Abstract
Platelets regulate vascular integrity by secreting a host of molecules that promote hemostasis and its sequelae. Given the importance of platelet exocytosis, it is critical to understand how it is controlled. The t-SNAREs, SNAP-23 and syntaxin-11, lack classical transmembrane domains (TMDs), yet both are associated with platelet membranes and redistributed into cholesterol-dependent lipid rafts when platelets are activated. Using metabolic labeling and hydroxylamine (HA)/HCl treatment, we showed that both contain thioester-linked acyl groups. Mass spectrometry mapping further showed that syntaxin-11 was modified on cysteine 275, 279, 280, 282, 283, and 285, and SNAP-23 was modified on cysteine 79, 80, 83, 85, and 87. Interestingly, metabolic labeling studies showed incorporation of [3H]palmitate into the t-SNAREs increased although the protein levels were unchanged, suggesting that acylation turns over on the two t-SNAREs in resting platelets. Exogenously added fatty acids did compete with [3H]palmitate for t-SNARE labeling. To determine the effects of acylation, we measured aggregation, ADP/ATP release, as well as P-selectin exposure in platelets treated with the acyltransferase inhibitor cerulenin or the thioesterase inhibitor palmostatin B. We found that cerulenin pretreatment inhibited t-SNARE acylation and platelet function in a dose- and time-dependent manner whereas palmostatin B had no detectable effect. Interestingly, pretreatment with palmostatin B blocked the inhibitory effects of cerulenin, suggesting that maintaining the acylation state is important for platelet function. Thus, our work shows that t-SNARE acylation is actively cycling in platelets and suggests that the enzymes regulating protein acylation could be potential targets to control platelet exocytosis in vivo.
Keywords: cardiovascular, posttranslational modification (PTM), lipid raft, membrane fusion, acyltransferase, cerulenin, palmostatin B, SNAP-23, syntaxin, thrombosis
Introduction
Platelet exocytosis is critical for hemostasis; however, it is increasingly obvious that platelets secrete components with wide-ranging effects on vascular microenvironments (1, 2). The diverse platelet secretome suggests that platelets can contribute to angiogenesis, inflammation, innate immunity, and atherosclerosis (3–5). Thus, understanding the mechanics and regulation of platelet exocytosis is critical. Previous work has shown that platelets use soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE)3–mediated fusion of granule and plasma membranes for granule cargo release. SNAREs assemble into a trans-bilayer, four-helix bundle that mediates fusion between target membranes and cargo-containing vesicles (6, 7). Genetic and biochemical studies have demonstrated roles for the vesicle/granule (v- or R-) SNAREs, vesicle-associated membrane protein (VAMP) -7 and -8, and the target membrane (t- or Q) SNAREs, SNAP-23 (Qbc) and syntaxin-8 and -11 (Qa), in mediating cargo release from all three classes of platelet granules (8–14). Further studies defined specific SNARE regulators, which control when and where the v- and t-SNAREs interact. Aside from the role that IκB kinase–mediated phosphorylation plays in controlling SNAP-23/syntaxin-11 association (15), little is known about other posttranslational modifications and their effects on the platelet exocytosis.
Despite their importance to secretion, neither SNAP-23 nor syntaxin-11 contains a classical transmembrane domain (TMD), nor any identifiable CAAX motifs. Instead, both proteins contain cysteine-rich regions: human syntaxin-11 has eight cysteines with six at its C terminus, and SNAP-23 contains six cysteines with five in a central domain. These cysteine-rich regions have been shown, in some cells, to be important for membrane association and are potential sites for S-acylation (16–20). Protein S-acylation is a reversible posttranslational modification that affects a protein's ability to interact with membranes. S-palmitoylation is the addition of a C16 fatty acid to cysteines via a thioester linkage. Palmitoyl acyl transferases (PATs) are a large family of enzymes, with a characteristic Asp-His-His-Cys (DHHC) motif, that uses fatty acyl-CoAs as a substrate (21–23). Platelets contain at least 13 PAT isoforms (24). The S-acyl linkage can be cleaved by a smaller family of acyl-protein thioesterases (APT); platelets have four isoforms (25, 26), setting up a potential enzymatic cycle that can control the extent and dynamics of this protein modification, much like kinases and phosphatases. The physiological significance of such a reversible modification is seen in RAS localization, which is, in part, controlled by such a cycle. Several different PAT and thioesterase inhibitors have been generated in an attempt to modulate RAS function and dysfunction (27, 28).
Past studies have shown that acylation occurs in platelets (29). Dowal et al. (30) characterized over 200 proteins in the platelet “palmitoylome,” many of which appear to be involved in platelet signaling pathways. Consistently, Sim et al. (25) showed that PAT-inhibitor treatment blocked platelet activation in response to several hemostatic agonists. This group also showed that treatment of permeabilized platelets with a recombinant thioesterase released SNAP-23 from platelet membranes. Despite these observations, the dynamics and potential importance of t-SNARE acylation, specifically of syntaxin-11 and SNAP-23, is largely undefined in platelets. In this manuscript, we showed that acylation SNAP-23 and syntaxin-11 are reversibly acylated in platelets and that acyl turnover is important for platelet secretion. We mapped the acylation sites in both t-SNAREs to specific cysteines in their conserved, cysteine-rich regions. Consistent with their acylation, we showed that both t-SNAREs are enriched in detergent-resistant, cholesterol-dependent membrane fractions or rafts, which may be the sites for membrane fusion because the primary v-SNARE, VAMP-8, also localizes to these fractions upon platelet activation. Finally, using PAT (cerulenin) and thioesterase (palmostatin B) inhibitors, we show that the cycling of the acyl groups is important for platelet function because palmostatin B pretreatment prevented the inhibitory effect of cerulenin. Given the dynamic nature of t-SNARE acylation in platelets, it may be possible to repurpose the acylation-directed drugs, originally formulated to modulate RAS and RAS-related proteins in tumors, for use as anti-thrombotic therapeutics.
Results
Syntaxin-11 and SNAP-23 are associated with membranes and lipid rafts
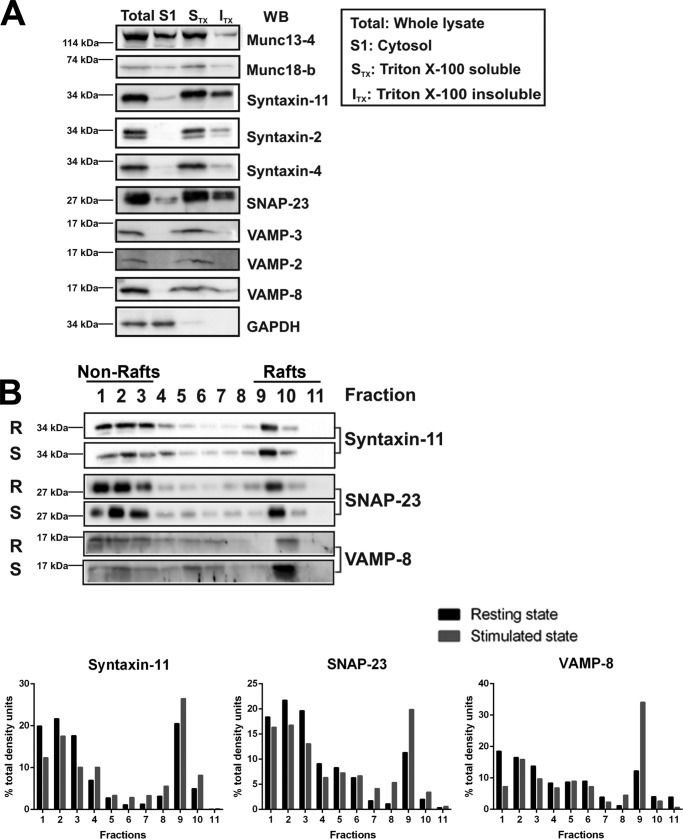
Given their lack of identifiable TMDs, we first sought to determine the extent to which syntaxin-11 and SNAP-23 associate with platelet membranes. Disrupting the platelets by freeze-thaw cycling, we generated a cytosolic and membrane fraction by centrifugation. In Fig. 1A, cytosolic proteins, e.g. glyceraldehyde 3-phosphate dehydrogenase (GAPDH), were present in the supernatant (S1) and membrane proteins (VAMP-2, -3, -8) in the pelleted fraction (STX + ITX). The two syntaxins (syntaxin-2 and -4) with TMDs were pelleted, as were syntaxin-11 and SNAP-23. Two peripheral membrane proteins, Munc18b and Munc13–4, were distributed between the supernatant and pellet. The pelleted membranes were treated with Triton X-100, which solubilized (STX) the TMD-containing syntaxins, but syntaxin-11 and SNAP-23 were found in both Triton X-100 soluble and insoluble fractions (ITX). The Triton X-100 insoluble fraction most likely contains actin cytoskeleton and lipid rafts (31, 32). Because SNAP-23 has been found in detergent-resistant lipid rafts in platelets (33), we fractionated the ITX material on a sucrose density gradient. The sample was loaded into the denser bottom of the gradient and the rafts were floated to their lighter density during centrifugation. A significant portion of SNAP-23 and syntaxin-11 from resting (R) and thrombin-stimulated (S) platelets did migrate into these lighter fractions (Fig. 1B, fractions 9–11). Interestingly, upon stimulation, more VAMP-8 was found in the raft fractions. This primary v-SNARE pairs with syntaxin-11 and SNAP-23 and is important for platelet exocytosis (10, 34).
Figure 1.
