Abstract
Recently, the binding kinetics of a ligand–target interaction, such as the residence time of a small molecule on its protein target, are seen as increasingly important for drug efficacy. Here, we investigate these concepts to explain binding and proton blockage of rimantadine variants bearing progressively larger alkyl groups to influenza A virus M2 wild type (WT) and M2 S31N protein proton channel. We showed that resistance of M2 S31N to rimantadine analogues compared to M2 WT resulted from their higher koff rates compared to the kon rates according to electrophysiology (EP) measurements. This is due to the fact that, in M2 S31N, the loss of the V27 pocket for the adamantyl cage resulted in low residence time inside the M2 pore. Both rimantadine enantiomers have similar channel blockage and binding kon and koff against M2 WT. To compare the potency between the rimantadine variants against M2, we applied approaches using different mimicry of M2, i.e., isothermal titration calorimetry and molecular dynamics simulation, EP, and antiviral assays. It was also shown that a small change in an amino acid at site 28 of M2 WT, which does not line the pore, seriously affects M2 WT blockage kinetics.
Keywords: Influenza M2, S31N mutation, rimantadine, rimantadine enantiomers, isothermal titration calorimetry, electrophysiology, binding kinetics, synthesis, antiviral assay, molecular dynamic simulations
Novel approaches are necessary in early drug discovery for optimal drug design and improved therapy. Recently, the kinetics of a ligand–target interaction, such as the residence time of a small molecule on its protein target, are seen as increasingly important for in vivo efficacy and safety.1
The antiviral agents amantadine (1) and rimantadine (2) (Scheme 1) are well-established to be blockers of proton transport by the influenza A virus (IAV).2,3 The primary binding site of 1 and 2 is the transmembrane domain lumen (TM, amino acids 22–46) in the four-helix bundle of tetrameric M2, which forms the proton transport path.2 Since 2008, high-resolution structures have become available for complexes of M2TM wild type (WT) with 1 or 2 (Figure S1).4−9
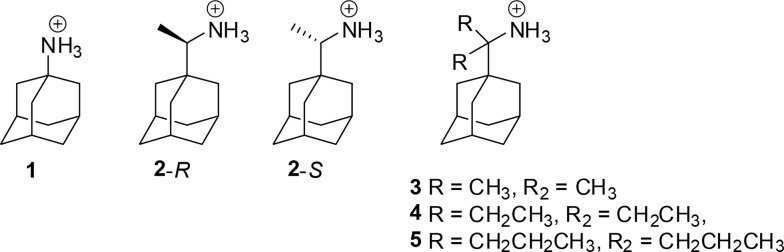
Scheme 1. Structures of Aminoadamantane Derivatives 1–5.
Compounds 1 and 2 are effective prophylactics and therapeutics against IAVs, provided they contain the M2TM WT such as A/Udorn/72 H3N2 (Udorn) and A/Hong Kong/68 H3N2 (HK), but not those containing M2 S31N such as A/WSN/33 H1N1 (WSN) (Figure S2). Since 2005, the amantadine (1)-insensitive Ser-to-Asn mutation at position 31 in M2 (S31N) has become globally prevalent, abrogating the clinical usefulness of 1.10
Compound 2 is ranked among the best binders to M2TM WT11,12 and most potent anti-IAV agents among the aminoadamantane derivatives.17,18 Thus, the synthesis of symmetrical analogues of 2 with the addition of two methyl (3), ethyl (4), and n-propyl (5) groups on the carbon bridge was accomplished (Scheme 1) aiming at filling progressively from 3 to 5 the extra space between the ligand and the walls in M2 WT or M2 S31N with a few alkyl groups. Binding affinities of 1, 2, 2-R, 2-S, and 3–5 were measured by ITC against the M2TM WT and its S31N variant in their closed form at pH 8. Furthermore, we measured the antiviral activity of the rimantadine analogues against IAV strains and the blocking effect of the compounds against full length Udorn M2, Udorn M2 S31N, and Udorn M2 V28I using electrophysiology (EP), and the kinetics of binding were compared. Molecular dynamics (MD) simulations of ligand binding to M2TM WT and its S31N variant in their closed form were performed for investigation of the binding mode interactions.
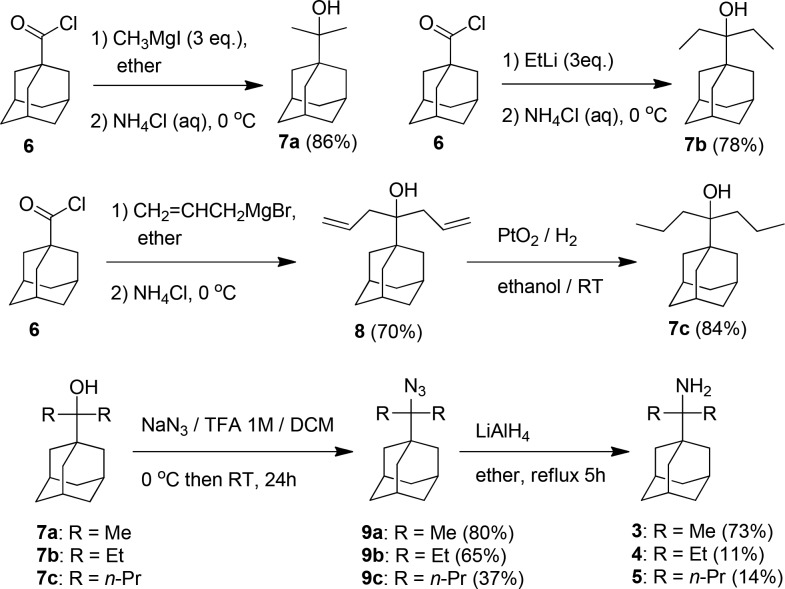
For the synthesis of primary tert-alkyl amines 3–5, the raw tert-alkyl alcohols 7a–c were prepared according to Scheme 2 from the reaction between 1-adamantanecarbonyl chloride 6 and an organometallic reagent (see Supporting Information). It was reported that the reaction of 1-adamantanecarbonitrile with ethyl magnesium bromide and titanium tetraisopropoxide afforded 4 in 52% yield.19 We tested twice this procedure with 1-adamantanecarbonitrile and methylmagnesium bromide yielding in our hands 3 with only 10% yield, which is lower than the ∼50% yield starting from 6 or 1-adamantanecarboxylic acid (Scheme 2).
