Abstract
Staphylococcus epidermidis is one of the most prevalent pathogens implicated in catheter-related bloodstream infections (CRBSI) at an academic hospital in Pretoria, South Africa. Little is known about the clonality and the prevalence of antibiotic resistance and virulence genes in S. epidermidis (e.g., icaAB, IS256, mecA, and qacA/B). A total of 508 intravascular catheters (IVCs) from 331 patients were submitted for culture from May to October 2013. Only 50% (n = 253/508) of the IVCs were accompanied by blood cultures (BCs) taken within 48 h. Forty-four percent (44%; n = 112/253) of IVCs were colonised, of which 26% (n = 65/253) were associated with a CRBSI. We identified S. epidermidis as the causal agent in 31% (n = 20/65) of the CRBSI cases. Fifty-nine S. epidermidis isolates were obtained, 23 isolates were cultured from 22 IVCs and 36 isolates were cultured from 36 BCs. All S. epidermidis isolates were resistant to β-lactams (100%; n = 59/59), followed by high levels of resistance toward erythromycin (86%; n = 51/59) and gentamicin (81%; n = 49/59). The mecA gene was prevalent in all the (100%, n = 59/59) isolates. Isolates contained the IS256 element (83%, n = 49/59), the icaAB gene (81%, n = 48/59) and, the qacA/B gene (81%, n = 48/59). All 48 isolates were qacA positive upon restriction enzyme digestion of the qacA/B amplicons. Phenotypic resistance toward 0.5% (m/v) chlorhexidine was not observed. Staphylococcal Cassette Chromosome (SCC) mec typing showed that SCCmec type IV (31%; n = 18/59) was the most prevalent. The remaining SCCmec elements were highly diverse. Pulsed-field gel electrophoresis (PFGE) showed that S. epidermidis isolates from individual patients were mostly clonal. Multilocus sequencing typing (MLST) of 10 sequenced isolates showed that sequence type (ST) 2 (40%; n = 4/10) was the most frequently detected, followed by ST54 (20%; n = 2/10), ST28 (10%; n = 1/10), ST59 (10%; n = 1/10) and ST490 (10%; 1/10). One isolate was newly assigned to ST596. These S. epidermidis infections can be attributed to patients' skin microflora or to poor infection control practices. Currently, S. epidermidis strains circulating in the studied hospital are multidrug-resistant and highly adaptive to environmental changes.
Keywords: Staphylococcus epidermidis, catheter-related bloodstream infections, South Africa, SCCmec typing, PFGE, MLST, ST596, ST2
Introduction
Catheter-related bloodstream infections are defined by the Infectious Diseases Society of America (IDSA) as bacteraemia or fungaemia in a catheterised febrile patient, with at least a positive blood culture (BC) and intravascular catheter (IVC) culture, yielding the same microorganism, with no other possible source of infection besides the IVC (Mermel et al., 2009). If the isolated microorganism is a common commensal, at least two positive blood cultures are required to exclude the possibility of contamination (Mermel et al., 2009). Staphylococcus epidermidis is a classic example of a common commensal and is an ever-present coloniser of the human skin (Percival et al., 2012). However, S. epidermidis can become an opportunistic pathogen if the external barrier of the skin surface is breached, such as during the insertion of an IVC (Schoenfelder et al., 2010).
Biofilm formation, encoded by the icaADBC operon, is central to the pathogenesis of S. epidermidis (Otto, 2009). The biofilm allows the bacterium to colonise the IVC and subsequently seed into the bloodstream (Otto, 2008). Certain S. epidermidis strains may have an evolutionary advantage due to methicillin-resistance facilitated by the mecA gene, phenotypic variation caused by insertion sequence IS256 and genotypic resistance to antiseptics, such as chlorhexidine, mediated by efflux pumps encoded by the qacA/B genes (Otto, 2009; Schoenfelder et al., 2010; Horner et al., 2012). As a result, certain strains may be resistant to infection prevention practices, able to evade host defences and subsequently cause disease (Otto, 2009).
Previously, S. epidermidis was identified as the most common CRBSI pathogen at an academic hospital in Pretoria, South Africa (Strasheim et al., 2015). This study molecularly characterised and genotyped S. epidermidis isolates implicated in CRBSI events and described S. epidermidis isolates from South Africa for the first time.
Materials and methods
Study setting and isolate collection
The IVCs and BCs were submitted from an 832-bed academic hospital in Pretoria, South Africa, over a six-month period (May to October 2013) to the diagnostic division of the Department of Medical Microbiology [Tshwane Academic Division, National Health Laboratory Service (NHLS)]. The laboratory processes specimens from academic hospitals as well as district hospitals and various clinics as part of standard care. Routine analysis was performed as requested by a medical practitioner on the laboratory requisition form and according to sample type. A S. epidermidis CRBSI was defined as the simultaneous isolation of the bacterium from an IVC and one or more BCs, which were submitted for culture within 48 h of each other (Mermel et al., 2009; Conrick-Martin et al., 2013). Even though a single positive BC yielding S. epidermidis was likely to present contamination, we included these samples due to the lack of local BC practices. In practice, compliance to guidelines were suboptimal in the studied hospital, since catheters were rarely accompanied by BCs. If a BC accompanied a catheter, often only one BC was submitted. Patient demographics [i.e., age, sex, ward, underlying conditions, length of hospitalisation, antimicrobial exposure and the presence of specific risk factors (HIV-status, diabetes, chronic renal failure, malnutrition, loss of skin integrity, neutropenia and total parenteral nutrition administration) for CRBSI] and catheter details (i.e., length of catheterisation, catheter type, vessel occupied, insertion site, pathway followed from insertion site to vessel and the number of lumens) were collected.
Staphylococcus epidermidis isolation, identification, and susceptibility testing
Intravascular catheter tips were cultured according to the semi-quantitative roll-plate method (Maki et al., 1977). Blood cultures were processed with the BacT/ALERT 3D system (bioMérieux, France). Identification and antimicrobial susceptibility testing of S. epidermidis isolates were performed with the VITEK® 2 automated system (bioMérieux, France). The minimum inhibitory concentration (MIC) was recorded according to the 2013 Clinical Laboratory Standards Institute (CLSI) guidelines (Clinical Laboratory Standards Institute, 2013). Identification of S. epidermidis isolates was confirmed by matrix-assisted laser desorption/ionisation time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, USA) along with a conventional identification (ID) polymerase chain reaction (PCR) amplification.
Total DNA extraction
Total DNA of the S. epidermidis isolates was extracted with the ZR Fungal/Bacterial DNA MiniPrep™ kit (Zymo Research, USA), with some modifications to the manufacturer's instructions. A volume of 2 mL of overnight Brain Heart Infusion broth (Merck, Germany) was used as starting material for the extractions. Beta-mercaptoethanol [0.5% (v/v)] (Merck, Germany) was added to the DNA binding buffer (Zymo Research, USA). A volume of 600 μL of Lysis solution (Zymo Research, USA), instead of the recommended 750 μL of Lysis solution (Zymo Research, USA), was used in the ZR BashingBead™ Lysis Tubes (Zymo Research, USA). The DNA obtained was used in all downstream PCR applications.