Syntaxin-11 and SNAP-23 are associated with membranes and lipid rafts. A, total lysates were prepared from washed platelets by five freeze-thaw cycles and were subjected to centrifugation to separate membrane and cytosol (S1) fractions. The membrane fractions were treated with 1% Triton X-100 to generate Triton X-100-soluble (STX) and insoluble (ITX) fractions, which were separated by centrifugation. Equivalent amounts of each fraction were analyzed by SDS-PAGE and probed by Western blotting with the indicated antibodies. B, resting (R) or thrombin-stimulated (S) platelets were lysed in 2× raft lysis buffer and layered underneath a sucrose gradient. The samples were centrifuged and the gradients were fractionated from the bottom. The fractions were probed for the indicated proteins by Western blotting. Quantification of the specific proteins in each fraction is shown. Data are representative of three independent experiments.
Cholesterol depletion affects t-SNARE localization
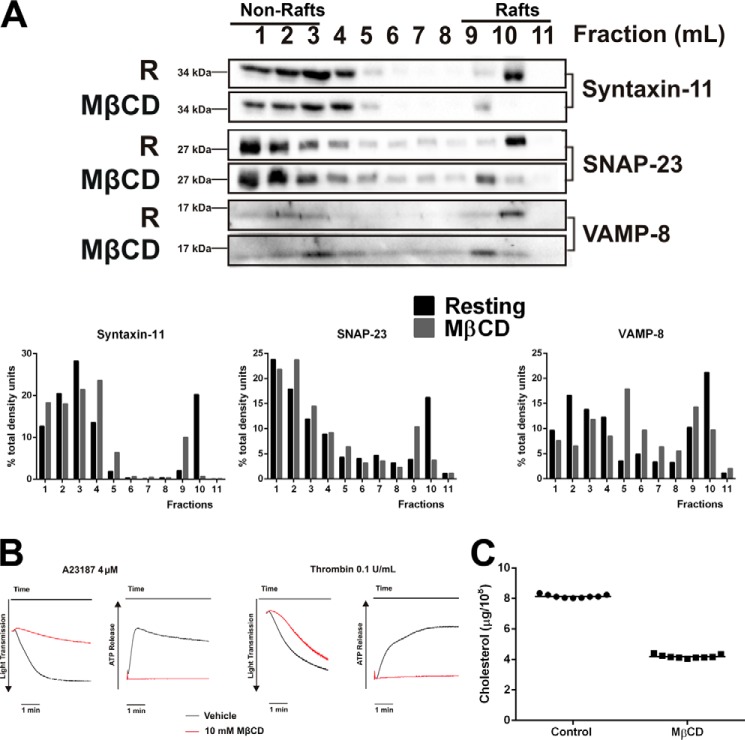
Because cholesterol is one of the major components in lipid rafts, we sought to determine whether altering platelet cholesterol affected syntaxin-11 and SNAP-23 distribution (Fig. 2). Cholesterol was reduced by more than 50% (Fig. 2C) using methyl-β-cyclodextrin (MβCD) and, consistent with previous studies (35, 36), MβCD treatment negatively affected dense granule secretion and platelet aggregation in response to both thrombin and A23187 (Fig. 2B). MβCD-treated platelets showed a decrease in syntaxin-11 and SNAP-23 in the lighter density fractions of the sucrose gradients (Fig. 2A, fractions 9–11), further supporting a lipid raft localization for the two t-SNAREs. These data imply that the cholesterol-rich lipid rafts in platelets serve as a SNARE-organizing structure that is important for exocytosis.
Figure 2.
Cholesterol depletion affects t-SNARE localization and platelet function. A, washed platelets were treated with 10 mm MβCD for 15 min at 37 °C. Untreated (R) or MβCD-treated platelets were lysed in 2× raft lysis buffer and lipid rafts were separated as in Fig. 1. The gradients were fractionated and probed for the indicated proteins by Western blotting. Quantification of specific proteins in each fraction is shown. B, platelets were treated with MβCD and their thrombin-stimulated aggregation and ATP-release were measured by lumi-aggregometer. C, the lipids from washed human platelets were extracted with chloroform:isopropanol:Nonidet P-40 (7:11:0.1) and cleared by centrifugation. Platelet cholesterol was determined using a cholesterol oxidase assay and converted into μg/109 platelets. Data are statistically significant (p < 0.0001) as determined using an unpaired Student's t test.
Acylation of syntaxin-11 and SNAP-23
Given the membrane distribution and raft association of the two TMD-deficient t-SNAREs and their lack of obvious CAAX-box prenylation signals, we next sought to determine whether they were acylated in platelets using both a chemical, acyl-biotin exchange (ABE) and a metabolic labeling method. In Fig. 3A, immunoprecipitated syntaxin-11 and SNAP-23 were labeled with the maleimido-biotin compound, 1-biotinamido-4-[4′-(maleimidomethyl) cyclohexanecarboxamido] butane (BMCC)-biotin, only after neutral hydroxylamine (HA) treatment. Because at neutral pH, HA specifically cleaves thioester bonds, this labeling pattern was consistent with both t-SNAREs being S-acylated. Note that free cysteines or those involved in disulfide bonds were capped with N-ethylmaleimide (NEM) prior to HA treatment and BMCC-biotin labeling. To further characterize this modification, we next used metabolic labeling with [3H]palmitic acid (Fig. 3B). Both syntaxin-11 and SNAP-23 showed time-dependent incorporation of radiolabel (Fig. 3B). The radiolabel was sensitive to neutral HA treatment (Fig. 3C). Because anucleated platelets show only limited protein synthesis under resting conditions (37), the time-dependent increase in [3H]palmitate incorporation implies that t-SNARE acylation is posttranslational.
Figure 3.
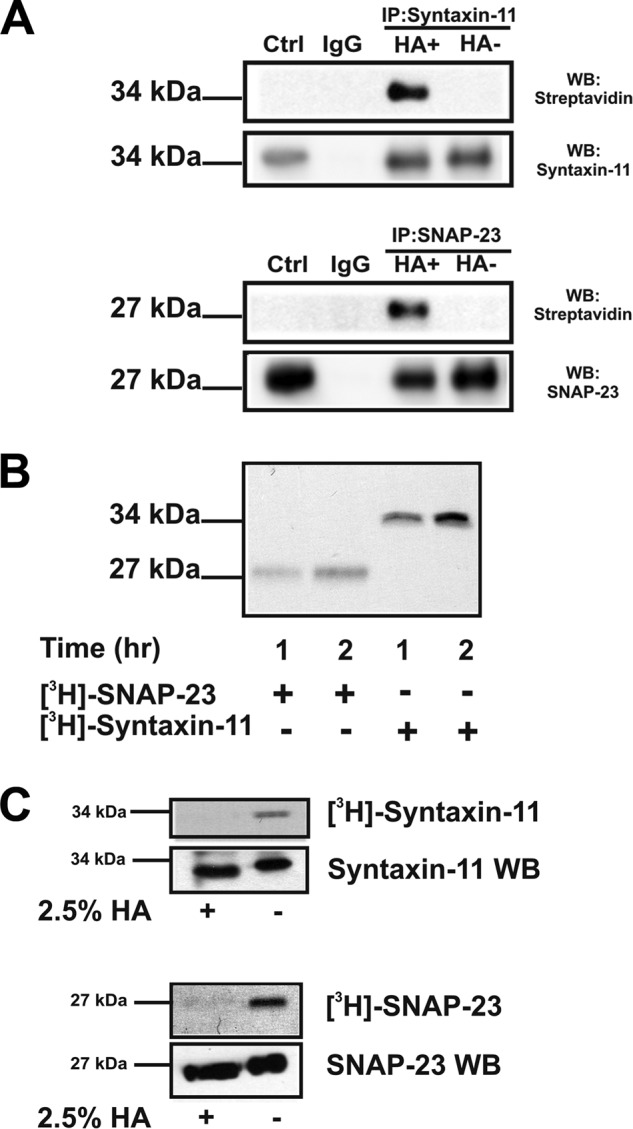
Syntaxin-11 and SNAP-23 are acylated in platelets. A, washed platelets (1 × 109) were lysed with cold 1× lysis buffer containing 50 mm NEM for 60 min. Anti–syntaxin-11 and anti–SNAP-23 antibodies were added and the immunoprecipitated proteins were treated with 100 mm HA for 30 min prior to biotinylation with BMCC-biotin. The omission of HA was used as the background control. The samples were separated by SDS-PAGE and probed for syntaxin-11 and SNAP-23 by Western blotting and for biotinylation with a streptavidin-HRP conjugate. B, washed human platelets were incubated with [3H]palmitate for 1 or 2 h and platelet lysates were subjected to immunoprecipitation with anti–SNAP-23 and anti–syntaxin-11 antibodies. The immunoprecipitated proteins were visualized by autoradiography. C, labeled samples were incubated with HA prior to separation by SDS-PAGE and visualization by autoradiography. The HA-treated samples were also probed by Western blotting with anti–syntaxin-11 and anti–SNAP-23 antibodies.