Scheme 2. Synthetic Scheme for the Preparation of Compounds 3–5.
Table 1 includes thermodynamic parameters of binding against M2TM WT and M2TM S31N. Binding affinities were determined by ITC for M2TM–ligand systems in dodecylphosphocholine (DPC) micelles at pH 8, where M2TM fragments form stable tetramers (see also Supporting Information).20 Compound 1 has a Kd of 2.17 μM. As depicted in Table 1, enantiomers 2-R and 2-S have the same Kd values21 against M2TM WT (Kd = 0.34 and 0.32 μM, respectively). Compound 3, having two methyl groups instead of one methyl group in 2, has the smallest Kd = 0.13 μM, i.e., the highest binding affinity of all studied aminoadamantane compounds, suggesting that polar and lipophilic characteristics are well balanced in its structure. The diethyl derivative 4 and dipropyl derivative 5 exhibit lower binding affinities against M2TM WT (Kd = 4.59 and 3.43 μM, respectively). A balance between enthalpy and entropy determines the free energy of binding as shown in Table 1. The entropy presumably changes significantly from 1, 2 to 3, 4 on binding because the ordered clathrate water surrounding the ligand is dispersed as the ligand enters the water-poor channel cavity. This is more prominent for 3 and 4, as expected due to their larger hydrophobic surfaces. Presumably, it would have gone up even more for 5, but this clathrate effect was probably countered by a reduced ligand entropy in the channel due to restricted rotation inside the receptor binding area. Compounds 1–3 did not bind efficiently to M2TM S31N according to isothermal titration calorimetry (ITC) and previous surface plasmon resonance measurements for 1,22 while 5, with a larger adduct connected to adamantane, binds weakly to M2TM S31N compared to M2TM WT according to ITC.
Table 1. Binding Constant, Free Energy, Enthalpy, and Entropy of Binding at 300 K Derived from ITC Measurements for M2TM WT (from Udorn, Upper Table) and the M2TM S31N (Lower Table).
| liganda | Kdb | ΔGc | ΔHd | –TΔSe |
|---|---|---|---|---|
| M2TM WT | ||||
| 1 | 2.17 ± 0.52 | –7.77 ± 0.14 | –6.66 ± 0.50 | –1.11 ± 0.52 |
| 2 | 0.51 ± 0.26 | –8.64 ± 0.30 | –7.60 ± 0.28 | –1.04 ± 0.41 |
| 2-R | 0.32 ± 0.16 | –8.97 ± 0.26 | –7.54 ± 0.34 | –1.42 ± 0.43 |
| 2-S | 0.34 ± 0.12 | –8.88 ± 0.21 | –7.73 ± 0.28 | –1.15 ± 0.35 |
| 3 | 0.13 ± 0.12 | –9.30 ± 0.43 | –4.19 ± 0.28 | –5.12 ± 0.51 |
| 4 | 4.59 ± 2.21 | –7.33 ± 0.28 | –3.29 ± 0.62 | –4.03 ± 0.68 |
| 5 | 3.43 ± 1.05 | –7.50 ± 0.18 | –6.23 ± 0.45 | –1.27 ± 0.48 |
| M2TM S31N | ||||
| 1–3 | f | f | f | f |
| 5 | >10 | f | f | f |
Binding constant Kd in μM 2.
Free energy of binding in kcal mol–1.
Binding enthalpy in kcal mol–1.
Entropy of binding in kcal mol–1.
Values could not be determined reliably due to the limitations of the methods in the area of very weak binding (see also SI for definition of quantities).
The cytopathic effect (CPE) inhibition assay was used23 to compare the antiviral potency of 1–5 against HK, Udorn, WSN, and WSN M2 N31S (generated by reverse genetics from WSN) in MDCK cells (Table 2). The amino acid sequences of M2 WT in Udorn and HK are identical, not just in the TM region but in the full length protein. There was no potency against the amantadine-resistant WSN with the compound concentrations used. All compounds showed low micromolar activity against Udorn, HK, and WSN M2 N31S with 3 being the most potent agent exhibiting submicromolar potency. Inhibition of replication of Udorn was further confirmed with plaque-reduction assay (results not shown). It is of note that 5 only reduced the plaque size but not the number of plaques. The cytotoxicity data (Table 2) showed that 1–4 are nontoxic with CC50 values >100 μM, but 5 is mildly toxic with CC50 ≈ 57 μM.
