Abstract
Iron is universally important to cellular metabolism, and mitoferrin-1 and -2 have been proposed to be the iron importers of mitochondria, the cell's assembly plant of heme and iron–sulfur clusters. These iron-containing prosthetic groups are critical for a host of physiological processes ranging from oxygen transport and energy consumption to maintaining protein structural integrity. Mitoferrin-1 (Mfrn1) belongs to the mitochondrial carrier (MC) family and is atypical given its putative metallic cargo; most MCs transport nucleotides, amino acids, or other small- to medium-size metabolites. Despite the clear importance of Mfrn1 in iron utilization, its transport activity has not been demonstrated unambiguously. To bridge this knowledge gap, we have purified recombinant Mfrn1 under non-denaturing conditions and probed its metal ion–binding and transport functions. Isothermal titration calorimetry indicates that Mfrn1 has micromolar affinity for Fe(II), Mn(II), Co(II), and Ni(II). Mfrn1 was incorporated into defined liposomes, and iron transport was reconstituted in vitro, demonstrating that Mfrn1 can transport iron. Mfrn1 can also transport manganese, cobalt, copper, and zinc but discriminates against nickel. Experiments with candidate ligands for cellular labile iron reveal that Mfrn1 transports free iron and not a chelated iron complex and selects against alkali divalent ions. Extensive mutagenesis identified multiple residues that are crucial for metal binding, transport activity, or both. There is a clear abundance of residues with side chains that can coordinate first-row transition metal ions, suggesting that these could form primary or auxiliary metal-binding sites during the transport process.
Keywords: iron, iron metabolism, membrane protein, membrane transport, membrane transporter reconstitution, metal ion-protein interaction, mitochondria, mitochondrial transport, metal transport, mitochondrial carrier
Introduction
The reactivity of iron is exploited across all kingdoms of life. The physiological roles that iron takes on are many. Iron readily participates in redox reactions where it can donate or accept electrons, depending on its oxidation state. Iron plays essential roles in gas transport and sensing and can also act as a non-redox–active metal ion cofactor in enzymes. Additionally, iron-containing cofactors can impart stability to folded proteins (1). Iron naturally occurs in the 2+ and 3+ oxidation states, but because of the insolubility of Fe(III), the bioavailable form of iron is almost exclusively the 2+ form. The main route of cellular iron uptake is via the transferrin receptor pathway whereby transferrin-bound iron is targeted to endosomes and subsequently released from transferrin during endosomal acidification. Fe(II) is transported out of endosomes by a divalent metal ion transporter (DMT1) and becomes available for incorporation into iron-containing proteins and cofactors (2).
In eukaryotes, mitochondria are the hot spots for iron utilization: these organelles house the heme assembly and the majority of the Fe–S cluster biosynthesis apparatus (3, 4). Mitochondria contain respiratory complexes that comprise many iron-containing cofactors. Not surprisingly, mitochondrial iron homeostasis is a tightly regulated process that affects overall cellular iron homeostasis (5). The importance of the metazoan mitoferrins was first demonstrated by analysis of the anemic zebrafish mutant frascati, leading to the discovery that Mfrn12 and -2 play essential roles in providing iron for heme and Fe–S cluster biosyntheses in animal mitochondria (6). Concordant to their proposed role in iron trafficking, pathological iron accumulation in mitochondria has been tied to up-regulation of mitoferrins (7). Studies in yeast had previously led to the discovery of putative iron transporters in the mitochondrial inner membrane, Mrs3/4, which are yeast orthologs of mitoferrins (8, 9). Deletions of Mrs3 and Mrs4 in yeast, however, result in a phenotype evident only in iron-starved conditions, whereas deletion of Mfrn1 is embryonically lethal (6, 10). Because the activity of Mfrn1/2 has not yet been validated by a reconstituted in vitro assay, the fundamental question of whether they can transport iron, or some other substrate that underlies their roles in iron trafficking and homeostasis, remains unanswered.
Mfrn1/2 belong to the SLC25 family of transporters, also known as the mitochondrial carrier (MC) family (11). Members of this family, which is the largest family of transporters, are also the smallest transporters known, encompassing only six transmembrane helices. However, they transport a wide array of small- and medium-size organic molecules and metabolites with a large stereochemical diversity (12). The conserved topology is a tripartite repeat of about 100 amino acids folded into two transmembrane helices connected by an amphipathic helix at the membrane-aqueous interface in the mitochondrial matrix (Fig. 1A) (13). All MCs contain the family's signature motif, PX(D/E)X2(K/R)X20–30(D/E)GX4(W/Y/F)(K/R)G, which is present, with some divergence, in all three repeats of each carrier (Fig. S1) (14). Additionally, on the cytosolic side of the even-numbered transmembrane helices in MCs are conserved (Y/F)(D/E)X2(K/R) motifs. The two (D/E)X2(K/R) motifs, one on each side of the membrane, are hypothesized to constitute two salt-bridge networks that alternate in gating the MCs during their transport cycles (13, 15, 16). The conserved topology, in combination with the diversity of the structures of molecules transported, pose intriguing questions about the mechanism of transport used by the MCs. Besides these universally conserved motifs across the entire family, there are residues that are strictly conserved in mitoferrins but not in the other members (Fig. S2). In the absence of a purification scheme for mitoferrins and a reconstituted in vitro assay, the individual importance of these residues in the function of mitoferrin-1/2 has not been addressed. Previously, conserved histidines were identified as requisite for iron uptake by Mrs3/4 (17); however, the specific role of these residues in the transport mechanism is unclear.
Figure 1.
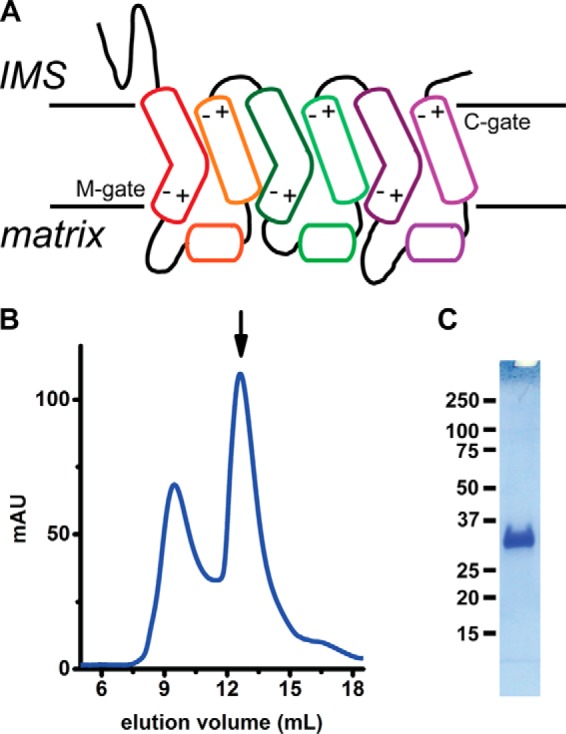
Purification of O. niloticus mitoferrin-1 (TMfrn1). A, putative topology of mitoferrin-1. +/− symbols depict locations of polar residues that form the salt bridges on the cytoplasmic and matrix sides. B, size-exclusion chromatogram of TMfrn1 as the final preparative step. The arrow indicates the TMfrn1 peak. C, SDS-PAGE of TMfrn1. IMS, intermembrane space; C-gate, cytosolic gate; M-gate, matrix gate; mAU, milli-absorbance units.
Detailed mechanistic understanding of the MC transport cycle has been hampered by a lack of structural information. The only structures solved to date are crystal structures of ADP/ATP carriers complexed with the inhibitor carboxyatractyloside, which enforces the cytosol-facing state of these transporters (13, 15), and NMR structures of mitochondrial uncoupling protein (18). Carboxyatractyloside has been shown to dramatically enhance ADP/ATP carrier stability, and there are no structures of the carrier without it (19). Nevertheless, the limited structural data have been integrated with mutational analyses of carrier-mediated substrate uptake, leading to proposals for contact sites within the MC cavity responsible for substrate recognition (20, 21). Iron is chemically quite different, however, from the typical cargoes of MCs; thus, it is virtually impossible to reliably deduce information about the transport mechanism of mitoferrins and the residues involved in the process from the scarce existing structural information.
