Abstract
Background
Tuberculosis (TB) caused by Mycobacterium tuberculosis (MTB) is one of the most common chronic infectious amphixenotic diseases worldwide. Prevention and control of TB are greatly difficult, due to the increase in drug-resistant TB, particularly multidrug-resistant TB. We speculated that there were some differences between drug-sensitive and drug-resistant MTB strains and that mazEF3,6,9 toxin–antitoxin systems (TASs) were involved in MTB viability. This study aimed to investigate differences in viability, biofilm formation, and MazEF expression between drug-sensitive and drug-resistant MTB strains circulating in Xinjiang, China, and whether mazEF3,6,9 TASs contribute to MTB viability under stress conditions.
Materials and methods
Growth profiles and biofilm-formation abilities of drug-sensitive, drug-resistant MTB strains and the control strain H37Rv were monitored. Using molecular biology experiments, the mRNA expression of the mazF3, 6, and 9 toxin genes, the mazE3, 6, and 9 antitoxin genes, and expression of the MazF9 protein were detected in the different MTB strains, H37RvΔmazEF3,6,9 mutants from the H37Rv parent strain were generated, and mutant viability was tested.
Results
Ex vivo culture analyses demonstrated that drug-resistant MTB strains exhibit higher survival rates than drug-sensitive strains and the control strain H37Rv. However, there was no statistical difference in biofilm-formation ability in the drug-sensitive, drug-resistant, and H37Rv strains. mazE3,6 mRNA-expression levels were relatively reduced in the drug-sensitive and drug-resistant strains compared to H37Rv. Conversely, mazE3,9 expression was increased in drug-sensitive strains compared to drug-resistant strains. Furthermore, compared with the H37Rv strain, mazF3,6 expression was increased in drug-resistant strains, mazF9 expression was increased in drug-sensitive strains, and mazF9 exhibited reduced expression in drug-resistant strains compared with drug-sensitive strains. Protein expression of mazF9 was increased in drug-sensitive and drug-resistant strains compared to H37Rv, while drug-resistant strains exhibited reduced mazF9 expression compared to drug-sensitive strains. Compared to H37Rv, H37RvΔmazEF3,6,9-deletion mutants grew more slowly under both stress conditions, and their ability to survive in host macrophages was also weaker. Furthermore, the host macrophage-apoptosis rate was higher after infection with any of the H37RvΔmazEF3,6,9 mutants than with the H37Rv strain.
Conclusion
The increased viability of MTB drug-resistant strains compared with drug-sensitive strains is likely to be related to differential MazEF mRNA and protein expression. mazEF3,6,9 TASs contribute to MTB viability under stress conditions.
Keywords: Mycobacterium tuberculosis, toxin–antitoxin system, viability, biofilm, apoptosis, drug resistance
Background
Tuberculosis (TB) caused by Mycobacterium tuberculosis (MTB) is one of the most common chronic infectious amphixenotic diseases worldwide. The incidence of TB in China ranks second only to that in India. The prevalence of TB in west China is significantly higher than in central or east China, and Xinjiang harbors the highest incidence of TB in the country.1 In recent years, following the emergence of drug-resistant TB strains, the global prevalence of TB has dramatically increased. China is one of 27 countries listed as having serious problems in relation to multidrug-resistant TB.2 Therefore, strategies to facilitate TB prevention and control are of utmost importance to prevent the further spread of this disease.
Toxin–antitoxin system (TAS) genes are found in low-copy-number plasmids, and these genes are required to allow bacteria to stabilize host plasmids. Recent studies have revealed that some bacteria, such as MTB, also harbor TAS genes on chromosomes.3 TASs are composed of two overlapping genes, one of which encodes a stable toxin protein, while the other encodes a labile antitoxin protein. Under stress conditions, such as nutritional deficiency or reduced oxygen levels,4 MazE degradation results in the synthesis of toxins that cause the release of MazF proteins.5 The two TAS proteins interact to form a toxin–antitoxin complex. The formation of the complex inhibits potentially lethal effects pertaining to the toxin proteins in bacteria.6 Many TASs have been identified, and the associated complexes are classified into six categories.7 MazEF was the first TAS found on the Escherichia coli chromosome. This complex involves the coexpression of the mazF and mazE genes, the former encoding the stable toxin protein MazF and the latter encoding the labile antitoxin protein MazE.8 MazEF TASs are capable of promoting programmed cell death, bacterial growth inhibition, persistence of infections, latent TB, and bacterial adaptability to the surrounding environment.9–11
Biofilms are specialized cellular groups caused by the formation of polymer-matrix package structures that attach to the surface of the microorganism; these complexes are produced by the bacteria themselves.7 More than 80% of infectious human diseases are mediated by bacterial biofilm formation.12 Biofilms confer resistant properties on bacteria, and are usually generated when the bacteria are exposed to conditions of adversity, including nutrient depletion and the presence of microbial inhibitors.7 Because biofilm systems confer resistance against drugs and host immune systems,13 resultant infections can be difficult to cure. Some studies have shown that biofilm formation is influenced by the action of multiple TAs.7
Materials and methods
Bacterial strains and identification
The control MTB strain H37Rv was provided by the Beijing Tuberculosis and Thoracic Tumor Research Institute. Both the drug-resistant strains and the drug-sensitive strains were collected from the sputum of TB patients in Xinjiang. Informed consent was obtained from all of the patients that were recruited as part of this study. The murine macrophage cell line RAW264.7 was purchased from the cell bank of the Committee on Type Culture Collection of the Chinese Academy of Sciences. The study was performed with the approval of the ethics committee of the First Hospital of Shihezi University. Culture, identification, and drug-susceptibility tests of all strains were performed according to the TB-diagnosis bacteriology-test criteria of the China Antituberculosis Association,14 and were performed at the Key Laboratory of Xinjiang Endemic and Ethnic Diseases. MTB resistance was investigated using the proportion method, as per our previous report.15 Drug-resistant strains were defined as those that were resistant to any first-line anti-TB drug (isoniazid, rifampin, streptomycin, and ethambutol), while drug-sensitive strains referred to strains that were sensitive to first-line anti-TB drugs (isoniazid, rifampin, streptomycin, and ethambutol).
Bacterial culture
MTB strains were grown in Middlebrook 7H9 broth (BD Biosciences, San Jose, CA, USA) supplemented with glycerol and Tween 80. The strains were subsequently cultured at 37°C for 7 days. After 7 days of culture, bacterial culture suspensions were adjusted to the same turbidity using PBS. Next, the bacterial suspensions were transferred to culture media at a final concentration of 106 CFU/mL. Next, bacterial strains were cultured in several different environmental conditions, including standard environmental conditions, whereby the strains were inoculated into 7H9 medium and the culture vessels stoppered with breathable silicone plugs.
