Abstract

The molecular determinants for the activities of the reported benzoic acid (SH4–54), salicylic acid (BP-1–102), and benzohydroxamic acid (SH5–07)-based STAT3 inhibitors were investigated to design optimized analogues. All three leads are based on an N-methylglycinamide scaffold, with its two amine groups condensed with three different functionalities. The three functionalities and the CH2 group of the glycinamide scaffold were separately modified. The replacement of the pentafluorobenzene or cyclohexylbenzene, or replacing the benzene ring of the aromatic carboxylic or hydroxamic acid motif with heterocyclic components (containing nitrogen and oxygen elements) all decreased potency. Notably, the Ala-linker analogues, 1a and 2v, and the Pro-based derivative 5d, all with (R)-configuration at the chiral center, had improved inhibitory activity and selectivity against STAT3 DNA-binding activity in vitro, with IC50 of 3.0 ± 0.9, 1.80 ± 0.94, and 2.4 ± 0.2 μM, respectively. Compounds 1a, 2v, 5d, and other analogues inhibited constitutive STAT3 phosphorylation and activation in human breast cancer and melanoma lines, and blocked tumor cell viability, growth, colony formation, and migration in vitro. Pro-based analogue, 5h, with a relatively polar tetrahydropyranyl (THP) ring, instead of the cyclohexyl, showed improved permeability. In general, the (R)-configuration Pro-based analogs showed the overall best profile, including physicochemical properties (e.g., microsomal metabolic stability, Caco-2 permeability), and in particular, 5d showed improved tumor-cell specificity.
Keywords: STAT, signal transducer and activator of transcription, BP-1−102, SH5−07, SH4−54, small-molecule inhibitors, breast cancer, melanoma, antitumor cell effects
Signal transducer and activator of transcription (STAT) family of cytoplasmic transcription factors have important roles in many cellular processes, including cell growth and differentiation, inflammation, and immune responses.1−4 Classically, STAT proteins are activated by tyrosine (Tyr) kinases, such as Janus kinases (JAKs) and Src family kinases, in response to the binding of cytokine and growth factors to their cognate receptors. The Tyr phosphorylation (pTyr) promotes dimerization between two activated STAT:STAT monomers through reciprocal pTyr-Src homology SH2 domain interactions. Active STAT:STAT dimers translocate to the nucleus to induce gene transcription by binding to specific DNA-response elements in the promoters of target genes.1,2,4
Aberrant activation of the family member, STAT3, occurs in a wide spectrum of human solid and hematological malignancies and promotes tumorigenesis and cancer progression.1,3−6 The mechanisms of constitutively active STAT3 in oncogenesis include the dysregulation of gene expression, resulting in uncontrolled cell proliferation and survival, enhanced tumor angiogenesis and metastasis, and the suppression of antitumor immunity. The critical role of STAT3 in cancer progression has made it an attractive target for anticancer drug discovery and anticancer therapy.3−5 One of the STAT3 anticancer drug discovery strategies is to target the key pTyr:SH2 domain interaction and hence the STAT3:STAT3 dimerization, thereby modulating STAT3 signaling.3,4,7−9 A number of small-molecule STAT3 inhibitors have been reported, none of which has gone beyond the preclinical stage because of potency and pharmacokinetic issues, except for OPB31121, although its exact mechanism of action is presently unclear and likely involves the inhibition of Tyr kinases.10−18
We have previously reported on BP-1–102, SH4–54, and SH5–07 as STAT3 dimerization disrupters8,19 (structures presented in Supplementary Table S6). All three lead compounds are based on the N-methylglycinamide scaffold, with its two amine moieties condensed with three different functionalities. To further improve the potency and physicochemical properties of the lead compounds and to furnish more potent and drug-like analogues, while maintaining selectivity, we took an iterative medicinal chemistry approach. The representative analogues, 1a, 5d, and 2v (Figure 1A), strongly inhibited STAT3 DNA-binding activity in vitro, with an IC50 of 3.0 ± 0.9, 2.4 ± 0.2, and 1.80 ± 0.94 μM, respectively.
Figure 1.