Interrogating the function of Mfrn1 in vitro required overcoming technical challenges pertaining to the instability of eukaryotic membrane proteins and redox activity of iron. The substrate specificities of mitochondrial carriers have been primarily established by protein expression in Escherichia coli, refolding from inclusion bodies, and reconstitution into proteoliposomes followed by in vitro transport assays (12). Although this method has been useful for discovering substrates of uncharacterized carriers, the biochemical integrity of the preparations has been called into question (19, 22). There have been very few cases of eukaryotic membrane proteins successfully purified from bacterial hosts for biochemical or structural studies (23). Moreover, eukaryotic membrane proteins are fragile molecules once extracted from the membrane, and the harsh detergents used for solubilization and purification of the carriers using this method cast doubt about the integrity of the proteins throughout such preparations. Accordingly, biophysical data on interactions between the carriers and their respective substrates exist for only a few MCs (24–27).
Iron is a troublesome substrate in a reconstituted transport assay in proteoliposomes due to 1) its oxidation from 2+ to 3+ and subsequent precipitation and 2) its ability to catalyze lipid peroxidation, leading to leaky liposomes (28). Many of the common buffers used for biochemistry also coordinate iron, thus significantly affecting the molecular identity of the species available for transport (29). These hurdles have likely contributed to a dearth of in vitro validation of iron transport by mitoferrins and any detailed biophysical and mutational analyses.
In this study, we report an in vitro reconstitution and detailed biochemical and biophysical investigation of the transport function of Mfrn1. We have developed a heterologous overexpression method in Pichia pastoris for purifying Mfrn1 in the natively folded state with mild detergents. Mfrn1 binds with ∼102 micromolar affinity to Fe(II), Mn(II), Co(II), and Ni(II). We have developed a robust proteoliposome-reconstituted transport assay for investigating the iron transport activity of Mfrn1 and show that, besides iron, it also transports manganese, cobalt, copper, and zinc but discriminates against nickel. We have investigated several residues that are specifically conserved in Mfrn1 and its orthologs to assess their importance in transport activity. Our results identify histidine, cysteine, and methionine residues that are critical for iron transport. A subset of these are also important for substrate binding by Mfrn1, making them strong contenders for forming metal ion–binding sites during the transport process. Iron transport by Mfrn1 is altered by the pH gradient across the proteoliposome membrane, but substrate translocation itself is not proton-coupled. Addition of putative in vivo low-molecular-weight ligands of iron has little effect on transport, indicating that Mfrn1 can uniport iron in ionic, unchelated form.
Results
Expression and purification of natively folded mitoferrin-1 from P. pastoris
An essential prerequisite for in vitro biochemical and biophysical characterization is a stable, biochemically well behaved protein construct amenable to heterologous overexpression and purification. At the outset, we elected not to use refolded material from inclusion bodies by overexpression in E. coli as has been done for various other MCs. Hence, we evaluated a number of Mfrn1 orthologs using fluorescence size-exclusion chromatography (FSEC) for expression yield in Pichia and behavior during membrane extraction and size-exclusion chromatography using mild detergents (30). Mfrn1 from Oreochromis niloticus, henceforth referred to as TMfrn1, was a suitably behaved candidate with high expression and a monodisperse chromatographic profile, indicating that the protein would be satisfactory for in vitro functional characterization. Importantly, TMfrn1 could be extracted in mild detergents. However, supplementation with lipids was essential during chromatographic purification steps. As shown in Fig. 1B, TMfrn1 runs as a symmetric, monodisperse peak at 12.5 ml with greater than 95% purity by SDS-PAGE (Fig. 1C). Molecular identity was confirmed by tryptic digestion and mass spectrometry (not shown).
Microcalorimetric measurement of transition metal ion binding by TMfrn1
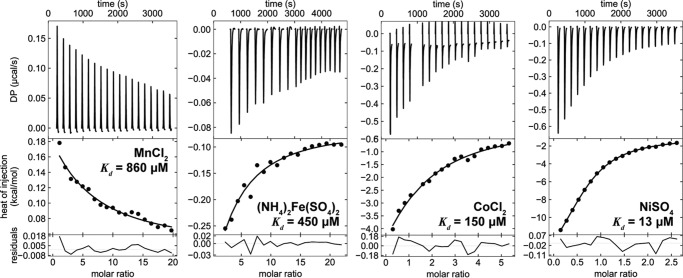
Substrate binding is an essential first step for most transporters as part of the transport cycle. Although Mfrn1 is a putative iron transporter, its binding to iron has not been studied (9, 31). Mitochondria require multiple transition metals, manganese, iron, copper, and zinc, which must be imported into the matrix for incorporation into their intended enzymes. To interrogate whether this range of transition metals could serve as substrates for TMfrn1, we used isothermal titration calorimetry (ITC). As depicted in Fig. 2, TMfrn1 binds various first-row transition metal ions with the order of affinities resembling the Irving-Williams series. The measured dissociation constants for Co2+ and Fe2+ are 150 and 450 μm, respectively, values similar to the affinities of other transporters for their substrates, whereas TMfrn1 affinity for Mn2+ is considerably weaker at 860 μm. Interestingly, TMfrn1 binds Ni2+ with an order of magnitude tighter affinity, 13 μm. This is consistent with nickel being an effective inhibitor of iron transport (see “Metal ion transport by TMfrn1” below). Unfortunately, titrations with Cu2+ and Zn2+ proved unfeasible because of rapid precipitation of either the metal ion or detergent-solubilized TMfrn1. Similarly, titrations with TMfrn1 at pH 6.5 failed due to protein precipitation.
Figure 2.
Transition metal binding to TMfrn1 assessed by isothermal titration calorimetry. Binding isotherms were fit to a single-site model. DP, differential power.
Development of an in vitro iron and copper transport assay
The next step was to verify whether TMfrn1 is a bona fide iron transporter. Other metal ions such as cobalt and manganese have been used as surrogates for iron in studying transition metal transporters in vitro (32, 33), and the use of iron as a transporter substrate is quite scarce. We sought without success a robust reconstituted proteoliposomal iron transport assay that worked reproducibly in our hands. One of the difficulties is illustrated in Fig. S3A, which shows an experiment to optimize conditions with protein-free control liposomes encapsulating the fluorescent probe Phen Green SK (PGSK), a sensor of first-row transition metal ions. We found substantial spurious quenching in the absence of added metal ions (Fig. S3A). As shown, removal of trace divalent ions from assay buffers with Chelex resin mitigates the background fluorescence quench.
Another problem is illustrated by Fig. S3B, which again shows a protein-free liposome control experiment. Addition of iron results in quenching of PGSK fluorescence, presumably through the formation of leaky vesicles likely caused by oxidation of lipids by Fenton-derived reactive oxygen species. This background signal would preclude accurate detection of protein-mediated metal transport. We tested a variety of additives to lower this background leakage signal. One important consideration was not to generate a stable complex of iron that would not release iron to TMfrn1. We decided to use sorbitol, which is a known scavenger of reactive oxygen species and was protective against lipid peroxidation in our assay but is not known to form strong complexes with divalent first-row transition metal ions in the pH range of our study (34).
Similarly, to investigate potential copper transport activity of TMfrn1, we surveyed the literature without success for a reconstituted proteoliposomal assay. As depicted, Cu2+ is also highly destabilizing to our protein-free liposome controls, thus rendering it impossible to delineate nonspecific leak from protein-mediated copper transport (Fig. S3C). We tested a variety of additives, sorbitol, glycine, methionine, and glutathione (not shown), to circumvent this nonspecific leak. The best results were obtained from addition of histidine, which protected the liposomes from copper-induced damage.