Mapping of the acylation sites in syntaxin-11 and SNAP-23
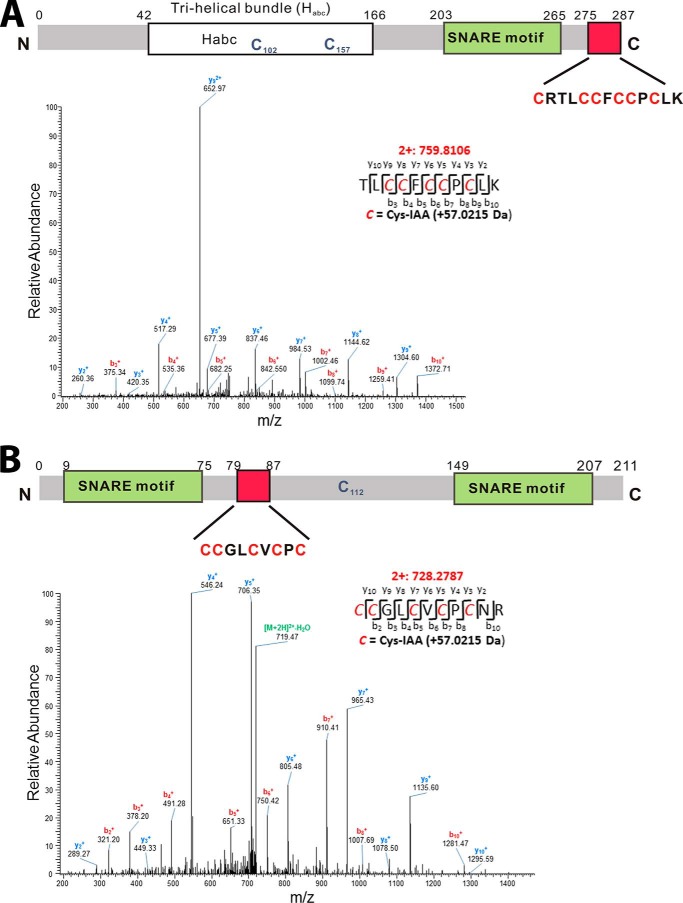
To determine the potential sites of S-acylation in syntaxin-11 and SNAP-23, we used two alkylating reagents, iodoacetic acid (IAA) and NEM, in combination with HA treatment and tryptic peptide mass spectrometry (Fig. 4). Immunopurified proteins were first treated with tris(2-carboxyethyl)phosphine (TCEP) to reduce disulfide bonds and the freed cysteines were exhaustively modified with NEM. The proteins were subsequently treated with neutral HA to cleave thioester-linked moieties. Those exposed cysteines were modified with IAA. Finally, the proteins were cleaved with trypsin and subjected to liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. In our analyses of syntaxin-11 from human platelets, we found evidence of NEM modifications on Cys-102 (GEVIHNEMC102K; monoisotopic m/z = 455.7301 Da) and Cys-157 (QRDNNEMC157KIR; monoisotopic m/z = 579.3026 Da). No other NEM-modified peptides were detected. After neutral HA treatment and modification with IAA, we detected two tryptic peptides that had been modified: KAVQYEEKNPIAAC275R (monoisotopic m/z = 507.9244 Da, data not shown) and TLIAAC279IAAC280FIAAC282IAAC283PIAAC285LK (monoisotopic m/z = 759.8106 Da) (Fig. 4A). These data are consistent with Cys-102 and -157 being either as free sulfhydryls or in disulfide bonds and Cys-275, -279, -280, -282, -283, and -285 being modified by an HA-sensitive, thioester-linked moiety. For SNAP-23, we found evidence of a NEM modification on Cys-112 (TTWGDGGENSPNEMC112NVVSK; monoisotopic m/z = 938.4195 Da). No other NEM modified peptides were detected. After neutral HA treatment and modification with IAA, we detected one tryptic peptide that had been modified with IAA: IAAC79IAAC80GLIAAC83VIAAC85PIAAC87NR (monoisotopic m/z = 728.2787 Da) (Fig. 4B). These data are consistent with Cys-112 being a free sulfhydryl and Cys-79, -80, -83, -85, and -87 being modified by an HA-sensitive, thioester-linked moiety.
Figure 4.
Detection of acylation sites by mass spectrometry. LC-MS/MS was used to detect the palmitoylation sites of syntaxin-11 and SNAP-23 and the peptides' identities were confirmed by tandem MS/MS fragmentation patterns. A, a schematic of syntaxin-11 and its features: Habc domain, SNARE motif, and cysteine-rich domain. The MS/MS of the IAA-modified tryptic peptide from syntaxin-11 is shown to demonstrate acylation sites: TLC279C280FC282C283PC285LK. B, a schematic of SNAP-23 and its features: SNARE motifs and cysteine-rich domain. The MS/MS spectrum of the IAA-modified peptide from SNAP-23 is shown to demonstrate acylation sites: C79C80GLC83VC85PC87NR of SNAP-23. Modification sites were identified based on MS/MS fragmentation patterns. For clarity, only y and b ions are labeled, with y ions in blue and b ions in red. This analysis was performed twice and identical results were obtained.
[3H]palmitate incorporation into syntaxin-11 and SNAP-23 from activated and passivated platelets
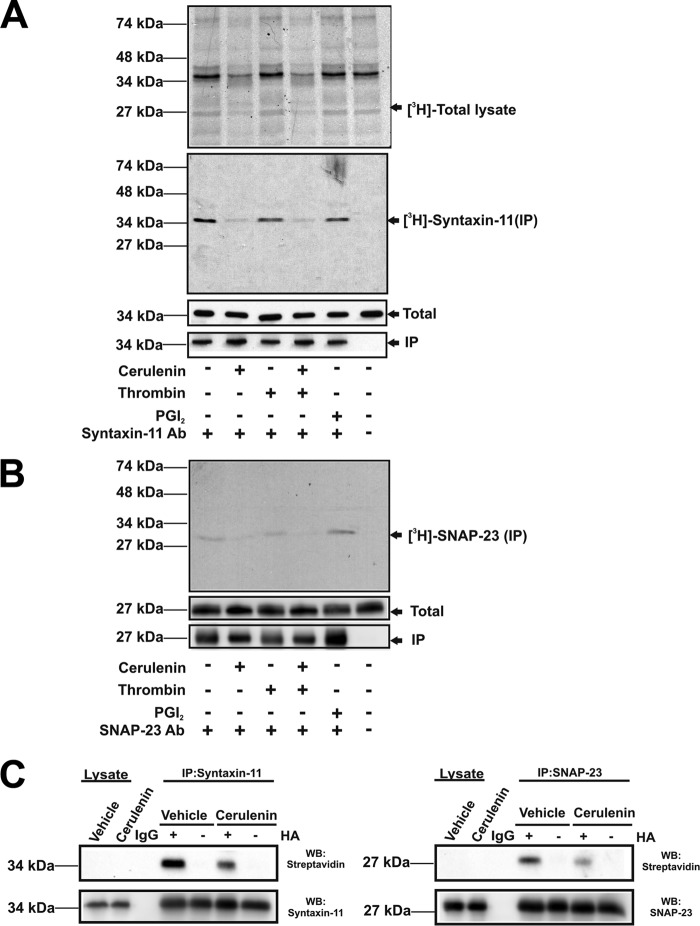
We further examined the incorporation of [3H]palmitate into platelet proteins under both passivated (treated with prostaglandin I2 (PGI2)) and activated (treated with thrombin) conditions. Fig. 5, A and B, show that [3H]palmitate incorporated into syntaxin-11 and SNAP-23 in the presence of PGI2. [3H]palmitate incorporation was not affected upon platelet activation with 0.1 units/ml of thrombin. Treatment of platelets with diethylamine NONOate, to generate nitric oxide (NO), also did not affect t-SNARE acylation as determined with the ABE assay (data not shown). This suggests that cysteine nitrosylation and acylation were not competitive in resting platelets.
Figure 5.
Acylation of t-SNARE in activated and passivated human platelets. A and B, platelets were incubated with [3H]palmitate in the presence or absence of cerulenin. Some were stimulated with thrombin and others were treated with PGI2. Lysates were prepared for immunoprecipitation with anti–syntaxin-11 (A) or anti-SNAP-23 antibodies (B). Total protein (top) or immunoprecipitated proteins (middle) were separated by SDS-PAGE and visualized by autoradiography. Total protein and immunoprecipitated samples were also probed by Western blotting with anti–syntaxin-11 or anti–SNAP-23 antibodies. C, washed platelets (1 × 109) were incubated in the presence or absence of cerulenin for 2 h and then lysed. The immunoprecipitation/ABE assay was used to detect acylation of syntaxin-11 and SNAP-23.
Treatment of platelets with a general PAT inhibitor, cerulenin, significantly reduced [3H]palmitate incorporation into platelet proteins in general as well as the two t-SNAREs. Total levels of syntaxin-11 and SNAP-23 were unchanged, as detected by Western blotting, consistent with no new protein being made and no adverse effect on protein levels being caused by cerulenin. To further demonstrate the effects of cerulenin, the ABE method was used to assess acylation (Fig. 5C). Compared with vehicle-control, cerulenin treatment reduced the level of BMCC-biotinylation of syntaxin-11 and SNAP-23, consistent with reduced acylation of the two t-SNAREs. Again, there was no significant change in total levels of the two proteins. Taken together, these data support the conclusion that the acyl groups on the t-SNAREs turn over, suggesting that dynamic remodeling of their hydrophobicity is occurring in platelets, in absence of new protein synthesis.