Multiplex PCR assay to identify Staphylococcus epidermidis
This first multiplex-PCR (M-PCR) assay detected five staphylococcal species simultaneously. The genes targeted in the M-PCR assay included an internal fragment of S. epidermidis [124 base pair (bp)] (Martineau et al., 1996), the nuclease (nuc) gene of S. aureus [359 bp], S. capitis [525 bp], and S. hominis [177 bp] (Hirotaki et al., 2011), and the mevalonate pathway (mvaA) gene of S. haemolyticus [271 bp] (Pereira et al., 2010). The 16S ribosomal (rRNA) gene for the Staphylococcus genus [597 bp] was included as an internal control (Al-Talib et al., 2009). A final concentration of 0.2 μM was used for each primer in the reaction mixture. The M-PCR reaction mixture consisted of 12.5 μL of 2 × QIAGEN® Multiplex PCR Master Mix (QIAGEN®, Netherlands), 2.5 μL of the 10 × primer mixture, 2.0 μL of template DNA and 8 μL of nuclease-free water. Amplification was done in a G-STORM™ thermocycler (Somerton Biotechnology Centre, UK). The cycling conditions included an initial Taq polymerase activation step (95°C for 15 min), followed by 30 cycles of denaturation (94°C for 30 s); annealing (58°C for 90 s) and extension (72°C for 90 s) and a final extension step (68°C for 15 min). The PCR amplicons were visualised on a 1.8% (m/v) MetaPhor™ agarose (Lonza, USA) gel, stained with 5 μL of ethidium bromide (10 μg/mL) (Sigma-Aldrich, USA). The amplicons were visualised under ultraviolet (UV) light (Transilluminator, Ultraviolet Products Incorporated, USA) and all visible bands were manually compared to a 50 bp DNA ladder (ThermoScientific, USA).
Multiplex PCR assay for the detection of is256, icaA/B, mecA, and qacA/B genes
S. epidermidis isolates were tested for the presence of the IS256 [762 bp] (Koskela et al., 2009), icaAB [546 bp] (Iorio et al., 2011), mecA [310 bp] (McClure et al., 2006) and qacA/B [220 bp] (Sekiguchi et al., 2004) genes, using a second M-PCR assay. Primer concentrations for the IS256, icaAB, mecA and qacA/B genes were 0.2, 2.0, 1.0, and 0.2 μM in the 10 × primer mix, respectively, which was prepared according to the recommendations of the manufacturer (QIAGEN®, USA). Each 25 μL reaction mixture contained 12.5 μL of QIAGEN® Multiplex PCR Master Mix, 2.5 μL of the 10 × primer mix, 2.5 μL of template DNA and 7.5 μL of nuclease-free water. Amplification was done in a G-STORM™ thermocycler (Somerton Biotechnology Centre, UK). The cycling conditions included an initial Taq polymerase activation step (95°C for 15 min), followed by 35 cycles of denaturation (94°C for 30 s); annealing (57°C for 90 s) and extension (72°C for 90 s) and a final extension step (72°C for 10 min). The detection and visualisation steps were repeated as described in the previous section, except for this assay a 2% (m/v) SeaKem® LE agarose (Lonza, USA) gel was used. A 50 bp DNA ladder (ThermoScientific, USA) was used as a marker. One positive amplicon for each of the screened genes was sequenced in both forward and reverse directions by Inqaba Biotechnical Industries, Pretoria, South Africa. These confirmed positive genes were used as positive controls in the M-PCR assays. A negative control [nuclease-free water (Qiagen, Germany)] was included for all M-PCR assays.
Enzyme digestion to distinguish between the qacA and-B gene
The qacA/B primers used in the M-PCR assays did not distinguish between the qacA and -B genes, since these genes are highly homologous and differ only at seven nucleotides (Sekiguchi et al., 2004; Horner et al., 2012). The method published by Sekiguchi et al. (2004) was followed to distinguish between the qacA [220 bp] and -B [44 bp and 176 bp] genes. The following modifications were made: (i) Instead of using the prescribed 5 U of AluI, 0.25 μl of the AluI restriction enzyme (New England Biolabs, USA) and 1 μl of the cut-smart buffer (New England Biolabs, USA) were added to 5 μl of the amplified products (a singleplex PCR assay was performed to detect the qacA/B genes. The cycling conditions was the same as described in the previous section); (ii) a 3.5% (m/v) MetaPhor™ agarose gel (Lonza, USA) was prepared instead of the suggested 15–25% polyacrylamide gel. The amplified products were digested at 36 ± 1°C for 90 min. The bands were visualised and captured as described elsewhere. A 50 bp DNA ladder (ThermoScientific, USA) was used as a marker. One positive amplicon form each of the screened genes were sequenced in both forward and reverse directions by Inqaba Biotechnical Industries, Pretoria, South Africa. The confirmed positive gene was used as a positive control. A negative control [nuclease-free water (Qiagen, Germany)] was included for all M-PCR assays. The identity of the genes was confirmed using BLAST after sequencing of the products.
Staphylococcal cassette chromosome mec typing
The M-PCR assays for the ccr and the mec gene complexes was performed as described by Kondo et al. (2007). Three additional primers (ccrCU1; α4U and β4U) were added to the ccr gene complex M-PCR assay to detect all known ccrC allotypes, as well as the ccrAB4 gene complex (Ruppé et al., 2009). The ccr and the mec gene complexes were classified as recommended by the guidelines from the International Working Group on the Classification of Staphylococcal Cassette Chromosome (SCC) elements (IWG-SCC) (Ito et al., 2009). SCC mec types were assigned based on the Roman numeral system, however the latter was not assigned to novel ccr and mec gene complex combinations if it was discovered in other methicillin-resistant staphylococci. These SCCmec types were reported using the raw combination of the ccr and mec gene complexes.