Metal ion transport by TMfrn1
TMfrn1 shows robust iron transport activity using the assay conditions described above (Fig. 3). When the iron concentration is varied, fitting of the initial quenching rates yields an apparent Km of 4 μm (Fig. 3, A and B). This Km value for iron lies within the measured chelatable iron in the cellular labile iron pool (35). TMfrn1 readily transports cobalt, copper, and zinc, whereas manganese is a poorer substrate (Fig. 3C). Intriguingly, TMfrn1 does not transport nickel. Given that TMfrn1 effectively binds nickel, we tested whether nickel can impede iron transport by TMfrn1. Indeed, nickel potently blocks iron transport with an IC50 of 1 μm (Fig. 3, D and E).
Figure 3.
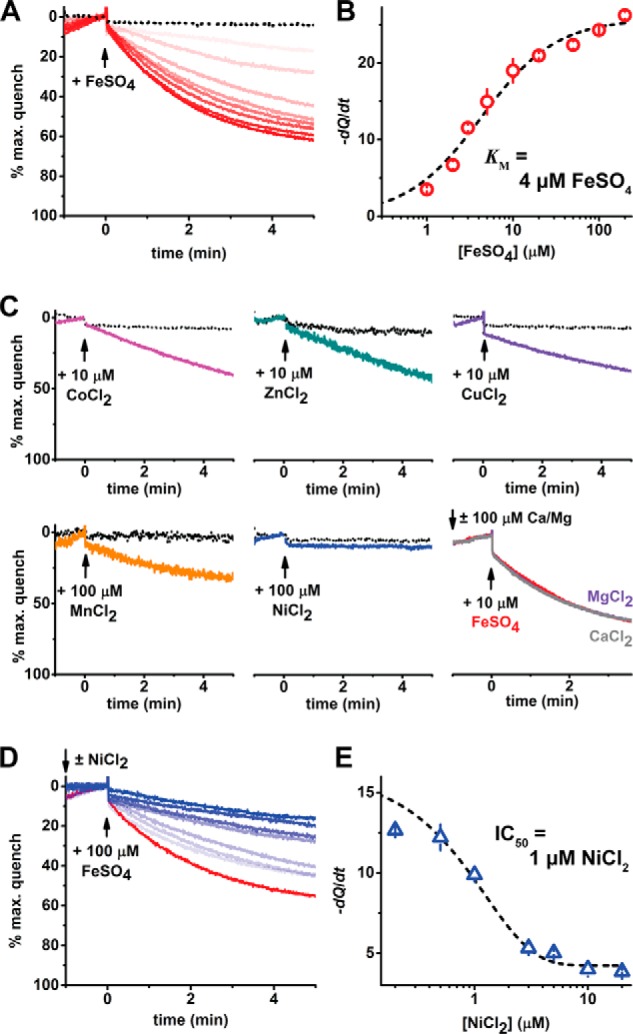
Proteoliposome-reconstituted TMfrn1 transports iron and other transition metals. A, representative PGSK quenching curves upon addition of iron to (proteo)liposomes. Red traces are escalating concentrations of iron (1–200 μm) applied to TMfrn1-containing liposomes. Black dotted traces show protein-free liposomes exposed to 100 μm iron. Traces are plotted as a percentage of maximal PGSK quenching after addition of the ionophore pyrithione (2 μm). B, concentration dependence of TMfrn1 iron transport. Depicted in red are initial quenching rates after iron addition, and points were fit to the Michaelis-Menten equation (black line). C, TMfrn1 transports cobalt, zinc, copper, and manganese but not nickel. Colored traces depict TMfrn1 liposomes; dotted black traces are protein-free liposomes. D, nickel blocks TMfrn1 iron transport. Shown are representative traces of 0–20 μm nickel applied prior to 100 μm iron; the red trace shows 0 nickel with blue traces showing escalating [nickel]. E, concentration dependence of nickel on TMfrn1 iron transport. Depicted in blue are initial quenching rates after iron addition; the black line shows a sigmoidal fit of initial quenching rates. In B and E, error bars depict S.D. of triplicates.
We also performed similar transport competition assays with the alkali metal ions magnesium (Mg2+) and calcium (Ca2+) (Fig. 3C, bottom right). Unlike with nickel, large excesses of magnesium or calcium had no effect on iron uptake, indicating that TMfrn1 efficiently discriminates between alkali and transition divalent metals. Curiously, the apparent Km of iron transport is 2 orders of magnitude lower than the affinity measured by ITC (4 versus 450 μm, respectively). This disparity could possibly arise from the differing milieus (detergent versus lipid bilayer) and the low titration temperature required for maintaining TMfrn1 stability in detergent.
pH dependence of iron transport
Mitochondria typically maintain a membrane potential (ΔΨm) across the inner membrane, in part composed by the pH gradient, which drives ATP synthesis. To test the effect of pH on the transport activity of TMfrn1, we systematically varied the pH of the external solution while holding the liposome internal pH at 6.5 or 7.4. The iron transport activity of TMfrn1 is substantially faster at alkaline pH inside the liposomes, which is consistent with the physiological ΔpH in mitochondria (Fig. 4A). Notably, the transport rate is unaffected by increasing the pH gradient across the membrane between external pH values of 7.4 and 6.5. Below an external pH of 6.5, however, iron transport slows, revealing that TMfrn1 activity is not dependent on a pH gradient per se but rather suggests that protonatable residue(s) with a pKa between 6 and 6.5 are important for metal translocation. This finding agrees with our TMfrn1 mutational analysis (see “Mutational analysis of TMfrn1 transport activity”), which shows several histidines playing critical roles in substrate recognition and transport.
Figure 4.
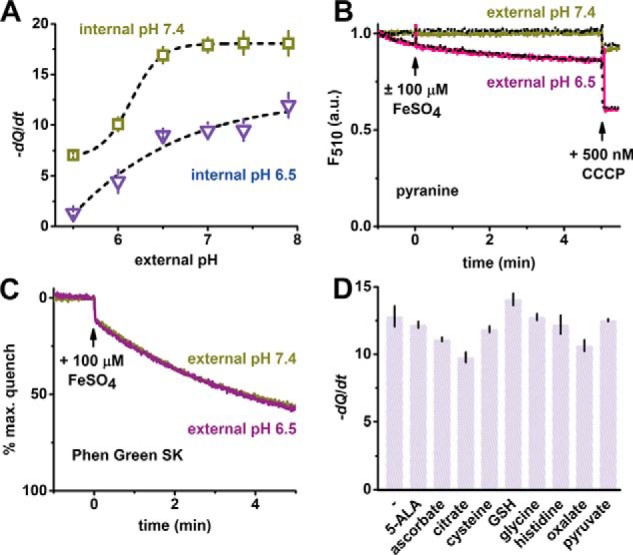
Modulation of TMfrn1 iron transport by pH and ligands. A, pH dependence of TMfrn1 iron transport. Points depict initial quenching rates after iron addition. Black lines are sigmoidal fits of quenching rates at internal pH 7.4 and 6.5. B, TMfrn1 iron transport does not alter intraliposome pH. Measurement of pH-dependent pyranine fluorescence in the presence or absence of iron. Solid colored lines show addition of iron to TMfrn1 proteoliposomes; dotted black lines lack iron. Liposomes contain 50 μm pyranine and contents are weakly buffered with 2 mm MOPS, pH 7.4, 0.5 mm EDTA, 140 mm KCl. Carbonyl cyanide m-chlorophenyl hydrazine (CCCP) was added as indicated to dissipate proton gradient. C, weakly buffered TMfrn1 proteoliposomes retain iron uptake. Liposome internal contents are similar to A except PGSK (instead of pyranine) was used, and no EDTA was added. D, labile iron pool candidates have negligible effect on TMfrn1 iron transport. A 100 μm concentration of each iron ligand was added to the TMfrn1 proteoliposome suspension followed by 10 μm iron, and PGSK fluorescence quenching was monitored. Plotted are initial PGSK quenching rates. Assays were performed with internal pH 7.4 TMfrn1 proteoliposomes diluted into external pH 6.5 buffer. 5-ALA, 5-aminolevulinic acid; GSH, glutathione; a.u., arbitrary units. In A and D, error bars depict S.D. of triplicates.