PAT and APT inhibitors affect human platelet secretion
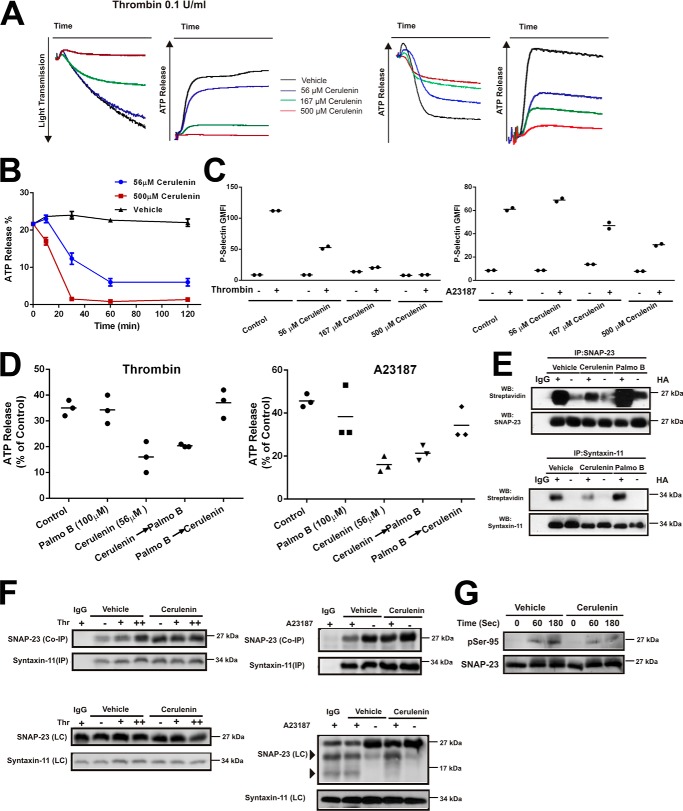
Previous reports showed that cerulenin treatment affected platelet activation (25). To further define these observations and focus on the t-SNAREs, platelets were pretreated with cerulenin, then aggregation, ATP release, and P-selectin exposure were monitored as metrics of dense and α-granule release. Platelets treated with cerulenin for 1 h showed a dose-dependent decrease in aggregation, ATP release (Fig. 6A), and P-selectin exposure (Fig. 6C) in response to thrombin, which signals through the PAR receptors, and A23187, which circumvents platelet signaling steps and directly increases intraplatelet calcium to trigger exocytosis. Cerulenin treatment did affect IκB kinase (IKK)-dependent phosphorylation of SNAP-23 at Ser-95 in response to thrombin (Fig. 6G) and A23187; however, it did not affect the calpain cleavage of SNAP-23 induced by A23187 stimulation (Fig. 6F; data not shown). Our previous studies (15) showed that this modification is critical for ternary SNARE complex formation and exocytosis in platelets. Cerulenin treatment had no effect on t-SNARE heterodimer formation (Fig. 6F) nor was there any effect on intracellular calcium increases seen in response to thrombin (data not shown). Although cerulenin treatment has been shown to affect platelet signaling (25), the similar inhibition of A23187-induced dense and α-granule exocytosis suggests that cerulenin treatment directly affects the secretory machinery.
Figure 6.
PAT and APT inhibitors affect human platelet secretion and activation. A, platelet suspensions (4 × 108/ml, 500 μl) were treated with cerulenin (0, 56, 167, or 500 μm) for 2 h at 37 °C. Aggregation traces and ATP release were monitored. Tracings are representative of three separate experiments. B, platelets were treated with cerulenin (56 or 500 μm) for the indicated times and aggregation in response to thrombin was measured by light transmission/aggregometry. Data are expressed as mean ± S.D. (n = 3 for each group). C, washed platelets (1 × 108/ml) were treated with cerulenin at 37 °C for 2 h and stimulated with thrombin (0.1 units/ml) or A23187 (4 μm) for 3 min. FITC-conjugated, anti–P-selectin antibody was added for 10 min. Fluorescent intensities were measured by FACS. The data were graphed using geometric mean fluorescence intensity: P-selectin. Data are representative of two different experiments. D, washed platelets were treated with cerulenin or palmostatin B alone or treated with both drugs sequentially: cerulenin- or palmostatin B–treated for 1 h then the other drug for an additional 1 h. ATP release in response to A23187 or thrombin was monitored. Quantification of the ATP release was expressed as mean ± S.E. (n = 3). E, washed platelets (1 × 109) were incubated in presence or absence of cerulenin or palmostatin B and then lysed with lysis buffer. The ABE assay was used to detect palmitoylation of the immunoprecipitated syntaxin-11 and SNAP-23. F, washed platelets (1 × 109) were incubated in presence or absence of cerulenin, stimulated with thrombin (0.1 units/ml (+) and 1 units/ml (++)) or A23187 (4 μm) and the t-SNARE heterodimer was immunoprecipitated (IP) with anti–syntaxin-11 antibody and the immunoprecipitates were analyzed by Western blotting with the indicated antibodies (Co-IP). Total extracts were also probed by Western blotting (LC) and the bands indicated by arrowheads in the right panel indicate calpain-mediated SNAP-23 cleavage products. G, washed platelets (1 × 109) were incubated in presence or absence of cerulenin, stimulated with thrombin and then probed by Western blotting with anti-SNAP-23 and anti–phospho-Ser-95 SNAP-23 antibodies.
Cerulenin's effects were time-dependent (Fig. 6B), suggesting that acyl cycling, in the absence of protein turnover, is occurring in platelets and is functionally relevant. To examine the role of deacylation, we examined the effects of an APT inhibitor, palmostatin B. A second APT inhibitor, bromopalmitate, had significant off-target effects on resting platelets and was not used further (data not shown). Treatment with palmostatin B had no effect on platelet exocytosis (Fig. 6D; data not shown) but significantly increased the acylation of both syntaxin-11 and SNAP-23, as measured with the ABE assay (Fig. 6E). These data confirm that deacylation is occurring and suggest that the acylated state of the t-SNAREs is important for exocytosis. To further address this point, we performed a staging experiment in which the two inhibitors were added in different sequences. When cerulenin was added first (before palmostatin B), the level of inhibition of ATP release was approximately equal to that seen when only cerulenin was added (Fig. 6D). When palmostatin B was added first, subsequent cerulenin addition had no inhibitory effect. Thus, pretreatment with palmostatin B counteracted the effects of cerulenin, but cerulenin pretreatment could not be reversed. These data are consistent with an active acylation/deacylation cycle in platelets and point to its functional relevance in platelet exocytosis. Because part of cerulenin's inhibitory effect is likely because of t-SNAREs deacylation without reacylation, these data suggest that the acylated form is the one needed for platelet exocytosis.
Exogenous fatty acids affect [3H]palmitate incorporation in platelets
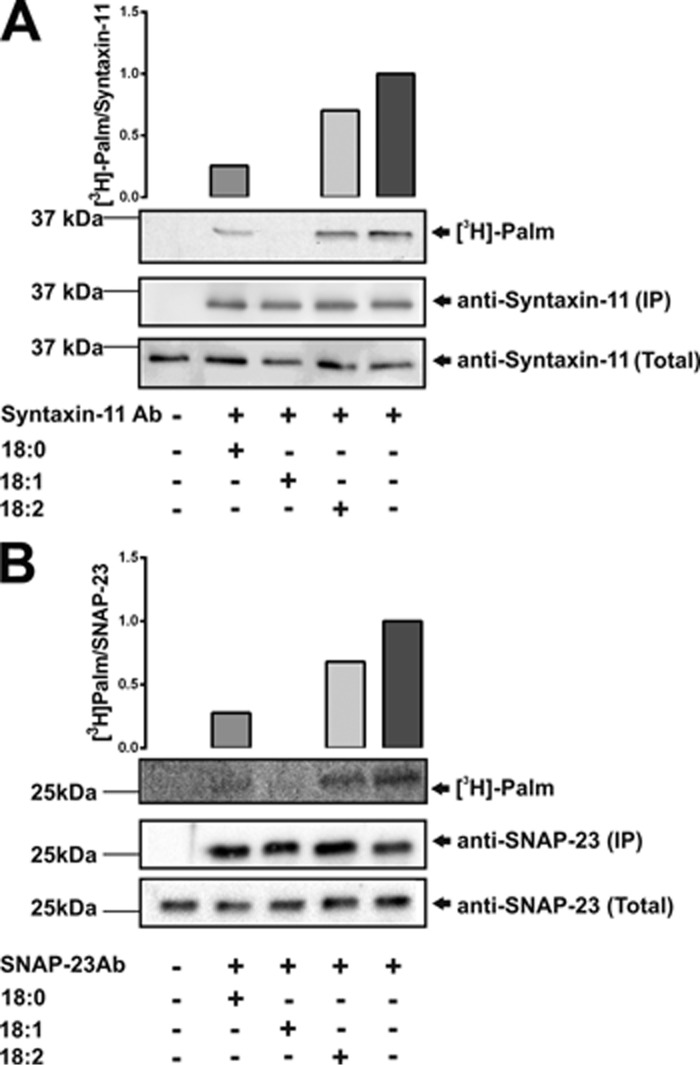
Given the evidence for an acylation/deacylation cycle in platelets, we asked whether the addition of exogenous fatty acids might compete with [3H]palmitate. Three dietary fatty acids (stearate, C18:0; oleate, C18:1; and linoleate, C18:2) were added into metabolic labeling reactions and the [3H] incorporated into the two t-SNAREs was evaluated by immunoprecipitation and autoradiography. The saturated stearate and monounsaturated oleate did reduce [3H]palmitate into both t-SNAREs with the oleate being more effective (Fig. 7). The diunsaturated linoleate had only a limited effect. These data show competition for incorporation into the t-SNAREs, but at this stage the mechanism is unclear. Some fatty acids may be directly incorporated into the t-SNAREs replacing palmitate; others may reduce the pools of available CoA to produce palmitoyl-CoA or inhibit the relevant PATs, thereby reducing the overall acylation of the t-SNAREs. Further examination of the acyl-CoA pools present in platelets and the fatty acids attached to the t-SNAREs will be required before this dynamic process can be completely understood.
Figure 7.