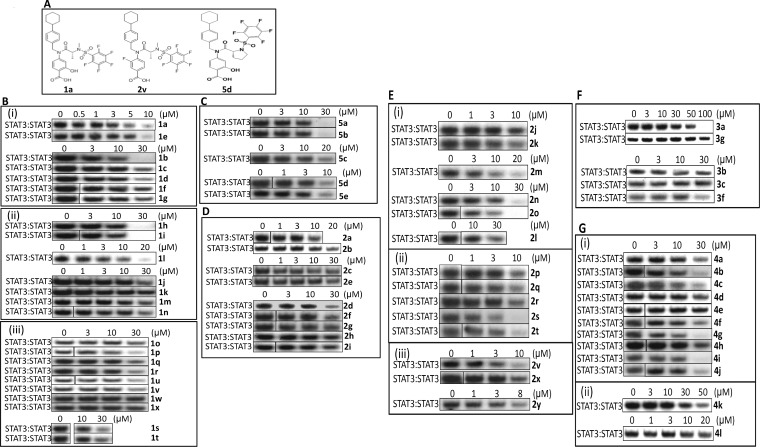
Structures of 1a, 2v, and 5d, and the effects of new analogues on STAT3 DNA-binding activity in vitro. (A) Analogues 1a, 2v, and 5d; (B–G) EMSA analysis of STAT3 DNA-binding activity in nuclear extracts of equal total protein prepared from v-Src-transformed fibroblasts containing activated STAT3 preincubated with 0–100 μM of the indicated compounds for 30 min prior to incubation with the radiolabeled hSIE probe that binds STAT3. Positions of STAT3:DNA complexes in gel are labeled; control lanes (0) represent nuclear extracts pretreated with 0.05% DMSO. Data are representative of 2–3 independent determinations.
Substitution of the glycine scaffold: alanine motif shows improved activity in disrupting STAT3 DNA-binding activity in vitro. The structure of the Gly-based initial lead compounds, BP-1–102, SH4–54, and SH5–07, was systematically optimized to simultaneously improve potency and physicochemical properties (e.g., lowering lipophilicity from the initial lead, SH5–07, cLogP of 5.6), while maintaining or lowering molecular weight (MW). In order to optimize the core scaffold of the lead compounds to furnish analogues with improved potency and better physicochemical properties, several groups of the pharmacophore were modified systematically (Supplementary Tables S1–S4). Supplementary Table S1 shows substitution at the methylene group of the glycine linker with small alkyl, hydroxyalkyl, aminomethyl, or carboxymethyl groups with the goal of improving potency or finding additional binding interactions. The inhibitory activities of the representative analogues against in vitro STAT3 DNA-binding activity, as measured by electrophoretic mobility shift assay (EMSA), are shown in Figure 1. Replacement of the Gly-linker by Ala (X = CHMe) to give the salicylic acid (R)-1a derivative (IC50 of 3.0 μM) moderately improved the STAT3-inhibitory activity over the parent compound, BP-1–102 (IC50 6.8 μM)13 (Figure 1B(i), Table S1). The corresponding enantiomer (S)-1e gave somewhat lower potency (IC50 of 5.0 μM) than its antipode (R)-1a.
Interestingly, contrary to the increase in potency observed with the salicylic acid-based analogs, the replacement of Gly-linker for Ala-linker in the benzoic acid-based and benzohydroxamic acid-based compounds did not improve potency (cf., SH4–54, IC50 4.4 μM with 1c and 1f, IC50 9.3 and 10.0 μM, respectively; and SH5–07, IC50 3.9 μM, with 1d and 1g, IC50 5.3 and 6.8 μM, respectively: Figure 1B(i), Table S1). Further substitutions at the methylene bridge of the salicylic acid-based BP-1–102 Gly-linker to further probe the structure–activity relationship (SAR), including ethyl (1h–i), bis-methylene to form a cyclopropyl (1j), dimethyl (1l), Ser-based linker (1m–p), hydroxyethyl (1q–r), Thr-based linker (1s–t), aminomethyl (1u–v), and Asp-based linker (1w–x, Table S1), led to compounds with reduced affinity compared to the Ala-linker analogues 1a and 1c (Table S1, Figure 1B(ii), (iii)). In general the (R)-configuration analogues consistently showed stronger potency compared to that of the corresponding enantiomeric (S)-analogues (Table S1). Based on this observation, therefore, only compounds with the (R)-configuration were considered for further optimization.
Modification of Benzoic Acid-Based Analogues. Aryl Ring Critical for the STAT3-Inhibitory Activity of Compounds
To further improve the potency of the lead compounds, we focused on optimizing the benzoic acid and benzohydroxamic acid moieties (Table S2). Interestingly, replacing the aromatic system with heterocyclic rings such as pyridine, pyridine-N-oxide, or oxazole (2b, 2c, 2e, and 2f) all greatly reduced the activity of the new analogues in vitro (Figure 1D, Table S2). We also noted that isosteric replacement of the standard 4-substituted benzoic acid system with a 3-substituted phenylacetic acid remarkably decreased the activity of the new analogue (2d) in vitro (Figure 1D, Table S2). Similarly, the isoindolinone and indazoles (2g, and 2i) also showed much weaker in vitro activity, compared to the parent compound (Figure 1D, Table S2). In addition, previous results from our lab also showed that the corresponding tetrazole system, instead of the carboxyl group, does not improve the STAT3-inhibitory activity.20 These data so far suggest that the carboxyl group can only be replaced with the hydroxamic acid moiety if STAT3-inhibitory activity is to be maintained. In addition, we noted that the sodium benzoate analogs (2a, 4c, and 1b) showed slightly decreased activity in inhibiting STAT3 DNA-binding ability in vitro, compared to their free acid forms (SH4–54, 4b, and 1a, respectively) (Figure 1B(i), 1b; Figure 1D, 2a; Figure 1G, 4c).
