Abstract
Derepression of chromatin-mediated transcriptional repression of paternal and maternal genomes is considered the first major step that initiates zygotic gene expression after fertilization. The histone variant H3.3 is present in both male and female gametes and is thought to be important for remodeling the paternal and maternal genomes for activation during both fertilization and embryogenesis. However, the underlying mechanisms remain poorly understood. Using our H3.3B-HA–tagged mouse model, engineered to report H3.3 expression in live animals and to distinguish different sources of H3.3 protein in embryos, we show here that sperm-derived H3.3 (sH3.3) protein is removed from the sperm genome shortly after fertilization and extruded from the zygotes via the second polar bodies (PBII) during embryogenesis. We also found that the maternal H3.3 (mH3.3) protein is incorporated into the paternal genome as early as 2 h postfertilization and is detectable in the paternal genome until the morula stage. Knockdown of maternal H3.3 resulted in compromised embryonic development both of fertilized embryos and of androgenetic haploid embryos. Furthermore, we report that mH3.3 depletion in oocytes impairs both activation of the Oct4 pluripotency marker gene and global de novo transcription from the paternal genome important for early embryonic development. Our results suggest that H3.3-mediated paternal chromatin remodeling is essential for the development of preimplantation embryos and the activation of the paternal genome during embryogenesis.
Keywords: chromatin remodeling, development, embryo, epigenetics, fertilization, gene expression, histone, oocyte, sperm, chromatin remodeling, embryonic development, epigenetic reprogramming, histone variant H3.3, paternal genome activation, embryogenesis, transcriptional repression
Introduction
Embryogenesis begins when two haploid genomes fuse to form a diploid zygotic genome after the sperm enter the oocyte (1, 2). At the time of fertilization, both the sperm and oocyte genomes are transcriptionally repressed while the maternal stored factors in the oocyte support and control the process of early embryogenesis. Maternal-to-zygotic transition is an embryonic development stage under the exclusive control of the newly formed zygotic genome. This developmental process requires zygotic genome activation (ZGA)4 to allow the transition from specified germ cells to a totipotent embryo in which both paternal and maternal genomes undergo dramatic epigenetic reprogramming regulated by maternal factors (3). Overcoming chromatin-mediated transcriptional repression of paternal and maternal genomes is thought to be the first major step to initiate zygotic gene expression after fertilization (4–9).
The mammalian sperm genome is packaged into highly condensed chromatin consisting primarily of protamine but 5–15% residual histones. Following fertilization, ZGA occurs first in the paternal genome (male pronucleus) at the one-cell stage embryo, whereas activation of the maternal genome is usually delayed and occurs at the two-cell stage in mice (10–13). Soon after fertilization, protamine is removed from the sperm genome. The paternal genome subsequently undergoes chromatin remodeling through a massive and highly regulated exchange of canonical and variant histones including H1FOO, H3.3, microH2A, and H2A.Z. The incorporation of histone variants into the paternal genome is suggested to be necessary for the acquisition of totipotency and ZGA (14–18), but the function and mechanism for each histone variant in this process are still poorly understood. The repressive H2A variant macroH2A is found preferentially in the mouse female pronucleus and appears to contribute to its transcriptional silence (15), whereas H1FOO and H3.3 can incorporate into both the paternal and maternal genomes and are probably associated with the transcriptional activation of zygotic genomes (8, 9, 16–19).
Chromatin is composed of histones and DNA. Each 147 base pairs of DNA wraps around eight core histone proteins, consisting of two copies each of the core histones H2A, H2B, H3, and H4, to form the basic chromatin unit defined as nucleosome. Packaging of DNA into nucleosomes not only helps store genetic information but also creates diverse means for regulating DNA-templated processes (6). Interplay between transcriptional machinery and chromatin in the early embryo likely regulates the timing of ZGA (20). Chromatin accessibility by transcriptional machinery is regulated through nucleosome positioning and configuration, heavily influenced by histone variants as well as post-transcriptional modifications of histone tails (14). Histone exchange is a general mechanism for chromatin remodeling during embryogenesis as gamete-specific variants are replaced by somatic versions to regulate chromatin accessibility (15–19). It has been observed that the epigenetic reprogramming of the paternal and maternal genomes is asymmetric and involves different maternal factors (15, 16, 18, 21, 22), indicating distinct mechanisms for the activation of paternal and maternal genomes during early embryonic development.
Histone variant H3.3 in mammals is encoded by two different genes (H3f3a and H3f3b), which give rise to an identical protein product (23, 24). H3.3 is constitutively expressed in cells and incorporated into the chromatin through a DNA synthesis–independent pathway during and outside of S phase (25). H3.3 has been a subject of increasing interest in the field of developmental biology because of its distinguishing roles in remodeling both the male and female genomes during fertilization and early embryonic development. H3.3 protein is enriched in both the genomes of sperm (sperm-derived H3.3 (sH3.3)) and oocyte (oocyte-derived H3.3 (oH3.3)). Notably, mature oocytes contain abundant H3.3 mRNA that gives rise to maternal H3.3 (mH3.3) upon activation (Fig. 1A) (26). H3.3 has been found in the decondensing sperm nucleus at fertilization and is important for male pronucleus formation (6, 8, 9, 27–29). However, the fate of these differently originated H3.3 proteins and their possible functions in regulation of paternal and maternal genomes during development remain unclear. In this study, we tracked the sH3.3 and mH3.3 in the paternal genome during fertilization and embryogenesis by taking advantage of our H3.3B-HA–tagged mouse model (26). By applying H3.3-specific siRNA in oocytes, we investigated the effects of mH3.3 knockdown on the transcriptional activation of the paternal genome in mouse embryos.
Figure 1.