Pulsed-field gel electrophoresis
Plug preparation of S. epidermidis isolates were done according to the unified PFGE protocol for Gram-positive bacteria developed by the Division of Health Care Quality Promotion from the Centers for Disease Control and Prevention (CDC) (http://www.cdc.gov/hai/pdfs/labSettings/Unified_PFGE_Protocol.pdf) with some modifications. The modifications included an overnight lysis step of the plugs at 51°C, instead of the 2-h lysis step at 54°C as described by the protocol. The SmaI restriction digested plugs were separated by PFGE using the Rotaphor® system 6.0 (Biometra, Germany) in a 1.2% (m/v) SeaKem LE (Lonza, Rockland, USA) agarose gel in 0.25 × TBE (Sigma-Aldrich, USA) buffer. The programmed parameters for electrophoresis were set at 220 V changing linearly to 200 V at a constant angle of 120°. The switch time was set at 5 s linearly to 40 s (McDougal et al., 2003). The gel ran for 25 h at 13°C. The gel was stained with ethidium bromide (0.25 μg/mL) (Sigma-Aldrich, USA) for 15 min and destained in ultrapure water for 30 min. The destained gel was visualised under UV light (Transilluminator, Ultra-violet Products Incorporated, USA) and photographed with the UVP GelDoc-It system (Transilluminator, Ultra-violet Products Incorporated, USA). Pulsed-field gel electrophoresis patterns were analysed with the GelCompar II system (Applied Maths, Belgium) and the percentage of relatedness was determined by the Dice Coefficient. Isolates from IVC tips and BCs were considered genetically-related if the pulsotype's banding patterns showed ≥ 80% similarity (Applied Maths, Belgium), which corresponds to the Tenover criteria (possibly related with 4–6 band differences) (Tenover et al., 1995; Peirano et al., 2014). Representatives from each major PFGE pulsotype (≥ 5 isolates), minor pulsotypes (< 5 isolates) with ≥ 80% similarity and selected singletons were chosen for MLST analyses.
Multilocus sequence typing
Multilocus sequence typing was performed as described by Thomas et al. (2007). Ten representative S. epidermidis isolates were selected from three major pulsotypes (n = 4), three minor pulsotypes (n = 3) and three singletons (n = 3). Sequencing of the seven housekeeping genes (arcC, aroE, gtr, mutS, pyrR, tpi, and yqiL) was done by Inqaba Biotechnical Industries (Pretoria, South Africa). Originally, the S. epidermidis MLST database (http://sepidermidis.mlst.net/) was used, [hosted now by pubMLST (https://pubmlst.org/sepidermidis/)] provided allelic profiles and the subsequent STs. A comparative electronic Based Upon Related Sequence Types (eBURST) v3 analysis (http://eburst.mlst.net/) was run to infer a hypothetical pattern of evolutionary descent. All STs were submitted to the curator (Jonathon Thomas, email address: miragaia@itqb.unl.pt) of the S. epidermidis database.
Results
Prevalence of Staphylococcus epidermidis catheter-related bloodstream infection cases
A total of 508 IVCs from 331 patients were submitted for culture during the study period. Only 50% (n = 253/508) of the IVCs were accompanied by BCs taken within 48 h. Forty-four percent (44%; n = 112/253) of these IVCs accompanied by a BC were colonised, of which 25,6% (n = 65/253) of the colonised IVCs were associated with a CRBSI. S. epidermidis was implicated as the aetiological agent in 31% (n = 20/65) of the CRBSI cases of which 59 S. epidermidis were obtained, 23 isolates were cultured from 22 IVCs and 36 isolates were cultured from 36 BCs. Nine patients (four adults and five paediatric patients) had a single positive BCs for S. epidermidis. The pulsotypes obtained from the IVC and the BC for these 9 patients were compared.
Clinical details of patients with a Staphylococcus epidermidis catheter-related bloodstream infection
Patients with an S. epidermidis CRBSI were mostly male (65%; n = 13/20). Adult patients (75%; 15/20) were more often affected than paediatric patients (25%; n = 5/20). Adult patients were on average 43 years [standard deviation (SD) = ± 14 years] old, whereas paediatric patients were on average 31 days (SD = ±16 days, outlier of 1 year) old. Sixty percent (60%; n = 12/20) of S. epidermidis CRBSI cases were from patients in an intensive care unit (ICU) and 40% (n = 8/20) were from patients located in other wards throughout the hospital. Thirty-five percent (40%; n = 8/20) of these patients had an underlying gastroenterological condition, followed by 30% (n = 6/20) of patients with either an underlying nephrological condition or a traumatic incident. The clinical condition of one patient was unknown. The remaining patients had a diverse range of underlying conditions [i.e., cardiovascular (5%; n = 1/20), dermatology (5%; n = 1/20), neurologic (5%; n = 1/20), surgery (5%; n = 1/20) and respiratory (5%; n = 1/20)].
On average, patients were exposed to two or more antimicrobials. Fifty-five percent (55%; n = 11/20) of patients with S. epidermidis CRBSI had meropenem exposure, followed by 30% (n = 6/20) for both vancomycin and colistin, and 15% (n = 3/20) to tazobactam. The presence of specific risk factors for the development of a CRBSI was unknown for a single patient (n = 19). Fifty-eight percent (58%; n = 11/19) of patients had previously received total parenteral nutrition and 16% (n = 3/19) of patients had HIV, diabetes or renal failure as a risk factor. Twenty-one percent (21%; n = 4/19) of patients were either malnourished or had lost the integrity of their skin. A single patient (5%; n = 1/19) had neutropaenia as a risk factor. Patients were hospitalised on average for 20 days (SD = ± 14 days), prior to the development of an S. epidermidis CRBSI.
Characteristics of the intravascular catheters implicated in a Staphylococcus epidermidis catheter-related bloodstream infection
A total of 22 IVC catheters were involved in the S. epidermidis CRBSI events. All IVCs were short non-cuffed lines and were not impregnated with antimicrobials. Seventy-three percent (73%; n = 16/22) of the IVCs submitted were central venous pressure (CVP) tips, followed by 14% (n = 3/22), 9% (n = 2/22), and 5% (n = 1/22) of IVCs being VasCath lines, arterial catheters and Broviac lines, respectively.
The duration of catheterisation, the vessel occupied, the insertion site, the pathway followed from the insertion site to the vessel and the number of lumens was unknown for a single CVP and thus was excluded (n = 15). The CVPs were placed either in the subclavian vein (67%; n = 10/15) or the internal jugular vein (33%; n = 5/15). All CVPs placed in the subclavian vein were non-tunnelled. Two of the subclavian vein CVPs (20%; n = 2/10) had a double-lumen, whereas the rest (80%; n = 8/10) had a triple-lumen. All the CVPs placed in the internal jugular vein had a triple-lumen and the pathway followed under the skin was non-tunnelled. The remaining CVPs were in place for an average of 12 days (SD = ± 6 days).
All VasCath lines were placed centrally in the internal jugular vein. The two triple-lumen VasCath lines were non-tunnelled, whereas the double-lumen VasCath line was tunnelled. The duration of placement was unknown for a single VasCath line, whereas the other two VasCath lines were in place for an average of 7 days. Both arterial catheters were short, single-lumen, non-tunnelled and placed in a radial vein. One of the arterial catheters was simultaneously in place with a CVP. The one arterial catheter was in place for 10 days prior to the development of S. epidermidis CRBSI. The short, double-lumen, non-tunnelled Broviac line was in place for 14 days centrally in the subclavian vein.