Exogenous fatty acids affect [3H]palmitate incorporation into syntaxin-11 and SNAP-23. A and B, human platelets (1 × 109) were incubated with [3H]palmitate in the presence or absence of the indicated fatty acids: C18:0, stearic acid; C18:1, oleic acid; C18:2, linoleic acid or cerulenin. Lysates were prepared for immunoprecipitation with anti-syntaxin-11 (A) or anti-SNAP-23 (B) antibodies. Total proteins (total) or immunoprecipitated proteins (IP) were separated by SDS-PAGE and visualized by autoradiography.
Discussion
In this manuscript, we built on previous work (25, 30) and examined the modification of two platelet t-SNAREs, syntaxin-11 and SNAP-23, which lack typical TMDs. Both behave as membrane proteins and partition into cholesterol-dependent lipid rafts. Both are S-acylated in their cysteine-rich domains. Based on mapping studies, all the available cysteines in these domains can be modified. A PAT inhibitor blocked [3H]palmitate incorporation into the two t-SNAREs and blocked platelet secretion from dense and α-granules. An APT inhibitor had no effect on platelet secretion but did reverse the inhibitory effects of the PAT inhibitor. These data imply that t-SNARE acylation is highly dynamic in platelets and cycles because of the actions of PATs and APTs, much like phosphorylation is controlled by kinases and phosphatases. Although the cycling of acyl groups has been reported on other proteins in other cells (38–40), our data are the first to demonstrate its importance in platelets.
Acylation's effects on t-SNAREs
Previous studies clearly demonstrated roles for syntaxin-11 and SNAP-23 in the membrane fusion events required for platelet granule release (10, 12, 15). It is unclear if SNAP-23 acylation is required for fusion. Studies with model proteoliposome systems clearly show that unmodified SNAP-23 can mediate membrane fusion (15) and similarly, acylation of SNAP-25 is not required in vivo (7). Alternatively, syntaxin-type t-SNAREs typically need their TMDs to progress from hemifusion of the outer leaflets to complete the bilayer fusion (41). However, longer, more hydrophobic moieties, such as artificially added C45 and C55 prenyl groups, are sufficient to promote full fusion in absence of a TMD. In contrast, C15 and C20 groups are not (42, 43). Acylation of SNAP-23 and syntaxin-11 has been shown to be important for exocytosis in other nucleated cell types, but the contribution to fusion versus a membrane targeting or trafficking role has been debated (20, 44–46). It seems possible that acylation of these two t-SNAREs must impart sufficient hydrophobicity to compensate for the loss of the syntaxin TMD. Full or even partial deacylation would be expected to limit fusogenicity, which perhaps accounts for part of the secretion defect we observed in the cerulenin-treated platelets (Fig. 6).
Acylation may also promote t-SNARE partitioning into lipid raft-like structures (47, 48). Clustering of t-SNAREs has been reported to be important for promoting membrane fusion and these clusters are cholesterol-dependent (49). Consistently, the Haynes group (50) showed that platelet membrane cholesterol directly affects cargo release from platelets by affecting fusion pore formation, dilation, and full fusion. The precise roles of lipid rafts in platelet exocytosis remains to be determined; however, it should be noted that stimulation with thrombin did increase the levels of VAMP-8 in platelet rafts (Fig. 1B). In mast cells, these lipid subdomains are sites of SNARE complex formation and perhaps membrane fusion. IKK/SNAP-23 association and SNAP-23 phosphorylation occur in rafts (51) as does the formation of ternary SNARE complexes (52) IKK is also found in the rafts of T cells (53). Interestingly, in platelets, cerulenin treatment not only inhibited secretion but also reduced the IKK-dependent phosphorylation of SNAP-23 (Fig. 6G). These data are suggestive of a mechanism by which deficient t-SNARE acylation, in platelets, would affect raft partitioning, ternary complex formation, and membrane fusion. Future super-resolution microscopy analysis will be needed to define the roles of rafts relative to the sites of granule/plasma membrane fusion and cargo release.
Extent of t-SNARE acylation
Our mass spectrometry analysis (Fig. 4) clearly detects all the cysteines in both t-SNAREs and distinguishes those which are free or in reducible, disulfide bonds (NEM modified) from those which are exposed upon HA treatment (IAA modified). These latter cysteines are likely the sites of acylation, because HA treatment completely removed metabolically incorporated [3H]palmitate (Fig. 3C). The acylated sites from syntaxin-11 were found on two peptides from the C-terminal domain. All were labeled with IAA, suggesting that six cysteines were modified. Despite exhaustive analysis of the MS/MS spectra, we did not detect any partially modified peptides (i.e. those containing both IAA and NEM). Similarly, for SNAP-23, one IAA-modified peptide was detected, which accounted for all five potential acylation sites in the central cysteine-rich domain. Again, no partially modified peptides were detected. Studies with a PEG-derivatized maleimide (54) were inconclusive, in part because the cysteines may be too close together, sterically impeding modification with the bulky PEG derivatives (data not shown). Click chemistry methods (55) using platelets incubated with 17-octadecynoic acid (ODYA) were also ineffective, perhaps because the platelet PATs failed to use the C17 fatty acid. Because no partially modified peptides were detected, our data argue that most of the two t-SNARE molecules are maximally acylated under steady-state conditions. However, treatment with palmostatin B did increase total acylation (Fig. 6E), so it is likely that there is a population of t-SNAREs that is dynamically underacylated in resting platelets. Shifts in the steady-state acylation of the two t-SNAREs could alter their hydrophobicity and thus their fusogenicity or their partitioning into lipid rafts. In pathogenic states, such as dyslipidemia, increased fatty acid availability could shift the balance toward greater acylation making the t-SNARE more hydrophobic.
Exogenous dietary fatty acids did modulate the acylation of the two t-SNAREs (Fig. 7), suggesting that they could compete with [3H]palmitate for incorporation into the SNAREs. The nature of the competition is likely complex; however, two potential mechanisms are plausible, but not mutually exclusive. The excess exogenous fatty acids could affect the CoA charging step skewing the platelet pools of available acyl-CoAs and thus reducing the effective concentrations of palmitoyl-CoA. Fatty acid binding proteins and fatty acid–CoA ligase/synthetases are detectible in platelets, by mass spectrometry, thus the machinery to convert free fatty acids into acyl-CoAs is present (56, 57). Unfortunately, our attempts to monitor platelet acyl-CoA pools by mass spectrometry have so far proved inconclusive. Alternatively, some fatty acids may generate acyl-CoAs that are preferred substrates for the t-SNARE–modifying PATs. Given that 13 different PATs have been detected in platelets (24), it is difficult to even guess which transferases modify the two t-SNAREs. Adding to the complexity, there could be acyl-CoA substrate preference overlaps between the PATs. Thus, further experimentation and technology development are required before we completely understand the dynamic nature of these substrate pools in platelets and the effects that pathologies such as dyslipidemia have on them.
Therapeutic value of platelet acylation
Our work (58) clearly shows that modulating secretion can be an effective tool to control occlusive thrombosis. Genetic studies suggest that the useful therapeutic window is ∼50–70% inhibition to control thrombus growth without causing bleeding.4 Targeting enzymes with critical activities is a more effective drug-development strategy than trying to selectively disrupt SNARE complexes. IκB kinase inhibitors, which block the phosphorylation of SNAP-23 and thus secretion, are an example of such a strategy (15). Can acylation be targeted to control platelet function? Perhaps. Our studies describe a combination of platelet function inhibitor (cerulenin) and antidote (palmostatin B) that could be valuable in managing hemostasis. Cerulenin-treated platelets showed a marked defect in their ability to form thrombi in vivo (25). However, neither drug is sufficiently specific for effective use at present. To realize the potential of targeting acylation, further studies to determine which PATs acylate the two t-SNAREs in platelets are needed to develop and test specific therapeutics.
Experimental procedures
Antibodies
Anti–syntaxin-11 rabbit polyclonal antibody was from Synaptic Systems GmbH (Goettingen, Germany). Anti-syntaxin-2 and -4 and SNAP-23 polyclonal antibodies were generated in our laboratory (13). Anti-Munc18b antibody (sc-14563) was purchased from Santa Cruz Biotechnology (Dallas, TX). The polyclonal rabbit–anti-human VAMP-8 and anti-Rab GDI polyclonal antibodies were as described previously (10, 34, 59). The anti–SNAP-23 phospho–Ser-95 antibody was described in Ref. 15. Fluorescein isothiocyanate (FITC)–conjugated PAC-1 and phycoerythrin (PE)-conjugated anti-human CD41a monoclonal antibodies were from BD Biosciences. Alkaline phosphatase–conjugated secondary anti-mouse, anti-rabbit, anti-sheep, and anti-goat IgG were from Sigma. Horseradish peroxidase (HRP)–conjugated anti-rabbit IgG was from Sigma. HRP-conjugated streptavidin was purchased from R&D Systems (Minneapolis, MN). Anti-GAPDH was from GeneTex (Irvine, CA).
General reagents
Acid citrate dextrose (ACD) blood collection tubes were from BD Vacutainer®. Apyrase, cerulenin (2,3 epoxy-4-oxo-7,10 dodecadienoylamide), urea, and NEM were purchased from Sigma. BCATM Protein Assay Kit, EZ-link® BMCC-biotin, HA, iodoacetamide, and Pierce® ECL plus Western blotting substrate were from Thermo Fisher Scientific. Complete, EDTA-free protease inhibitor mixture was obtained from Roche. A23187, the calcium ionophore, was from Calbiochem. Palmostatin B [(3S,4S)-3-decyl-4-[2-(3,4-dimethoxyphenyl)ethyl]oxetan-2-one] was purchased from EMD Millipore. PGI2 was from Cayman Chemical Company (Ann Arbor, MI). [9,10-3H(N)]palmitic acid was purchased from Perkin Elmer.