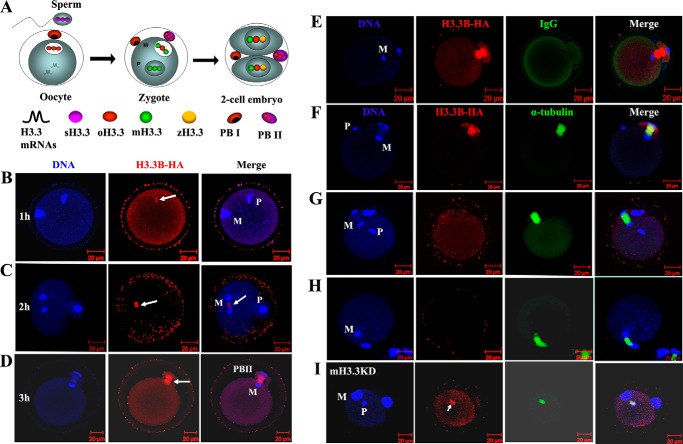
Sperm-derived H3.3 is removed from the genome upon fertilization and extruded from the zygote. A, dynamic distribution of H3.3 protein in the genomes of sperm (sH3.3) and oocyte (oH3.3). After oocyte activation, mH3.3 is produced from H3.3 mRNA originally stored in mature oocytes and deposited into both paternal and maternal genomes (zygote). At the early two-cell stage, de novo expressed zygotic H3.3 (zH3.3) is produced and deposited into the zygotic genome as well. The fates of these different origins of H3.3 protein during fertilization and embryogenesis are not clear. In this experiment, we aimed to track sH3.3 in the embryos during fertilization and embryogenesis. PBI, the first polar body. B, confocal image of B6D2F1 oocyte fertilized with a H3.3B-HA–tagged sperm 1 h post-sperm injection. A sperm H3.3 (arrow), detected by anti-HA staining, is found in the decondensing sperm genome (P) and not in the oocyte genome (M). C, B6D2F1 oocyte fertilized with H3.3B-HA sperm 2 h post-sperm injection. Both the sperm (P) and oocyte (M) genomes are negative for HA staining. Strong HA staining (arrow) is observed in the middle of the meiosis II spindle between the segregating maternal genome, which is not overlapped with the DNA staining. D, single plate confocal image of the B6D2F1 oocyte fertilized with H3.3B-HA sperm 3 h post-sperm injection and the extrusion of the PBII. HA staining is positive in the spindle of the PBII (arrow) but negative for the genome DNA. E, B6D2F1 oocyte fertilized with H3.3B-HA sperm 3 h post-sperm injection. HA and IgG antibodies were used. Only HA staining is positive in the PBII; IgG staining was negative in the whole zygote. F, B6D2F1 oocyte fertilized with H3.3B-HA sperm 3 h post-sperm injection. HA staining for H3.3B (red) and α-tubulin staining for meiosis II spindle (green) are shown; Hoechst 33348 was used for DNA staining. HA staining is positive in the PBII. G, B6D2F1 oocyte fertilized with WT sperm 3 h post-sperm injection. HA staining is negative both in the paternal and maternal genomes as well as in the PBII; α-tubulin staining shows the meiosis II spindle. H, B6D2F1 oocyte artificially activated with SrCl2 for 3 h. HA staining was negative both in the DNA and spindle. I, the removal of sH3.3 is impaired when mH3.3 is depleted in the oocyte. B6D2F1 oocytes were injected with siH3.3 to knock down mH3.3 following activation with SrCl2 for 10 h to achieve the maximum depletion of mH3.3. H3.3B-HA sperm were injected into activated oocytes, and oocytes were fixed 3 h after the sperm injection. Strong signal for HA staining was found in the decondensing sperm genome (arrow).
Results
Sperm-derived H3.3 is removed from the paternal genome and extruded from the zygote during fertilization
We have detected H3.3 in the mature sperm genome using our H3.3B-HA–tagged mice (26). However, the fate of sH3.3 in the embryo is not clear. To track sH3.3 during fertilization, we injected H3.3B-HA sperm into wild-type (WT) oocytes and performed immunofluorescence (IF) staining with anti-HA antibody in the embryos collected 1, 2, and 3 h after sperm injection. The α-tubulin antibody was used to display the meiosis II spindles and the second polar bodies (PBII). As expected, sH3.3 was detected in the decondensing sperm genome in the embryo collected 1 h postinjection (Fig. 1B). However, sH3.3 was undetectable in the male pronucleus in any of the embryos collected at 2 and 3 h post-intracytoplasmic sperm injection (ICSI) (Fig. 1C). Female pronuclei remained negative for HA staining throughout (Fig. 1, B–D). To our surprise, HA staining was positive in meiosis II spindles and PBII in these embryos (Fig. 1, C, D, and F). To exclude nonspecific staining for the HA antibody in organelles, we used IgG as the control. We found that only the HA antibody staining was positive, whereas the IgG staining was negative in these embryos (Fig. 1E), suggesting that the HA antibody is specific. To determine whether the protein recognized by HA antibody in the meiosis II spindle and the PBII is from H3.3B-HA sperm and is H3.3B-HA sperm–specific, we generated embryos using WT sperm for ICSI and the parthenogenetic embryos from artificially activated WT oocytes as the controls. Indeed, the staining was positive only in the embryos obtained by ICSI with H3.3B-HA–tagged sperm (Fig. 1F); embryos obtained with WT sperm (Fig. 1G) or parthenogenetically activated embryos (Fig. 1H) were all negative. Our results showed that the HA staining in the meiosis II spindles and the PBII is H3.3B-HA sperm–specific, suggesting that the protein recognized by HA antibody is from the H3.3B-HA sperm. This result further supports our observation that sH3.3 is removed from the sperm genome postfertilization. Collectively, our results suggest that sH3.3 is removed from the sperm genome and excluded from the zygotes through PBII extrusion during embryogenesis.
In somatic cell nuclear transfer embryos, we have shown that donor nucleus–derived H3.3 is removed from the genome after activation, and the removal of donor nucleus–derived H3.3 requires the incorporation of mH3.3 (19). To determine whether the removal of sH3.3 requires the incorporation of mH3.3, we depleted mH3.3 by injection of siH3.3 into the MII oocytes. As shown in our previous study, over 80% of the mH3.3 is depleted 10 h after oocyte activation (19). To achieve the maximum depletion of mH3.3 in the oocytes, we injected H3.3B-HA sperm into the H3.3 knockdown (mH3.3KD) oocytes 10 h after activation and collected the embryos for IF staining of H3.3B-HA 3 h post-sperm injection. We found that sH3.3 was retained in the paternal genome in these embryos (Fig. 1I). This result suggests that mH3.3 is required for the removal of sH3.3 in the embryos during fertilization.
Maternal H3.3 is incorporated into the paternal genome and is detectable in the genome of preimplantation embryos until the morula stage
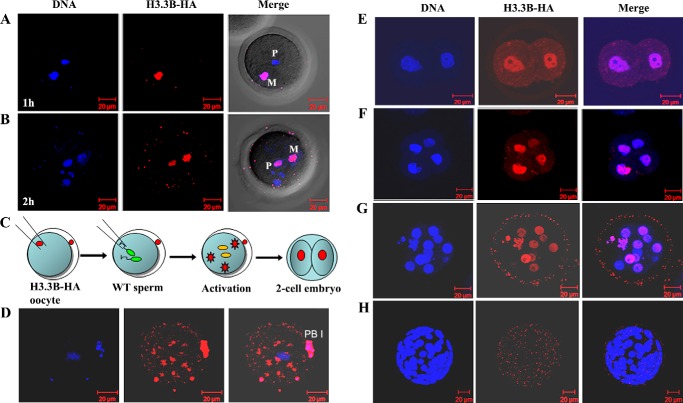
Maternal H3.3 can be deposited to the sperm genome after fertilization (22, 28). To track the deposition of mH3.3 into the paternal genome, we injected WT sperm into H3.3B-HA oocytes and monitored the incorporation of mH3.3 in the paternal genome in embryos recovered at different time points after sperm injection. At 1 h postfertilization, mH3.3 was hardly detectable in the sperm genome, whereas a strong signal was detected in the maternal genome (oH3.3) (Fig. 2A). Then mH3.3 was clearly detected in the decondensing paternal genome in embryos 2 h postfertilization (Fig. 2B), approximately the same time when sH3.3 was removed from the paternal genome (Fig. 1C). To track the deposition of mH3.3 in the paternal genome after the first embryonic division, we generated androgenetic diploid embryos by injecting two WT sperm into an enucleated H3.3B-HA oocyte (Fig. 2C) and monitored the incorporation of mH3.3 into the paternal genome. We could detect mH3.3 in the cytoplasm of the oocytes 1–2 h post-sperm injection (Fig. 2D). Strong signals were detected in the paternal genome in the two- and four-cell stage embryos, and remained detectable in the early morula embryos (Fig. 2, E–G). However, mH3.3 was not detected in androgenetic blastocysts (Fig. 2H), suggesting that mH3.3 is either removed from the genome or diluted out at the blastocyst stage. Altogether, our results demonstrated that mH3.3 is deposited into the paternal genome shortly after fertilization and retained in the paternal genome until the early morula stage.