A number of mitochondrial carriers symport protons with their substrates, exploiting the energy of the pH gradient. To more explicitly address whether TMfrn1 uses a similar symport with H+, we tested transport in the presence of a pH-sensitive fluorophore, pyranine, entrapped within TMfrn1-containing proteoliposomes. At pH 7.4 inside and two different external pH values, 6.5 and 7.4, no iron-dependent detectable change in pH was observed, indicating that TMfrn1 does not cotransport H+ along with iron (Fig. 4B).
Effect of candidate physiological ligands on TMfrn1 transport
Iron has been postulated to be sequestered in the cytosol by a “labile iron pool” that refers to iron chelated by cellular small-molecule ligands (35). We tested various such putative ligands of iron in the transport assay to investigate the possibility of Mfrn1 transporting iron in a ligated form. Although in a few cases we saw a small decrease in the transport rate, we did not observe any of the ligands noticeably enhancing uptake (Fig. 4D). The decrease in transport rates in the presence of citrate, oxalate, and ascorbate most likely stems from these multidentate ligands coordinating and thus shrinking the pool of available iron.
Mutational analysis of TMfrn1 transport activity
Equipped with a reconstituted in vitro transport assay, we pursued a mutagenesis-based analysis of Mfrn1 function. In choosing the residues, we used the following considerations: 1) residues that are conserved in orthologs of Mfrn1 but not necessarily across all mitochondrial carriers (36) and 2) residues with side chains that might serve as a coordinating ligand to a soft metal ion (Fig. S2). Also, a small subset of mitochondrial carriers, including Mfrn1/2, has N-terminal extensions (Fig. S1) that the ADP/ATP carrier and most other members of the family lack. Of these, the N-terminal extensions of glutamate/aspartate carriers AGC1/2 and ATP-Mg/Pi carriers APC1/2/3 encompass EF-hand domains, which bind calcium (37, 38). However, the functions of the N-terminal extensions in other members of this subset, including mitoferrins, are largely unknown. We examined the effect of truncating the N terminus on transport activity by TMfrn1.
Interestingly, TMfrn1 is particularly sensitive to truncation of both C and N termini. Deletion of 18 residues from the N terminus or 10 residues from the C terminus drastically reduces iron transport (Fig. 5A). Of the residues that were most disruptive to function, Glu-30 and Tyr-31 are on the N-terminal side, putatively in the intermembrane space. Mutation of Tyr-31 to Ser retains function, pointing to the importance of the hydroxyl group in the side chain at this position. Mutation of this conserved EYE motif to AAA nearly abolishes TMfrn1 expression, implying a role in the protein's folding (Fig. S4A). Two other highly conserved residues, Asn-222 and Tyr-227, show little influence on iron uptake. Mutation of these residues, present in intermembrane space loop 2, results in poorer protein expression, again suggesting a role in biogenesis and/or stability of TMfrn1. We also examined three positions not conserved in mitoferrins, Met-41, Met-44, and Ser-243. As anticipated, Met-41 and Ser-243 had very little effect on transport. Conversely, M44A retained only 60% of wildtype activity and was rescued by M44C, pointing to a possible utility of a coordinating side chain with sulfur at this position. Although Met-44 is not conserved in lower eukaryotes, it is more conserved in metazoans (Fig. S2).
Figure 5.
Mutational analysis of TMfrn1 substrate transport and binding. A, relative iron uptake of the indicated TMfrn1 mutants reconstituted into proteoliposomes. Assays were performed as in Fig. 3 with symmetric pH 7.4 buffer inside and outside liposomes. The extent of PGSK quenching was measured 5 min after addition of iron and normalized to wildtype. Shown is a topology diagram of TMfrn1 with mutations color-coded according to the lower panel; the diagram was adapted from Protter output (53). Error bars depict S.D. from five or six replicates across two independent experiments. B, ITC measurement of TMfrn1 mutant affinities for cobalt. Measurements were performed and fitted similarly to Fig. 2 and plotted for comparison with iron uptake efficiencies from A. IMS, intermembrane space.
Because Fe2+ is a borderline soft metal ion, it is presumably coordinated by amino acid side chains with soft coordinating atoms during the mitoferrin-1 transport cycle. This supposition is clearly borne out in the residues that are important in our mutagenesis study: all the histidines we investigated perturb transport function of TMfrn1; His-55 and His-210 have more severe effects than His-113 and His-152. Interestingly, the H55C and H113S mutants fully recover transport function. Although the hydroxyl group of serine is chemically different from the imidazole group of histidine, these results argue for the importance of coordinating side chains at these two positions. Two methionines, Met-156 and Met-202, also appear to be important for TMfrn1 activity; the M156C mutant retains almost full function, whereas the M156A mutant is severely compromised, suggesting that the Met-156 side chain aids in iron coordination and is recapitulated by cysteine. The T149S mutant is only about 50% as efficient as wildtype, indicating that the coordinating capability of threonine at this position is probably dispensable.
Because cysteines are known to coordinate soft transition metal ions in proteins (39), we examined the importance of the cysteines in TMfrn1 for metal transport activity. TMfrn1 has four cysteines of which the N-terminal Cys-4 is not conserved; of the remaining three, all are highly conserved. When we made a quadruple cysteine → serine mutant, TMfrn1 stability was severely diminished, and the protein precipitated during the purification process (Fig. S5A). We then broadly examined orthologous Mfrn1 sequences to choose better replacements than serine for cysteine using sequence alignment and engineered a cysteine-free version of TMfrn1, C4S/C119T/C179T/C253I, that had reasonable expression yield and stability. Although this mutant was amenable to large-scale purification, it was not competent for iron transport. Of the individual mutants, C119T and C253I were severely compromised in transport activity, whereas C179T retained about 50% of transport activity, implying that Cys-119 and/or Cys-253 may be important for coordination of the substrate.
MCs have two sets of highly conserved polar residues, one on the matrix side and the other on the cytosolic side (Figs. 1 and S1). These residues are thought be important for stabilizing the matrix- or cytosol-facing conformations of the carriers (16). Ablating one, two, or three cytosolic gate (C-gate) salt bridges in TMfrn1 by replacing charged Lys or Asp/Glu with uncharged Ala results in essentially non-expressing protein (Fig. S4B). Mutations of the matrix-gate residues, however, have very different effects: mutating all three bridges by replacing Lys with Ala yields a chromatographically well behaved construct. However, transport activity of the mutant is severely compromised, thus pointing to the importance of these bridge residues for the function of Mfrn1/2. Abolition of even a single putative matrix-gate salt bridge, mutant K254A, results in a loss of about 50% activity.
Microcalorimetry of cobalt binding by TMfrn1 mutants
The effect of a mutation on transport activity can manifest through multiple mechanisms. We decided to test the metal ion–binding properties of selected mutants to investigate whether the specific mutants affected substrate binding as well. We hypothesized that a combination of decreased transport and binding would be a stronger indicator of that residue being involved in metal ion coordination during the transport cycle. Three of the four His → Gln mutants, H55Q, H113Q, and H210Q, have substantially weakened affinity for cobalt, whereas H152Q retains wildtype binding (Fig. 5B). Proximal to His-152 are Asp-153 and Met-156, and mutations to Asn and Val, respectively, dramatically lessen TMfrn1 affinity for cobalt. Unexpectedly, TMfrn1 CysΔ retains wildtype binding of cobalt and nickel, which contrasts with the mutant's severely diminished transport activity (Figs. 5B and S5B).