Table 2. Cytotoxicity (CC50) and Antiviral Activity (EC50) of Compounds 1–5 against IAVs HK, Udorn, WSN, and WSN M2 N31S in Madin–Darby Canine Kidney Cells.
| EC50 (μM)a |
|||||
|---|---|---|---|---|---|
| HK | Udorn | WSN |
|||
| ligand | M2 (V28; S31) | M2 (V28; S31) | M2 (I28; N31S) | M2 (I28; N31) | CC50 (μM)a |
| 1 | NDc | 0.78 ± 0.44 | 0.48 ± 0.05 | >100 | >100 |
| 2 | 0.05 ± 0.04 | 0.09 ± 0.03 | 0.04 ± 0.02 | >100 | >100 |
| 2-R | NDc | 0.05 ± 0.01 | 0.04 ± 0.01 | >100 | >100 |
| 2-S | NDc | 0.06 ± 0.02 | 0.02 ± 0.01 | >100 | >100 |
| 3 | 0.012 ± 0.003 | 0.01 ± 0.001 | 0.03 ± 0.02 | >100 | >100 |
| 4 | 0.46 ± 0.25 | 0.41 ± 0.23 | 1.01 ± 0.13 | >100 | >100 |
| 5 | 0.45 ± 0.34 | 1.07 ± 0.31b | 1.06 ± 0.23 | >100 | 57.3 ± 11.3 |
| Oseltamivir | 0.002 ± 0.001 | 0.001 | 0.02 ± 0.01 | 0.03 ± 0.01 | >100 |
Mean and standard deviations of the 50% inhibitory concentration (EC50) and the 50% cytotoxic concentration (CC50) of at least three independent assays.
Inhibition of plaque size without reduction of plaque number.
ND: Not determined.
The EC50 values for 1–5 (Table 2) prioritize the same derivative for M2 WT virus inhibition, i.e., 3, in agreement with the results from the Kd values from ITC experiments based on M2TM WT binding (Table 1). Compound 3 is almost equal in structure with rimantadine (2) without having a chiral center. Compound 3 has also a promising selectivity index based on the in vitro cytotoxicity data.
The inhibitors were tested with a two-electrode voltage clamp (TEVC) assay using X. laevis frog oocytes microinjected with RNA expressing the M2 protein as in previous reports.17,21 The blocking effect of the aminoadamantane derivatives against M2 was investigated with EP experiments using Udorn M2 and Udorn M2 S31N. Because WSN has the V28I substitution in M2, Udorn M2 V28I was generated and studied in parallel to examine whether small changes in WSN in the side chains of amino acids that do not line the pore (Figure S2) affect aminoadamantane blocking properties. The blocking effect of the inhibitors was expressed as the inhibition percentage of the M2 current observed after 2, 5, and/or 10 min of incubation with 100 μM compound (Tables 3 and S1).
Table 3. Blocka of Inward Currents in Oocytesb Transfected with Full-Length Udorn M2 by Selected Compounds.
| ligand | Udorn M2a,b |
||||
|---|---|---|---|---|---|
| % block (2 min) | % block (5 min) | % block (10 min) | IC50 (2 min) (μM) | IC50 (5 min) (μM) | |
| 1 | 90 ± 2 | 95 ± 1 | NDc | 12.5 | 4.7 |
| 2 | 96 ± 1 | 96 ± 1 | NDc | 10.8 | NDc |
| 2-R | 95 ± 1 | 96 ± 1 | NDc | NDc | NDc |
| 2-S | 93 ± 1 | 95 ± 1 | NDc | NDc | NDc |
| 3 | 90 ± 2 | 96 ± 1 | NDc | 9.3 | 4.0 |
| 4 | 78 ± 2 | 91 ± 1 | NDc | 24.3 | 13.2 |
| 5 | 27 ± 1.2 | 38 ± 1.6 | 61 ± 2.3 | NDc | NDc |
| ligand | kon (M–1 s–1) | koff (s–1) | Kd (μM)d |
|---|---|---|---|
| 1 | 327 | 0.003 | 9 |
| 2 | 416 | 0.003 | 7 |
| 2-R | 412 | 0.0013 | 3.2 |
| 2-S | 407 | 0.0016 | 3.9 |
| 3 | 230 | 0.003 | 13 |
| 4 | NDc | NDc | NDc |
| 5 | 34 | 0.003 | 88 |
For each compound, percent block of pH-dependent M2 current at listed concentrations (±SEM) and IC50 (μM) are shown.
Three replicates were used for measurements at 100 μM.
ND: Not determined.
Kd (TEVC) = koff/kon.
After 5 min, 3 and 4 block Udorn M2 and Udorn M2 V28I as well as 1 (about 90% and 80%, respectively). Generally, after 2 and 5 min, the percentage of current inhibition was progressively increased for 3 and 4. It is noteworthy that 5 against Udorn M2 exhibited 27% blocking at 2 min, 38% at 5 min, and 61% at 10 min (Table 3). The IC50 values of 3 and 4 for Udorn M2 and Udorn M2 V28I were reduced from 2 to 5 min time points (Tables 3 and S1). These measurements at 2, 5, or 10 min are made prior to the establishment of equilibrium25 due to very slow on- and off-rates for entry (see the kon and koff rate values in Tables 3 and 4), especially of the bulky ligands like 5, together with the difficulty of maintaining cells at low pH for extended periods. Thus, the very slow binding of 5 (Table 3) should not be viewed as inconsistent with the high antiviral potency (submicromolar EC50) against WT (V28; S31) viruses (Table 2), the latter representing much longer exposure times than EP experiments.