A hypoxic environment was generated by culturing bacterial suspensions in rubber-plugged vessels containing 7H9 medium. These vessels were subsequently sealed with parafilm. A nutrient-starved environment was established by resuspending the cells in PBS instead of 7H9 medium. The air:medium volume ratio was 1:2 in all of the different culture environments that were tested. Bacterial suspensions were cultured at 37°C, and turbidimetry was used to detect turbidity at the different time points (2, 4, 6, 8, and 10 days).
Biofilm formation
Bacterial suspensions in the logarithmic phase of growth in 7H9 medium were collected and diluted to an OD600 of 0.4. The suspensions were subsequently inoculated into PVC plastic 96-well (200 μL/well) microtiter plates. Next, plates were incubated at 37°C in a bacteriological incubator for 15 days. A total of 25 μL 1% crystal violet solution (crystal violet was solubilized in 95% ethanol) was subsequently added to each well. Next, the plates were incubated at room temperature for 15 minutes, washed gently four times with water, and patted dry using paper towels. Biofilm formation was detected by enzyme-linked immunosorbent assay using a wavelength of 570 nm.16
RNA extraction and reverse-transcription reactions
Total RNA was isolated from exponentially growing bacterial cultures using an RNeasy Plus universal minikit and Qiazol lysis reagent (Qiagen, Venlo, Netherlands) according to the manufacturer’s recommendations. Ultraviolet spectrophotometry was used to determine RNA concentration and purity. First-strand cDNA synthesis was performed using a TianScript cDNA synthesis kit (Tiangen, Beijing, China). Reverse transcription was carried out using 1–5 μg total RNA, 2 μL oligo(dT)1, 2 μL super-pure deoxynucleoside triphosphate, and RNase-free ddH2O to a final volume of 14.5 μL. Each reaction was performed at 70°C for 5 minutes. This was followed by incubation on ice for 2 minutes. Next, 4 μL 5× first-strand buffer (containing dithiothreitol), 0.5 μL RNasin, and 1 μL TianScript M-MLV were added and the associated tubes gently mixed. The reaction mixtures were subsequently incubated at 42°C for 50 minutes in a water bath. Reactions were terminated by incubation at 95°C for 5 minutes. The resultant cDNA was stored at –20°C until required.
Analysis of mRNA by quantitative real-time polymerase chain reaction
Specific primers for mazE3, mazE6, mazE9, mazF3, mazF6, mazF9, and the reference gene, sigA, were designed using Primer 5.0 software. These primers were used for quantitative real-time polymerase chain reaction (Table 1). Reaction mixtures were made to a final volume of 20 μL and included 2 μL cDNA (60 ng/mL) template, 1 μL each primer, 10 μL SYBR Select Master Mix (Thermo Fisher Scientific, Waltham, MA, USA), and 6 μL ddH2O. Samples were analyzed in a 7500 fast fluorescence immunoassay detector (Thermo Fisher Scientific). Quantification was performed in triplicate for each cDNA sample. Reaction conditions utilized to facilitate quantification were 50°C for 2 minutes, 95°C for 2 minutes, 45 cycles of 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 1 minute.
Table 1.
Primers for quantitative real-time polymerase chain reaction
| Gene | Sequence (5′–3′) | Gene | Sequence (5′–3′) |
|---|---|---|---|
| mazE3 | F: CCAGCGTATCCAGATCACC | mazF3 | F: TATGACACCACCCAATCG |
| R: GCGGGTGCATACCAAACT | R: ACCTATCCACTACGCACAGC | ||
| mazE6 | F: TCACCACTCATCGTCCTG | mazF6 | F: GGTCGGTGAGGTCAGTCTTG |
| R: ATGAAGACAGCTATTTCTCTGCC | R: GGTGATTAGTCGTGCCGAGAT | ||
| mazE9 | F: CATGCGTTGGCATAGTCATC | mazF9 | F: TCAAAGCCTCATCGAGCTG |
| R: TATGTGAAACGAGCGGGATT | R: GAGGTAGCGAAGCGAACAAC | ||
| sigA | F: TCGAGGTGATCAACAAGCTG | ||
| R: CTGCAGCAAAGTGAAGGACA |
Synthesis and preparation of MazF3, 6, and 9 antibodies
The pET28a vector was used as the expression vector for MazF3, 6, and 9. The bases marked in red in Figure 1 are representative of the restriction-enzyme-recognition sites (former BamHI, latter XhoI). The first line represents the target DNA sequence of mazF3, 6, and 9, the second line the sequencing results. The sequences before and after the restriction-enzyme-recognition site are vector sequences. Sequence alignment results show that the coincidence rate was 100%. After successful construction, the plasmid was transformed into DH5α E. coli cells. MazF3, 6 and 9 recombinant protein molecular weights inclusive of His-tag molecular weight were 15, 16, and 16 kDa, respectively.
Figure 1.
Sequence results of mazF3,6,9.
Notes: (A) mazF3; (B) mazF6; (C) mazF9.