Figure 2.
Maternal H3.3 is incorporated into the paternal genome and remains until morula stage. A, anti-HA staining of an H3.3B-HA–tagged oocyte fertilized with WT sperm. HA staining, performed 1 h post-sperm injection, is positive in the oocyte genome and negative in the sperm genome. B, anti-HA staining of fertilized H3.3B-HA oocyte is positive in both the paternal (P) and maternal (M) genomes 2 h post-sperm injection. C, schematic illustration of androgenetic diploid embryo derivation for tracking mH3.3 in the paternal genome during embryogenesis. H3.3B-HA MII oocytes were enucleated by removal of the oocyte nuclei with a pipette followed by injection of two WT sperm heads. Embryos were collected for HA staining at two-cell, four-cell, morula, and blastocyst stages. D, confocal image of the H3.3B-HA oocyte after activation with a sperm. HA staining shows newly produced H3.3 protein accumulated in the ooplasm before deposition to the genome. E–G, strong signals for HA staining in the genomes of androgenetic embryos at two-cell, four-cell, and morula (12-cell) stages. H, maternal H3.3 disappearance in androgenetic blastocysts. PBI, first polar body.
Maternal H3.3 is required for early embryonic development and the activation of paternal Oct4 pluripotent gene
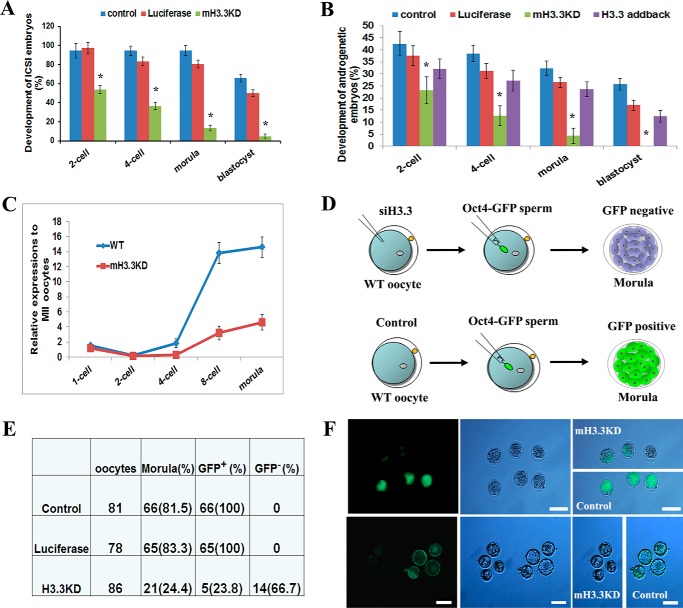
We reported previously that mH3.3 is required for development of somatic cell nuclear transfer embryos and for the activation of pluripotency-associated genes during oocyte reprogramming (19). We hypothesize that mH3.3 is also required for the development of fertilized embryos and for the activation of paternal pluripotent genes. To test this hypothesis, we knocked down mH3.3 in oocytes by injecting siH3.3 followed by ICSI fertilization. Untreated fertilized embryos and embryos injected with an siRNA against luciferase served as controls. In a total of 82 siH3.3-injected oocytes, 53.6, 36.6, 13.4, and 4.8% of them reached the two-cell, four-cell, morula, and blastocyst stages, respectively (Fig. 3A and Table 1). The developmental potential of H3.3KD embryos was significantly decreased compared with both control embryos (p < 0.01, χ2 test), suggesting that mH3.3 is essential for early embryogenesis for fertilized embryos.
Figure 3.
Maternal H3.3 is required for early embryonic development and activation of paternal Oct4. A, developmental potentials of fertilized embryos injected with siH3.3 and luciferase and control siRNAs (χ2 test; *, p < 0.01). B, developmental potentials of androgenetic haploid embryos injected with siH3.3 and luciferase and control siRNAs. H3.3-addback embryos were supplemented with exogenous H3.3 mRNA (χ2 test; *, p < 0.01). C, Oct4 down-regulation by mH3.3 depletion in androgenetic haploid embryos. D, schematic illustration of the experiment to test the effect of mH3.3 depletion on paternal Oct4 gene activation in embryos fertilized with OCT4-GFP sperm. E and F, impaired paternal OCT4-GFP expression in mH3.3KD morula embryos and blastocysts. Error bars represent S.D. Scale bar: 100 μm.
Table 1.
Compromised developmental potential for mH3.3KD fertilized embryos
Control, ICSI embryos; mH3.3KD, ICSI embryos injected with siRNAs against both H3f3a and H3f3b in oocytes (4 μm each set; n = 4 sets; total concentration, 16 μm); luciferase, ICSI embryos injected with siRNAs against luciferase (16 μm) serving as H3.3 siRNA control. Percentages are based on the number of oocytes. *, p < 0.01, χ2 test (statistical significance of differences was determined by comparison versus luciferase).
| No. of oocytes | No. of two-cell (%) | No. of four-cell (%) | No. of morula (%) | No. of blastocyst (%) | |
|---|---|---|---|---|---|
| mH3.3KD | 82 | 44 (53.6)* | 30 (36.6)* | 11 (13.4)* | 4 (4.8)* |
| Luciferase | 36 | 35 (97.2) | 30 (83.3) | 29 (80.5) | 18 (50.0) |
| Control | 73 | 69 (94.5) | 69 (94.5) | 69 (94.5) | 48 (65.8) |
Previous studies have demonstrated that androgenetic haploid embryos generated by injection of one sperm into an enucleated oocyte can develop to blastocysts and give rise to embryonic stem cell lines (30, 31), providing a simple model to study the reprogramming of the paternal genome by oocytes. To investigate the role of mH3.3 in remodeling the paternal genome without considering the effect of the maternal genome on the developing embryos, we generated androgenetic haploid embryos by injecting one sperm head into an enucleated oocyte after mH3.3 was depleted by siH3.3. In rescue experiments, exogenous H3.3 mRNA was injected back (H3.3-addback) into the oocytes. Untreated and luciferase siRNA–injected androgenetic haploid embryos were used as controls. For luciferase-injected embryos, 25.6 and 16.3% developed to morula and blastocyst stages, respectively, similar to what was observed for untreated androgenetic haploid embryos (30.8 and 19.2%). In contrast, only 3.7% of H3.3KD embryos developed to morula stage, and none of the 82 mH3.3KD embryos developed to blastocysts (Fig. 3B and Table 2); H3.3-addback significantly increased the percentage of morula (23.2%) and blastocyst (12.5%) embryos (p < 0.01, χ2 test). Our results demonstrated that knockdown of mH3.3 resulted in compromised embryonic development for both the fertilized embryos and androgenetic haploid embryos, suggesting that mH3.3 plays an essential role for reprogramming of the paternal genome during embryogenesis.