Table 4. Block of Full-Length Udorn M2 S31N-Dependent Current 2 by Selected Compoundsa,b.
| Udorn
M2 S31N |
||||||
|---|---|---|---|---|---|---|
| ligand | % block after 2 min | % block after 5 min | % block after 10 min | kon (M–1 s–1) | koff (s–1) | Kde |
| 1 | 35 ± 2 | 36 ± 1 | 36.3 ± 1 | 143 | 0.03 | 210 μM |
| 2c | 1.0 ± 0.2 | 1.5 ± 0.4 | NDd | 22 | 0.9 | >10 mM |
| 3 | 21 ± 2 | 30 ± 3 | 33 ± 1 | 18 | 0.008 | 444 μM |
| 5 | 7.0 ± 0.4 | 7.6 ± 0.2 | 8.0 ± 0.4 | 79 | 0.14 | 1.8 mM |
For each compound, percent block of pH-dependent M2 current at listed concentrations (±SEM) and IC50 (μM) are shown.
Three replicates were used for measurements at 100 μM.
Racemic.
ND: Not determined.
Kd = koff/kon.
In a very recent paper,24 the authors showed that when Kd(TEVC) = koff/kon was smaller than a threshold, an in vitro antiviral activity was exhibited. For amantadine (1), koff/kon = 10–6 M (1 μM) was measured against M2 WT, which correlates with good in vitro antiviral potency. When koff/kon ∼100 μM or higher, antiviral potency was not observed even for quick binders. For example, 4-(2-adamantyl)piperidine (compound 3 in ref (24)), although a quick blocker against Udorn M2 and the amantadine resistant Udorn M2 V27A, was ineffective against the corresponding influenza A strains. The authors also showed that 2-(1-adamantyl)piperidine (compound 8 in ref (24)) was a slow binder against Udorn M2 (48% at 2 min, but 90% at 6 min) but still has good antiviral efficacy, possibly because, although kon is low, koff is really low. This is the case with compound 5 against Udorn M2 WT in the present study (see Table 3). It has a reduced onset of block compared to 1, 2, and 3, but also has a low dissociation rate constant, so it still has micromolar efficacy against infections of cell cultures by viral strains with M2 WT.
In studies focusing on the development of aminoadamantane ligands against IAV, derivatives are often initially tested in TEVC assays at 100 μM concentration at 2 min, and only the most potent compounds are then tested using whole cell assays.25 If the same procedure had been applied here, 5 would not have been tested, even though it proved to be a low micromolar inhibitor according to Kd values from ITC experiments with M2TM WT (Table 1) and CPE assay (Table 2) results. Thus, TEVC percent block for 100 μM at 2 min in MDCK cells underestimated the potential of 5. Similarly 4 would not have been tested based on percent block at 2 min in TEVC with Udorn M2 V28I (Table S1). Slow block could be associated with tight block, and this phenomenon should not be overlooked in short-lasting experiments. The results suggested that TEVC results, when used for compound filtering, need careful interpretation for compounds having low association rate constant for binding to the full length M2, which also depends on the M2 pore. In this regard, ITC measurements represent an important additional tool for clarifying the binding energies of novel derivatives to M2TM given their capacity for sufficient relaxations of equilibrium between titration injections. Nevertheless, it is clear from the ITC, CPE (all strains with M2 WT), that 5 (and 4 where tested) are ∼10-fold less active than 3.
For Udorn M2 V28I, the percentage of current inhibition is lower; compounds 3 and 4 inhibit Udorn M2 more rapidly than Udorn M2 V28I (Table S1). A small change in an amino acid at site 28 (V28I) of M2, which does not line the pore, seriously affects M2 blockage kinetics. The inhibition of 3 and 4 on both Udorn M2 and Udorn M2 V28I are irreversible in our experimental time frame, as was also observed for 1 with both proteins (data not shown).
After the aforementioned results highlighted the importance of kon, koff values on ligand behavior, we were intrigued to further investigate the block the block of rimantadine enantiomers against Udorn M2 WT protein in EP, considering the differences in resonances seen in ssNMR studies of 2-R or 2-S bound to the full-length Udorn M2 protein.26 We previously showed21 that 2-R and 2-S showed similar channel blockage against Udorn M2 WT when tested in EP at 100 μM at the 2- or 5 min time point, and this result is consistent with that from ITC measurements (see ref (21) and Table 1) and antiviral assays (see ref (21) and Table 2). In our present work we seek to further investigate the binding kinetics 2-R, 2-S by measuring the respective kon, koff, Kd (TEVC) values. The EP measurements showed a koff = 0.0013 s–1 for 2-R and a koff = 0.0016 s–1 for 2-S (Table 3), i.e., the two enantiomers had very similar binding kinetics. Thus, 2-R has a bit longer residence time inside the receptor than 2-S, as reflected by its slightly lower koff and Kd values (2.4 vs 3.2 μM). In ref (26), the first ssNMR study of the full length M2 in complex with rimantadine enantiomers was published. Compound 2-R was argued to have a higher affinity than 2-S based on differences in peak intensities and position restrained MD simulations. The results published in ref (26) are in qualitative agreement with those reported here, but not in quantitative agreement, as here we see no statistically significant (ITC and EC50) or meaningful (EP) difference. Perhaps this is a consequence of the different methodologies applied, i.e., EP vs ssNMR spectroscopy. Chemical shifts differences and peak intensities do not provide an accurate quantitative estimate of binding affinity values. The EP results, antiviral assays, and ITC results showed clearly that the two rimantadine enantiomers have similar binding free energies, channel blockage, kon and koff rate constants, and antiviral potencies. We conclude that they form equally stable complexes and have the same residence time inside M2 WT.