Detection and purification of recombinant MazF3, 6, and 9 protein
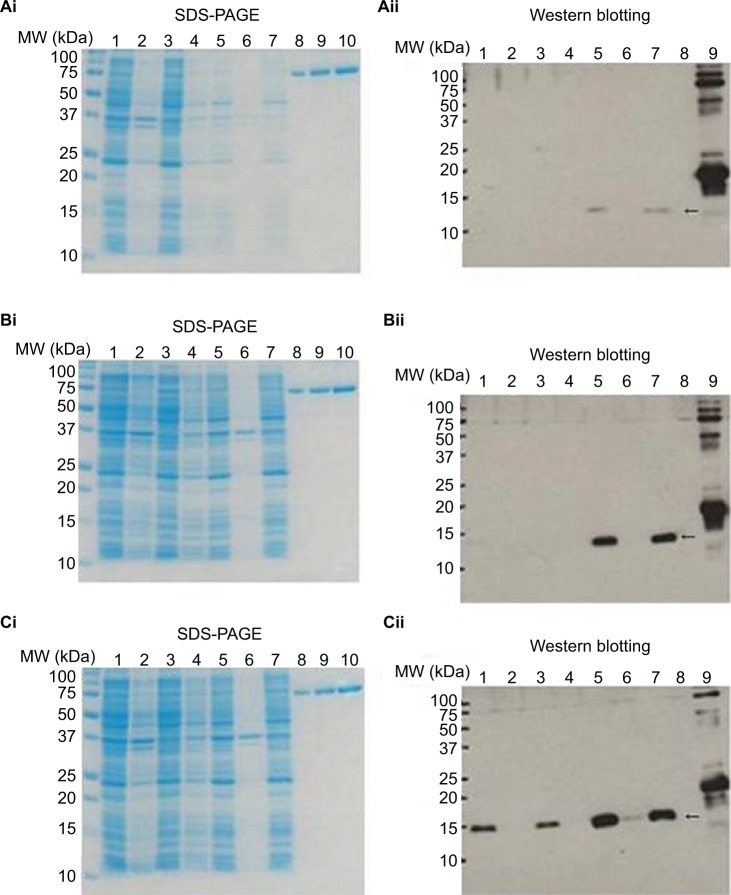
Figure 2 shows sodium dodecyl sulfate polyacrylamide-gel electrophoresis (SDS-PAGE) and Western blot images of MazF3, 6, and 9 proteins. No objective protein bands were observed following SDS-PAGE. Western blot analysis revealed minimal MazF3 expression. MazF6 and 9 protein expression was induced in large quantities. However, MazF6 was toxic to the host bacterium, and during induction transformant bacteria died. MazF9 was subsequently purified and the recombinant protein used to prepare monoclonal antibodies.
Figure 2.
SDS-PAGE and Western blot analyses to facilitate detection of recombinant protein expression.
Notes: SDS-PAGE for detection of MazF3, 6, and 9 (i); Western blot for detection of MazF3, 6, and 9 (ii). SDS-PAGE: 1, 37°C O/N whole; 2, 37°C O/N ppt; 3, 37°C O/N sup; 4, IPTG– whole; 5, IPTG+ whole; 6, IPTG+ ppt; 7, IPTG+ sup; 8, BSA 0.5 μg; 9, BSA 1.0 μg; 10, BSA 2 μg. Western blot: 1, 37°C O/N whole; 2, 37°C O/N ppt; 3, 37°C O/N sup; 4, IPTG– whole; 5, IPTG+ whole; 6, IPTG+ ppt; 7, IPTG+ sup; 8, negative control; 9, positive control. The arrows show recombinant protein MazF3 (Aii), MazF6 (Bii), and MazF9 (Cii), respectively.
Abbreviations: SDS-PAGE, sodium dodecyl sulfate polyacrylamide-gel electrophoresis; IPTG, isopropyl β-d-1-thiogalactopyranoside; MW, molecular weight; ppt, precipitation; sup, supernatant; BSA, bovine serum albumin.
Protein extraction
MTB colonies growing exponentially on Löwenstein–Jensen medium were aseptically scraped off culture plates and resuspended in 500 μL distilled water. Bacterial cells were subsequently inactivated at 85°C for 30 minutes. Inactivated cells were centrifuged at 8,000 rpm for 5 minutes and the supernatant discarded. The resultant pellets were collected and stored at –80°C. Total bacterial protein was extracted using a Qproteome bacterial protein prep kit. Briefly, the frozen bacterial pellets were thawed for 15 minutes on ice and resuspended in 1 mL native lysis buffer. Resuspensions were subsequently incubated on ice for a further 30 minutes. Next, the chilled suspensions were mixed two to three times by gentle swirling. The resultant lysates were centrifuged at 14,000× g for 30 minutes at 4°C, in order to collect the cellular debris. The cell-lysate supernatants containing the soluble fractions of the bacterial cell proteins were retained, and the concentration and purity of total proteins were determined using a nucleic acid/protein detector.
Analysis of protein by Western blot analysis
Samples were separated by SDS-PAGE using 5% spacer gel and 15% separation gel. GAPDH was utilized as a loading control. GAPDH monoclonal antibody was purchased from Bioworld Technology (Minneapolis, MN, USA; the antibody was diluted 1:1,000). The MTB MazF monoclonal antibody (host: mouse) was prepared by MBL (Nagoya, Japan). The HRP-conjugated secondary antibody was obtained from MBL. The latter antibody was raised in goat against mouse IgG to generate goat antimouse IgG HRP. The dilution rate for the GAPDH monoclonal antibody was 1:10,000.
Construction and identification of mazEF3,6,9-deletion mutants from H37Rv
Construction and identification of deletion mutants of TAS mazEF3,6,9 of MTB H37Rv was conducted by splicing-overlap extension polymerase chain reaction. H37Rv, H37RvΔmazEF3, H37RvΔmazEF6, and H37RvΔmazEF9 were grown at 37°C in Middlebrook 7H9 liquid medium supplemented with 10% oleic albumin dextrose catalase or on Löwenstein–Jensen slants. To determine what role the mazEF3,6,9 TASs play in MTB growth and survival, we compared cell growth between the parent H37Rv strain and H37RvΔmazEF3,6,9-deletion mutants under both normal and stressful culture conditions, including hypoxia and nutrient starvation or after in vitro intracellular infection of host macrophages.
Infection model, viability, and apoptosis rate of MTB in RAW264.7 macrophages
An MTB infection model in RAW264.7 macrophages was established, making a final ratio of ten bacilli to one cell. At different time points after time 0 (0.5, 1, 1.5, 2, 3, or 5 days later), bacterial cells were collected by adding and thoroughly mixing 250 μL radioimmunoprecipitation-assay lysis buffer containing 1% phenylmethylsulfonyl fluoride to the infected macrophages for 30 minutes to ensure complete lysis of the host cells. Equivalent amounts of bacteria were then cultured on Löwenstein–Jensen slants for 3–4 weeks to observe growth, and colonies were counted after tenfold dilution.
The apoptosis rate of infected macrophages was assessed based on the manufacturer’s instructions for the annexin V–fluorescein isothiocyanate–propidium iodide apoptosis-detection kit (BioVision, Milpitas, CA, USA). Briefly, host macrophage cells infected with H37Rv andH37RvΔmazEF3,6,9 were harvested by centrifugation at different time points after time 0 of bacterial infection (1, 6, 12, or 24 hours later); uninfected macrophages were used as control cells. Cells were then resuspended in 500 μL binding buffer, and 5 μL annexin V–fluorescein isothiocyanate plus 5 μL PI (50 μg/mL) were added before incubation at room temperature for 5 minutes in the dark. The apoptosis rate was evaluated within 1 hour of staining by flow cytometry.