We next substituted the aromatic ring with methyl, chlorine, or fluorine for SAR evaluation (Table S2). Aromatic methyl substitutions of Gly-linker analogues (2j, 2l, and 2n) significantly reduced the ability to disrupt STAT3 DNA-binding activity in vitro, compared to the lead compound, SH4–54 (Figure 1E(i), Table S2). However, their corresponding Ala analogues (2k, 2m, and 2o) showed comparable (2k and 2m, IC50 of 5.7, 6.6, respectively) or slightly improved (2o, IC50 4.2 μM) in vitro STAT3-inhibitory activity compared to the lead compound, SH4–54 (IC50 4.4 μM) (Figure 1E(i), Table S2). The ortho- or meta-substitution with chlorine decreased the in vitro activity of the new analogues (2p and 2s, IC50 8.6–14.4 μM) (Figure 1E(ii), Table S2), compared to SH4–54, whereas their corresponding Ala analogues (2q and 2t, IC50 4.1 and 3.6 μM, respectively), and the benzohydroxamic analogue of 2q (2r, IC50 3.5 μM), showed moderately improved activity, compared to the parental lead compound (Figure 1E(ii), Table S2). Notably, contrary to chlorine, a single fluorine substitution at the 3-position (2v, IC50 1.8 μM) led to a 2-fold improvement in potency against STAT3 activity in vitro (Figure 1E(iii), Table S2). Similar substitution of a fluorine at the 3-position of the indazole also led to improvement (Table S2, entry 26 vs entry 9). By contrast, other fluorine replacements, including substitution at the 2-position or 3,5-positions (2y and 2x), showed no improvement in activity compared to SH4–54 (Figure 1E(iii), Table S2).
Substitutions on pentafluorophenyl or cyclohexylbenzyl group decrease the potency of analogues in vitro. We have previously reported that the replacement of the 4-toluene-sulfonamide moiety of S3I-201.1066 with pentafluoro-benzenesulfonamide generated the first orally bioavailable small-molecule inhibitor of STAT3, BP1–102,8 which substantially improved the STAT3-inhibitory potency in vitro (IC50 of 6.8 μM) over the parental compound (IC50 of 35 μM). Our new strategy focused on the pentafluorophenyl ring for SAR evaluation (Table S3) and for addressing whether this motif is critical for the potency of the lead compounds. Interestingly, reducing the number of fluorine atoms in a variety of different ways on the aromatic ring (analogues 3a, 3b, 3c, 3d and 3e), or replacing the pentafluorophenyl group with 3,5-bis(trifluoromethyl)phenyl or pyridyl (3f, and 3g) all significantly suppressed the activity of the new analogues against STAT3 DNA-binding activity in vitro (IC50 higher than 26 μM) (Figure 1F, Table S3). For SAR studies, the sulfonamide linker was replaced with an amide or a methylene. However, the modifications of the pentafluorobenzenesulfonamide group to a pentafluorobenzamide or a pentafluorobenzylamide were similarly not tolerated (3h–i, Table S3).
In our previous study, the incorporation of the (4-cyclohexyl)benzyl moiety into the compound S3I-201 generated S3I-201.1066,7 which showed a 3-fold improvement in activity, with an IC50 of 35 μM. This important substructure is also incorporated in the three lead compounds (BP1–102, SH4–54, and SH5–07). In order to improve the physicochemical properties of the scaffold, for example, reduce the hydrophobicity, and improve the solubility of the new analogues, we substituted the cyclohexyl group with heterocycles containing oxygen or nitrogen (Table S4). Interestingly, the replacement of cyclohexane by tetrahydro-4H-pyran (THP) (4a, 4e, and 4d) or piperidine (4l and 4f) decreased the activity of the new analogues (Figure 1G, Table S4). However, the THP analogue, 4b (IC50 5.0 μM, Table S4), which incorporates an Ala-linker, shows a moderately improved potency against STAT3 DNA-binding activity in vitro compared to the parental compound, BP-1–102 (IC50 6.9 μM), indicating that the incorporation of alanine can compensate for the loss of activity caused by oxygen substitution in the cyclohexane moiety. Other analogues also designed and prepared include replacing the cyclohexane with cycloheptane (4i), cycloheptene (4j), or 4,4-difluorocyclohexane (4g, 4h), or replacing the benzyl system with a pyridylmethyl moiety (4k), but all resulted in compromised STAT3-inhibitory potency (Figure 1G, Table S4).