The compounds did not bind to Udorn M2TM S31N according to ITC and did not exhibit antiviral potency against WSN virus, which contains both the S31N and the V28I mutations. We showed that a valuable parameter to explain the resistance of M2 S31N viruses to rimantadine analogues compared to M2 WT is a higher koff rate (i.e., a smaller residence time inside M2 S31N). According to our previous results, this is due to the fact that, in M2 S31N, the loss of the V27 pocket for the adamantyl cage11 resulted in low residence time inside M2TM and a lack of antiviral potency; but for 5, the sizable adducts resulted in a weak binding, which is albeit not sufficient for antiviral potency.11 It is the high dissociation rate constants that render aminoadamantanes useless against S31N viruses like WSN leading to Kd (TEVC) in the millimolar range compared to the micromolar range for M2 WT binding.
In the S31N variants, TEVC (Table 4) shows very high exit rate constants, especially for 2 (0.9 s–1) and 5 (0.14 s–1), consistent with the unmeasurably high Kd in ITC (Table 1, lower) and EC50 in CPE (Table 2, WSN). Interestingly, in these two cases, 2 and 5 have low % block of inward currents at 2, 5, and 10 min in Udorn M2 S31N (Table 4), and somewhat similar kon rates to 1 and 3, albeit lower compared to M2 WT, (Table 3), demonstrating that mutations can have complex, ligand-dependent effects on entry and exit rates.
M2TM WT complexes were simulated using an experimental structure of M2TM WT (PDB ID 2KQT(4,8)) determined at pH 7.5 in the presence of 1 (see ref (11) for details). No significant differences in measures were detected between trajectories with production times of 4 and 80 ns (Tables S2 and S3). To ensure that the measures were meaningful, the equilibration of the membranes was tested. To verify this, the average area per lipid headgroup was measured in the simulation of the different lipids and compared with experimental results.27 The calculated values approached the experimental ones of pure lipid bilayers (see Figure S4 and Table S5). The center of mass between the four V27 residues and the adamantane cage of the ligand stabilized as explained in Figure 1 varies between 4.1 and 4.5 Å on average (Tables S2 and S3). Hydrogen bond interactions for 2-R and 2-S and geometric measures, which reflect van der Waals contacts, were found to be similar for the two enantiomers, suggesting equal binding interactions as previously discussed (Figure S3).21 In diethyl and di-n-propyl derivatives (4 and 5), the alkyl groups seem to better fill the space between the ligand and the pore walls; but in these cases, restricted motion and the resulting entropy cost of binding may be significant and decrease the binding affinities compared to 3 (Tables 1–3 and S1). Configurations from the simulations of ligands 3 and 5 are depicted in Figure 1. In all cases, in the region located above the adamantane core (i.e., toward the N-terminus) no water molecules were found, which is consistent with the proton blocking effect of the aminoadamantane derivatives.2,11,26,28
Figure 1.
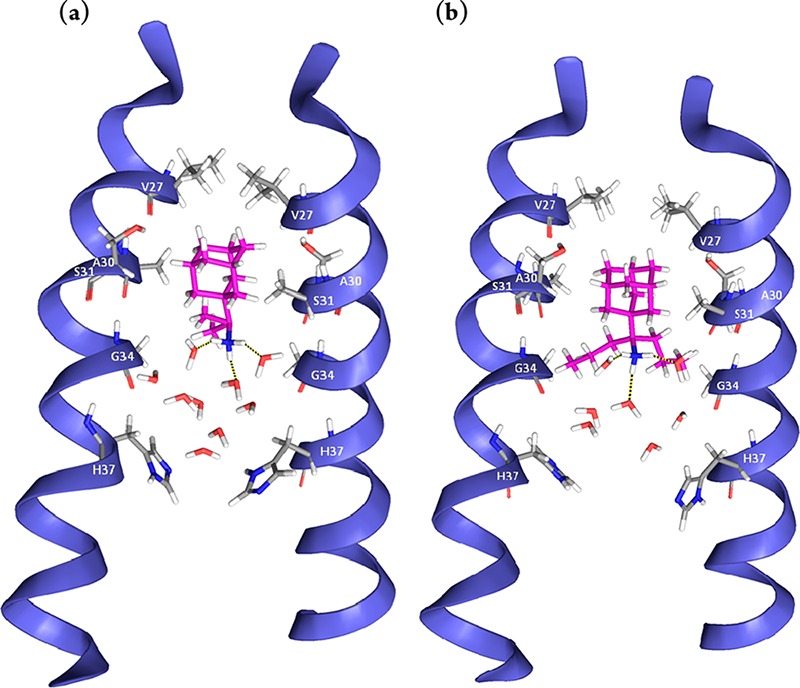
Representative snapshots from the simulation of ligands 3 (a) and 5 (b) bound to M2TM WT. Nine and seven water molecules are shown between the ligand and H37 residues for 3 and 5, respectively. Three hydrogen bonds between the ammonium group of the ligand and three water molecules are shown. Hydrogen bonding with water molecules and van der Waals interactions of the adamantane core with V27 and A30 side chains stabilize the ligand inside the pore with its ammonium group oriented towards the C-terminus of the channel.