Statistics
Differences between groups were analyzed using Student’s t-test or analysis of variance. Data are represented as means ± SEM for three replicates. These data are representative of results obtained from at least three independent experiments. Data were analyzed using GraphPad Prism 5.0 software, and P<0.05 was considered to have statistical significance.
Results
Growth profiles of MTB strains under different culture conditions
MTB strains were cultured under standard conditions in 7H9 medium in culture vessels containing breathable silicone plugs (Figure 3A). OD600 values for each of the bacterial cultures that were analyzed increased gradually following inoculation. However, differences in growth ability among the different strains become apparent after 2 days of culture. Under standard conditions, the drug-resistant strains appeared to grow more rapidly than drug-sensitive strains and H37Rv. However, growth-profile differences between drug-resistant strains and H37Rv were observed 8 and 10 days after inoculation, and these differences were significant.
Figure 3.
Growth profiles of MTB in standard, hypoxic and nutrient starvation conditions at different culture time points.
Notes: (A) MTB incubated under standard conditions in 7H9 medium. Difference in growth ability became apparent on the eighth day, and resistant strains grew more rapidly than H37Rv (P<0.05). (B) Strains incubated under hypoxic conditions. Difference in growth ability became apparent at 2 days, 8 days, and 10 days, and resistant strains grew more rapidly than H37Rv (P<0.05). (C) Strains incubated under starvation conditions. Difference in growth ability became apparent on the sixth day. Resistant strains grew more rapidly than sensitive strains and H37Rv (P<0.05). #Comparison of drug-sensitive strains with H37Rv (P<0.05); *comparison of drug-resistant strains with H37Rv (P<0.05). **Comparison of drug-resistant strains with H37Rv (P<0.01).
Abbreviation: MTB, Mycobacterium tuberculosis; MCF, McFarland turbidity standards.
MTB strains were also cultured in hypoxic conditions in culture vessels containing 7H9 medium (Figure 3B). The vessels were stoppered with rubber plugs and sealed with Parafilm. OD600 values for each of the bacterial cultures that were analyzed increased gradually following inoculation. However, differences in growth ability among the different strains became apparent at 2, 8, and 10 days following inoculation. Overall, drug-resistant strains grew more rapidly than drug-sensitive strains and H37R. However, the only significance difference was between drug-resistant strains and H37Rv.
As shown in Figure 3C, MTB strains were cultured in conditions that simulated nutrient depletion. This involved resuspension of bacterial cultures in PBS in culture vessels stoppered with breathable silicone plugs. OD600 values for each of the bacterial cultures that were analyzed increased gradually following inoculation. However, differences in growth ability among the different strains became apparent at 6, 8, and 10 days following inoculation. Overall, drug-resistant strains grew more rapidly than drug-sensitive strains and H37Rv. At 6 days after inoculation, drug-sensitive strains were observed to grow more rapidly than H37Rv, and the differences were statistically significant.
Biofilm formation of drug-sensitive and drug-resistant MTB strains
As shown in Figure 4, absorbance values were measured using spectrophotometry at a wavelength of 570 nm. This analysis permitted us to determine the different biofilm-formation abilities of drug-sensitive strains, drug-resistant strains, and the control H37Rv. We observed that drug-sensitive strains exhibited greater biofilm-formation ability than H37Rv. The H37Rv strain displayed greater biofilm-formation ability than drug-resistant strains; however, none of these differences was statistically significant (P>0.05).
Figure 4.

Biofilm-formation ability of drug-sensitive and drug-resistant MTB strains.
Note: There were no differences among drug-resistant strains, drug-sensitive strains, and control strain H37Rv (P>0.05).
Abbreviation: MTB, Mycobacterium tuberculosis.
mRNA expression of mazE3,6,9 and mazF3,6,9 in drug-sensitive and drug-resistant MTB strains relative to H37Rv
The antitoxin genes mazE3 and mazE6 exhibited reduced expression in drug-sensitive strains and drug-resistant strains compared with control strain H37Rv (P<0.01). mazE9 exhibited increased expression in drug-sensitive strains compared with drug-resistant strains (P<0.05) (Figure 5). The toxin genes mazF3 and mazF6 exhibited increased expression in drug-resistant strains compared with control strain H37Rv (P<0.01), and only mazF9 exhibited increased expression in drug-sensitive strains compared with control strain H37Rv (P<0.01). Upon comparison of drug-resistant strains with drug-sensitive strains, mazF9 exhibited reduced expression, and the difference was statistically significant (P<0.01) (Figure 6).
Figure 5.
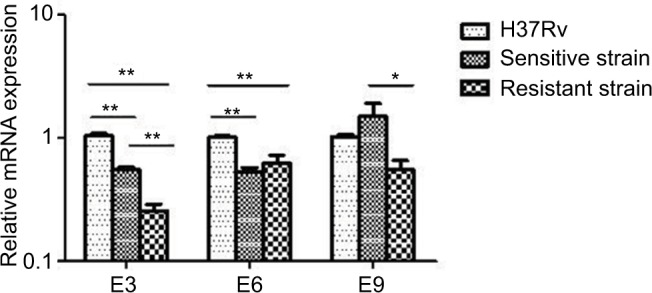
mRNA expression of mazE3,6,9 in drug-sensitive and drug-resistant MTB strains and the control strain H37Rv.
Notes: *P<0.05; **P<0.01.
Abbreviation: MTB, Mycobacterium tuberculosis.
Figure 6.
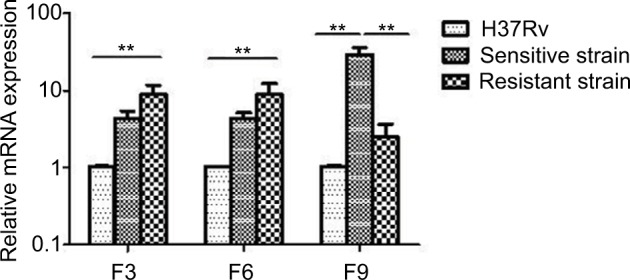
mazF3,6,9 mRNA expression in drug-sensitive and drug-resistant MTB strains and the control strain H37Rv.
Note: **P<0.01.
Abbreviation: MTB, Mycobacterium tuberculosis.