Identification, antimicrobial susceptibility profiles, distribution of the icaAB, IS256, mecA, and qacA/B genes, restriction enzyme digestion of the qacA/B genes and chlorhexidine susceptibility testing
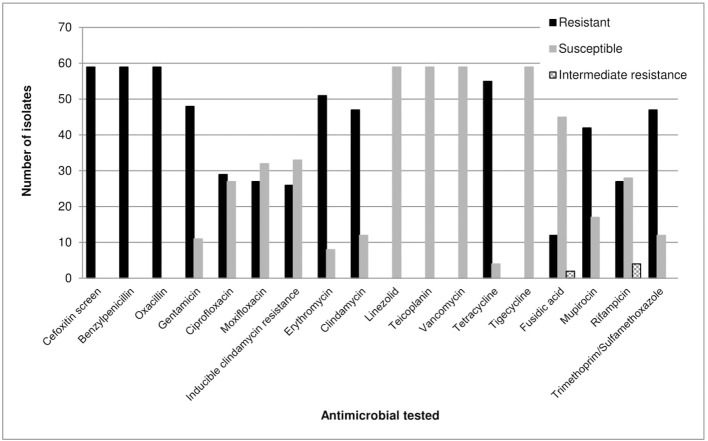
The VITEK® 2 automated system, MALDI-TOF MS analysis and the M-PCR assay results were in agreement and all isolates were identified as S. epidermidis. All isolates (100%) were positive for the cefoxitin screen and showed resistance to benzylpenicillin and oxacillin. A total of 86% (n = 51/59) and 81% (n = 48/59) of isolates were resistant to erythromycin and gentamicin, respectively (Figure 1). All isolates (100%; n = 59/59) were susceptible to linezolid, teicoplanin, vancomycin and tigecycline (Figure 1).
Figure 1.
Antimicrobial susceptibility profiles of 59 S. epidermidis isolates as determined by the VITEK® 2 system (bioMérieux, France) (AST-P603) according to the CLSI guidelines (Clinical Laboratory Standards Institute, 2013).
The icaAB (n = 48/59) and qacA/B (n = 48/59) genes were both present in 81% of the isolates (Table 1). All isolates (n = 59/59) carried the mecA gene, whereas 83% (n = 49/59) of isolates carried the IS256 element. Seventy-six percent (76%; n = 45/59) of the isolates harboured all four genes. The amplified qacA/B products (81%; n = 48/59) were digested with AluI (New England Biolabs, USA). All isolates (100%; n = 48/48) harboured the qacA gene (220 bp product obtained after digestion).
Table 1.
Molecular characterisation of Staphylococcus epidermidis isolates involved in CRBSIs.
| Patientnumber | Isolate number | Type ofspecimen | IS256 | icaAB | mecA | qacA/B | mecAcontrol | ccrgroup | mecgroup | Combination of the ccr group and the mec group | SCCmectype | MLST |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 7.2 | CVP | + | – | + | + | + | ND | ND | ND | ||
| 7.3 | + | + | + | + | + | 4, 5 | C | 4,5C | ||||
| 7.4 | BC | + | + | + | + | + | 3,5 | A | 3,5A | |||
| 7.5 | BC | + | + | + | + | + | 2 | B | 2B | IV | ||
| 7.6 | BC | + | – | + | + | + | 2,5 | B | 2,5B | |||
| 9 | 9.1 | CVP | + | + | + | + | + | 5 | A | 5A | ||
| 9.2 | BC | + | + | + | + | + | 2 | B | 2B | IV | ||
| 12 | 12.1 | CVP | + | + | + | + | + | 1 | A | 1A | ||
| 12.3 | BC | + | + | + | + | + | 1 | A | 1A | |||
| 12.4 | BC | + | + | + | + | + | 1 | A | 1A | |||
| 12.5 | BC | + | + | + | + | + | 1 | A | 1A | ST54 | ||
| 14 | 14.1 | CVP | + | + | + | + | + | 4 | A | 4A | ||
| 14.2 | AC | + | + | + | + | + | 3 | A | 3A | III | ||
| 14.3 | BC | + | + | + | + | + | 3 | A | 3A | III | ST2 | |
| 14.4 | BC | + | + | + | + | + | 3,4 | A | ND | |||
| 14.5 | BC | + | + | + | + | + | ND | A | mec class A | |||
| 18 | 18.1 | BC | – | + | + | – | + | 4,5 | A | 4,5A | ||
| 18.2 | BC | – | + | + | – | + | ND | A | mec class A | ST490 | ||
| 18.4 | CVP | – | – | + | – | + | 2 | B | 2B | IV | ||
| 20 | 20.1 | CVP | + | + | + | + | + | ND | A | mec class A | ||
| 20.2 | BC | + | + | + | + | + | 1,2,5 | A | 1,2,5A | |||
| 22 | 22.1 | CVP | + | + | + | + | + | 2 | B | 2B | IV | |
| 22.2 | BC | + | + | + | + | + | 2 | B | 2B | IV | ||
| 26 | 26.1 | CVP | + | + | + | + | + | 2,5 | B,C | ND | ||
| 26.2 | BC | + | + | + | + | + | 2 | B | 2B | IV | ST28 | |
| 27 | 27.1 | CVP | + | + | + | + | + | 3 | A | 3A | III | |
| 27.2 | + | + | + | + | + | 2,5 | C | 2,5C | ||||
| 27.3 | BC | + | + | + | + | + | 3 | A | 3A | III | ST2 | |
| 28 | 28.2 | CVP | + | + | + | + | + | 2,4 | B | 2,4B | ||
| 28.5 | BC | + | + | + | + | + | 2 | B | 2B | IV | ||
| 30 | 30.1 | CVP | + | + | + | + | + | ND | B | mec class B | ||
| 30.3 | BC | + | + | + | + | + | ND | B | mec class B | ST2 | ||
| 36 | 36.1 | AC | + | + | + | + | + | 2,4,5 | B,C | ND | ||
| 36.2 | BC | + | + | + | + | + | 4,5 | C | 4,5C | |||
| 36.3 | BC | + | + | + | + | + | 2 | B | 2B | IV | ||
| 38 | 38.1 | CVP | + | – | + | – | + | 1,5 | B | 1,5B | ||
| 38.2 | BC | – | – | + | – | + | 1,5 | B | 1,5B | ST596 | ||
| 38.3 | BC | + | – | + | – | + | 1,5 | B | 1,5B | |||
| 40 | 40.1 | CVP | + | + | + | + | + | 4,5 | C | 4,5C | ||
| 40.2 | BC | + | + | + | + | + | 2 | B | 2B | IV | ||
| 40.3 | BC | + | + | + | + | + | 2 | B | 2B | IV | ||
| 42 | 42.1 | CVP | + | + | + | + | + | 2,5 | B,C | ND | ||
| 42.5 | BC | – | – | + | + | + | 2 | B | 2B | IV | ||
| 43 | 43.2 | CVP | + | + | + | + | + | 2 | B | 2B | IV | |
| 43.3 | VC | + | + | + | + | + | 3 | A | 3A | III | ||
| 43.4 | VC | + | + | + | + | + | 2 | B | 2B | IV | ||
| 43.5 | BC | + | + | + | + | + | 2 | B | 2B | IV | ST2 | |
| 43.6 | BC | + | + | + | + | + | 4,5 | C | 4,5C | |||
| 43.7 | BC | + | + | + | + | + | 2 | B | 2B | IV | ||
| 44 | 44.1 | CVP | + | + | + | + | + | 1 | A | 1A | ||
| 44.2 | BC | + | + | + | + | + | 1 | A | 1A | ST54 | ||
| 53 | 53.1 | CVP | – | – | + | – | + | 5 | C | 5C | V | |
| 53.2 | BC | – | – | + | – | + | 5 | C | 5C | V | ST59 | |
| 53.3 | BC | – | – | + | – | + | 5 | C | 5C | V | ||
| 59 | 59.1 | VC | + | + | + | + | + | 2 | B | 2B | IV | |
| 59.2 | BC | + | + | + | + | + | 2 | B | 2B | IV | ||
| 60 | 60.1 | CVP | + | + | + | + | + | 2 | B | 2B | IV | |
| 60.2 | BC | – | + | + | – | + | 2,5 | B | 2,5B | |||
| 60.3 | BC | – | – | + | – | + | 5 | C | 5C | V |
The VITEK® 2 automated system (bioMérieux, France), MALDI-TOF MS (matrix-assisted laser deabsorption time of flight mass spectrometry) and ID PCR all identified the collected isolates as Staphylococcus epidermidis; AC, arterial catheter; BC, blood culture; CVP, central venous catheter; MLST, multilocus sequence typing; M-PCR assay, multiplex polymerase chain reaction assay; ND, no data; SCCmec, Staphylococcal Chromosome Cassette mec; ST, sequence type; VC, VasCath line; +, detected by M-PCR assay; –, not detected by the M-PCR assay. The shaded areas under the SCCmec type column indicate that certain isolates (n = 32/59) had multiple or unusual ccr and mec gene complex combinations and in these cases the raw ccr and mec gene combinations, instead of the SCCmec type, were reported. A Roman numeral was therefore not assigned to novel ccr and mec gene complex combinations if it is discovered in other methicillin-resistant staphylococci.
