1186/s12866-017-1012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T. (1984). Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12, 19–36. 10.1016/0147-619X(84)90063-5 [DOI] [PubMed] [Google Scholar]
- Kumarasamy K. K., Toleman M. A., Walsh T. R., Bagaria J., Butt F., Balakrishnan R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Logan L. K. (2012). Carbapenem-resistant enterobacteriaceae: an emerging problem in children. Clin. Infect. Dis. 55, 852–859. 10.1093/cid/cis543 [DOI] [PubMed] [Google Scholar]
- Logan L. K., Renschler J. P., Gandra S., Weinstein R. A., Laxminarayan R., Centers for Disease Control Prevention Epicenters Program (2015). Carbapenem-resistant Enterobacteriaceae in children, United States, 1999–2012. Emerg. Infect. Dis. 21, 2014–2021. 10.3201/eid2111.150548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan L. K., Weinstein R. A. (2017). The epidemiology of carbapenem resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215, S28–S36. 10.1093/infdis/jiw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Freijo P., Fluit A. C., Schmitz F. J., Grek V. S., Verhoef J., Jones M. E. (1998). Class I integrons in gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J. Antimicrob. Chemother. 42, 689–696. 10.1093/jac/42.6.689 [DOI] [PubMed] [Google Scholar]
- Mshana S. E., Imirzalioglu C., Hossain H., Hain T., Domann E., Chakraborty T. (2009). Conjugative IncFI plasmids carrying CTXM- 15 among Escherichia coli ESBL producing isolates at a University hospital in Germany. BMC Infect. Dis. 9:97 10.1186/1471-2334-9-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Poirel L., Dortet L. (2012). Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 18, 1503–1507. 10.3201/eid1809.120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál T., Ghazawi A., Darwish D., Villa L., Carattoli A., Hashmey R., et al. (2017). Characterization of NDM-7 carbapenemase-producing Escherichia coli isolates in the arabian peninsula. Microb. Drug Resist. 23, 871–878. 10.1089/mdr.2016.0216 [DOI] [PubMed] [Google Scholar]
- Pavel A. B., Vasile C. I. (2012). PyElph-a software tool for gel images analysis and phylogenetics. BMC Bioinformatics 13:9. 10.1186/1471-2105-13-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperell C., Kus J. V., Gardam M. A., Humar A., Burrows L. L. (2002). Low-virulence citrobacter species encode resistance to multiple antimicrobials. Antimicrob. Agents Chemother. 46, 3555–3560. 10.1128/AAC.46.11.3555-3560.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J. D., Naqvi S. H., Mirza I. A., Alizai S. A., Hussain A., Ghirardi S., et al. (2011). Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J. Antimicrob. Chemother. 66, 2288–2294. 10.1093/jac/dkr299 [DOI] [PubMed] [Google Scholar]
- Pesesky M. W., Hussain T., Wallace M., Wang B., Andleeb S., Burnham C. A., et al. (2015). KPC and NDM-1 genes in related Enterobacteriaceae strains and plasmids from Pakistan and the United States. Emerg. Infect. Dis. 21, 1034–1037. 10.3201/eid2106.141504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Morfin S., Bocanegra-Ibarias P., Morfin-Otero R., Garza-González E., Perez-Gomez H. R., González-Diaz E., et al. (2017). New Delhi Metallo-Beta-Lactamase (NDM-1)-producing Klebsiella pneumoniae isolated from a burned patient. Am. J. Case Rep. 18, 805–809. 10.12659/AJCR.903992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo N., Vranić-Ladavac M., Feudi C., Villa L., Fortini D., Barišić N., et al. (2016). Spread of Enterobacter cloacae carrying blaNDM-1, blaCTX-M-15, blaSHV-12 and plasmid-mediated quinolone resistance genes in a surgical intensive care unit in Croatia. J. Glob. Antimicrob. Resist. 4, 44–48. 10.1016/j.jgar.2015.09.008 [DOI] [PubMed] [Google Scholar]
- Pitout J. D., Nordmann P., Poirel L. (2015). Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59, 5873–5884. 10.1128/AAC.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Dortet L., Bernabeu S., Nordmann P. (2011). Genetic features of blaNDM-1 positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55, 5403–5407. 10.1128/AAC.00585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Zhou M., Zhang Q., Tao H., Ye Y., Chen H., et al. (2016). First identification of NDM-4-producing Escherichia coli ST410 in China. Emerg. Microb. Infect. 5:e118. 10.1038/emi.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., Ray C. G. (ed.). (2004). Sherris Medical Microbiology: An Introduction to Infectious Diseases, 4th Edn. New York, NY: McGraw-Hill. [Google Scholar]
- Sartor A. L., Raza M. W., Abbasi S. A., Day K. M., Perry J. D., Paterson D. L., et al. (2014). Molecular epidemiology of NDM-1-producing Enterobacteriaceae and Acinetobacter baumannii isolates from Pakistan. Antimicrob. Agents Chemother. 58, 5589–5593. 10.1128/AAC.02425-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh M., Tam A., Steinberg D. (2012). Expression of biofilm-associated genes of Streptococcus mutans in response to glucose and sucrose. J. Med. Microbiol. 56, 1528–1535. 10.1099/jmm.0.47146-0 [DOI] [PubMed] [Google Scholar]
- Versalovic J., Koeuth P., Lupski R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 24, 6823–6831. 10.1093/nar/19.24.6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. R., Weeks J., Livermore D. M., Toleman M. A. (2011). Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11, 355–362. 10.1016/S1473-3099(11)70059-7 [DOI] [PubMed] [Google Scholar]
- Wang J., Yuan M., Chen H., Chen X., Jia Y., Zhu X., et al. (2017). First report of Klebsiella oxytoca strain simultaneously producing NDM-1, IMP-4 and KPC-2 carbapenemases. Antimicrob. Agents Chemother. 61, 00877–00817. 10.1128/AAC.00877-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D., Toleman M. A., Giske C. G., Cho H. S., Sundman K., Lee K., et al. (2009). Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi A. K., Huskins W. C., Thaver D., Bhutta Z. A., Abbas Z., Goldmann D. A. (2005). Hospital-acquired neonatal infections in developing countries. Lancet 365, 1175–1188. 10.1016/S0140-6736(05)71881-X [DOI] [PubMed] [Google Scholar]
- Zhang F., Xie L., Wang X., Han L., Guo X., Ni Y., et al. (2016). Further Spread of blaNDM-5 in Enterobacteriaceae via IncX3 Plasmids in Shanghai, China. Front. microbial. 7:424 10.3389/fmicb.2016.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lou D., Xu Y., Shang Y., Li D., Huang X., et al. (2013). First identification of coexistence of blaNDM-1 and blaCMY-42 among Escherichia coli ST167 clinical isolates. BMC Microbiol. 13:282. 10.1186/1471-2180-13-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. Q., Zhao J. Y., Xu C., Zhao H., Jia N., Li Y. N. (2016). Identification of an NDM-5-producing Escherichia coli sequence type 167 in a neonatal patient in China. Sci. Rep. 6:29934. 10.1038/srep29934 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.