The MD simulations of the complex of 3 or 5 with M2TM S31N showed that the ligand cannot bind tightly to M2TM S31N because significant favorable van der Waals interactions are missing (Figure 2). The S31N mutation of M2TM results in a shift of the hydrophobic adamantyl ring toward the C-terminus, due to the enhanced repulsive forces of the asparagine amide side chains to the adamantyl ring and attraction to water molecules. As a consequence, the stabilizing hydrophobic interactions of the V27 isopropyl groups with the adamantyl ring that are present in the M2TM WT are lost in M2TM S31N.11 The bulky Val27 and N31 side chains are oriented toward the N-terminus the latter forming hydrogen bonding interactions with water molecules; the ammonium group of the ligands are also turned toward the N-terminus, allowing significant hydrogen bonding interactions with the polar N31 side chains and the nearby water molecules.11 The hydrogen bonding interactions with N31 are consistent with our magic angle spinning (MAS) experimental data for spiro[pyrrolidine-2,2′-adamantane]–M2TM S31N complex. With the adamantane compound present, there was a chemical shift perturbation for N31 and G34 compared to the apo M2TM S31N.11 In the M2TM WT, the adamantyl ring is well accommodated by the V27 and A30 side chains, and sizable adducts such as ligands 4 and 5 additionally fill the region between A30 and G34 (Figure 1); but in M2TM S31N, the adamantyl ring is between A30 and G34 (Figure 2), due to the lack of a favorable hydrophobic pocket. We have also found11 an absence of chemical shift perturbations for V27 in M2TM S31N in presence of a bulky ligand, in comparison with the apo M2TM S31N, contrasting with the significant chemical shift changes at V27, S31, and G34 relative to the apo state reported when rimantadine is bound to M2TM WT.21,26
Figure 2.
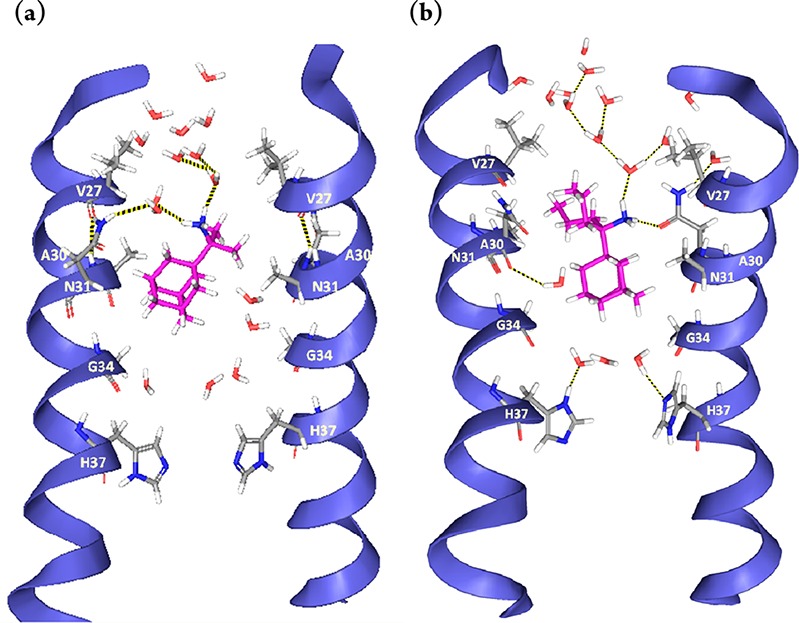
Representative snapshots from the simulations of ligand 3 (a) and 5 (b) bound to M2TM S31N. Five and three water molecules are shown between ligand 3 and 5, respectively, and the H37 residues and ten and twelve water molecules, respectively, between N31 and the mouth of the pore. The ammonium group of the ligand is oriented towards the N-terminus, where it forms hydrogen bonds with water molecules or the carbonyl group of the N31 amide side chain. The loss of the V27 pocket for the adamantyl cage would be expected to lead to weak binding of aminoadamantane ligands.