Extension of the Alanine SAR
Proline analogues show further activity improvement in disrupting STAT3 DNA-binding in vitro. A further modification to the Ala linker was to join the two methyl groups to become a novel Pro-linker system. Notably, the (R)-enantiomer Pro analogue 5d (IC50 of 2.4 μM, Figure 1C, Table S5) showed 3-fold improvement in potency in inhibiting STAT3 DNA-binding activity in vitro, compared to that of BP-1–102 (IC50 6.8 μM). Consistent with what was observed before, the (S)-5c showed lower activity than that of the (R)-5d (IC50 of 7.2 μM vs 2.4 μM, respectively) [Figure 1C]. Interestingly, an isomeric modification of the Pro pyrrolidine ring, such as in compounds 5a and 5b (IC50 6.5–6.9 μM, Table S5, Figure 1C), led to lower potency compared to that of the Pro-linker 5d (IC50 of 2.4 μM) [Figure 1C], but still similar to that of the original lead compound BP-1–102 (IC50 6.8 μM). Unlike alanine, the increase in potency seen in the case of the meta-fluorine analogue 2v (IC50 1.8 μM, Table S5 vs SH4–54, IC50 4.4 μM) was not realized for the Pro-based analogue, 5e (IC50 5.3 μM). Substitution of a chlorine in the 2-position or conversion of the salicylic acid to the benzohydroxamic acid showed no improvement (5k, IC50 3.8 μM; 5l, IC50 5.5 μM). In addition, the substitution of the cyclohexyl group of 5d with a THP provided analogue 5g (IC50 5.4 μM, Table S5), which although showed moderately improved potency in disrupting STAT3 DNA-binding activity in vitro when compared to their parent compound, BP-1–102 (IC50 6.9 μM), it was less potent than 5d. However, further substitutions of the cyclohexane group such as tetrahydrofuran (THF) 5j (IC50 4.5 μM, Table S5) or a cyclopentane 5i (IC50 3.6 μM, Table S5) showed promise in the proline series. Select variations of the pentafluorophenylsulfonyl group in the proline series were not tolerated as observed with the alanine series (Table S3; Table S5, entries 13–16).
On selectivity, we note that select analogues disrupt the DNA-binding activities of both STAT3 and STAT5, but not that of STAT1 in vitro (Supplementary Figure S1). These are discussed in detail in the Supporting Information.
New analogues inhibited constitutive STAT3 activation in cancer cells, with minimal effects on other signaling proteins. Select analogues were investigated for their inhibitory activities against constitutively active STAT3 in tumor cells. The human breast cancer cells, MDA-MB-231, were treated with 10 μM 1e or 1a for 1, 3, or 24 h, or the human breast cancer line, MDA-MB-468, was treated with 5 μM 1a, 2j, 2p, 2k, or 2q for 3 or 24 h. Nuclear extracts were prepared and samples of equal total protein were subjected to STAT3 DNA-binding activity/EMSA analysis to determine effects on intracellular STAT3 activity. Results show variable degrees of inhibition of STAT3 activity, which are cell-type dependent. STAT3 DNA-binding activity in MDA-MB-231 cells was inhibited by both 1e and 1a following 3 h treatment, with little change at 1 h treatment (Supplementary Figure S2A(i)). At the 24 h time point, the treatment with 1e, but not with 1a, continued to inhibit STAT3 DNA-binding activity (Supplementary Figure S2A, 24 h), suggesting the inhibitory effect of 1e is sustained up to 24 h. Moreover, EMSA analysis shows STAT3 DNA-binding activity in MDA-MB-468 cells is inhibited by treatment with 5 μM 1a for 3 or 24 h, 5 μM 2k for 3 h, and with 5 μM 2q for 24 h (Supplementary Figure S2A(ii)). Other compounds tested (2j and 2p) showed minimal effect on STAT3 DNA-binding activity in cells (Supplementary Figure S2A(ii)).