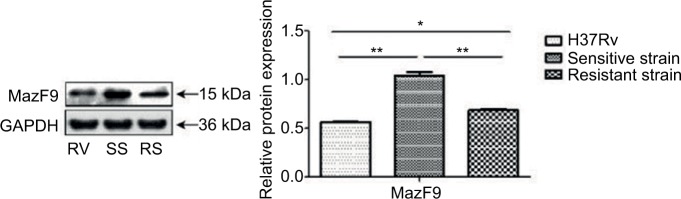
Protein expression of MazF9 in drug-sensitive and drug-resistant MTB strains
Increased MazF9 expression was observed in both drug-sensitive and drug-resistant strains compared with H37Rv (Figure 7). Differences in expression between these strains were statistically significant in each case. Reduced MazF9 expression was observed the drug-resistant strains compared with drug-sensitive strains, and the differences were statistically significant. These results are consistent with those generated during the mazF9 mRNA-expression analysis in drug-sensitive and drug-resistant MTB strains.
Figure 7.
MazF9 protein expression in drug-sensitive and drug-resistant MTB strains.
Notes: MazF9 exhibited increased expression both in drug-sensitive strains (**P<0.01) and drug-resistant strains (*P<0.05) compared with the control strain H37Rv. The drug-sensitive strains exhibited increased expression compared with the drug-resistant strains (**P<0.01).
Abbreviations: MTB, Mycobacterium tuberculosis; SS, sensitive strain; RS, resistant strain; RV, H37Rv.
mazEF3,6,9 TASs promote MTB growth under stressful culture conditions
In terms of bacterial cell survival, H37Rv and H37RvΔmazEF3,6,9 strain viability showed no significant differences at days 2, 4, 6, 8, and 10 under normal conditions (P>0.05). However, the viability of the three deletion mutants was significantly reduced compared to the H37Rv parent strain at day 10 in hypoxic conditions (P<0.05), although no significant differences were observed among the three deletion mutant strains. H37Rv and H37RvΔmazEF3,6,9 bacterial cell growth in nutrient-starved conditions showed a downward trend, and the viability of the deletion mutants was significantly reduced on days 6, 8, and 10 compared to H37Rv (P<0.01), but no significant differences were observed among the three deletion mutant strains (Table 2). Taken together, these results suggested that mazEF3,6,9 functioned to promote MTB growth when bacteria were exposed to stress conditions.
Table 2.
The number of living bacteria in various conditions over time
| Day | Logarithm of counted colony numbers (log10 CFU/mL)
|
|||||
|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | ||
| Control | H37Rv | 5.74±0.05 | 5.96±0.07 | 6.48±0.03 | 6.76±0.1 | 6.90±0.03 |
| H37RvΔmazEF3 | 572±0.04 | 5.94±0.07 | 6.44±0.04 | 6.72±0.4 | 6.86±0.05 | |
| H37RvΔmazEF6 | 5.71±0.03 | 5.91±0.04 | 6.43±0.04 | 6.7±0.03 | 6.84±0.04 | |
| H37RvΔmazEF9 | 5.72±0.03 | 5.91±0.03 | 6.44±0.02 | 6.71±0.03 | 6.87±0.04 | |
| Compare result | F=0.361 | F=0.578 | F=1.07 | F=0.679 | F=0.869 | |
| P>0.05 | P>0.05 | P>0.05 | P>0.05 | P>0.05 | ||
| Hypoxia | H37Rv | 5.27±0.06 | 5.81±0.05 | 5.61±0.09 | 5.1±0.03 | 4.69±0.09 |
| H37RvΔmazEF3 | 5.22±0.04 | 6.77±0.02 | 5.55±0.06 | 5.06±0.03 | 4.42±0.03 | |
| H37RvΔmazEF6 | 5.23±0.06 | 5.75±0.04 | 5.53±0.05 | 5.03±0.05 | 4.4±0.03 | |
| H37RvΔmazEF9 | 5.21±0.04 | 5.76±0.03 | 5.54±0.03 | 5.04±0.03 | 4.41±0.03 | |
| Compare result | F=0.716 | F=1.645 | F=1.014 | F=2.207 | F=22.849 | |
| P>0.05 | P>0.05 | P>0.05 | P>0.05 | P<0.01 | ||
| Nutrient starvation | H37Rv | 4.6±0.06 | 4.35±0.06 | 4.15±0.07 | 4.07±0.05 | 3.62±0.1 |
| H37RvΔmazEF3 | 4.53±0.02 | 4.32±0.06 | 4.11±0.07 | 3.84±0.03 | 3.41±0.02 | |
| H37RvΔmazEF6 | 4.52±0.035 | 4.27±0.061 | 4.09±0.087 | 3.78±0.04 | 3.37±0.031 | |
| H37RvΔmazEF9 | 4.54±0.051 | 4.28±0.082 | 4.1±0.053 | 3.81±0.03 | 3.39±0.032 | |
| Compare result | F=2.141 | F=0.904 | F=138.179 | F=40.388 | F=426.488 | |
| P>0.05 | P>0.05 | P<0.01 | P<0.01 | P<0.01 | ||
Notes: Three deletion strains compared with H37Rv in various growth conditions. Data presented as mean ± SD.
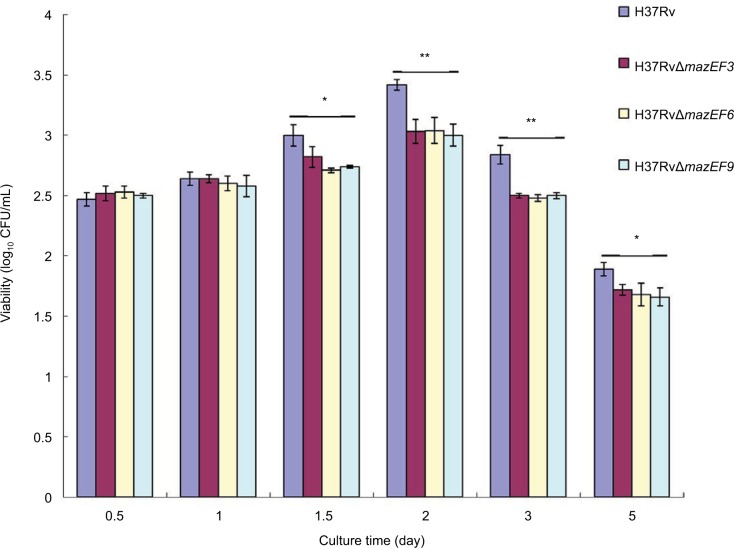
mazEF3,6,9 promote MTB viability in macrophages during intracellular infection
The H37Rv strain had significantly higher viability in macrophages than the three deletion mutant strains H37RvΔmazEF3,6,9 (P<0.05) on days 1.5, 2, 3, and 5, and no significant differences in viability were observed among the three deletion mutant strains (P>0.05) (Figure 8). These results suggested that mazEF3,6,9 also promoted bacterial cell survival of MTB within the host macrophage.