Compound 5 has sizeable adducts in addition to the adamantyl ring that can fill the region between A30 and G34, and the interactions needed for binding are slightly improved, resulting in weak binding to M2TM S31N according to ITC compared to no binding for 1–3 (Table 1). This can be observed from the snapshot for the complex of 5 with M2TM S31N in Figure 2b. The results showed no binding of 1 and similar in size analogues to M2 S31N and M2TM S31N, and only possible weak binding for sizable adducts in the region between A30 and G34, which is reflected by the high koff values. The last point is in agreement with the results from MAS and OS ssNMR spectra.11
In summary, in this work, we compared the potency between the rimantadine analogues against M2 using four different M2 mimicry methods, i.e., ITC, MD simulations, EP, and antiviral assays. We investigated the binding kinetics of rimantadine analogues with M2 WT and M2 S31N and how they influenced the outcome of potency. We provided a kinetic perspective to explain rimantadine variant binding, proton transport blockage, and antiviral potency against influenza M2 WT and M2 S31N. According to this study, aminoadamantane variants bearing a polar head should exhibit a kinetic profile of small koff rates (i.e., long residence time inside the M2 S31N protein channel pore) resulting in Kd = koff/kon values at the low micromolar region, for them to exhibit inhibitory potency against M2 S31N protein.29
Acknowledgments
We are grateful to Chiesi Hellas for supporting this research and the Ph.D. work of C.T.; J.W. thanks the support from NIH AI119187 and PhRMA Foundation 2015 Research Starter Grant in Pharmacology and Toxicology; A.H. and M.S. thank Andreas Sauerbrei for continuous kind support; A.K. and M.S. thank Professor Adolfo Garcia-Sastre group and the CEIRS program (NIAD Centers of Excellence for Influenza Research and Surveillance) for providing the Udorn virus.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.7b00458.
Experimental and MD simulations additional material (PDF)
Author Present Address
A.D.: Pharmaceutical and Medicinal Chemistry, Institute of Pharmacy and Food Chemistry, Julius-Maximilians-Universität Würzburg, Am Hubland, 97074 Würzburg, Germany.
Author Contributions
A.K. designed this research project. A.D. and C.T. contributed equally. A.D. and C.T. did ligand synthesis. C.T., A.D., Ath.K., and D.K. did the MD simulations. C.M. in J.W. group did the EP experiments against the Udorn M2 and WSN M2 N31S and the plague reduction assay against Udorn virus. K.M. in D.B. group did the EP experiments against the Udorn M2 and kinetic measurements. K.F. and J.H. in G.G. group did the ITC measurements. A.H. in the M.S. group performed CPE inhibitory assays with WSN M2 N31S and HK virus and plague reduction assay against Udorn virus as well as the cytotoxicity determinations. A.K. wrote the manuscript, and J.W., M.S., A.H., A.D., and D.B. revised it. This research includes part of the Master thesis work of A.D. and part of the Ph.D. work of C.T.
The authors declare no competing financial interest.
Supplementary Material
References
- Ferruz N.; De Fabritiis G. Binding Kinetics in Drug Discovery. Mol. Inf. 2016, 35, 216–226. 10.1002/minf.201501018. [DOI] [PubMed] [Google Scholar]
- Wang C.; Takeuchi K.; Pinto L. H.; Lamb R. A. Ion Channel Activity of Influenza A Virus M2 Protein: Characterization of the Amantadine Block. J. Virol. 1993, 67, 5585–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden F. G. Clinical Applications of Antiviral Agents for Chemophrophylaxis and Therapy of Respiratory Viral Infections. Antiviral Res. 1985, 5, 229–239. 10.1016/S0166-3542(85)80033-4. [DOI] [PubMed] [Google Scholar]
- Hu J.; Asbury T.; Achuthan S.; Li C.; Bertram R.; Quine J. R.; Fu R.; Cross T. A. Backbone Structure of the Amantadine-Blocked Trans-Membrane Domain M2 Proton Channel from Influenza A Virus. Biophys. J. 2007, 92, 4335–4343. 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell J. R.; Chou J. J. Structure and Mechanism of the M2 Proton Channel of Influenza A Virus. Nature 2008, 451, 591–595. 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer A. L.; Acharya R.; Salom D.; Levine A. S.; Di Costanzo L.; Soto C. S.; Tereshko V.; Nanda V.; Stayrook S.; DeGrado W. F. Structural Basis for the Function and Inhibition of an Influenza Virus Proton Channel. Nature 2008, 451, 596–599. 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady S. D.; Mishanina T. V.; Hong M. Structure of Amantadine-Bound M2 Transmembrane Peptide of Influenza A in Lipid Bilayers from Magic-Angle-Spinning Solid-State NMR: The Role of Ser31 in Amantadine Binding. J. Mol. Biol. 2009, 385, 1127–1141. 10.1016/j.jmb.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady S. D.; Schmidt-Rohr K.; Wang J.; Soto C. S.; DeGrado W. F.; Hong M. Structure of the Amantadine Binding Site of Influenza M2 Proton Channels in Lipid Bilayers. Nature 2010, 463, 689–692. 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady S. D.; Wang J.; Wu Y.; Degrado W. F.; Hong M. Specific Binding of Adamantane Drugs and Direction of Their Polar Amines in the Pore of the Influenza M2 Transmembrane Domain in Lipid Bilayers and Dodecylphosphocholine Micelles Determined by NMR Spectroscopy. J. Am. Chem. Soc. 2011, 133, 4274–4284. 10.1021/ja102581n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High Levels of Adamantane Resistance among Influenza A (H3N2) Viruses and Interim Guidelines for Use of Antiviral Agents--United States, 2005–06 Influenza Season. MMWR. Morb. Mortal. Wkly. Rep. 2006, 55, 44–46. [PubMed] [Google Scholar]