Table 2.
Compromised developmental potential for mH3.3KD androgenetic haploid embryos
Control, androgenetic haploid embryos; mH3.3KD, androgenetic haploid embryos injected with siRNAs against both H3f3a and H3f3b in oocytes (4 μm each set; n = 4 sets, total concentration, 16 μm); H3.3-addback, androgenetic haploid embryos injected with exogenous H3.3 mRNA (30 ng/μl) into H3.3KD (4 μm each set, n = 4 sets) oocytes; luciferase, androgenetic haploid embryos injected with siRNAs against luciferase (16 μm) serving as H3.3 siRNA control. Percentages are based on the number of oocytes. *, p < 0.01, χ2 test (statistical significance of differences was determined by comparison versus luciferase).
| No. of oocytes | No. of two-cell (%) | No. of four-cell (%) | No. of morula (%) | No. of blastocyst (%) | |
|---|---|---|---|---|---|
| mH3.3KD | 82 | 20 (24.4)* | 10 (12.2)* | 3 (3.7)* | 0 (0)* |
| H3.3-addback | 56 | 19 (33.4) | 16 (28.6) | 13 (23.2) | 7 (12.5) |
| Luciferase | 43 | 17 (39.5) | 13 (30.2) | 11 (25.6) | 7 (16.3) |
| Control | 52 | 23 (44.2) | 19 (36.5) | 16 (30.8) | 10 (19.2) |
Expression of Oct4 (Pou5f1) is the hallmark for pluripotency establishment and the activation of the zygotic genome during embryonic development (32). To determine whether mH3.3 is involved in the activation of paternal pluripotent genes, we first checked the effect of mH3.3 on expression of Oct4 in androgenetic haploid embryos using RT-qPCR. Because Oct4 mRNA is largely stockpiled in the MII oocyte cytoplasm, we used the maternal Oct4 mRNA level in MII oocytes as the baseline to normalize our RT-qPCR data. Initially, Oct4 transcript levels from one-cell (8 h after fertilization) to two-cell stages were decreased, indicating degradation of maternal Oct4 mRNA after fertilization. Expression of Oct4 then increased from the four-cell to eight-cell stages and peaked at the morula stage. In mH3.3KD embryos, Oct4 levels were significantly lower at the eight-cell and morula stages (Fig. 3C). This result suggests that the expression of paternal Oct4 gene is significantly down-regulated when mH3.3 is depleted in the oocytes.
The expression of Oct4 can be visualized by GFP in morulae and blastocysts using the GFP transgene driven by the Oct4 promoter (OCT4-GFP) (33). To validate whether knockdown of mH3.3 in oocytes affects the expression of paternal Oct4 protein in fertilized embryos, we injected OCT4-GFP sperm into WT oocytes and monitored GFP expression with fluorescence microscopy (Fig. 3D). As expected, strong GFP fluorescence was observed in morula and blastocyst control embryos, whereas the majority of mH3.3KD embryos did not show GFP fluorescence at either stage (Fig. 3, E and F). Our study demonstrated that depletion of mH3.3 in oocytes resulted in down-regulation of the Oct4 level in androgenetic haploid embryos and failure to activate the paternal OCT4-GFP gene in fertilized embryos. This result indeed suggests that mH3.3 is required for the activation of paternal Oct4 gene during embryogenesis.
Maternal H3.3 facilitates the paternal genome activation during embryogenesis
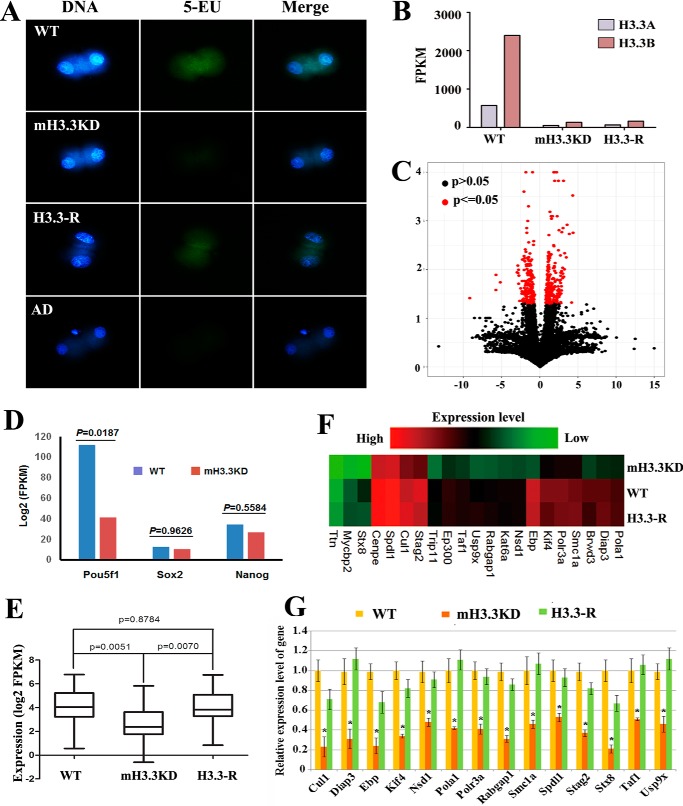
As shown above, mH3.3 is required for the activation of paternal Oct4 gene during embryogenesis. We further investigated whether mH3.3 is involved in the global activation of the paternal genome during development. The earliest zygotic transcription in mouse embryos begins at the one-cell stage as a minor wave of ZGA involving as many as 800 genes; the second wave of ZGA occurs at the two-cell stage and involves about 3500 genes (11–14). We thus used 5-EU to label newly synthesized transcripts in two-cell androgenetic embryos and indeed observed robust transcriptional activity that was markedly reduced in H3.3KD embryos. Notably, 5-EU incorporation was rescued by H3.3-addback in all mH3.3-depleted embryos. Embryos treated with actinomycin D, which inhibits transcription in general, served as a negative control (Fig. 4A). Our results suggest that mH3.3 is required for de novo transcription of the paternal genome in androgenetic haploid embryos, at least at the two-cell stage.