Select analogues were evaluated for their inhibitory effects on pY705STAT3, relative to nonspecific effects on pJak2, pSrc, and pERK1/2 induction in the human breast cancer, MDA-MB-231 and MDA-MB-468 cells, and the metastatic melanoma, C8161, 1205LU, and UACC903 cells. Results, which are discussed in detail in the Supporting Information, show that at concentrations that inhibit pY705STAT3 and STAT3 DNA-binding activity (Supplementary Figure S2), the analogues have little effect on other signaling proteins (Supplementary Figure S3). Altogether, the new analogues show strong and variable inhibitory effects against constitutive STAT3 DNA-binding activity and Tyr phosphorylation in human tumor cells. The variable responses may reflect the complexities in the STAT3 signaling pathway that impacts small molecule inhibitors in cells.19 We note that the Ala analogues 2k and 2q are more potent compared to their Gly-based counterparts, 2j and 2p.
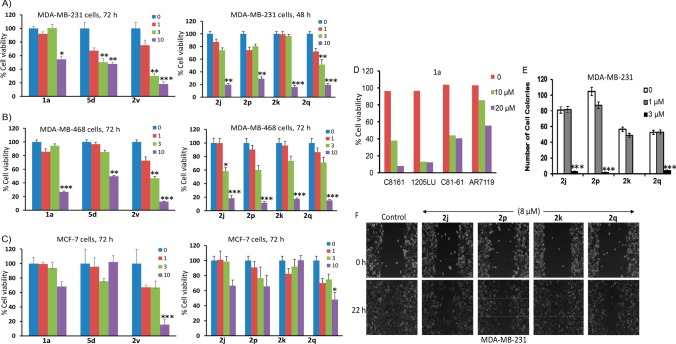
Novel active analogues decreased the growth, colony formation, and migration of human cancer cells harboring persistently active STAT3. Constitutive STAT3 activity promotes tumor cell growth and proliferation and survival.2 We have previously shown that the lead agents SH4–54 and SH5–07 inhibit human tumor cell growth.19 We determined the effects of the new active analogues on tumor cells harboring aberrantly active STAT3. Based on the activity of the compounds in inhibiting STAT3 DNA-binding in vitro, we selected potent analogues for further evaluation of their effects on inhibiting tumor cell phenotype. We note the variable responses of the compounds tested against tumor cells harboring constitutively active STAT3, compared to cells that do not. Treatment with 5d, 2v, 2j, 2p, 2k, and 2q variably inhibited the viability of MDA-MB-231 and MDA-MB-468 cells (Figure 2A,B). These results are consistent with the previous report19 on the lead agents. Analogue 2v further inhibited MDA-MB-231 cell growth (Supplementary Figure S4), which is discussed in detail in the Supporting Information. By contrast, 72 h treatment with 5d, 2v, 2j, 2p, 2k, or 2q of MCF-7 cells that do not harbor aberrantly active STAT3 led to relatively weaker inhibitory effects on cell viability and growth, as measured by CyQuant viability assay (Figure 2C). Overall, 10 μM concentration of these analogues showed relatively stronger effects on MDA-MB-231 and MDA-MB-468, compared to the generally weak-to-moderate effects on cells (MCF-7) that do not harbor constitutively active STAT3 (Figure 2), suggesting preferential effects on STAT3-dependent tumor cells. Intracellular activity overall are moderately improved, which in part reflects the enhancement in physicochemical factors that influence cell membrane permeability, metabolic stability, and hence increased intracellular concentrations. Importantly, the Pro-based analogue, 5d, improved tumor-cell specificity and preferentially inhibited the growth of MDA-MB-231 and MDA-MB-468 human breast cancer cells harboring constitutively active STAT3 over the MCF-7 line that does not.
Figure 2.
Effects of compounds on cell viability, growth, colony survival, and migration in vitro. Human breast cancer MDA-MB-231 (A) and MDA-MB-468 (B) cells harboring aberrantly active STAT3 or MCF-7 cells that do not (C) grow in 96-well plates were treated once with 0–10 μM of the indicated compounds for 72 h and subjected to CyQuant cell proliferation assay for viable cells, which are plotted as % viability against concentration. (D) Cultures of melanoma cells harboring aberrantly active STAT3 (C8161, 1205LU, and C81–61) and counterpart that does not (AR7119) were treated with 1a or 1c at the indicated concentrations for 72 h and subjected to MTT assay for viable cells, which were plotted as % viability. (E) Human breast cancer MDA-MB-231 cells were seeded as single-cell culture and treated once with 0–3 μM of the indicated compounds and allowed to culture until large colonies were visible, which were stained with crystal violet, counted, and plotted. (F) Human breast cancer MDA-MB-231 cells in culture were wounded and treated once with or without 8 μM of the indicated compounds and allowed to migrate to the denuded area over a 23 h period and imaged. Control (0) represents 0.05% DMSO-treated cells. Values, mean ± SD of three independent determinations. Data are representative of three independent determinations. *p < 0.01, **p < 0.005, ***p < 0.001.