Platelet preparation
Blood from healthy donors was collected into acid citrate dextrose collection tubes at the University of Kentucky Clinic. After collection, all were transferred into 50-ml conical tubes and centrifuged at 250 × g for 20 min at room temperature to generate platelet-rich plasma. The platelet-rich plasma was carefully transferred to a 15-ml tube containing 0.2 units/ml apyrase, 10 ng/ml PGI2 and after 10-min incubation at room temperature, was centrifuged at 900 × g for 10 min. The pelleted platelets were resuspended with HEPES-Tyrode buffer (pH 6.5, 20 mm HEPES/KOH) containing apyrase, PGI2 for 10 min at room temperature. Finally, washed platelets were obtained by centrifugation at 850 × g for 8 min and resuspended with HEPES-Tyrode buffer pH 7.4. Platelets were counted with a Z2 Coulter Particle Count and Size Analyzer (Beckman-Coulter) and adjusted to 1–2 × 109/ml.
Subcellular fractionation of platelets
Subcellular fractionation of platelets was performed as in Ref. 12. Briefly, washed human platelets (1 × 109) in HEPES-Tyrode buffer (pH 7.4) with protease inhibitors were disrupted by five freeze-thaw cycles. Membranes were recovered by centrifugation at 100,000 × g for 1 h at 4 °C. The supernatant (cytosol) was kept and the pellet was treated with 1% Triton X-100 on ice for 30 min. Triton X-100 soluble and insoluble material were separated by centrifugation at 100,000 × g. The fractions were analyzed by Western blotting.
Fractionation of lipid rafts
Washed platelets (1 × 109) were lysed with 2× lysis buffer (50 mm MES/NaOH, pH 6.5, 300 mm NaCl, 2% CHAPS, protease inhibitor mixture, and 4 mm Na3VO4) and mixed with 80% sucrose to make 40% sucrose-lysate mixture. The mixture was added to ultracentrifuge tubes (12 ml, 14 mm × 89 mm, Beckman), and then overlaid with 30% sucrose (6 ml) followed by 5% sucrose (2 ml). The gradients were centrifuged at 200,000 × g at 4 °C for 18 h in an SW Ti 41 rotor (Beckman). After centrifugation, fractions (1 ml) were collected, from the bottom, using peristaltic pump (Mettler-Toledo Rainin, Oakland CA). Trichloroacetic acid precipitation was used to concentrate each sample and they were subjected to Western blot analysis.
Lumi-aggregometry
Lumi-aggregometry was measured in a Model 460Vs Lumi-Dual aggregometer (Chrono-log, Havertown, PA). Washed platelets (500 μl, 4 × 108/ml) were stirred at 800 rpm at 37 °C in siliconized glass cuvettes (Chrono-log). ATP release from dense granules was measured by preincubating platelets with Chrono-Lume® reagent (Chrono-log) for 1 min at 37 °C. Agonists were added and the data were collected using a Model 810 Aggro/Link computer interface and analyzed with Aggro/Link software (Chrono-log).
Flow cytometry analysis
Washed human platelets were treated with inhibitors at 37 °C for 2 h. Calcium (1 mm) was added and the platelets were incubated for 30 min prior to assay. Poststimulation, FITC–anti-human CD62P (P-selectin), FITC-PAC1, or PE–anti-human CD41a were added for 10 min and the samples were transferred to tubes for analysis. Analysis was performed using a BD FACSCalibur flow cytometer (BD Biosciences) and geometric mean fluorescence intensity (GMFI) was measured. In the cholesterol-depletion studies, washed platelets were pretreated with methyl-β-cyclodextrin (10 mm) for 15 min at 37 °C, and then FACS analysis was performed.
Metabolic labeling with [3H]palmitate
The fatty acids and bovine serum albumin (BSA) were allowed to incubate prior to addition to the platelets. For metabolic labeling, washed platelets were incubated with 200 μCi/ml [9,10-3H(N)]palmitic acid for 1 or 2 h at 37 °C, in HEPES-Tyrode buffer, pH 7.4 containing 1% fatty acid–free BSA.
Measurement and manipulation of platelet cholesterol
To measure platelet cholesterol, washed human platelets (1 × 108) were resuspended in HEPES-Tyrode buffer (pH 7.4) and lipids were extracted with chloroform:isopropanol:Nonidet P-40 (7:10:0.1, 400 μl). The extracts were clarified by centrifugation at 15,000 × g at room temperature and dried to remove organic solvents. The cholesterol assay was performed with the Amplex Red Cholesterol Assay Kit (Thermo Fisher Scientific) following manufacturer's instructions. The dried lipids were dissolved in 1× reaction buffer containing 300 μm Amplex Red reagent, 2 units/ml HRP, 2 units/ml cholesterol oxidase, and 0.2 units/ml cholesterol esterase. After incubation for 30 min 37 °C, the fluorescent reaction product was measured at excitation of 544 nm and emission at 590 nm.
For cholesterol depletion, washed platelets were incubated with MβCD (10 mm) at 37 °C for 30 min. After incubation, platelets were washed and resuspended at 4 × 108/ml in HEPES-Tyrode buffer (pH7.4). The treated platelets were used for aggregation and subcellular fractionation studies.
Immunoprecipitation
Washed unlabeled or [3H]palmitate–labeled platelets (500 μl) were warmed at 37 °C for 5 min and held resting or stimulated with thrombin. Platelets were lysed with 2× lysis buffer (40 mm Tris-HCl (pH 7.5), 150 mm NaCl, 2 mm EDTA, 2 mm EGTA, 2% Triton X-100, 2% sodium deoxycholate, 5 mm Na4P2O7, 2 mm Na3VO4, and protease inhibitor mixture) for 45 min on ice. The lysates were clarified by centrifugation at 13,000 rpm, for 5 min and the supernatants were incubated with rabbit IgG and Protein A Sepharose 4 Fast Flow (GE Healthcare). After preclearing, the lysate was incubated with anti–syntaxin-11 or –SNAP-23 antibodies were added for 2 h at 4 °C. Protein A Sepharose 4 Fast Flow was added for 1 h at 4 °C, and then washed thrice. The bound proteins were eluted with 2× SDS loading buffer for 8 min at 95 °C. For [3H]-labeled platelets, proteins were eluted with 2× SDS-PAGE loading buffer (no β-mercaptoethanol) at 95 °C. The immunoprecipitates were analyzed by SDS-PAGE and either immunoblotting or autoradiography using EN3HANCE (PerkinElmer) following manufacturer's instructions.
Acyl-biotin exchange analysis
The degree of S-acylation was measured using the ABE method as described in Ref. 60. Washed platelets (1 × 109) were incubated in presence or absence of cerulenin or palmostatin B and washed in HEPES-Tyrode buffer (pH 7.4). The platelets were lysed with cold 1× lysis buffer (with protease inhibitors, 1 mm PMSF, 2 mm Na3VO4, 2% Triton X-100, 2% sodium deoxycholate, 50 mm Tris-HCl pH7.5, 150 mm NaCl) containing 50 mm NEM. After clearing, anti–syntaxin-11 and SNAP-23 antibodies were added and the two t-SNAREs were immunoprecipitated. The precipitated proteins were treated with 100 mm HA in lysis buffer (pH 7.4) for 30 min at 37 °C and the exposed cysteines were biotinylated with the maleimido-biotin compound, BMCC-biotin (1.5 μm) for 1 h at 4 °C. Omission of HA treatment was used as the negative control. The samples were then separated by SDS-PAGE, transferred to polyvinylidene fluoride (PVDF) membranes, and probed with streptavidin-HRP.
Mass spectrometry analysis
Immuno-purified syntaxin-11 and SNAP-23 were first treated with tris(2-carboxyethyl)phosphine and NEM treatment to reduce the cysteines involved in disulfide linkages. The t-SNAREs were then sequentially treated with HA and IAA to modify acylated cysteines. After trypsin digestion, peptides were extracted, concentrated, and analyzed as described in Yang et al. (61).
LC-MS/MS analysis was performed using a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled with a cHiPLC™-nanoflex system (Eksigent, Dublin, CA) through a nano-electrospray ionization source. The peptide samples were separated with a reversed phase cHiPLC column (75 μm × 150 mm) at a flow rate of 300 nl/min. Mobile phase A was water with 0.1% (v/v) formic acid whereas B was acetonitrile with 0.1% (v/v) formic acid. A 50-min gradient was applied: initial 3% mobile phase B was increased linearly to 40% in 24 min and further to 85 and 95% for 5 min each before it was decreased to 3% and re-equilibrated. The MS/MS analysis method consisted of one segment with eight scan events. The first scan event was an Orbitrap MS scan (300–1800 m/z) with 60,000 resolution for parent ions followed by data-dependent MS/MS for fragmentation of the seven most intense ions with collision-induced dissociation method.
The LC-MS/MS data were submitted to a local MASCOT server for MS/MS protein identification via Proteome Discoverer (version 1.3, Thermo Fisher Scientific) against a customized database containing syntaxin-11 or SNAP-23. Typical parameters used in the MASCOT MS/MS ion search were trypsin digestion with a maximum of two miscleavages, cysteine carbamidomethylation, cysteine N-ethylmaleimide modification, methionine oxidation, a maximum of 10 ppm MS error tolerance, and a maximum of 0.8 Da MS/MS error tolerance. Modifications on cysteines of interest in the tryptic peptides of syntaxin-11 or SNAP-23 proteins were manually inspected and confirmed based on the ladder of the b- and y-type fragment ions in the MS/MS spectra.