Figure 4.
Maternal H3.3 facilitates activation of paternal genome during embryogenesis. A, de novo transcription of the paternal genome in androgenetic haploid embryos at the two-cell stage. De novo transcription of the paternal genome labeled with 5-EU was detected in the WT embryos but was hardly detectable in the mH3.3KD embryos. De novo transcription is rescued in H3.3-addback embryos. Embryos treated with actinomycin D (AD) were used as a negative control. B, efficient knockdown of H3f3a and H3f3b transcripts in mH3.3KD and H3.3-addback (H3.3-R) embryos revealed by RNA-seq. C, Pou5f1 (Oct4) is significantly down-regulated in mH3.3KD embryos 50 h after fertilization (p < 0.05); the expression levels of Nanog and Sox2 are only slightly decreased (p > 0.05). D, RNA-seq analysis of differentially expressed genes between mH3.3KD and WT embryos. E, bar plot of average expression levels of the 21 genes down-regulated in mH3.3KD but rescued by H3.3-addback (H3.3-R). F, heat map of the expression levels of the 21 genes. G, qRT-PCR analysis confirmed the down-regulation of the 14 genes in mH3.3KD embryos and up-regulation in H3.3-addback (H3.3-R) embryos (t test; *, p < 0.01). Error bars represent S.D.
We further performed high-throughput RNA sequencing (RNA-seq) on androgenetic haploid embryos. We collected embryos 50 h after ICSI (eight-cell or 16-cell stage), which are expected to have robust zygotic transcription with maternal mRNAs being largely depleted. RNA-seq results confirmed excellent knockdown of both H3f3a and H3f3b mRNAs in mH3.3KD and H3.3-addback embryos (Fig. 4B). A total of 460 genes were found significantly (p < 0.05) differentially expressed between WT and mH3.3KD embryos including Oct4 gene, and 176 of them were down-regulated at least 1.5-fold in mH3.3KD embryos (Fig. 4, C and D, and Table S1). From a total of 333 genes significantly differentially expressed between mH3.3KD and H3.3-addback (p < 0.05), 165 genes were increased in expression in H3.3-addback embryos (Table S2). We also identified 21 genes that were down-regulated in mH3.3KD embryos and significantly up-regulated by H3.3-addback (Fig. 4, E and F; p < 0.05). RT-qPCR analysis further confirmed the expression patterns of these genes (Fig. 4G). Pathway analysis revealed that many of these genes were involved in cell cycle, mitotic nuclear division, and cell division (Table S3). Collectively, our study revealed that mH3.3 is essential for the expression of many paternal genes in androgenetic haploid embryos and plays a critical role in regulating paternal genome activation during embryogenesis.
Discussion
Histone variant H3.3 has drawn particular interest in the fields of chromatin and developmental biology for its distinctive characteristics. First, unlike replication-coupled deposition of its canonical counterparts H3.1/2 proteins, H3.3 incorporation into chromatin is replication-independent. Second, H3.3 has been consistently associated with an active state of chromatin, which is expected to be causally involved in the regulation of gene expression or to have downstream consequences for the structure and function of chromatin (34–39). Third, we and others have shown that H3.3 is enriched in both the genomes of male (sH3.3) and female gametes (oH3.3) and that H3.3 mRNA is abundant in the cytoplasm of mature oocytes (mH3.3) (19, 26, 28, 29). H3.3 plays a critical role in regulating the formation of the pronucleus in zygotes and the development of early embryos (8, 9, 19, 22, 27–29). As H3.3 differs from its canonical counterparts H3.1/2 in only four to five amino acids, it has been difficult to generate highly specific H3.3 antibodies for in vivo studies. To monitor H3.3 deposition in vivo, we have generated H3.3B-HA-IRES-EYFP/mCherry knock-in mice (26), which allow us to track H3.3 protein in vivo and, importantly, to distinguish the different origins of H3.3 proteins (sH3.3, mH3.3, and oH3.3) during fertilization and embryogenesis. In this study, we used H3.3B-HA sperm to fertilize WT oocytes to track sH3.3 in embryos by HA antibody staining. We found that sH3.3 is removed from the sperm genome shortly after fertilization and that sH3.3 removal is tightly associated with the incorporation of mH3.3. Notably, we further demonstrated that mH3.3 is essential for early embryonic development and for paternal genome transcriptional activation. Our results suggest a multifaceted role of H3.3 in paternal chromatin remodeling during fertilization and embryogenesis.
The mammalian spermatozoon is packaged largely with protamine and 5–15% of histones (40, 41). Global analysis on both mouse and human spermatozoa shows that retained histones are mainly enriched at embryonic developmental promoters coupled with transcription repression–associated histone modifications. This pattern is suggested to serve as an epigenetic memory mark that poises the genes in the sperm genome for activation upon fertilization (1, 22, 40–44). During the transition from transcriptional quiescence to ZGA, the sperm nucleus undergoes massive changes in chromatin composition including protamine removal from the sperm genome and loading of maternally derived histones (34, 36, 40, 45, 46). Although protamine is known to be quickly removed from the sperm genome, the fate of retained histones in the sperm genome during fertilization remains unclear, in part due to the difficulty in distinguishing sperm-derived from maternally derived histones in fertilized embryos. Using our H3.3B-HA–tagged mouse sperm, we found that sH3.3 is removed from the paternal genome shortly after fertilization, a process that requires the presence of mH3.3. This finding is unexpected as H3.3 enrichment in the sperm genome is thought to serve as an epigenetic signature for gene activation and to be transmitted to the embryo through fertilization (1, 40–44, 47). Our observation underlines the need to further investigate the function of H3.3 in the sperm genome. Indeed, H3.3 is enriched at the promoter of many genes in the sperm genome whose transcriptional activation depends on H3.3 during embryogenesis (Fig. S1). We speculate that H3.3 enrichment at the promoter together with specific histone modifications in the sperm genome marks developmentally important genes for activation during ZGA. H3.3 replacement (newly synthesized H3.3 replacing nucleosomal canonical H3.1/2 and H3.3) is a general mechanism for pluripotent gene activation during reprogramming (17, 19, 48). Interestingly, H3.3 replacement is also closely associated with the removal of repressive histone modifications at the promoter such as histone H3 lysine 27 trimethylation.5 We hypothesize that sH3.3 carrying repressive histone modifications is replaced by mH3.3 to create a permissive chromatin state for transcriptional activation during ZGA. Although the results from this study support extensive sH3.3 removal during fertilization, it remains possible that a fraction of sH3.3 with special modifications is retained and transmitted to the zygote after fertilization (1, 43, 44). Using the recently established technology for ChIP-sequencing on embryos (49, 50) and our H3.3B-HA–tagged mice, identification of the dynamic location of mH3.3 and sH3.3 as well as histone modifications in the paternal genome during ZGA would provide more important insights.