Molecular typing of Staphylococcus epidermidis isolates
SCC mec type IV (2B) was prevalent (31%; n = 18/59), followed by SCCmec type 1A (10%; n = 6/59) and SCCmec type III (3A) (9%; n = 5/59) (Table 1). Four isolates (7%; n = 4/59) harboured either SCCmec type V (5C2) or SCCmec type VII (5C1). The M-PCR assay by Kondo et al. (2007) did not distinguish between these two mec class C allotypes (i.e. mec class C1 and mec class C2) and it is therefore reported as SCCmec type 5C. Other ccr and mec gene classes detected included type VIII (4A) (3%; n = 2/59) and 5A (2%; n = 1/59). Only the mec gene complex could be detected in five isolates [mec class A (n = 3/5) and mec class B (n = 2/5)]. Three isolates harboured two mec groups (B,C) and multiple ccr groups [2,4,5B,C (n = 1/3) and 2,5B,C (n = 2/3)]. The remaining isolates (25%; n = 15/59) harboured multiple ccr gene complexes associated with a single mec gene complex [i.e., 4,5C (n = 4/15); 1,5B (n = 3/15); 2,5B (n = 2/15); 2,4B (n = 1/15); 2,5C (n = 1/15); 3,4A (n = 1/15); 3,5A (n = 1/15); 4,5A (n = 1/15), and 1,2,5A (n = 1/15)]. A single isolate was untypeable, even though it harboured the mecA gene. Only 46% (n = 27/59) of the isolates were typeable. The remaining 54% (n = 32/59) of the isolates had mec-ccr combinations that did not agree to the current classification scheme according to Kondo et al. (2007) and Ruppé et al. (2009). More than one ccr complex were detected in 31% (n = 18/59) of isolates.
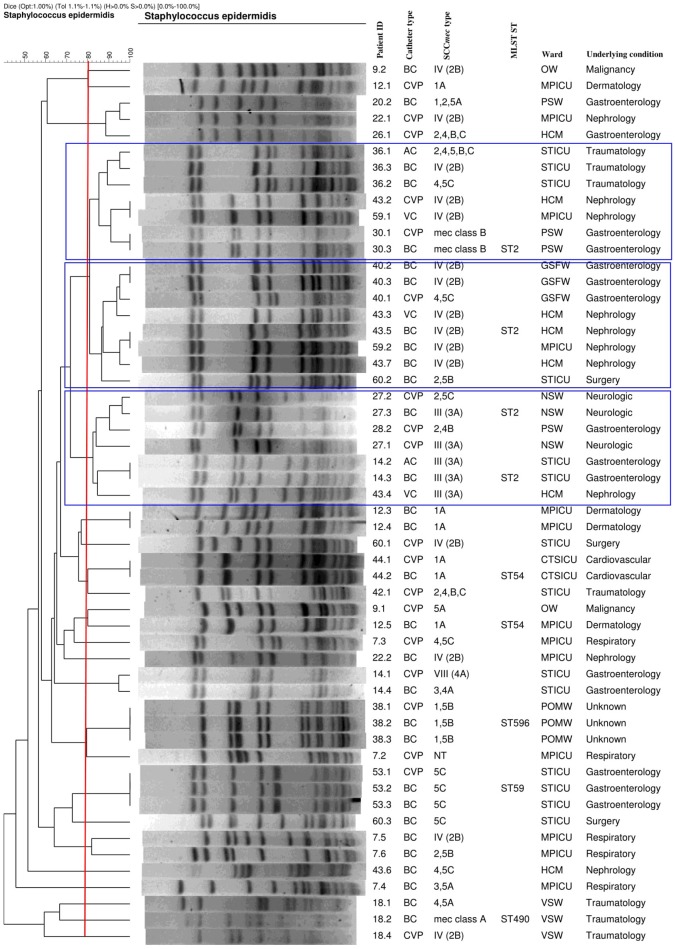
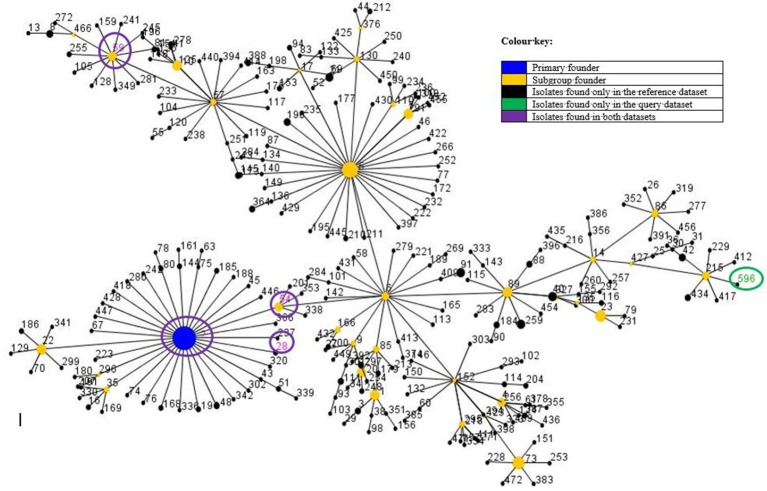
The PFGE results showed three major pulsotypes, three minor pulsotypes and several singletons (Figure 2). The dominant ST among the 10 sequenced isolates was ST2 (40%, n = 4/10), followed by ST54 (20%; n = 2/10), ST28 (10%; n = 1/10), ST59 (10%; n = 1/10), and ST490 (10%, 1/10) (Table 1). A new ST was assigned to isolate 38.3 as ST596. A population snapshot of S. epidermidis is represented in Figure 3. The majority of isolates belonged to a single clonal complex [CC2], using eBURST analysis.