- Tzitzoglaki C.; Wright A.; Freudenberger K.; Hoffmann A.; Tietjen I.; Stylianakis I.; Kolarov F.; Fedida D.; Schmidtke M.; Gauglitz G.; Cross T. A.; Kolocouris A. Binding and Proton Blockage by Amantadine Variants of the Influenza M2 WT and M2 S31N Explained. J. Med. Chem. 2017, 60, 1716–1733. 10.1021/acs.jmedchem.6b01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis H.; Drakopoulos A.; Tzitzoglaki C.; Homeyer N.; Kolarov F.; Gkeka P.; Freudenberger K.; Liolios C.; Gauglitz G.; Cournia Z.; Gohlke H.; Kolocouris A. Alchemical Free Energy Calculations and Isothermal Titration Calorimetry Measurements of Aminoadamantanes Bound to the Closed State of Influenza A/M2TM. J. Chem. Inf. Model. 2016, 56, 862–876. 10.1021/acs.jcim.6b00079. [DOI] [PubMed] [Google Scholar]
- Kolocouris A.; Tzitzoglaki C.; Johnson F. B.; Zell R.; Wright A. K.; Cross T. A.; Tietjen I.; Fedida D.; Busath D. D. Aminoadamantanes with Persistent in Vitro Efficacy against H1N1 (2009) Influenza A. J. Med. Chem. 2014, 57, 4629–4639. 10.1021/jm500598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolocouris N.; Foscolos G. B.; Kolocouris A.; Marakos P.; Pouli N.; Fytas G.; Ikeda S.; De Clercq E. Synthesis and Antiviral Activity Evaluation of Some Aminoadamantane Derivatives. J. Med. Chem. 1994, 37, 2896–2902. 10.1021/jm00044a010. [DOI] [PubMed] [Google Scholar]
- Fan Z.; Shu S.; Ni J.; Yao Q.; Zhang A. Ligand-Promoted Pd(II)-Catalyzed Functionalization of Unactivated C (sp3)–H Bond: Regio- and Stereoselective Synthesis of Arylated Rimantadine Derivatives. ACS Catal. 2016, 6, 769–774. 10.1021/acscatal.5b02483. [DOI] [Google Scholar]
- Salom D.; Hill B. R.; Lear J. D.; DeGrado W. F. pH-Dependent Tetramerization and Amantadine Binding of the Transmembrane Helix of M2 from the Influenza A Virus. Biochemistry 2000, 39, 14160–14170. 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakopoulos A.; Tzitzoglaki C.; Ma C.; Freudenberger K.; Hoffmann A.; Hu Y.; Gauglitz G.; Schmidtke M.; Wang J.; Kolocouris A. Affinity of Rimantadine Enantiomers against Influenza A/M2 Protein Revisited. ACS Med. Chem. Lett. 2017, 8, 145–150. 10.1021/acsmedchemlett.6b00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M. R.; Casarotto M. G. Coexistence of Two Adamantane Binding Sites in the Influenza A M2 Ion Channel. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 13866–13871. 10.1073/pnas.1002051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtke M.; Schnittler U.; Jahn B.; Dahse H.-M.; Stelzner A. A Rapid Assay for Evaluation of Antiviral Activity against Coxsackie Virus B3, Influenza Virus A, and Herpes Simplex Virus Type 1. J. Virol. Methods 2001, 95, 133–143. 10.1016/S0166-0934(01)00305-6. [DOI] [PubMed] [Google Scholar]
- Barniol-Xicota M.; Gazzarrini S.; Torres E.; Hu Y.; Wang J.; Naesens L.; Moroni A.; Vázquez S. Slow but Steady Wins the Race: Dissimilarities among New Dual Inhibitors of the Wild-Type and the V27A Mutant M2 Channels of Influenza A Virus. J. Med. Chem. 2017, 60, 3727–3738. 10.1021/acs.jmedchem.6b01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. J.; Wu Y.; Ma C.; Fiorin G.; Pinto L. H.; Lamb R. A.; Klein M. L.; Degrado W. F. Structure and Inhibition of the Drug-Resistant S31N Mutant of the M2 Ion Channel of Influenza A Virus. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 1315–1320. 10.1073/pnas.1216526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A. K.; Batsomboon P.; Dai J.; Hung I.; Zhou H. X.; Dudley G. B.; Cross T. A. Differential Binding of Rimantadine Enantiomers to Influenza A M2 Proton Channel. J. Am. Chem. Soc. 2016, 138, 1506–1509. 10.1021/jacs.5b13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T.; Ogushi F.; Sugita Y. Analysis of Lipid Surface Area in Protein-Membrane Systems Combining Voronoi Tessellation and Monte Carlo Integration Methods. J. Comput. Chem. 2012, 33, 286–293. 10.1002/jcc.21973. [DOI] [PubMed] [Google Scholar]
- Andreas L. B.; Eddy M. T.; Pielak R. M.; Chou J.; Griffin R. G. Magic Angle Spinning NMR Investigation of Influenza A M2 18–60: Support for an Allosteric Mechanism of Inhibition. J. Am. Chem. Soc. 2010, 132, 10958–10960. 10.1021/ja101537p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Hu Y.; Xu S.; Zhang Y.; Musharrafieh R.; Hau R.-K.; Ma C.; Wang J. In Vitro Pharmacokinetic Optimizations of AM2-S31N Channel Blockers Led to the Discovery of Slow-Binding Inhibitors with Potent Antiviral Activity against Drug-Resistant Influenza A Viruses. J. Med. Chem. 2018, 10.1021/acs.jmedchem.7b01536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.