Although sH3.3 was found to be removed from the paternal genome during fertilization, our results also showed that mH3.3 is incorporated into the paternal genome approximately at the time as sH3.3 is removed and that incorporated mH3.3 remains detectable in the paternal genome in developing embryos until the morula stage. This result indicates that mH3.3 may play an important role in regulating paternal genome activation during embryogenesis. Indeed, depletion of mH3.3 in oocytes resulted in significantly compromised embryonic development of both normal fertilized embryos and androgenetic haploid embryos. Mechanistically, we found that activation of pluripotent Oct4 gene in the paternal genome was impaired and that the de novo transcription from the paternal genome was significantly affected when mH3.3 was depleted in oocytes. Furthermore, analysis of high-throughput RNA sequencing of androgenetic haploid embryo revealed that hundreds of embryo-expressed genes (at eight-cell and 16-cell stages) were transcriptionally altered by mH3.3 depletion in oocytes. Thus, mH3.3 supports early embryonic development at least partially through paternal genome activation. The next step will be to identify the exact location of mH3.3 in the paternal genome during fertilization and embryogenesis. Our system in combination with the recently established low input ChIP-seq technology on mouse embryos (49, 50) would provide a practical model for such future studies.
Our findings may also be relevant to human-assisted reproduction technologies as improper zygotic genome activation is one of the major reasons that fertilized embryos failed to develop (51). It may prove useful to determine whether in such cases the oocyte has a proper level of H3.3 mRNA and whether the sperm genome has incorporated mH3.3 adequately after fertilization.
Experimental procedures
Animals and oocytes
Animals were housed and prepared according to the protocol approved by the Institutional Animal Care and Use Committee of Weill Cornell Medical College (protocol number 2014-0061). B6D2F1 and ICR mice were purchased from Taconic Farms (Germantown, NY). Females were superovulated at 6–8 weeks with 5 IU of pregnant mare serum gonadotrophin (Sigma-Aldrich) and 5 IU of human chorionic gonadotrophin (hCG; Sigma-Aldrich) at intervals of 48 h. MII oocytes from superovulated female mice were recovered 14–16 h after hCG injection.
Injection of siRNA and mRNA into MII oocytes
MII oocytes from superovulated B6D2F1 females with PMSG (Pregnant mare serum gonadotropin, Sigma, G4527) were recovered 14–16 h after hCG injection. siRNAs or mRNAs were injected into the oocytes with a piezo-operated microcapillary pipette (3–5 μm inner diameter). After injection, oocytes were kept in the room temperature for 10 min and then moved into the incubator for at least another 30min before enucleation and ICSI or parthenogenetic activation. siRNA sequences for H3f3a and H3f3b and cDNA sequence for H3.3-addback construct are provided previously (26).
Enucleation and ICSI
Oocytes were transferred into a droplet of HEPES containing 5 μg ml−1 cytochalasin B, which had previously been placed in the operation chamber on the microscope stage. Oocytes undergoing micromanipulation were held with a holding pipette, and the metaphase II chromosome-spindle complex was aspirated into the pipette with a minimal volume of oocyte cytoplasm. After enucleation, oocytes were transferred into cytochalasin B–free KSOM and returned to the incubator for at least 1 h for recovery. The sperm head was picked up with the injection pipette, and each enucleated oocyte was injected with one sperm head. Following ICSI, oocytes were kept at room temperature for 10 min and moved to KSOM to culture in the incubator at 37 °C under 5% (v/v) CO2 in air.
Fluorescence microscopy, immunohistochemistry, and confocal imaging
Expression of fluorescence was detected in live embryos using a fluorescence inverted microscope (Nikon, TE2000-U). Images were captured with a digital camera and merged in NIS-Elements D software (Nikon).
For detecting newly synthesized RNA of paternal genes in androgenetic embryos, an iClickTM EU Andy Fluor 488 Imaging kit (GeneCopoeia, A009) was used. Briefly, two-cell stage embryos were incubated with 1 mm EU, which can incorporate into nascent RNA, for 5 min and then in iClick reaction mixture for 30 min. Embryos treated with 0.1 μg/ml actinomycin D for 30 min served as a negative control.
For immunohistochemistry staining (IF), oocytes or embryos were fixed (4% paraformaldehyde), permeabilized (0.5% Triton X-100 in PBS), blocked (10% normal donkey serum and 0.5% Triton X-100 in PBS), and incubated in working dilutions of the antibodies. As primary antibodies, anti-HA goat IgG (Abcam, ab9134; 1:100) and anti-α-tubulin-FITC (Sigma, F2168; 1:300) were used. We used anti-goat IgG conjugated with Alexa Fluor 647 (Invitrogen, A-21245) as a secondary antibody. Imaging was performed with a Zeiss 710 confocal imaging system. Z-stack images with 20 sequential sections for each embryo were taken.
RT-qPCR
Primers were designed to span an exon–exon junction (Table S4). RT-qPCR was performed using an Applied Biosystems StepOnePlus system and Power SYBR Green PCR Master Mix. RNA from three to 10 embryos was isolated using TRIzol, and cDNA was made using SuperScript III (Invitrogen). cDNA was treated with RNase H and diluted 1:10 in H2O with 8 μl used per PCR. Gapdh was used as a control. Experiments were performed in biological triplicate and technical duplicate with data represented as the mean and error represented as S.D.
RNA sequencing of embryos
Five to 10 embryos were transferred into a PCR tube with 100 μl of lysate buffer by mouth pipette, and RNA was prepared using an Arcturus PicoPure kit (Life Technologies, KIT0204). Library preparation was done according to a published protocol (52). Briefly, purified RNA was used for first-strand synthesis, second-strand synthesis, and PCR amplification (×2). 200 ng of resulting DNA was sonicated to ∼100–300 bp and then used to construct a sequencing library according to standard Illumina protocols. Libraries were sequenced using the Illumina HiSeq2000 platform (single end, 51 bp) multiplexed at three samples per lane.
Data analysis
All data are presented as mean ± S.D. Differences between groups were tested for statistical significance using Student's t test or χ2 test. Statistical significance was set at p < 0.05 or p < 0.01. For RNA-seq analysis, RNA-seq reads were aligned to the mouse genome (mm10) using TopHat. Expression levels were obtained using CuffLinks using upper quartile and GC normalization. Genes were considered expressed if their FPKM value was greater than 5. Differentially expressed genes were identified using a 2-fold cutoff and the requirement that one of the expression values (FPKM) was equal to or greater than 5. Pathway analysis was performed using mouse gene ontology pathways. All data have been deposited to the Gene Expression Omnibus (GEO) under accession number GSE108769.