Figure 2.
Genetic relatedness of catheter and blood culture S. epidermidis isolates implicated in CRBSI events (with the blue blocks representing the major pulsotypes). AC, arterial catheter; BC, blood culture; CTSICU, cardiothoracic surgery ICU; CVP, central venous catheter; GSFW, general surgery female ward; HCM, high care, multidisciplinary; MPICU, medical and pulmonology ICU; MLST, multilocus sequence typing; NSW, neurosurgery ward; NT, not typeable; OW, oncology ward; PSW, paediatric surgery ward; POMW, plastic/maxillofacial ward; ST, sequence type; STICU, surgery and trauma ICU; VC, VasCath; VSW, vascular surgery ward. Banding patterns of the catheter culture and BC isolates showing ≥80% similarity were clonal. Please note that isolates 14.5, 20.1, 26.2, 28.5, and 42.5 were untypeable and not included in the dendrogram.
Figure 3.
Comparative population snapshot of S. epidermidis STs detected in this study vs. STs in the S. epidermidis MLST database of clonal complex 2 as on the 16th of May 2017. Ten isolates were sequenced: ST2 (n = 4); ST54 (n = 2); ST28 (n = 1); ST59 (n = 1); ST490 (n = 1) and ST596 (n = 1). The sequence types (STs) found in this study are encircled, except for ST490 [not currently in the MLST database (last update 24-09-2015)]. The colour and meaning of each circle can be read according to the colour key.
Discussion
S. epidermidis is one of the most common isolated pathogens associated with CRBSI in this study. This is often due to the abundance of this opportunistic pathogen on the human skin, providing it with the ideal opportunity to contaminate IVC upon insertion. The molecular epidemiology and prevalence of the icaAB, IS256, mecA, and qacA/B genes in the S. epidermidis isolates (implicated in the CRBSI events) were described at a clinical setting in this study.
The level of resistance in S. epidermidis CRBSI isolates toward commonly prescribed antimicrobials, such as β-lactams, aminoglycosides, and macrolides, was high indicating multidrug resistance (MDR) as shown in Figure 1. The IS256, icaAB, and qacA/B genes were also highly prevalent among these isolates. All S. epidermidis isolates carried the mecA gene, which is similar to a study conducted in China (~96%) (Li et al., 2009), but higher compared to a study done in Portugal (~70%) (Rolo et al., 2012) and India (~67%) (Jena et al., 2017). The high degree of antimicrobial resistance and the presence of the mecA gene may indicate the presence of methicillin-resistant S. epidermidis (MRSE), which is more often resistant to other antimicrobials compared to methicillin-sensitive S. epidermidis (Otto, 2009). MRSE can also serve as a mecA gene donor to methicillin-sensitive S. aureus, which can lead to the emergence of methicillin-resistant S. aureus (MRSA) (Bloemendaal et al., 2010).
Schoenfelder et al. (2010) suggested that controlling MRSE may reduce the prevalence of MRSA. Infection prevention strategies, designed to decrease the incidence of CRBSI, include the use of antiseptics, such as chlorhexidine skin preparations prior to catheter insertion (Marschall et al., 2014). The qacA/B genes in S. epidermidis encode an efflux pump, which is associated with reduced susceptibility toward quaternary ammonium compounds and chlorhexidine (Horner et al., 2012; Wassenaar et al., 2015). According to the literature the qacA/B genes may indicate chlorhexidine failure but other unknown genes could be involved (Sekiguchi et al., 2004; Horner et al., 2012). The QacA and QacB efflux pumps have different substrate specificities (Wassenaar et al., 2015). The QacA efflux pump can transport a broad range of substrates, which include QACs, intercalating dyes (e.g., ethidium bromide) and cationic biocides (e.g., chlorhexidine), whereas the QacB efflux pump can transport only a limited range of substrates, which include QACs and intercalating dyes, but not cationic biocides (Wassenaar et al., 2015). Additional efflux pumps may also play a role (Otter et al., 2013).
The SCCmec types detected in the study were diverse, which are in agreement with other studies done in Finland and Western China (Ibrahem et al., 2008; Zong et al., 2011). The hospital environment is known to play a fundamental role in the amplification and the diversification of SCCmec elements in S. epidermidis (Rolo et al., 2012). S. epidermidis harbouring multiple ccr and mec gene complexes have been extensively reported in the literature (Ibrahem et al., 2008; Garza-González et al., 2010; Svensson et al., 2011; Zong et al., 2011; Rolo et al., 2012; Jena et al., 2017), but none of these studies have addressed the possibility of polyclonality. Polyclonality among S. epidermidis isolates has been reported in prosthetic joint infections (Galdbart et al., 1999), prosthetic valve endocarditis (Van Wijngaerden et al., 1997; Jena et al., 2017), healthy ocular surfaces (Ueta et al., 2007), and CRBSI (Rijnders et al., 2001). The overlapping of SCCmec types in isolates obtained from different specimens (patient 14 and patient 36) suggests the presence of a polyclonal S. epidermidis infection, due to the fusion of elements or due to the separate integration of various structural components (Ito et al., 2009; Shore and Coleman, 2013).
The PFGE results showed that the two STs (ST2 and ST54) were detected from different patients residing in different wards. These results were in agreement with a study done in Belgium at a clinical setting, by Cherifi et al. (2013) who also reported ST2 and ST54 as the most frequently detected clones. Du et al. (2013) conducted a study in China among patients, healthy volunteers and healthcare workers, who reported that the ST2 S. epidermidis clone was the most frequently detected among participants, while ST54 was only detected among healthcare workers. To the best of the authors knowledge, no literature could be found regarding S. epidermidis ST54 in Africa or South Africa. S. epidermidis isolates, obtained from different specimens (i.e., IVC and BCs), but from the same patient, were mostly clonal. However, some of the isolates collected from the same patient (patients 7, 9, 12, 20, 22, 26, 28, 43, and 60) were diverse (Figure 2). This could be explained by the introduction of multiple organisms residing on the skin as several patients had multiple catheters inserted or due to poor infection control measures at this setting (Table 1). The majority of ST2 strains harboured SCCmec IV with a single ccr group (2). This was similar to the results of Cherifi et al. (2013) who also detected SCCmec IV as the most dominant SCCmec type; however, multiple ccr complexes were reported by the authors. A new ST (596) was detected and assigned to one isolate [ID = 1031; isolate = UP2; country = South Africa] (https://pubmlst.org/bigsdb?db=pubmlst_sepidermidis_isolates&page=profiles).