Moreover, treatment with 1a for 48 h strongly inhibited the viability of human metastatic melanoma C8161, 1205LU, and C81–61 cell lines that harbor constitutively active STAT3, compared to the relatively weaker activity against the immortalized human normal melanocyte, AR7119 line that does not harbor aberrant STAT3 activation (Figure 2D). We surmise that at concentrations (<10 μM) that inhibit STAT3 activity in tumor cells, the tested compounds showed relative preferential effects against the viability of tumor cells that harbor constitutive STAT3 activation over cells that do not.
Colony survival assay in which single-cell cultures of MDA-MB-231 line were treated once with 0–20 μM 2j, 2p, 2k or 2q and allowed to culture until colonies were visible showed that all four analogues strongly inhibited the formation of colonies at 3 μM, with the minimum effect at 1 μM (Figure 2E, Supplementary Figure S4C). In vitro cell migration scratch assay showed that treatment of MDA-MB-231 cells with 8 μM of the analogues variably suppressed cell migration, with the strongest inhibition caused by 2j, 2p, and 2q, while 2k had moderate to weak effect (Figure 2F).
Initial Pharmacokinetic Evaluation
We were interested in understanding how the physicochemical properties of these compounds influence their pharmacokinetic parameters and initially focused on reducing lipophilicity by introducing heteroatoms at different sites of the pharmacophore structure. For example, replacement of the pentafluorophenyl group with a 3-pyridyl ring (3g, Table S3), replacement of the benzoic acid group with different heterocyclic systems (2b-c, 2e–f, 2zTable S2), modification of the 4-cyclohexylbenzyl group to 4-cyclohexyl-3-pyridylmethyl (4k, Table S4), or the modification of the cyclohexyl ring to 1- and 4-piperidyl systems (4f, 4l, Table S4) were explored, which all gave weaker in vitro STAT3-inhibitory activity compared to BP-1–102. Although the replacement of the cyclohexyl ring with 4-tetrahydropyranyl (THP) also tended to give compounds with relatively lower activity (4a, 4c-e, Table S4), some other THP analogues gave comparable potency with the parent BP-1–102 (e.g., 4b, IC50 5.0 μM, Table S4, vs BP-1–102, IC50 6.8 μM). This finding was exploited to improve the physicochemical properties of the compounds (vide supra). Preliminary in vitro assessment of aqueous solubility, cell membrane permeability (Caco-2 cells assay), and mouse and human liver microsomal (MLM and HLM) metabolic stability studies were conducted on selected analogues and compared to the lead compounds, SH4–54 and SH5–07 (Table S6).
Results show that solubility in both pH 7.4 PBS and pH 6.8 buffered simulated intestinal fluid (SIF, Table S6) was improved by creating sodium salts, as observed for 1b (127 μg/mL both PBS and SIF), 4c (132 and 127 μg/mL, respectively), and 5h (123 and 117 μg/mL, respectively), compared to the lead agent, SH4–54, which had only limited solubility as a carboxylic acid (7.6 and 14 μg/mL, respectively), possibly as a result of enhanced dissolution rate.21 However, the benzohydroxamic acids, SH5–07 and 2w, present good solubility in SIF (74 and 72 μg/mL, respectively), but not in PBS pH 7.4 (0.3 and 4.7 μg/mL, respectively), a result likely due to the emulsifying effects on dissolution at SIF.22
Ala-based analogues show lower metabolic stability compared to Gly-based compounds (e.g., 1b and 4c, RLM/HLM t1/2 ≤ 5 min vs SH4–54 and SH5–07 RLM/HLM t1/2 ≥ 17 min, Table S6). Hypothesizing that the Ala-based compounds are more prone to metabolic N-demethylation,23 Pro-linker compounds were designed with the goal to improve metabolic stability and maintain potency. Indeed, the Pro-based analogues 5f (HLM and MLM t1/2 = 54 and 26 min, respectively) and 5h (HLM and MLM t1/2 = 15 and 18 min, respectively) showed much improved microsomal metabolic stability (Table S6).