Figure 8.
Viability in parent and mazEF-deletion mutant H37Rv strains in host macrophages after infection.
Notes: Viable H37Rv, H37RvΔmazEF3, H37RvΔmazEF6, and H37RvΔmazEF9 bacteria after being phagocytosed by macrophages. Mean values from duplicates of three independent experiments are shown. *P<0.05, **P<0.01 compared to H37Rv group.
mazEF3,6,9 TASs in MTB play a role in inhibiting macrophage apoptosis during infection
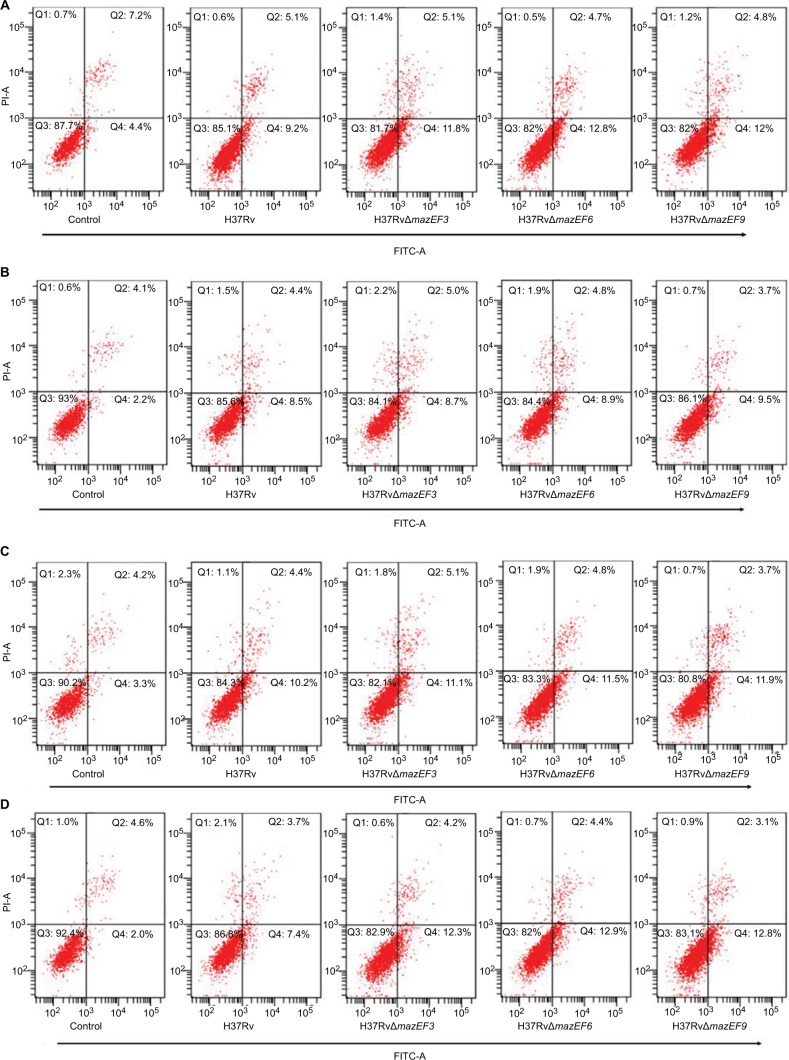
The macrophage-apoptosis rate was significantly higher in H37Rv-, H37RvΔmazEF3-, H37RvΔmazEF6-, and H37RvΔmazEF9-infected macrophages than in uninfected control macrophages (P<0.05); moreover, the apoptosis rate of macrophages infected with any of the MTB strains had increased significantly at 1, 12, and 24 hours after infection (P<0.05). Among the strains, macrophages infected with any of the deletion mutant strains exhibited significantly increased apoptosis over H37Rv-infected macrophages (P<0.05) (Figure 9). These results indicated that the presence of these mazEF3,6,9 TASs in MTB could inhibit macrophage apoptosis, potentially prolonging the life of MTB.
Figure 9.
Host-macrophage apoptosis over time after infection with H37Rv, H37RvΔmazEF3, H37RvΔmazEF6, or H37RvΔmazEF9.
Notes: Flow-cytometry analysis of macrophage apoptosis after (A) 1, (B) 6, (C) 12, and (D) 24 hours after infection by annexin V and propidium iodide staining. Control group represents uninfected macrophages. (E) Kinetics of macrophage-apoptosis rate after infection or in uninfected controls (P<0.05).
Abbreviations: FITC, fluorescein isothiocyanate; PI, propidium iodide.
Discussion
TB represents a continued global health problem. The vast majority of MTB infections are caused by emerging multidrug-resistant and extensively drug-resistant strains. This phenomenon is exacerbated by the lack of effective drugs available for treatment.17 Gene-encoded drug resistance predominantly arises in MTB through the occurrence of chromosomal mutations;18,19 however, research suggests that drug resistance may also be related to TASs.
TASs are commonly found in bacterial genomes and can affect cell function in many different ways, including protein synthesis, cell division, peptidoglycan biosynthesis, and ribosome assembly.7 It is likely that these occurrences contribute to the formation of biofilms following exposure to adverse conditions.7 TASs are induced under different adverse environmental conditions and occurrences, including starvation, hypoxia, drug targeting, and macrophage-mediated immunoreactions.20–22 The MTB genome harbors an abundance of TASs, most of which belong to the MazEF, RelBE, and ParDE families.20 The present study was conducted to investigate mazEF expression in drug-sensitive, drug-resistant, and control MTB strains. The study also investigated the growth profiles of the different strains following exposure to different growth conditions. We also analyzed the biofilm-forming ability of the different strains.
It was observed that expression of the antitoxin genes mazE3 and mazE6 was comparatively downregulated in drug-sensitive and drug-resistant strains compared with the control strain H37Rv. The expression levels of mazE3 were lowest in drug-resistant strains, and mazE9 was downregulated in drug-resistant strains compared with drug-sensitive strains. Upon analysis of the toxin genes, increased expression of mazF3 and mazF6 was observed in drug-resistant strains compared with H37Rv, and mazF9 expression was higher in drug-sensitive strains, but relatively lower in drug-resistant strains. Toxin and antitoxin expression varied between the different strains that were analyzed. Whether the latter phenomenon is related to characteristics that are specific to drug-resistant strains remains to be elucidated. Following growth analyses using different culture conditions, we observed that drug-resistant strains exhibited higher expression of toxin genes following exposure to hypoxic and nutrition-depleted conditions. However, accumulation of MazF, which is both a restriction ribonuclease that cleaves single-stranded RNA at ACA8,23,24 and a toxin protein, causes inhibition of bacterial growth and even cell death.