Some of the study limitations included, that the results cannot be extrapolated to the rest of South Africa, since the study was done in a single centre, compliance toward the IDSA guidelines, in regards to BC submissions, is lacking, catheter cultures were not always accompanied by one or more BCs, which could have led to the inclusion of possible S. epidermidis contaminants. Due to limited funding all 59 S. epidermidis isolates could not be subjected to MLST.
Conclusion
The isolated S. epidermidis isolates were MDR and all isolates carried the mecA gene. The high prevalence of the mecA gene and other virulence factors is worrisome since S. epidermidis can serve as a genetic reservoir to the more pathogenic S. aureus. The predominant ST in the studied hospital was ST2, followed by ST54 but a new ST (596) was also discovered. It is recommended that infection control and prevention strategies are intensified to limit the spread of S. epidermidis in the hospital environment. Further investigation is needed to determine resistance determinants toward chlorhexidine.
Ethics statement
The study was approved by the Research Ethics Committee, Faculty of Health Sciences, University of Pretoria (Protocol number: 118/2013) and informed individual patient consent was waivered, since the study was observational and patient care was not influenced at any stage.
Author contributions
ME, MK, and WS: conceived and designed the study; WS: collected all the clinical isolates; WS and ML: performed all the laboratory analyses; ME, ML, and WS: wrote the manuscript with critical appraisal and contributions received from all of the authors. All authors read and approved the final version of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the Department of Microbiology and Plant Pathology, Faculty of Natural and Agricultural Sciences, University of Pretoria, in particular Ms. A. Lombard and Ms. Z. Zulu, for performing MALDI-TOF MS (Bruker Daltonics, USA) analysis. The authors would like to thank Dr. Cheryl A. Tosh (Faculty of Health Sciences, University of Pretoria) for editing the manuscript before submission. The authors would like to thank Dr. A. Smith, coordinator for PulseNet Africa for the donation of the Salmonella strain (ATCC BAA-664). This work was previously reported in part as an Oral ePoster presentation (EP084) at the 25th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID 2015) in Copenhagen, Denmark from the 25–28 April 2015.
Footnotes
Funding. The authors hereby acknowledge the NRF and the National Health Laboratory Service (NHLS) for financial support. This work is based on the research supported in part by the National Research Foundation (NRF) of South Africa [Grant specific unique reference number (UID) 74426]. Opinions expressed and conclusions arrived at are those of the authors and are not necessarily to be attributed to the NRF.
References
- Al-Talib H., Yean C. Y., Al-Khateeb A., Hassan H., Singh K. K., Al-Jashamy K., et al. (2009). A pentaplex assay for the rapid detection of methicillin-resistant Staphylococcus aureus and Panton-valentine leucocidin. BMC Microbiol. 9:113. 10.1186/1471-2180-9-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemendaal A. L., Brouwer E. C., Fluit A. C. (2010). Methicillin resistance transfer from Staphylococcus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS ONE 5:e11841. 10.1371/journal.pone.0011841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherifi S., Byl B., Deplano A., Nonhoff C., Denis O., Hallin M. (2013). Comparative epidemiology of Staphylococcus epidermidis isolates from patients with catheter-related bacteremia and from healthy volunteers. J. Clin. Microbiol. 51, 1541–1547. 10.1128/JCM.03378-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute (CLSI) (2013). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-third Informational Supplement M100-S23. Wayne, PA: CLSI. [Google Scholar]
- Conrick-Martin I., Foley M., Roche F. M., Fraher M. H., Burns K. M., Morrison P., et al. (2013). Catheter-related infection in Irish intensive care units diagnosed with HELICS criteria: a multi-centre surveillance study. J. Hosp. Infect. 83, 238–243. 10.1016/j.jhin.2012.11.020 [DOI] [PubMed] [Google Scholar]
- Du X., Zhu Y., Song Y., Li T., Luo T., Sun G., et al. (2013). Molecular analysis of Staphylococcus epidermidis strains isolated from community and hospital environments in China. PLoS ONE 8:e62742. 10.1371/journal.pone.0062742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdbart J. O., Morvan A., Desplaces N., El Solh N. (1999). Phenotypic and genomic variation among Staphylococcus epidermidis strains infecting joint prostheses. J. Clin. Microbiol. 37, 1306–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-González E., Morfín-Otero R., Llaca-Díaz J. M., Rodriguez-Noriega E. (2010). Staphylococcal cassette chromosome mec (SCCmec) in methicillin-resistant coagulase-negative staphylococci. A review and the experience in a tertiary-care setting. Epidemiol. Infect. 138, 645–654. 10.1017/S0950268809991361 [DOI] [PubMed] [Google Scholar]
- Hirotaki S., Sasaki T., Kuwahara-Arai K., Hiramatsu K. (2011). Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR. J. Clin. Microbiol. 49, 3627–3631. 10.1128/JCM.00488-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner C., Mawer D., Wilcox M. (2012). Reduced susceptibility to chlorhexidine in staphylococci: is it increasing and does it matter? J. Antimicrob. Chemother. 67, 2547–2559. 10.1093/jac/dks284 [DOI] [PubMed] [Google Scholar]
- Ibrahem S., Salmenlinna S., Lyytikäinen O., Vaara M., Vuopio-Varkila J. (2008). Molecular characterization of methicillin-resistant Staphylococcus epidermidis strains from bacteraemic patients. Clin. Microbiol. Infect. 14, 1020–1027. 10.1111/j.1469-0691.2008.02080.x [DOI] [PubMed] [Google Scholar]
- Iorio N. L., Azevedo M. B., Frazão V. H., Barcellos A. G., Barros E. M., Pereira E. M., et al. (2011). Methicillin-resistant Staphylococcus epidermidis carrying biofilm formation genes: detection of clinical isolates by multiplex PCR. Int. Microbiol. 14, 13–17. 10.2436/20.1501.01.130 [DOI] [PubMed] [Google Scholar]
- Ito T., Hiramatsu K., Oliveira D. C., De Lencastre H., Zhang K., Westh H., et al. (2009). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53, 4961–4967. 10.1128/AAC.00579-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena S., Panda S., Nayak K. C., Singh D. V. (2017). Identification of major sequence types among multidrug-resistant Staphylococcus epidermidis strains isolated from infected eyes and healthy conjunctiva. Front. Microbiol. 8:1430. 10.3389/fmicb.2017.01430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y., Ito T., Ma X. X., Watanabe S., Kreiswirth B. N., Etienne J., et al. (2007). Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51, 264–274. 10.1128/AAC.00165-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela A., Nilsdotter-Augustinsson A., Persson L., Söderquist B. (2009). Prevalence of the ica operon and insertion sequence IS256 among Staphylococcus epidermidis prosthetic joint infection isolates. Eur. J. Clin. Microbiol. Infect. Dis. 28, 655–660. 10.1007/s10096-008-0664-6 [DOI] [PubMed] [Google Scholar]