Discussion
Mitoferrin-1 and -2 are the only reported major carriers of iron into mitochondria, and this process is crucial for high-throughput biosynthesis of iron-containing cofactors and maintenance of cellular iron homeostasis. The importance of mitoferrins and their function as iron transporters have only been inferred by genetic and cell-based studies. Moreover, deletion of Mrs3/4, which are the yeast orthologs of mitoferrin-1/2, results in very different phenotypes compared with deletion of Mfrn1 in animals, making it difficult to infer the function of mitoferrin-1/2 from studies on Mrs3/4. In any case, the only study reporting on the functional reconstitution of Mrs3 used refolded proteins from E. coli inclusion bodies, a method whose biochemical integrity has been called into question. Here we resolve various aspects about the biochemistry of iron transport by mitoferrins. 1) We present a strategy for native purification of TMfrn1 from a heterologous expression host, P. pastoris, using mild detergents for extraction and a combination of affinity and size-exclusion chromatographic steps. 2) We show, using a fluorescence-based assay that, when reconstituted in proteoliposomes, TMfrn1 has robust iron transport activity. Reminiscent of other transporters of divalent first-row transition metal ions (40–42), TMfrn1 is promiscuous and can transport divalent iron, manganese, cobalt, copper, and zinc. 3) We investigate the importance of residues that are conserved in mitoferrins and identify a number of candidates that are likely involved in iron binding and/or transport.
Our findings strongly support the premise that mitoferrins are high-affinity or -throughput iron transporters. Mitochondria are sites for utilization of other transition metals as well, notably manganese, copper, and zinc, but the mechanisms by which these metal ions traverse the mitochondrial inner membrane are poorly understood, likely due to the presence of multiple and redundant transport pathways. Our data unambiguously demonstrate that TMfrn1 can transport each of these metals in their free ionic forms, although the means of metal delivery to mitoferrins in the cellular context is obscure. TMfrn1 transports the relatively soft first-row transition metal ions, but we could not detect any competition by the harder divalent alkali metal ions magnesium and calcium, arguing that TMfrn1 efficiently discriminates between these two classes. Given that there are dedicated transport systems in the mitochondrial inner membrane for alkali metal ions (43–46), mitoferrins do not appear to function redundantly for magnesium/calcium uptake. TMfrn1 substrate promiscuity within the first-row transition metal ions is reminiscent of other softer transition metal ion transporters (40–42).
Transition metal ions are closely sequestered and chaperoned during their trafficking in cells; thus, the metal ion selectivity in the physiological context of some of these transporters likely arises from the selectivity of the metal delivery to the transporter. We speculate that interaction with various chaperones leads, for instance, to differential copper selectivity between Mrs3 and -4 (47) because the residues we identified as important for transport are highly conserved among mitoferrins. Clearly, further study is needed to ascribe specific shuttles for delivering metal ions to mitoferrins at the mitochondrial inner membrane.
To demonstrate the transport activity of TMfrn1, we developed, separately, robust iron and copper transport assays in proteoliposomes. Typically, other transition metal ions have been used as a surrogate for iron in the biochemical investigation of iron transporters in vitro (32, 33). Our results suggest that the difficulty with using iron in a minimal proteoliposome-based assay is due to the formation of nonspecific leaks in the vesicles, presumably derived from lipid peroxidation. After an extensive survey, we found several factors, including Chelex treatment of solutions, appropriate choice of buffer, and addition of sorbitol or histidine, to be crucial for a successful and unambiguous reconstitution of transport function. Because iron and copper are ubiquitously important in biology, these assays will be valuable tools in studying other iron/copper transporters in vitro.
A part of cellular iron has been proposed to exist as a kinetically available pool, bound to low-molecular-weight ligands (48). Because the known substrates of members of the MC family are predominantly small organic molecules like nucleotides, keto acids, and vitamins, at the outset we hypothesized that the substrate for Mfrn1 could be a chelated form of iron that would resemble other canonical substrates of the MC family. For the battery of plausible intracellular ligands for iron that we tested, we did not measure any detectable increase in transport activity of TMfrn1 in their presence. This leads us to conclude that Mfrn1 transports iron in an unchelated form.
Iron transport by TMfrn1 shows a degree of sensitivity to acidic pH. Our experiments with pyranine, a pH-sensitive fluorophore, indicate that this is not due to H+ being a cosubstrate for Mfrn1 because we did not observe any detectable changes in intravesicular pH resulting from iron transport. The dependence of the transport rate on the external pH (when the internal pH is 7.4; Fig. 4A) implies that the protonation of a histidine residue is involved. This is consistent with our mutagenesis data that identified two histidines, His-55 and His-210, important for the transport activity of TMfrn1. Interestingly, the transport activity is otherwise not affected by a pH gradient mimicking the mitochondrial ΔpH. Our result contrasts with prior findings as iron uptake in isolated yeast mitochondria or submitochondrial particles was seen to be dependent on mitochondrial polarization (31, 49). The disparity perhaps reflects functional differences between a metazoan Mfrn1 and its fungal counterpart or, more likely, differences in our defined proteoliposomes versus complex organellar membranes.
The ion selectivity of TMfrn1 suggests that it uses soft coordinating ligands such as cysteine, histidine, and methionine for binding and transport. In agreement with that prediction, our mutational analyses of transport function show that the residues that have the highest effect predominantly contain side chains that can coordinate soft metal ions, His-55, Cys-119, Met-156, Met-202, His-210, and Cys-253. These residues could be involved either in forming the metal-binding site or in making important interactions during the transport process. Interestingly, nickel potently blocks iron transport by TMfrn1, and although this may not be physiologically relevant, it may prove to be a useful tool for biophysical studies of TMfrn1.
TMfrn1 binds metal ions in calorimetric studies with affinities that reflect the Irving-Williams series, implying that TMfrn1 does not have a substrate-binding site specifically tailored for iron. Surprisingly, the CysΔ version of TMfrn1 is inactive for metal transport but retains wildtype metal binding, indicating that none of the cysteines are involved in forming a stable metal ion–binding site, which the calorimetric data presumably report on. The residues we identified as important for substrate binding are dispersed along the first four transmembrane helices of TMfrn1 (Fig. 5A). Presumably, these residues are involved in forming more than one (i.e. primary and auxiliary) binding site for the metal or form important contacts in different conformational states during the transport process. Clarifying these questions will require further structural and functional interrogation.
Mitochondrial carriers have highly conserved sets of polar residues, one set on the matrix side and another on the cytosolic side, that have been proposed to form salt bridges stabilizing states facing alternate sides of the membrane (Fig. 1A). Our mutational data suggest that these interactions are important in the case of Mfrn1 as well. Thus, Mfrn1 seemingly shares some of the basic features of the transport mechanism with other MCs. Conversely, ionic Fe(II) is markedly disparate relative to substrates of the other MCs. Conformational exchange of other MCs appears to be instantiated by substrates contacting sites on even-numbered helices within the carriers' cavity (50). Our mutational analysis implies that TMfrn1 utilizes a somewhat dissimilar recognition motif with functionally important residues present on each of the first four transmembrane helices. Mfrn1 also diverges from other MCs due to the functional requirement of its extended N terminus. In the case of Mfrn1, perhaps the N-terminal extension promotes conformational exchange after the initial engagement of the substrate metal ion. Future high-resolution structural studies in different states of the transport pathway will help unravel further detailed atomic insights into the chemical basis of mitoferrin function.
Experimental procedures
Materials
N-Dodecyl β-d-maltopyranoside (DDM) was purchased from Anatrace, and Triton X-100 and Chelex resin were from Sigma-Aldrich. Lipids were purchased from Avanti Polar Lipids. Phen Green SK (dipotassium salt) was from Thermo Fisher Scientific. Zeocin was obtained from Invitrogen. Components for bacterial and yeast media were procured from Fisher; yeast nitrogen base was from Research Products International (RPI), Corp. Talon cobalt metal affinity resin was from Clontech. Other chemicals were obtained from Sigma-Aldrich.