Caco-2 cell permeability studies show that incorporation of the THP group instead of the lipophilic cyclohexyl ring gives better permeability (Table S6). For example, whereas compounds containing the lipophilic cyclohexyl group, instead of the THP ring, show no appreciable permeability (SH4–54, SH5–07, and 1b, Table S6), the THP compounds 5f and 5h (Table S6) show detectable apical (A) to basolateral (B) relative permeability (Papp 0.1 and 0.2 × 10–6 cm/s, respectively). Another THP compound, 4c (Table S6), shows increased B to A permeability (Papp 0.7 × 10–6 cm/s). That cell membrane permeability from the Caco-2 assay was notably higher for the analogues that incorporate the THP ring is likely due to the disruption of the amphiphilic character of the molecule, specifically the lipophilic cyclohexylphenyl and perfluorophenyl moieties at two different ends, and the polar carboxyl group at the third other end in the case of the leads BP-1–102, SH4–54, or SH5–07. Taken together, these studies demonstrate significant progress toward developing a more suitable STAT3 inhibitor with drug-like features.
In summary, Ala-linker analogues 1a and 2v and the Pro-based derivative 5d, all with (R)-configuration at the chiral center, had improved inhibitory activity and selectivity against STAT3 DNA-binding activity in vitro, with IC50 of 3.0 ± 0.9, 1.80 ± 0.94, and 2.4 ± 0.2 μM, respectively. All three analogues, 1a, 2v, and 5d, inhibited constitutive STAT3 phosphorylation and activation in human breast cancer and melanoma lines, and blocked tumor cell viability, growth, colony formation, and migration in vitro. However, in general, the (R)-configuration Pro-based analogues showed overall the best profile, including microsomal metabolic stability, Caco-2 permeability, and in particular, 5d showed improved tumor-cell specificity preferentially inhibiting the growth of MDA-MB-231 and MDA-MB-468 human breast cancer cells harboring constitutively active STAT3, with minimal effects at concentrations up to 10 μM on the growth of MCF-7 human breast cancer line that does not harbor persistent STAT3 activity compared to 2v. Additional efforts are currently focused on further optimizing potency and refining the physicochemical properties, thereby further improving the activity and efficacy to ultimately identify suitable STAT3-inhibiting candidates for clinical application.
Acknowledgments
We thank all colleagues and members of our laboratory for the stimulating discussions. Part of the services for this work was provided by the Chemical Biology Core, which is supported in part by grant 4P30CA071789 from the NIH/NCI to the University of Hawaii Cancer Center. This work was supported by the NIH/NCI Grant CA161931 (to J.T.), CA208851 (to J.T.), and University of Hawaii start-up funds.
Glossary
ABBREVIATIONS
- STAT
signal transducer and activator of transcription
- EMSA
electrophoretic mobility shift assay
- MAPK
mitogen-activated protein kinase
- ERK
extracellular signal-regulated kinases.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.7b00544.
Information on compound synthesis, chemical data, experimental procedures for nuclear extract preparation, gel shift assay, immunoblotting analysis, cell proliferation, viability assay, and trypan blue exclusion/phase-contrast microscopy, clonogenic survival assay, and scratch assay (PDF)
Author Contributions
∥ These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Darnell J. E. Validating Stat3 in cancer therapy. Nat. Med. 2005, 11, 595. 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- Yu H.; Jove R. The STATS of Cancer-New molecular targets come of age. Nat. Rev. Cancer 2004, 4, 97. 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- Yue P.; Turkson J. Targeting STAT3 in cancer: how successful are we?. Expert Opin. Invest. Drugs 2009, 18, 45. 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklossy G.; Hilliard T. S.; Turkson J. Therapeutic modulators of STAT signaling for human diseases. Nat. Rev. Drug Discovery 2013, 12, 611. 10.1038/nrd4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkson J. STAT proteins as novel targets for cancer drug discovery. Expert Opin. Ther. Targets 2004, 8, 409. 10.1517/14728222.8.5.409. [DOI] [PubMed] [Google Scholar]