The reason for higher expression of toxin encoding genes in drug-resistant strains remains to be elucidated; however, this phenomenon may be due to greater exposure to adverse conditions. Whether excess accumulation of toxins as a consequence of increased expression results in host death remains to be elucidated. It is possible that accumulation of these toxins, including MazF, can increase the drug-resistance capacity of bacteria upon exposure to adverse environments. Conversely, the production of higher concentrations of similar toxins might induce bacterial death. However the function of MazF homologues in stress adaptation and drug tolerance of MTB is unknown, and requires further study.
Ex vivo culture results demonstrated that drug-resistant MTB strains exhibited higher survival rates than drug-sensitive strains and the control strain H37Rv. Due to drug resistance, mutation occurs mainly on the necessary genes of MTB, such as rpoB (coding RNA polymerase) and gyrA (coding DNA gyrase). Mutation of these genes can reduce or lose corresponding protein/enzyme activity, which leads to a decline in environmental adaptability of MTB, that is, to produce “fitness cost”.25 The adaptive decline shows that the reproduction rate and survival capability of the resistant strains were less than that of the wild strains in the absence of antibiotic pressure in the environment.26 However, recent studies have shown that drug-resistant MTB strains’ adaptive evolution may have played an important role in the spread of drug-resistant TB, ie, this fitness cost can be ameliorated secondarily by compensatory mutations, which help restore the fitness of drug-resistant strains. The compensatory mutations increase the adaptability of drug-resistant strains, and promote the fixation and transmission of drug-resistant bacteria in the population.27–29 Borrel et al used M. smegmatis as a model for human TB and an in vitro competitive fitness assay to explore the combined fitness effects and interaction between mutations conferring resistance to rifampicin and ofloxacin.30 It was found that there were significant differences in the adaptive cost of different combinations of drug-resistant gene mutations.30 In 17 kinds of double-combination drug-resistance mutations, the adaptability of six kinds of mutation combinations was significantly higher than the adaptability of the single drug-resistant mutant strain, such as rpoBH526P + gyrAD94N and rpoBH526R + gyrAD94N. Among these, individual drug-resistant mutations (such as rpoBH526P + gyrAD94N) can even make the drug-resistance mutations more adaptable than wild ones.30
Biofilm formation is an important phenomenon in bacterial drug resistance. Studies have shown that the formation of biofilms is associated with MazF expression.7 However, we observed that higher mazF expression in drug-resistant strains compared with drug-sensitive strains and control strains did not affect relative biofilm-formation ability. This phenomenon is likely to be regulated by multiple genes, with differential expression of these genes affecting biofilm-formation ability. Mycobacterial biofilms contain microcolonies composed of numerous bacilli held together by extracellular material of proteins, lipids, polysaccharides, and extracellular DNA. These microcolonies are attached to the substratum by cellulose along with unidentified polysaccharides and extracellular DNA. Cellulose is an essential component of the extracellular matrix and a key component of MTB biofilms. Free mycolic acids are only a minor fraction of extracellular material. As cellulose is absent in humans, this polysaccharide could potentially be used as a biomarker for the detection of MTB biofilms in vivo.31 Formation of biofilm is influenced by the action of multiple TASs. The most penetrating insights into the role of TAs in biofilm organization come from research on E. coli biofilms. TASs related to biofilm formation are involved, eg, mazFE, mqsRA, hipAB, hhA–tomB, and yafQ–dinJ.7 In addition to TASs, some genes of MTB are associated with the formation of biofilms, such as groEL1, which modulates synthesis of mycolates – long-chain fatty-acid components of the mycobacterial cell wall – specifically during biofilm formation and physically associates with KasA;32 H37Ra_3775(H37Rv3737), which encodes conservative transmembrane protein, might be related to forming pellicle biofilm;33 the Rv1013 gene, which encodes a putative acyl-CoA synthase, is related to the maturation of biofilm;34 and the Rv2454c and Rv2455c genes, encoding 2-oxoglutarate dehydrogenase, are associated with promoting biofilm formation,34 pks1, a polyketide synthase gene, also contributes to the formation of MTB biofilm.35 MTB biofilm-formation genes merit further investigation to determine the relationship between biofilm formation and drug resistance fully. MTB biofilms are observed in primary lesion residual necrosis and coating cavities. Such biofilm formation by MTB and other mycobacteria has been shown to reduce their sensitivity to antimycobacterial agents. The biofilm architecture and the extracellular polymeric substance matrix limit drug diffusion, protecting individual cells.36 Biofilm MTB differs from planktonic MTB in many ways. Biofilms are known to protect pathogens from phagocytosis and to increase tolerance to drug treatment. Slower growth rates and lower metabolic activity are implicated in higher tolerance to drug treatment. However, biofilms have been shown to have higher metabolic activities than their planktonic counterparts.36
In this study, we found that deletion mutants without intact mazEF3,6,9 TASs exhibited slower growth than the mazEF-sufficient H37Rv strain under hypoxic or nutrient-starved conditions, suggesting that mazEF3,6,9 played a role in sustaining MTB survival during stress conditions. We tested whether mazEF3,6,9 TASs were also involved in bacterial growth after infecting murine RAW264.7 macrophages by comparing the ability of the different strains to survive within macrophages, and found that the number of viable H37Rv bacterial cells was higher than H37RvΔmazEF3,6,9 strains over time. This finding suggests that the presence of mazEF3,6,9 in MTB contributes to their ability to survive in macrophages. MTB mainly survives and reproduces in host macrophages after infecting the body. Macrophage apoptosis is one of the most important immunomechanisms that can kill MTB and limit its dissemination in vivo. However, MTB can sometimes evade immunodefenses by restraining and regulating macrophage apoptosis through a variety of mechanisms. Here, by monitoring the apoptosis rate of macrophages infected with various MTB strains, our findings showed that the apoptosis rate significantly increased 1, 12, and 24 hours after infection; moreover, the apoptosis rate of macrophages infected with the mazEF-sufficient H37Rv parent strain or any of the deletion mutant strains was significantly higher than uninfected macrophages, while the apoptosis rate for macrophages infected with any of the deletion mutant strains was significantly higher than that for H37Rv-infected macrophages. Therefore, these findings support a role for the tested mazEF3,6,9 TASs in immunoevasion mechanisms that inhibit macrophage apoptosis, thus potentially sustaining MTB survival within the host body.