Protein expression and purification
The cDNA encoding for O. niloticus Mfrn1 (and other orthologs) was codon-optimized for P. pastoris, synthesized, and cloned into expression plasmid pPICZ A. Protein expression was assessed by FSEC (30). The best behaving Mfrn1 construct was engineered as a fusion protein between N-terminal small ubiquitin-like modifier and C-terminal enhanced GFP tags with a C-terminal decahistidine affinity tag. Human rhinovirus 3C protease sites were included for proteolytic tag removal. Mutageneses were performed via PCR and confirmed by DNA sequencing. Constructs were electroporated into Pichia strain SMD1163H, and transformants were selected on yeast extract-peptone-dextrose-sorbitol plates supplemented with 400 μg/ml Zeocin. Individual Pichia colonies were screened for TMfrn1 expression using in-cell GFP fluorescence or FSEC. Pichia cultures were grown in buffered minimal glycerol medium at 30 °C to an A600 of ∼25, pelleted by centrifuging at 3000 × g for 5 min, resuspended into buffered minimal methanol medium, and shaken for 24 h at 25 °C with two total additions of methanol up to 0.5%. Pichia were again pelleted and flash frozen in liquid N2.
TMfrn1 was purified from isolated mitochondria. Mitochondria were harvested from Pichia by resuspending cells (100 g/150 ml of buffer) in lysis buffer (100 mm HEPES-Tris, pH 7.8, 650 mm sorbitol, 5 mm EDTA, 5 mm aminohexanoic acid, 5 mm benzamidine HCl, 5 mm EDTA, 10 mm DTT, 0.2% BSA) and then passing one time through a horizontal bead mill (Dynamill). Lysate was clarified by centrifugation at 3500 × g for 30 min. The supernatant was then spun at 24,000 × g for 90 min to harvest mitochondria. Pellets were resuspended in wash buffer (100 mm HEPES-Tris, pH 7.4, 650 mm sorbitol, 5 mm aminohexanoic acid, 5 mm benzamidine HCl, 10 mm β-ME) and spun again at 24,000 × g for 50 min. Mitochondrial pellets were then resuspended in storage buffer (100 mm HEPES-KOH, pH 7.4, 10% glycerol, 5 mm β-ME) and spun a final time at 24,000 × g for 50 min. Mitochondrial pellets were resuspended in storage buffer to a protein concentration of ≈20 mg/ml, aliquoted, and flash frozen in liquid N2. TMfrn1 mutants were purified from cell powder produced by cryomilling Pichia in liquid N2-cooled stainless-steel ball milling jars (Retsch).
TMfrn1 was extracted from mitochondria by adding KCl to 150 mm, TCEP to 2 mm, protease inhibitors, and DDM to 3% and then stirring for 3 h at 4 °C. Extraction from cell powder entailed resuspending the powder (10 g up to 50 ml) in extraction buffer (50 mm HEPES-KOH, pH 7.8, 150 mm KCl, 10 mm β-ME, 2 mm TCEP, protease inhibitors, 0.5 μg/ml DNase), then adding DDM to 3%, and stirring for 3 h at 4 °C. Extracts were clarified by centrifuging at 38,000 × g for 30 min, and then supernatants were supplemented with 2 mm imidazole, pH adjusted to ≈7.4 with 1 n KOH, and batch-bound to Talon affinity resin for 4–5 h. Talon resin was collected by gravity flow in a polypropylene column, washed with 10 bed volumes of wash buffer (20 mm MOPS-KOH, pH 7.4, 140 mm KCl, 10 mm imidazole, 5 mm β-ME, 2 mm TCEP, 1 mm DDM, 0.05 mg/ml toCL). The column was plugged; 1 bed volume of wash buffer was added; DDM and toCL were raised to 2 mm and 0.1 mg/ml, respectively; and human rhinovirus 3C protease was added to cleave TMfrn1 from fusion proteins. After rocking the resin slurry overnight at 4 °C, imidazole was added to 30 mm, and TMfrn1 was eluted from the column. TMfrn1 was concentrated and subjected to size-exclusion chromatography over Superdex 200 Increase 10/300 GL equilibrated in 20 mm MOPS-KOH, pH 7.4, 140 mm KCl, 2 mm TCEP, 1 mm DDM, 0.05 mg/ml toCL. TMfrn1 was finally concentrated to ≈2 mg/ml for downstream use.
Isothermal titration calorimetry
TMfrn1 was freshly purified, concentrated, and then dialyzed at 4 °C for 20–24 h against size-exclusion buffer (20 mm MOPS-KOH, pH 7.4, 140 mm KCl, 2 mm TCEP, 1 mm DDM, 0.05 mg/ml toCL). All titrations were performed at 6 °C to preserve TMfrn1 stability. Most experiments were performed on a MicroCal ITC200. Protein solutions were degassed for 15 min prior to placement into the calorimeter, and dialysis buffer was degassed for 1 h before addition of CoCl2, NiSO4, or MnCl2. (NH4)2Fe(SO4)2 titrations were performed on a TA Instruments Nano ITC placed inside an anaerobic glove bag (Coy Laboratory Products). Dialysis buffer for (NH4)2Fe(SO4)2 titrations was additionally sparged with argon for 30 min prior to dialysis, and plasticware was degassed overnight. After placement into the glove bag, TMfrn1 solutions were degassed for 1 h, and dialysis buffer was degassed for 4 h. Thermograms were analyzed using the software packages NITPIC, SEDPHAT, and GUSSI (51).
Proteoliposome preparation
TMfrn1 proteoliposomes were prepared via hydrophobic chromatography (52); protein-free liposomes were prepared in the same manner except with DDM-containing size-exclusion buffer replacing TMfrn1. Lipids in chloroform were dried with argon to a film and then further vacuum-dried overnight. Lipid composition was 55:40:5 1-palmitoyl-2-oleoylphosphatidylcholine:1-palmitoyl-2-oleoylphosphatidylethanolamine:toCL (w/w). All buffers were treated with Chelex resin to remove trace divalent ions. Lipid films were resuspended to 20 mg/ml in 10 mm MOPS, pH 7.4; layered with argon; and bath-sonicated to large unilamellar vesicles, and KCl was added up to 140 mm. Ten sequential additions of 10% Triton X-100 were added to destabilize the liposomes, and the final lipid:detergent ratio was 10:6 by mass. To the destabilized liposomes, freshly purified TMfrn1 was added (10 mg of lipid:100 μg of TMfrn1), and the suspension was rocked at 4 °C for 30 min. The proteoliposome suspension was then passed 15 times over 550 mg of Amberlite XAD-2 at room temperature, diluted to 6.5 mg/ml lipid with inside buffer (10 mm MOPS, pH 7.4, 140 mm KCl), and flash frozen in liquid N2. Phen Green SK (50 μm) was incorporated by freezing/thawing three times; liposomes were then extruded through a 400-nm filter, and free dye was removed by passing the vesicles over Sepharose CL-2B equilibrated in inside buffer.
For pH-dependence transport assays, large unilamellar vesicles were exchanged to internal pH 6.5 (and pH 7.4) by centrifuging pre-extruded proteoliposomes at 21,000 × g for 10 min at 4 °C. Vesicle pellets were resuspended in 10 mm MES-KOH, pH 6.5, 140 mm KCl (or pH 7.4 buffer); PGSK was added to 50 μm; and the samples were freeze/thawed three times, extruded, and passed over Sepharose CL-2B equilibrated in pH 6.5 (or 7.4) buffer.
For pyranine-based detection of intravesicular pH, non-extruded vesicle pellets were resuspended in 2 mm MOPS-KOH, pH 7.4, 140 mm KCl, 0.5 mm EDTA; pyranine was added to 50 μm; and the samples were freeze/thawed three times, extruded, and passed over Sepharose CL-2B equilibrated in 2 mm MOPS-KOH, pH 7.4, 140 mm KCl. PGSK (50 μm) was similarly incorporated, subtracting EDTA.
Proteoliposome metal uptake assays
Fluorescence quenching of Phen Green SK (λex = 506 nm, λem = 530 nm) and pyranine (λex = 450 nm, λem = 520 nm) was monitored using a Cary Varian Eclipse spectrofluorometer at room temperature. (Proteo)liposomes were diluted ≈30-fold to 20 μm [lipid] into Chelex-treated assay buffers (10 mm MOPS-KOH, pH 7.0/7.4/7.9, or 10 mm MES-KOH, pH 5.5/6.0/6.5, 140 mm KCl). Metal stock solutions were made to 100 mm Me2+ (FeSO4 in 20 mm H2SO4; CoCl2, CuCl2, MnCl2, NiCl2, and ZnCl2 in water) each experimental day. For Fe(II) uptake assays in buffers pH 7.0 and higher, assays were supplemented with 1 mm sorbitol. For copper uptake assays, 50 μm histidine was added to assay solutions. Maximal dye quenching was determined by addition of the ionophore calcimycin (for Cu2+ and Mn2+) or pyrithione (other metals) to liposomes to equilibrate internal and external divalent ions.