- Bowman T.; Garcia R.; Turkson J.; Jove R. STATs in oncogenesis. Oncogene 2000, 19, 2474. 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Yue P.; Fletcher S.; Zhao W.; Gunning P. T.; Turkson J. A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes. Biochem. Pharmacol. 2010, 79, 1398. 10.1016/j.bcp.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Yue P.; Page B. D.; Li T.; Zhao W.; Namanja A. T.; Paladino D.; Zhao J.; Chen Y.; Gunning P. T.; Turkson J. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 9623. 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkson J.; Ryan D.; Kim J. S.; Zhang Y.; Chen Z.; Haura E.; Laudano A.; Sebti S.; Hamilton A. D.; Jove R. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J. Biol. Chem. 2001, 276, 45443. 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- Chen J.; Bai L.; Bernard D.; Nikolovska-Coleska Z.; Gomez C.; Zhang J.; Yi H.; Wang S. Structure-Based Design of Conformationally Constrained, Cell-Permeable STAT3 Inhibitors. ACS Med. Chem. Lett. 2010, 1, 85. 10.1021/ml100010j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Nikolovska-Coleska Z.; Yang C.-Y.; Gomez C.; Gao W.; Krajewski K.; Jiang S.; Roller P.; Wang S. Design and synthesis of a new, conformationally constrained, macrocyclic small-molecule inhibitor of STAT3 via ’click chemistry’. Bioorg. Med. Chem. Lett. 2007, 17, 3939. 10.1016/j.bmcl.2007.04.096. [DOI] [PubMed] [Google Scholar]
- Zheng H.; Hong H.; Zhang L.; Cai X.; Hu M.; Cai Y.; Zhou B.; Lin J.; Zhao C.; Hu W. Nifuratel, a novel STAT3 inhibitor with potent activity against human gastric cancer cells. Cancer Manage. Res. 2017, 9, 565. 10.2147/CMAR.S146173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosenko I.; Yu Y.; Busker S.; Dyczynski M.; Liu J.; Haraldsson M.; Palm Apergi C.; Helleday T.; Tamm K. P.; Page B. D. G.; Grander D. Identification of novel small molecules that inhibit STAT3-dependent transcription and function. PLoS One 2017, 12, e0178844. 10.1371/journal.pone.0178844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath B.; Xu S.; Neamati N. Small molecule inhibitors of signal transducer and activator of transcription 3 (Stat3) protein. J. Med. Chem. 2012, 55, 6645. 10.1021/jm300207s. [DOI] [PubMed] [Google Scholar]
- Oh D. Y.; Lee S. H.; Han S. W.; Kim M. J.; Kim T. M.; Kim T. Y.; Heo D. S.; Yuasa M.; Yanagihara Y.; Bang Y. J. Phase I study of OPB-31121, an Oral STAT3 Inhibitor, in Patients with Advanced Solid Tumors. Cancer Res. Treat. 2015, 47, 607. 10.4143/crt.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendell J. C.; Hong D. S.; Burris H. A. 3rd; Naing A.; Jones S. F.; Falchook G.; Bricmont P.; Elekes A.; Rock E. P.; Kurzrock R. Phase 1, open-label, dose-escalation, and pharmacokinetic study of STAT3 inhibitor OPB-31121 in subjects with advanced solid tumors. Cancer Chemother. Pharmacol. 2014, 74, 125. 10.1007/s00280-014-2480-2. [DOI] [PubMed] [Google Scholar]
- Xiao H.; Bid H. K.; Jou D.; Wu X.; Yu W.; Li C.; Houghton P. J.; Lin J. A novel small molecular STAT3 inhibitor, LY5, inhibits cell viability, cell migration, and angiogenesis in medulloblastoma cells. J. Biol. Chem. 2015, 290, 3418. 10.1074/jbc.M114.616748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W.; Li C.; Zhang W.; Xia Y.; Li S.; Lin J. Y.; Yu K.; Liu M.; Yang L.; Luo J.; Chen Y.; Sun H.; Kong L. Discovery of an Orally Selective Inhibitor of Signal Transducer and Activator of Transcription 3 Using Advanced Multiple Ligand Simultaneous Docking. J. Med. Chem. 2017, 60, 2718. 10.1021/acs.jmedchem.6b01489. [DOI] [PubMed] [Google Scholar]
- Yue P.; Lopez-Tapia F.; Paladino D.; Li Y.; Chen C.-H.; Namanja A. T.; Hilliard T.; Chen Y.; Tius M.; Turkson J. Hydroxamic acid and benzoic acid-based Stat3 inhibitors suppress human glioma and breast cancer phenotypes in vitro and in vivo. Cancer Res. 2016, 76, 652. 10.1158/0008-5472.CAN-14-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Sun Y.; Pireddu R.; Yang H.; Urlam M. K.; Lawrence H. R.; Guida W. C.; Lawrence N. J.; Sebti S. M. A novel inhibitor of STAT3 homodimerization selectively suppresses STAT3 activity and malignant transformation. Cancer Res. 2013, 73, 1922. 10.1158/0008-5472.CAN-12-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serajuddin A. T. Salt formation to improve drug solubility. Adv. Drug Delivery Rev. 2007, 59, 603. 10.1016/j.addr.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Klein S. The use of biorelevant dissolution media to forecast the in vivo performance of a drug. AAPS J. 2010, 12, 397. 10.1208/s12248-010-9203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot M. J.; Ackland M. J.; Horne V. A.; Alex A. A.; Jones B. C. A novel approach to predicting P450 mediated drug metabolism. CYP2D6 catalyzed N-dealkylation reactions and qualitative metabolite predictions using a combined protein and pharmacophore model for CYP2D6. J. Med. Chem. 1999, 42, 4062. 10.1021/jm991058v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.