[Ca2+]i measurements
Intraplatelet calcium was measured using Fura-2-acetoxymethyl ester (Fura-2 AM, Thermo Fisher Scientific) as described (34). Washed platelets (4 × 108/ml) in HEPES-Tyrode buffer (pH 7.4) were incubated in presence or absence of cerulenin at 37 °C for 1 h. Then 1 μm Fura-2 AM was added to platelets at 37 °C for 1 h. After incubation, the Fura-2–loaded platelets were washed and resuspended in HEPES-Tyrode buffer (pH 7.4). The platelet concentration was adjusted to 2 × 108/ml. Calcium chloride (0.7 mm) and platelets (750 μl) were added to siliconized cuvettes and stimulated with 0.1 units/ml thrombin with stirring. Fluorescence was analyzed by excitation at 340 nm and 380 nm, and emission was measured at 509 nm using a model LS55 Luminescence Spectrometer (Perkin-Elmer). The ratio of emissions was calculated simultaneously using FL WinLab4.0 software (Perkin-Elmer) and used to calculate free calcium levels.
Author contributions
J. Z. and S. W. W. conceptualization; J. Z., Y. H., J. C., H. Z., and S. W. W. formal analysis; J. Z. and S. W. W. investigation; J. Z., Y. H., J. C., H. Z., and S. W. W. methodology; J. Z. and S. W. W. writing-original draft; H. Z. and S. W. W. writing-review and editing; S. W. W. resources; S. W. W. supervision; S. W. W. funding acquisition.
Acknowledgments
We thank members of the Whiteheart Laboratory: Dr. Meenakshi Banerjee, Smita Joshi, Dr. Harry Chanzu, and Laura Tichacek for their careful perusal of this manuscript. We thank Dr. Greg Bauman and Jennifer Strange, at the University of Kentucky Flow Cytometry Core Facility. We also thank the Kentucky Blood Center for their assistance and Ming Zhang for her technical assistance.
This work was supported by NHLBI, National Institutes of Health Grants HL56652 and HL138179; American Heart Association Grant-in-Aid AHA16GRNT27620001; and Veterans Affairs Merit Award (to S. W. W.). This study was also supported in part by NIGMS, National Institutes of Health Grant P30GM110787. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or reflect the position or policy of the Department of Veterans Affairs or the United States government.
S. Joshi, M. Banerjee, J. Zhang, A. Kesaraju, S. M. Dymecki, and S. W. Whiteheart, manuscript in preparation.
- SNARE
- soluble NSF attachment protein receptor
- MβCD
- methyl-β-cyclodextrin
- IP
- immunoprecipitation
- SNAP-23
- synaptosome associated protein 23
- PGI2
- prostaglandin I2
- PAT
- palmitoyl acyltransferase
- TMD
- transmembrane domain
- VAMP
- vesicle-associated membrane protein
- HA
- hydroxylamine
- APT
- acyl-protein thioesterases
- ABE
- acyl-biotin exchange
- NEM
- N-ethylmaleimide
- BMCC-biotin
- 1-biotinamido-4-[4′-(maleimidomethyl) cyclohexanecarboxamido] butane
- IAA
- iodoacetic acid
- IKK
- IκB kinase
- HEPES/KOH buffer
- N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid
- HEPES-Tyrode buffer
- pH 6.5, 20 mm HEPES/KOH, 128 mm NaCl, 2.8 mm KCl, 1 mm MgCl2, 5 mm d-glucose, 12 mm NaHCO3, 0.4 mm NaH2PO4
- CHAPS
- 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate.
References
- 1. Ren Q., Ye S., and Whiteheart S. W. (2008) The platelet release reaction: Just when you thought platelet secretion was simple. Curr. Opin. Hematol. 15, 537–541 10.1097/MOH.0b013e328309ec74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Joshi S., and Whiteheart S. W. (2017) The nuts and bolts of the platelet release reaction. Platelets 28, 129–137 10.1080/09537104.2016.1240768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Menter D. G., Kopetz S., Hawk E., Sood A. K., Loree J. M., Gresele P., and Honn K. V. (2017) Platelet “first responders” in wound response, cancer, and metastasis. Cancer Metastasis Rev. 36, 199–213 10.1007/s10555-017-9682-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mancuso M. E., and Santagostino E. (2017) Platelets: Much more than bricks in a breached wall. Br. J. Haematol. 178, 209–219 10.1111/bjh.14653 [DOI] [PubMed] [Google Scholar]
- 5. Gue Y. X., and Gorog D. A. (2017) Importance of endogenous fibrinolysis in platelet thrombus formation. Int. J. Mol. Sci. 18, E1850 10.3390/ijms18091850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brunger A. T. (2001) Structural insights into the molecular mechanism of calcium-dependent vesicle-membrane fusion. Curr. Opin. Struct. Biol. 11, 163–173 10.1016/S0959-440X(00)00186-X [DOI] [PubMed] [Google Scholar]
- 7. Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Söllner T. H., and Rothman J. E. (1998) SNAREpins: Minimal machinery for membrane fusion. Cell 92, 759–772 10.1016/S0092-8674(00)81404-X [DOI] [PubMed] [Google Scholar]
- 8. Golebiewska E. M., Harper M. T., Williams C. M., Savage J. S., Goggs R., Fischer von Mollard G., and Poole A. W. (2015) Syntaxin 8 regulates platelet dense granule secretion, aggregation, and thrombus stability. J. Biol. Chem. 290, 1536–1545 10.1074/jbc.M114.602615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koseoglu S., Peters C. G., Fitch-Tewfik J. L., Aisiku O., Danglot L., Galli T., and Flaumenhaft R. (2015) VAMP-7 links granule exocytosis to actin reorganization during platelet activation. Blood 126, 651–660 10.1182/blood-2014-12-618744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ye S., Karim Z. A., Al Hawas R., Pessin J. E., Filipovich A. H., and Whiteheart S. W. (2012) Syntaxin-11, but not syntaxin-2 or syntaxin-4, is required for platelet secretion. Blood 120, 2484–2492 10.1182/blood-2012-05-430603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arneson L. N., Brickshawana A., Segovis C. M., Schoon R. A., Dick C. J., and Leibson P. J. (2007) Cutting edge: Syntaxin 11 regulates lymphocyte-mediated secretion and cytotoxicity. J. Immunol. 179, 3397–3401 10.4049/jimmunol.179.6.3397 [DOI] [PubMed] [Google Scholar]
- 12. Chen D., Bernstein A. M., Lemons P. P., and Whiteheart S. W. (2000) Molecular mechanisms of platelet exocytosis: role of SNAP-23 and syntaxin 2 in dense core granule release. Blood 95, 921–929 [PubMed] [Google Scholar]
- 13. Chen D., Lemons P. P., Schraw T., and Whiteheart S. W. (2000) Molecular mechanisms of platelet exocytosis: Role of SNAP-23 and syntaxin 2 and 4 in lysosome release. Blood 96, 1782–1788 [PubMed] [Google Scholar]
- 14. Lemons P. P., Chen D., and Whiteheart S. W. (2000) Molecular mechanisms of platelet exocytosis: Requirements for alpha-granule release. Biochem. Biophys. Res. Commun. 267, 875–880 10.1006/bbrc.1999.2039 [DOI] [PubMed] [Google Scholar]
- 15. Karim Z. A., Zhang J., Banerjee M., Chicka M. C., Al Hawas R., Hamilton T. R., Roche P. A., and Whiteheart S. W. (2013) IκB kinase phosphorylation of SNAP-23 controls platelet secretion. Blood 121, 4567–4574 10.1182/blood-2012-11-470468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Valdez A. C., Cabaniols J. P., Brown M. J., and Roche P. A. (1999) Syntaxin 11 is associated with SNAP-23 on late endosomes and the trans-Golgi network. J. Cell Sci. 112, 845–854 [DOI] [PubMed] [Google Scholar]
- 17. Prekeris R., Klumperman J., and Scheller R. H. (2000) Syntaxin 11 is an atypical SNARE abundant in the immune system. Eur. J. Cell Biol. 79, 771–780 10.1078/0171-9335-00109 [DOI] [PubMed] [Google Scholar]
- 18. Offenhäuser C., Lei N., Roy S., Collins B. M., Stow J. L., and Murray R. Z. (2011) Syntaxin 11 binds Vti1b and regulates late endosome to lysosome fusion in macrophages. Traffic 12, 762–773 10.1111/j.1600-0854.2011.01189.x [DOI] [PubMed] [Google Scholar]
- 19. Pallavi B., and Nagaraj R. (2003) Palmitoylated peptides from the cysteine-rich domain of SNAP-23 cause membrane fusion depending on peptide length, position of cysteines, and extent of palmitoylation. J. Biol. Chem. 278, 12737–12744 10.1074/jbc.M208598200 [DOI] [PubMed] [Google Scholar]
- 20. Vogel K., and Roche P. A. (1999) SNAP-23 and SNAP-25 are palmitoylated in vivo. Biochem. Biophys. Res. Commun. 258, 407–410 10.1006/bbrc.1999.0652 [DOI] [PubMed] [Google Scholar]
- 21. Huang K., Yanai A., Kang R., Arstikaitis P., Singaraja R. R., Metzler M., Mullard A., Haigh B., Gauthier-Campbell C., Gutekunst C. A., Hayden M. R., and El-Husseini A. (2004) Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron 44, 977–986 10.1016/j.neuron.2004.11.027 [DOI] [PubMed] [Google Scholar]
- 22. Fukata M., Fukata Y., Adesnik H., Nicoll R. A., and Bredt D. S. (2004) Identification of PSD-95 palmitoylating enzymes. Neuron 44, 987–996 10.1016/j.neuron.2004.12.005 [DOI] [PubMed] [Google Scholar]
- 23. Roth A. F., Feng Y., Chen L., and Davis N. G. (2002) The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23–28 10.1083/jcb.200206120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Munday A. D., and López J. A. (2007) Posttranslational protein palmitoylation: Promoting platelet purpose. Arterioscler. Thromb. Vasc. Biol. 27, 1496–1499 10.1161/ATVBAHA.106.136226 [DOI] [PubMed] [Google Scholar]