Author contributions
A. B. conceived the study. A. B. and E. T. C. designed the study and wrote the paper. E. T. C. carried out most of the experiments. A. S. G. contributed to generation of point mutants and characterization of their activities. A. B. and E. T. C. analyzed data. A. B. and E. T. C. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Susan Buchanan (NIDDK) for use of the Dynamill, Yan Li (NINDS) for mass spectrometric analysis, and Caroline Philpott (NIDDK) for use of the anaerobic glove bag and microcalorimeter. We thank Caroline Philpott (NIDDK), Tracey Rouault (NICHD), and Gisela Storz (NICHD) for comments on the manuscript.
This work was supported by National Institutes of Health Intramural Research Program Grant ZIA HD008928-04 and by the Eunice Kennedy Shriver National Institute of Child Health and Human Development and the National Institute of Neurological Disorders and Stroke. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5.
- Mfrn
- mitoferrin
- MC
- mitochondrial carrier
- PGSK
- Phen Green SK
- toCL
- tetraoleoylcardiolipin
- FSEC
- fluorescence size-exclusion chromatography
- ITC
- isothermal titration calorimetry
- DDM
- N-dodecyl β-d-maltopyranoside
- β-ME
- β-mercaptoethanol
- TCEP
- tris (2-carboxyethyl)phosphine.
References
- 1. Pantopoulos K., Porwal S. K., Tartakoff A., and Devireddy L. (2012) Mechanisms of mammalian iron homeostasis. Biochemistry 51, 5705–5724 10.1021/bi300752r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen C., and Paw B. H. (2012) Cellular and mitochondrial iron homeostasis in vertebrates. Biochim. Biophys. Acta 1823, 1459–1467 10.1016/j.bbamcr.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lane D. J., Merlot A. M., Huang M. L., Bae D. H., Jansson P. J., Sahni S., Kalinowski D. S., and Richardson D. R. (2015) Cellular iron uptake, trafficking and metabolism: key molecules and mechanisms and their roles in disease. Biochim. Biophys. Acta 1853, 1130–1144 10.1016/j.bbamcr.2015.01.021 [DOI] [PubMed] [Google Scholar]
- 4. Rouault T. A., and Maio N. (2017) Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem. 292, 12744–12753 10.1074/jbc.R117.789537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mühlenhoff U., Hoffmann B., Richter N., Rietzschel N., Spantgar F., Stehling O., Uzarska M. A., and Lill R. (2015) Compartmentalization of iron between mitochondria and the cytosol and its regulation. Eur. J. Cell Biol. 94, 292–308 10.1016/j.ejcb.2015.05.003 [DOI] [PubMed] [Google Scholar]
- 6. Shaw G. C., Cope J. J., Li L., Corson K., Hersey C., Ackermann G. E., Gwynn B., Lambert A. J., Wingert R. A., Traver D., Trede N. S., Barut B. A., Zhou Y., Minet E., Donovan A., et al. (2006) Mitoferrin is essential for erythroid iron assimilation. Nature 440, 96–100 10.1038/nature04512 [DOI] [PubMed] [Google Scholar]
- 7. Rouault T. A. (2016) Mitochondrial iron overload: causes and consequences. Curr. Opin. Genet. Dev. 38, 31–37 10.1016/j.gde.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Foury F., and Roganti T. (2002) Deletion of the mitochondrial carrier genes MRS3 and MRS4 suppresses mitochondrial iron accumulation in a yeast frataxin-deficient strain. J. Biol. Chem. 277, 24475–24483 10.1074/jbc.M111789200 [DOI] [PubMed] [Google Scholar]
- 9. Mühlenhoff U., Stadler J. A., Richhardt N., Seubert A., Eickhorst T., Schweyen R. J., Lill R., and Wiesenberger G. (2003) A specific role of the yeast mitochondrial carriers Mrs3/4p in mitochondrial iron acquisition under iron-limiting conditions. J. Biol. Chem. 278, 40612–40620 10.1074/jbc.M307847200 [DOI] [PubMed] [Google Scholar]
- 10. Troadec M. B., Warner D., Wallace J., Thomas K., Spangrude G. J., Phillips J., Khalimonchuk O., Paw B. H., Ward D. M., and Kaplan J. (2011) Targeted deletion of the mouse mitoferrin1 gene: from anemia to protoporphyria. Blood 117, 5494–5502 10.1182/blood-2010-11-319483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wiesenberger G., Link T. A., von Ahsen U., Waldherr M., and Schweyen R. J. (1991) MRS3 and MRS4, two suppressors of mtRNA splicing defects in yeast, are new members of the mitochondrial carrier family. J. Mol. Biol. 217, 23–37 10.1016/0022-2836(91)90608-9 [DOI] [PubMed] [Google Scholar]
- 12. Palmieri F. (2013) The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol. Aspects Med. 34, 465–484 10.1016/j.mam.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 13. Pebay-Peyroula E., Dahout-Gonzalez C., Kahn R., Trézéguet V., Lauquin G. J., and Brandolin G. (2003) Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 10.1038/nature02056 [DOI] [PubMed] [Google Scholar]
- 14. Walker J. E., and Runswick M. J. (1993) The mitochondrial transport protein superfamily. J. Bioenerg. Biomembr. 25, 435–446 10.1007/BF01108401 [DOI] [PubMed] [Google Scholar]
- 15. Ruprecht J. J., Hellawell A. M., Harding M., Crichton P. G., McCoy A. J., and Kunji E. R. (2014) Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc. Natl. Acad. Sci. U.S.A. 111, E426–E434 10.1073/pnas.1320692111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. King M. S., Kerr M., Crichton P. G., Springett R., and Kunji E. R. (2016) Formation of a cytoplasmic salt bridge network in the matrix state is a fundamental step in the transport mechanism of the mitochondrial ADP/ATP carrier. Biochim. Biophys. Acta 1857, 14–22 10.1016/j.bbabio.2015.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brazzolotto X., Pierrel F., and Pelosi L. (2014) Three conserved histidine residues contribute to mitochondrial iron transport through mitoferrins. Biochem. J. 460, 79–89 10.1042/BJ20140107 [DOI] [PubMed] [Google Scholar]
- 18. Berardi M. J., Shih W. M., Harrison S. C., and Chou J. J. (2011) Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature 476, 109–113 10.1038/nature10257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crichton P. G., Lee Y., Ruprecht J. J., Cerson E., Thangaratnarajah C., King M. S., and Kunji E. R. (2015) Trends in thermostability provide information on the nature of substrate, inhibitor, and lipid interactions with mitochondrial carriers. J. Biol. Chem. 290, 8206–8217 10.1074/jbc.M114.616607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kunji E. R., and Robinson A. J. (2006) The conserved substrate binding site of mitochondrial carriers. Biochim. Biophys. Acta 1757, 1237–1248 10.1016/j.bbabio.2006.03.021 [DOI] [PubMed] [Google Scholar]
- 21. Monné M., Palmieri F., and Kunji E. R. (2013) The substrate specificity of mitochondrial carriers: mutagenesis revisited. Mol. Membr. Biol. 30, 149–159 10.3109/09687688.2012.737936 [DOI] [PubMed] [Google Scholar]
- 22. Zoonens M., Comer J., Masscheleyn S., Pebay-Peyroula E., Chipot C., Miroux B., and Dehez F. (2013) Dangerous liaisons between detergents and membrane proteins. The case of mitochondrial uncoupling protein 2. J. Am. Chem. Soc. 135, 15174–15182 10.1021/ja407424v [DOI] [PubMed] [Google Scholar]