Author contributions
Q. K., Z. L., K. S., G. D. P., C. D. A., S. R., Z. R., and D. W. resources; Q. K., L. A. B., F. G., X. Z., J. Z., P. Y., and D. W. data curation; Q. K., H. Z., C. L. O., P. Y., and Z. L. formal analysis; Q. K., L. A. B., X. Z., J. Z., and D. W. investigation; Q. K. visualization; Q. K., L. A. B., F. G., X. Z., J. Z., H. Z., P. Y., and D. W. methodology; Q. K. and D. W. writing-original draft; Q. K., F. G., C. L. O., K. S., G. D. P., S. R., Z. R., and D. W. writing-review and editing; H. Z. and P. Y. software; C. L. O. and P. Y. validation; Z. L., K. S., C. D. A., S. R., and D. W. supervision; C. D. A. and Z. R. funding acquisition; D. W. conceptualization; D. W. project administration.
Supplementary Material
This work was supported in part by a Starr Foundation Tri-Institutional Stem Cell Core and Derivation grant (to D. W., Z. R., and S. R.), the National Key Research and Development program of China-Stem cell and Translational Research Grant 2016YFA0100200 (to Q. K. and Z. L.), and “Academic Backbone” Project of Northeast Agricultural University Grant 15XG19 (to Q. K. and Z. L.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and Tables S1–S4.
Y. Liu, Q. Kong, and D. Wen, unpublished data.
- ZGA
- zygotic genome activation
- sH3.3
- sperm-derived H3.3
- mH3.3
- maternal H3.3
- oH3.3
- oocyte-derived H3.3
- IF
- immunofluorescence
- PBII
- second polar bodies
- ICSI
- intracytoplasmic sperm injection
- siH3.3
- H3.3 siRNA
- MII
- meiosis II
- KD
- knockdown
- qPCR
- quantitative PCR
- EU
- ethynyl uridine
- seq
- sequencing
- hCG
- human chorionic gonadotrophin
- KSOM
- potassium-supplemented simplex optimized medium
- FPKM
- fragments per kilobase of transcript per million mapped reads.
References
- 1. Arico J. K., Katz D. J., van der Vlag J., and Kelly W. G. (2011) Epigenetic patterns maintained in early Caenorhabditis elegans embryos can be established by gene activity in the parental germ cells. PLoS Genet. 7, e1001391 10.1371/journal.pgen.1001391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burton A., and Torres-Padilla M.-E. (2010) Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief. Funct. Genomics 9, 444–454 10.1093/bfgp/elq027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tadros W., and Lipshitz H. D. (2009) The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- 4. Farley B. M., and Ryder S. P. (2008) Regulation of maternal mRNAs in early development. Crit. Rev. Biochem. Mol. Biol. 43, 135–162 10.1080/10409230801921338 [DOI] [PubMed] [Google Scholar]
- 5. Ferg M., Sanges R., Gehrig J., Kiss J., Bauer M., Lovas A., Szabo M., Yang L., Straehle U., Pankratz M. J., Olasz F., Stupka E., and Müller F. (2007) The TATA-binding protein regulates maternal mRNA degradation and differential zygotic transcription in zebrafish. EMBO J. 26, 3945–3956 10.1038/sj.emboj.7601821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Inoue A., and Zhang Y. (2014) Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat. Struct. Mol. Biol. 21, 609–616 10.1038/nsmb.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng J., Xia X., Ding H., Yan A, Hu S., Gong X., Zong S., Zhang Y., and Sheng H. Z. (2008) Erasure of the paternal transcription program during spermiogenesis: the first step in the reprogramming of sperm chromatin for zygotic development. Dev. Dyn. 237, 1463–1476 10.1002/dvdy.21499 [DOI] [PubMed] [Google Scholar]
- 8. Lin C.-J., Conti M., and Ramalho-Santos M. (2013) Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development 140, 3624–3634 10.1242/dev.095513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin C.-J., Koh F. M., Wong P., Conti M., and Ramalho-Santos M. (2014) Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev. Cell 30, 268–279 10.1016/j.devcel.2014.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park S.-J., Komata M., Inoue F., Yamada K., Nakai K., Ohsugi M., and Shirahige K. (2013) Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev. 27, 2736–2748 10.1101/gad.227926.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aoki F., Worrad D. M., and Schultz R. M. (1997) Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 181, 296–307 10.1006/dbio.1996.8466 [DOI] [PubMed] [Google Scholar]
- 12. Hamatani T., Carter M. G., Sharov A. A., and Ko M. S. (2004) Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell 6, 117–131 10.1016/S1534-5807(03)00373-3 [DOI] [PubMed] [Google Scholar]
- 13. Clift D., and Schuh M. (2013) Re-starting life: fertilization and the transition from meiosis to mitosis. Nat. Rev. Mol. Cell Biol. 14, 549–562 10.1038/nrm3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang P., Wu W., and Macfarlan T. S. (2015) Maternal histone variants and their chaperones promote paternal genome activation and boost somatic cell reprogramming. BioEssays 37, 52–59 10.1002/bies.201400072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang C.-C., Ma Y., Jacobs S., Tian X. C., Yang X., and Rasmussen T. P. (2005) A maternal store of macroH2A is removed from pronuclei prior to onset of somatic macroH2A expression in preimplantation embryos. Dev. Biol. 278, 367–380 10.1016/j.ydbio.2004.11.032 [DOI] [PubMed] [Google Scholar]
- 16. Gao S., Chung Y. G., Parseghian M. H., King G. J., Adashi E. Y., and Latham K. E. (2004) Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Dev. Biol. 266, 62–75 10.1016/j.ydbio.2003.10.003 [DOI] [PubMed] [Google Scholar]
- 17. Jullien J., Astrand C., Szenker E., Garrett N., Almouzni G., and Gurdon J. B. (2012) HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin 5, 17 10.1186/1756-8935-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teranishi T., Tanaka M., Kimoto S., Ono Y., Miyakoshi K., Kono T., and Yoshimura Y. (2004) Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1foo in nuclear transfer. Dev. Biol. 266, 76–86 10.1016/j.ydbio.2003.10.004 [DOI] [PubMed] [Google Scholar]
- 19. Wen D., Banaszynski L. A., Liu Y., Geng F., Noh K. M., Xiang J., Elemento O., Rosenwaks Z., Allis C. D., and Rafii S. (2014) Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc. Natl. Acad. Sci. U.S.A. 111, 7325–7330 10.1073/pnas.1406389111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee M. T., Bonneau A. R., and Giraldez A. J. (2014) Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 30, 581–613 10.1146/annurev-cellbio-100913-013027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Santos F., Hendrich B., Reik W., and Dean W. (2002) Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241, 172–182 10.1006/dbio.2001.0501 [DOI] [PubMed] [Google Scholar]
- 22. van der Heijden G. W., Dieker J. W., Derijck A. A., Muller S., Berden J. H., Braat D. D., van der Vlag J., and de Boer P. (2005) Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 122, 1008–1022 10.1016/j.mod.2005.04.009 [DOI] [PubMed] [Google Scholar]