- Li M., Wang X., Gao Q., Lu Y. (2009). Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J. Med. Microbiol. 58, 456–461. 10.1099/jmm.0.007567-0 [DOI] [PubMed] [Google Scholar]
- Maki D. G., Weise C. E., Sarafin H. W. (1977). A semi-quantitative culture method for identifying intravenous catheter-related infection. N. Engl. J. Med. 296, 1305–1309. 10.1056/NEJM197706092962301 [DOI] [PubMed] [Google Scholar]
- Marschall J., Mermel L. A., Fakih M., Hadaway L., Kallen A., O'Grady N. P., et al. (2014). Strategies to prevent central line–associated bloodstream infections in acute care hospitals: 2014 update. Infect. Control Hosp. Epidemiol. 35, 753–771. 10.1086/676533 [DOI] [PubMed] [Google Scholar]
- Martineau F., Picard F. J., Roy P. H., Ouellette M., Bergeron M. G. (1996). Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J. Clin. Microbiol. 34, 2888–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure J. A., Conly J. M., Lau V., Elsayed S., Louie T., Hutchins W., et al. (2006). Novel multiplex PCR assay for detection of the staphylococcal virulence marker panton-valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from -resistant staphylococci. J. Clin. Microb. 44, 1141–1144. 10.1128/JCM.44.3.1141-1144.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal L. K., Steward C. D., Killgore G. E., Chaitram J. M., McAllister S. K., Tenover F. C. (2003). Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41, 5113–5120. 10.1128/JCM.41.11.5113-5120.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel L. A., Allon M., Bouza E., Craven D. E., Flynn P., O'Grady N. P., et al. (2009). Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 49, 1–45. 10.1086/599376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otter J. A., Patel A., Cliff P. R., Halligan E. P., Tosas O., Edgeworth J. D. (2013). Selection for qacA carriage in CC22, but no CC30, methicillin-resistant Staphylococcus aureus bloodstream infection isolates during a successful institutional infection control programme. J. Antimicrob. Chemother. 68, 992–999. 10.1093/jac/dks500 [DOI] [PubMed] [Google Scholar]
- Otto M. (2008). Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322, 207–288. 10.1007/978-3-540-75418-3_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2009). Staphylococcus epidermidis — the “accidental” pathogen. Nat. Rev. Microbiol. 7, 555–567. 10.1038/nrmicro2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano G., van der Bij A. K., Freeman J. L., Poirel L., Nordmann P., Costello M., et al. (2014). Travel-related carbapenemase-producing gram-negative bacteria in Alberta, Canada: the first 3 years. Antimicrob. Agents. Chemother. 58, 3762–3767. 10.1128/JCM.00162-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival S. L., Emanuel C., Cutting K. F., Williams D. W. (2012). Microbiology of the skin and the role of biofilms in infection. Int. Wound J. 9, 14–32. 10.1111/j.1742-481X.2011.00836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira E. M., Schuenck R. P., Malvar K. L., Iorio N. L., Matos P. D., Olendzki A. N., et al. (2010). Staphylococcus aureus, Staphylococcus epidermidis, and Staphylococcus haemolyticus: methicillin-resistant isolates are detected directly in blood cultures by multiplex PCR. Microbiol. Res. 165, 243–249. 10.1016/j.micres.2009.03.003 [DOI] [PubMed] [Google Scholar]
- Rijnders B. J., van Wijngaerden E., van Eldere J., Peetermans W. E. (2001). Polyclonal Staphylococcus epidermidis intravascular catheter-related infections. Clin. Microbiol. Infect. 7, 388–391. 10.1046/j.1198-743x.2001.00272.x [DOI] [PubMed] [Google Scholar]
- Rolo J., de Lencastre H., Miragaia M. (2012). Strategies of adaptation of Staphylococcus epidermidis to hospital and community: amplification and diversification of SCCmec. J. Antimicrob. Chemother. 67, 1333–1341. 10.1093/jac/dks068 [DOI] [PubMed] [Google Scholar]
- Ruppé E., Barbier F., Mesli Y., Maiga A., Cojocaru R., Benkhalfat M., et al. (2009). Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob. Agents Chemother. 53, 442–449. 10.1128/AAC.00724-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S. M., Lange C., Eckart M., Hennig S., Kozytska S., Ziebuhr W. (2010). Success through diversity - how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int. J. Med. Microbiol. 300, 380–386. 10.1016/j.ijmm.2010.04.011 [DOI] [PubMed] [Google Scholar]
- Sekiguchi J., Hama T., Fujino T., Araake M., Irie A., Saruta K., et al. (2004). Detection of the antiseptic- and disinfectant-resistance genes qacA, qacB, qacC in methicillin-resistant Staphylococcus aureus isolated in a Tokyo Hospital. Jpn. J. Infect. Dis. 57, 288–291. [PubMed] [Google Scholar]
- Shore A. C., Coleman D. C. (2013). Staphylococcal cassette chromosome mec: recent advances and new insights. Int. J. Med. Microbiol. 303, 350–359. 10.1016/j.ijmm.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Strasheim W., Kock M. M., Ueckermann V., Hoosien E., Dreyer A. W., Ehlers M. M. (2015). Surveillance of catheter-related infections: the supplementary role of the microbiology laboratory. BMC Infect. Dis. 15:5. 10.1186/s12879-014-0743-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson K., Hellmark B., Söderquist B. (2011). Characterization of SCCmec elements in methicillin-resistant Staphylococcus epidermidis isolated from blood cultures from neonates during three decades. APMIS 119, 885–893. 10.1111/j.1600-0463.2011.02801.x [DOI] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. C., Vargas M. R., Miragaia M., Peacock S. J., Archer G. L., Enright M. C. (2007). Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J. Clin. Microbiol. 45, 616–619. 10.1128/JCM.01934-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta M., Iida T., Sakamoto M., Sotozono C., Takahashi J., Kojima K., et al. (2007). Polyclonality of Staphylococcus epidermidis residing on the healthy ocular surface. J. Med. Microbiol. 56, 77–82. 10.1099/jmm.0.46810-0 [DOI] [PubMed] [Google Scholar]
- Van Wijngaerden E., Peetermans W. E., Van Lierde S., Van Eldere J. (1997). Polyclonal Staphylococcus endocarditis. Clin. Infect. Dis. 25, 69–71. 10.1086/514499 [DOI] [PubMed] [Google Scholar]
- Wassenaar T. M., Ussery D., Nielsen L., Ingmer H. (2015). Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur. J. Microbiol. Immunol. 5, 44–61. 10.1556/EuJMI-D-14-00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Z., Peng C., Lü X. (2011). Diversity of SCCmec Elements in methicillin-resistant coagulase-negative staphylococci clinical isolates. PLoS ONE 6:e20191. 10.1371/journal.pone.0020191 [DOI] [PMC free article] [PubMed] [Google Scholar]