- 23. He Y., Wang K., and Yan N. (2014) The recombinant expression systems for structure determination of eukaryotic membrane proteins. Protein Cell 5, 658–672 10.1007/s13238-014-0086-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brüschweiler S., Yang Q., Run C., and Chou J. J. (2015) Substrate-modulated ADP/ATP-transporter dynamics revealed by NMR relaxation dispersion. Nat. Struct. Mol. Biol. 22, 636–641 10.1038/nsmb.3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Run C., Yang Q., Liu Z., OuYang B., and Chou J. J. (2015) Molecular basis of MgATP selectivity of the mitochondrial SCaMC carrier. Structure 23, 1394–1403 10.1016/j.str.2015.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whittaker M. M., Penmatsa A., and Whittaker J. W. (2015) The Mtm1p carrier and pyridoxal 5′-phosphate cofactor trafficking in yeast mitochondria. Arch. Biochem. Biophys. 568, 64–70 10.1016/j.abb.2015.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sounier R., Bellot G., and Chou J. J. (2015) Mapping conformational heterogeneity of mitochondrial nucleotide transporter in uninhibited states. Angew. Chem. 54, 2436–2441 10.1002/anie.201408417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng Z., and Li Y. (2007) What is responsible for the initiating chemistry of iron-mediated lipid peroxidation: an update. Chem. Rev. 107, 748–766 10.1021/cr040077w [DOI] [PubMed] [Google Scholar]
- 29. Ferreira C. M., Pinto I. S., Soares E. V., and Soares H. M. (2015) (Un)suitability of the use of pH buffers in biological, biochemical and environmental studies and their interaction with metal ions—a review. RSC Adv. 5, 30989–31003 10.1039/C4RA15453C [DOI] [Google Scholar]
- 30. Kawate T., and Gouaux E. (2006) Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 10.1016/j.str.2006.01.013 [DOI] [PubMed] [Google Scholar]
- 31. Froschauer E. M., Schweyen R. J., and Wiesenberger G. (2009) The yeast mitochondrial carrier proteins Mrs3p/Mrs4p mediate iron transport across the inner mitochondrial membrane. Biochim. Biophys. Acta 1788, 1044–1050 10.1016/j.bbamem.2009.03.004 [DOI] [PubMed] [Google Scholar]
- 32. Ehrnstorfer I. A., Geertsma E. R., Pardon E., Steyaert J., and Dutzler R. (2014) Crystal structure of a SLC11 (NRAMP) transporter reveals the basis for transition-metal ion transport. Nat. Struct. Mol. Biol. 21, 990–996 10.1038/nsmb.2904 [DOI] [PubMed] [Google Scholar]
- 33. Taniguchi R., Kato H. E., Font J., Deshpande C. N., Wada M., Ito K., Ishitani R., Jormakka M., and Nureki O. (2015) Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin. Nat. Commun. 6, 8545 10.1038/ncomms9545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tadolini B. (1987) Iron autoxidation in Mops and Hepes buffers. Free Radic. Res. Commun. 4, 149–160 10.3109/10715768709088100 [DOI] [PubMed] [Google Scholar]
- 35. Hider R. C., and Kong X. (2013) Iron speciation in the cytosol: an overview. Dalton Trans. 42, 3220–3229 10.1039/C2DT32149A [DOI] [PubMed] [Google Scholar]
- 36. Robinson A. J., Overy C., and Kunji E. R. (2008) The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc. Natl. Acad. Sci. U.S.A. 105, 17766–17771 10.1073/pnas.0809580105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thangaratnarajah C., Ruprecht J. J., and Kunji E. R. (2014) Calcium-induced conformational changes of the regulatory domain of human mitochondrial aspartate/glutamate carriers. Nat. Commun. 5, 5491 10.1038/ncomms6491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harborne S. P., Ruprecht J. J., and Kunji E. R. (2015) Calcium-induced conformational changes in the regulatory domain of the human mitochondrial ATP-Mg/Pi carrier. Biochim. Biophys. Acta 1847, 1245–1253 10.1016/j.bbabio.2015.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andreini C., Bertini I., Cavallaro G., Najmanovich R. J., and Thornton J. M. (2009) Structural analysis of metal sites in proteins: non-heme iron sites as a case study. J. Mol. Biol. 388, 356–380 10.1016/j.jmb.2009.02.052 [DOI] [PubMed] [Google Scholar]
- 40. Pinilla-Tenas J. J., Sparkman B. K., Shawki A., Illing A. C., Mitchell C. J., Zhao N., Liuzzi J. P., Cousins R. J., Knutson M. D., and Mackenzie B. (2011) Zip14 is a complex broad-scope metal-ion transporter whose functional properties support roles in the cellular uptake of zinc and nontransferrin-bound iron. Am. J. Physiol. Cell Physiol. 301, C862–C871 10.1152/ajpcell.00479.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Illing A. C., Shawki A., Cunningham C. L., and Mackenzie B. (2012) Substrate profile and metal-ion selectivity of human divalent metal-ion transporter-1. J. Biol. Chem. 287, 30485–30496 10.1074/jbc.M112.364208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mitchell C. J., Shawki A., Ganz T., Nemeth E., and Mackenzie B. (2014) Functional properties of human ferroportin, a cellular iron exporter reactive also with cobalt and zinc. Am. J. Physiol. Cell Physiol. 306, C450–C459 10.1152/ajpcell.00348.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baughman J. M., Perocchi F., Girgis H. S., Plovanich M., Belcher-Timme C. A., Sancak Y., Bao X. R., Strittmatter L., Goldberger O., Bogorad R. L., Koteliansky V., and Mootha V. K. (2011) Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345 10.1038/nature10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Stefani D., Raffaello A., Teardo E., Szabò I., and Rizzuto R. (2011) A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340 10.1038/nature10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schindl R., Weghuber J., Romanin C., and Schweyen R. J. (2007) Mrs2p forms a high conductance Mg2+ selective channel in mitochondria. Biophys. J. 93, 3872–3883 10.1529/biophysj.107.112318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsai M. F., Jiang D., Zhao L., Clapham D., and Miller C. (2014) Functional reconstitution of the mitochondrial Ca2+/H+ antiporter Letm1. J. Gen. Physiol. 143, 67–73 10.1085/jgp.201311096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vest K. E., Wang J., Gammon M. G., Maynard M. K., White O. L., Cobine J. A., Mahone W. K., and Cobine P. A. (2016) Overlap of copper and iron uptake systems in mitochondria in Saccharomyces cerevisiae. Open Biol. 6, 150223 10.1098/rsob.150223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kakhlon O., and Cabantchik Z. I. (2002) The labile iron pool: characterization, measurement, and participation in cellular processes. Free Radic. Biol. Med. 33, 1037–1046 10.1016/S0891-5849(02)01006-7 [DOI] [PubMed] [Google Scholar]
- 49. Lange H., Kispal G., and Lill R. (1999) Mechanism of iron transport to the site of heme synthesis inside yeast mitochondria. J. Biol. Chem. 274, 18989–18996 10.1074/jbc.274.27.18989 [DOI] [PubMed] [Google Scholar]
- 50. Robinson A. J., and Kunji E. R. (2006) Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc. Natl. Acad. Sci. U.S.A. 103, 2617–2622 10.1073/pnas.0509994103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brautigam C. A., Zhao H., Vargas C., Keller S., and Schuck P. (2016) Integration and global analysis of isothermal titration calorimetry data for studying macromolecular interactions. Nat. Protoc. 11, 882–894 10.1038/nprot.2016.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Palmieri F., Indiveri C., Bisaccia F., and Iacobazzi V. (1995) Mitochondrial metabolite carrier proteins: purification, reconstitution, and transport studies. Methods Enzymol. 260, 349–369 10.1016/0076-6879(95)60150-3 [DOI] [PubMed] [Google Scholar]
- 53. Omasits U., Ahrens C. H., Müller S., and Wollscheid B. (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 10.1093/bioinformatics/btt607 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.