- 23. Frank D., Doenecke D., and Albig W. (2003) Differential expression of human replacement and cell cycle dependent H3 histone genes. Gene 312, 135–143 10.1016/S0378-1119(03)00609-7 [DOI] [PubMed] [Google Scholar]
- 24. Wellman S. E., Casano P. J., Pilch D. R., Marzluff W. F., and Sittman D. B. (1987) Characterization of mouse H3.3-like histone genes. Gene 59, 29–39 10.1016/0378-1119(87)90263-0 [DOI] [PubMed] [Google Scholar]
- 25. Szenker E., Ray-Gallet D., and Almouzni G. (2011) The double face of the histone variant H3.3. Cell Res. 21, 421–434 10.1038/cr.2011.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wen D., Noh K.-M., Goldberg A. D., Allis C. D., Rosenwaks Z., Rafii S., and Banaszynski L. A. (2014) Genome editing a mouse locus encoding a variant histone, H3.3B, to report on its expression in live animals. Genesis 52, 959–966 10.1002/dvg.22827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loppin B., Bonnefoy E., Anselme C., Laurençon A., Karr T. L., and Couble P. (2005) The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437, 1386–1390 10.1038/nature04059 [DOI] [PubMed] [Google Scholar]
- 28. Torres-Padilla M. E., Bannister A. J., Hurd P. J., Kouzarides T., and Zernicka-Goetz M. (2006) Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50, 455–461 10.1387/ijdb.052073mt [DOI] [PubMed] [Google Scholar]
- 29. Santenard A., Ziegler-Birling C., Koch M., Tora L., Bannister A. J., and Torres-Padilla M.-E. (2010) Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol. 12, 853–862 10.1038/ncb2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang H., Shi L., Wang B.-A., Liang D., Zhong C., Liu W., Nie Y., Liu J., Zhao J., Gao X., Li D., Xu G. L., and Li J. (2012) Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell 149, 605–617 10.1016/j.cell.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 31. Li W., Shuai L., Wan H., Dong M., Wang M., Sang L., Feng C., Luo G. Z., Li T., Li X., Wang L., Zheng Q. Y., Sheng C., Wu H. J., Liu Z., et al. (2012) Androgenetic haploid embryonic stem cells produce live transgenic mice. Nature 490, 407–411 10.1038/nature11435 [DOI] [PubMed] [Google Scholar]
- 32. Lee M. T., Bonneau A. R., Takacs C. M., Bazzini A. A., DiVito K. R., Fleming E. S., and Giraldez A. J. (2013) Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 10.1038/nature12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoshimizu T., Sugiyama N., De Felice M., Yeom Y. I., Ohbo K., Masuko K., Obinata M., Abe K., Schöler H. R., and Matsui Y. (1999) Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 41, 675–684 10.1046/j.1440-169x.1999.00474.x [DOI] [PubMed] [Google Scholar]
- 34. Henikoff S., McKittrick E., and Ahmad K. (2004) Epigenetics, histone H3 variants, and the inheritance of chromatin states. Cold Spring Harb. Symp. Quant. Biol. 69, 235–243 10.1101/sqb.2004.69.235 [DOI] [PubMed] [Google Scholar]
- 35. Ahmad K., and Henikoff S. (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 10.1016/S1097-2765(02)00542-7 [DOI] [PubMed] [Google Scholar]
- 36. Henikoff S., and Smith M. M. (2015) Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 7, a019364 10.1101/cshperspect.a019364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ng R. K., and Gurdon J. B. (2008) Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 10, 102–109 10.1038/ncb1674 [DOI] [PubMed] [Google Scholar]
- 38. Ng R. K., and Gurdon J. B. (2008) Epigenetic inheritance of cell differentiation status. Cell Cycle 7, 1173–1177 10.4161/cc.7.9.5791 [DOI] [PubMed] [Google Scholar]
- 39. Saade E., Pirozhkova I., Aimbetov R., Lipinski M., and Ogryzko V. (2015) Molecular turnover, the H3.3 dilemma and organismal aging (hypothesis). Aging Cell 14, 322–333 10.1111/acel.12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hammoud S. S., Nix D. A., Zhang H., Purwar J., Carrell D. T., and Cairns B. R. (2009) Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478 10.1038/nature08162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brykczynska U., Hisano M., Erkek S., Ramos L, Oakeley E. J., Roloff T. C., Beisel C., Schübeler D., Stadler M. B., and Peters A. H. (2010) Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17, 679–687 10.1038/nsmb.1821 [DOI] [PubMed] [Google Scholar]
- 42. Erkek S., Hisano M., Liang C.-Y., Gill M., Murr R., Dieker J., Schübeler D., van der Vlag J., Stadler M. B., and Peters A. H. (2013) Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat. Struct. Mol. Biol. 20, 868–875 10.1038/nsmb.2599 [DOI] [PubMed] [Google Scholar]
- 43. van der Heijden G. W., Derijck A. A., Ramos L., Giele M., van der Vlag J., and de Boer P. (2006) Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev. Biol. 298, 458–469 10.1016/j.ydbio.2006.06.051 [DOI] [PubMed] [Google Scholar]
- 44. van der Heijden G. W., Ramos L., Baart E. B., van den Berg I. M., Derijck A. A., van der Vlag J., Martini E., and de Boer P. (2008) Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev. Biol. 8, 34–34 10.1186/1471-213X-8-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Güne S., and Kulaç T. (2013) The role of epigenetics in spermatogenesis. Turk. J. Urol. 39, 181–187 10.5152/tud.2013.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Castillo J., Estanyol J. M., Ballescá J. L., and Oliva R. (2015) Human sperm chromatin epigenetic potential: genomics, proteomics, and male infertility. Asian J. Androl. 17, 601–609 10.4103/1008-682X.153302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Teperek M., and Miyamoto K. (2013) Nuclear reprogramming of sperm and somatic nuclei in eggs and oocytes. Reprod. Med. Biol. 12, 133–149 10.1007/s12522-013-0155-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wen D., Banaszynski L. A., Rosenwaks Z., Allis C. D., and Rafii S. (2014) H3.3 replacement facilitates epigenetic reprogramming of donor nuclei in somatic cell nuclear transfer embryos. Nucleus 5, 369–375 10.4161/nucl.36231 [DOI] [PubMed] [Google Scholar]
- 49. Liu X., Wang C., Liu W., Li J., Li C., Kou X., Chen J., Zhao Y., Gao H., Wang H., Zhang Y., Gao Y., and Gao S. (2016) Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 10.1038/nature19362 [DOI] [PubMed] [Google Scholar]
- 50. Zhang B., Zheng H., Huang B., Li W., Xiang Y., Peng X., Ming J., Wu X., Zhang Y., Xu Q., Liu W., Kou X., Zhao Y., He W., Li C., et al. (2016) Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 10.1038/nature19361 [DOI] [PubMed] [Google Scholar]
- 51. Neri Q. V., Lee B., Rosenwaks Z., Machaca K., and Palermo G. D. (2014) Understanding fertilization through intracytoplasmic sperm injection (ICSI). Cell Calcium 55, 24–37 10.1016/j.ceca.2013.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B. B., Siddiqui A., Lao K., and Surani M. A. (2009) mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 10.1038/nmeth.1315 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.