Acknowledgments
The authors acknowledge the financial support of the National Natural Science Foundation of China (81560264) and the “AIDS and viral hepatitis and other major infectious disease prevention and treatment” Special Major Science and Technology Project (2013ZX10003003-002). A small portion of this paper was presented at the Fourth European Congress of Immunology, Vienna, September 6–9, 2015, on the role of the mazEF toxin–antitoxin system in Mycobacterium tuberculosis macrophages as a poster presentation.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Technical guidance group of the fifth national TB epidemiological survey The fifth national tuberculosis epidemiological survey in 2010. Zhongguo Fang Lao Xie Hui. 2012;34:485–508. Chinese. [Google Scholar]
- 2.Xu Y, Zhang Z, Sun Z. Drug resistance to MTB: from the traditional Chinese view to modern systems biology. Crit Rev Microbiol. 2015;41:399–410. doi: 10.3109/1040841X.2013.860948. [DOI] [PubMed] [Google Scholar]
- 3.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiwari P, Arora G, Singh M, Kidwai S, Narayan OP, Singh R. MazF ribonucleases promote MTB drug tolerance and virulence in guinea pigs. Nat Commun. 2015;6:6059. doi: 10.1038/ncomms7059. [DOI] [PubMed] [Google Scholar]
- 5.Robson J, McKenzie JL, Cursons R, et al. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J Mol Biol. 2009;390:353–367. doi: 10.1016/j.jmb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Engelberg-Kulka H, Glaser G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu Rev Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- 7.Kedzierska B, Hayes F. Emerging roles of toxin-antitoxin modules in bacterial pathogenesis. Molecules. 2016;21:E790. doi: 10.3390/molecules21060790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 9.Chao MC, Rubin EJ. Letting sleeping does lie: does dormancy play a role in tuberculosis? Annu Rev Microbiol. 2010;64:293–311. doi: 10.1146/annurev.micro.112408.134043. [DOI] [PubMed] [Google Scholar]
- 10.Gengenbacher M, Kaufmann SH. MTB: success through dormancy. FEMS Microbiol Rev. 2012;36:514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol. 2012;66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 12.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 13.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 14.Chinese Antituberculosis Association The laboratory science procedure of diagnostic bacteriology in tuberculosis. Zhongguo Fang Lao Xie Hui. 1996;18:28–31. Chinese. [Google Scholar]
- 15.Yuan L, Huang Y, Mi LG, et al. There is no correlation between sublineages and drug resistance of Mycobacterium tuberculosis Beijing/W lineage clinical isolates in Xinjiang, China. Epidemiol Infect. 2015;143:141–149. doi: 10.1017/S0950268814000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter G, Wu M, Drummond DC, Bermudez LE. Characterization of biofilm formation by clinical isolates of Mycobacterium avium. J Med Microbiol. 2003;52:747–752. doi: 10.1099/jmm.0.05224-0. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization . Global Tuberculosis Report 2012. Geneva: WHO; 2012. [Google Scholar]
- 18.World Health Organization Totally drug-resistant TB: a WHO consultation on the diagnostic definition and treatment options. 2012. [Accessed December 5, 2017]. Available from: http://www.who.int/tb/challenges/xdr/Report_Meeting_total-lydrugresistantTB_032012.pdf.
- 19.World Health Organization . Multidrug and Extensively Drug-Resistant TB (M/XDR-TB): 2010 Global Report on Surveillance and Response. Geneva: WHO; 2010. [Google Scholar]
- 20.Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A. 2011;108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Q, Awano N, Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol. 2011;79:109–118. doi: 10.1111/j.1365-2958.2010.07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutschler H, Gebhardt M, Shoeman RL, Meinhart A. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 2011;9:e1001033. doi: 10.1371/journal.pbio.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem. 2005;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 24.Erental A, Sharon I, Engelberg-Kulka H. Two programmed cell death systems in Escherichia coli: an apoptotic-like death is inhibited by the mazEF-mediated death pathway. PLoS Biol. 2012;10:e1001281. doi: 10.1371/journal.pbio.1001281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melnyk AH, Wong A, Kassen R. The fitness costs of antibiotic resistance mutations. Evol Appl. 2015;8:273–283. doi: 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogwill T, MacLean RC. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl. 2015;8:284–295. doi: 10.1111/eva.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comas I, Borrell S, Roetzer A, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2011;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Vos M, Müller B, Borrell S, et al. Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob Agents Chemother. 2013;57:827–832. doi: 10.1128/AAC.01541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li QJ, Jiao WW, Yin QQ, et al. Compensatory mutations of rifampin resistance are associated with transmission of multidrug-resistant Myco-bacterium tuberculosis Beijing genotype strains in China. Antimicrob Agents Chemother. 2016;60:2807–2812. doi: 10.1128/AAC.02358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borrell S, Teo Y, Giardina F, et al. Epistasis between antibiotic resistance mutations drives the evolution of extensively drug-resistant tuberculosis. Evol Med Public Health. 2013;2013:65–74. doi: 10.1093/emph/eot003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trivedi A, Mavi PS, Bhatt D, Kumar A. Thiol reductive stress induces cellulose-anchored biofilm formation in Mycobacterium tuberculosis. Nat Commun. 2016;7:11392. doi: 10.1038/ncomms11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR, Jr, Hatfull GF. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell. 2005;123:861–873. doi: 10.1016/j.cell.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Guo QL, Wang RJ, Wang HH, Pei XY, Zhang XL. Screening and identification of Mycobacterium tuberculosis biofilm formation related genes. China Biotechnol. 2013;33:15–21. Chinese. [Google Scholar]
- 34.Ojha AK, Baughn AD, Sambandan D, et al. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol. 2008;69:164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pang JM, Layre E, Sweet L, et al. The polyketide Pks1 contributes to biofilm formation in Mycobacterium tuberculosis. J Bacteriol. 2012;194:715–721. doi: 10.1128/JB.06304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solokhina A, Brückner D, Bonkat G, Braissant O. Metabolic activity of mature biofilms of Mycobacterium tuberculosis and other non-tuberculous mycobacteria. Sci Rep. 2017;7:9225. doi: 10.1038/s41598-017-10019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]