- 25. Sim D. S., Dilks J. R., and Flaumenhaft R. (2007) Platelets possess and require an active protein palmitoylation pathway for agonist-mediated activation and in vivo thrombus formation. Arterioscler. Thromb. Vasc. Biol. 27, 1478–1485 10.1161/ATVBAHA.106.139287 [DOI] [PubMed] [Google Scholar]
- 26. Haudek V. J., Slany A., Gundacker N. C., Wimmer H., Drach J., and Gerner C. (2009) Proteome maps of the main human peripheral blood constituents. J. Proteome Res. 8, 3834–3843 10.1021/pr801085g [DOI] [PubMed] [Google Scholar]
- 27. De Vos M. L., Lawrence D. S., and Smith C. D. (2001) Cellular pharmacology of cerulenin analogs that inhibit protein palmitoylation. Biochem. Pharmacol. 62, 985–995 10.1016/S0006-2952(01)00739-0 [DOI] [PubMed] [Google Scholar]
- 28. Lawrence D. S., Zilfou J. T., and Smith C. D. (1999) Structure-activity studies of cerulenin analogues as protein palmitoylation inhibitors. J. Med. Chem. 42, 4932–4941 10.1021/jm980591s [DOI] [PubMed] [Google Scholar]
- 29. Huang E. M. (1989) Agonist-enhanced palmitoylation of platelet proteins. Biochim. Biophys. Acta 1011, 134–139 10.1016/0167-4889(89)90200-0 [DOI] [PubMed] [Google Scholar]
- 30. Dowal L., Yang W., Freeman M. R., Steen H., and Flaumenhaft R. (2011) Proteomic analysis of palmitoylated platelet proteins. Blood 118, e62–e73 10.1182/blood-2011-05-353078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kübler E., Dohlman H. G., and Lisanti M. P. (1996) Identification of Triton X-100 insoluble membrane domains in the yeast Saccharomyces cerevisiae. Lipid requirements for targeting of heterotrimeric G-protein subunits. J. Biol. Chem. 271, 32975–32980 10.1074/jbc.271.51.32975 [DOI] [PubMed] [Google Scholar]
- 32. Regula J. T., Boguth G., Görg A., Hegermann J., Mayer F., Frank R., and Herrmann R. (2001) Defining the mycoplasma “cytoskeleton”: the protein composition of the Triton X-100 insoluble fraction of the bacterium Mycoplasma pneumoniae determined by 2-D gel electrophoresis and mass spectrometry. Microbiology 147, 1045–1057 10.1099/00221287-147-4-1045 [DOI] [PubMed] [Google Scholar]
- 33. Choi W., Karim Z. A., and Whiteheart S. W. (2006) Arf6 plays an early role in platelet activation by collagen and convulxin. Blood 107, 3145–3152 10.1182/blood-2005-09-3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ren Q., Barber H. K., Crawford G. L., Karim Z. A., Zhao C., Choi W., Wang C. C., Hong W., and Whiteheart S. W. (2007) Endobrevin/VAMP-8 is the primary v-SNARE for the platelet release reaction. Mol. Biol. Cell 18, 24–33 10.1091/mbc.E06-09-0785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shiraishi M., Tani E., and Miyamoto A. (2010) Modulation of rabbit platelet aggregation and calcium mobilization by platelet cholesterol content. J. Vet. Med. Sci. 72, 285–292 10.1292/jvms.09-0385 [DOI] [PubMed] [Google Scholar]
- 36. Grgurevich S., Krishnan R., White M. M., and Jennings L. K. (2003) Role of in vitro cholesterol depletion in mediating human platelet aggregation. J. Thromb. Haemost. 1, 576–586 10.1046/j.1538-7836.2003.00087.x [DOI] [PubMed] [Google Scholar]
- 37. Zimmerman G. A., and Weyrich A. S. (2008) Signal-dependent protein synthesis by activated platelets: new pathways to altered phenotype and function. Arterioscler. Thromb. Vasc. Biol. 28, s17–s24 10.1161/ATVBAHA.107.160218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shipston M. J. (2014) Ion channel regulation by protein S-acylation. J. Gen. Physiol. 143, 659–678 10.1085/jgp.201411176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. el-Husseini Ael-D., and Bredt D. S. (2002) Protein palmitoylation: A regulator of neuronal development and function. Nat. Rev. Neurosci. 3, 791–802 10.1038/nrn940 [DOI] [PubMed] [Google Scholar]
- 40. Linder M. E., and Deschenes R. J. (2007) Palmitoylation: Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 10.1038/nrm2084 [DOI] [PubMed] [Google Scholar]
- 41. Laage R., Rohde J., Brosig B., and Langosch D. (2000) A conserved membrane-spanning amino acid motif drives homomeric and supports heteromeric assembly of presynaptic SNARE proteins. J. Biol. Chem. 275, 17481–17487 10.1074/jbc.M910092199 [DOI] [PubMed] [Google Scholar]
- 42. McNew J. A., Weber T., Parlati F., Johnston R. J., Melia T. J., Söllner T. H., and Rothman J. E. (2000) Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J. Cell Biol. 150, 105–117 10.1083/jcb.150.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu H., Zick M., Wickner W. T., and Jun Y. (2011) A lipid-anchored SNARE supports membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 108, 17325–17330 10.1073/pnas.1113888108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gonzalo S., and Linder M. E. (1998) SNAP-25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Mol. Biol. Cell 9, 585–597 10.1091/mbc.9.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hellewell A. L., Foresti O., Gover N., Porter M. Y., and Hewitt E. W. (2014) Analysis of familial hemophagocytic lymphohistiocytosis type 4 (FHL-4) mutant proteins reveals that S-acylation is required for the function of syntaxin 11 in natural killer cells. PLoS One 9, e98900 10.1371/journal.pone.0098900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vogel K., Cabaniols J. P., and Roche P. A. (2000) Targeting of SNAP-25 to membranes is mediated by its association with the target SNARE syntaxin. J. Biol. Chem. 275, 2959–2965 10.1074/jbc.275.4.2959 [DOI] [PubMed] [Google Scholar]
- 47. Salaün C., Gould G. W., and Chamberlain L. H. (2005) Lipid raft association of SNARE proteins regulates exocytosis in PC12 cells. J. Biol. Chem. 280, 19449–19453 10.1074/jbc.M501923200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Salaün C., Gould G. W., and Chamberlain L. H. (2005) The SNARE proteins SNAP-25 and SNAP-23 display different affinities for lipid rafts in PC12 cells. Regulation by distinct cysteine-rich domains. J. Biol. Chem. 280, 1236–1240 10.1074/jbc.M410674200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kreutzberger A. J., Kiessling V., and Tamm L. K. (2015) High cholesterol obviates a prolonged hemifusion intermediate in fast SNARE-mediated membrane fusion. Biophys. J. 109, 319–329 10.1016/j.bpj.2015.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ge S., White J. G., and Haynes C. L. (2010) Critical role of membrane cholesterol in exocytosis revealed by single platelet study. ACS Chem. Biol. 5, 819–828 10.1021/cb100130b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Suzuki K., and Verma I. M. (2008) Phosphorylation of SNAP-23 by IκB kinase 2 regulates mast cell degranulation. Cell 134, 485–495 10.1016/j.cell.2008.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jennings B. C., Nadolski M. J., Ling Y., Baker M. B., Harrison M. L., Deschenes R. J., and Linder M. E. (2009) 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. J. Lipid Res. 50, 233–242 10.1194/jlr.M800270-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thome M. (2004) CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat. Rev. Immunol. 4, 348–359 10.1038/nri1352 [DOI] [PubMed] [Google Scholar]
- 54. Percher A., Ramakrishnan S., Thinon E., Yuan X., Yount J. S., and Hang H. C. (2016) Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation. Proc. Natl. Acad. Sci. U.S.A. 113, 4302–4307 10.1073/pnas.1602244113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martin B. R. (2013) Nonradioactive analysis of dynamic protein palmitoylation. Curr. Protoc. Protein Sci. 73, 14.15: 14.15.1–14.15.9 10.1002/0471140864.ps1415s73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Burkhart J. M., Vaudel M., Gambaryan S., Radau S., Walter U., Martens L., Geiger J., Sickmann A., and Zahedi R. P. (2012) The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood 120, e73–e82 10.1182/blood-2012-04-416594 [DOI] [PubMed] [Google Scholar]
- 57. Grevengoed T. J., Klett E. L., and Coleman R. A. (2014) Acyl-CoA metabolism and partitioning. Annu. Rev. Nutr. 34, 1–30 10.1146/annurev-nutr-071813-105541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Graham G. J., Ren Q., Dilks J. R., Blair P., Whiteheart S. W., and Flaumenhaft R. (2009) Endobrevin/VAMP-8-dependent dense granule release mediates thrombus formation in vivo. Blood 114, 1083–1090 10.1182/blood-2009-03-210211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen D., and Whiteheart S. W. (1999) Intracellular localization of SNAP-23 to endosomal compartments. Biochem. Biophys. Res. Commun. 255, 340–346 10.1006/bbrc.1999.0173 [DOI] [PubMed] [Google Scholar]
- 60. Brigidi G. S., and Bamji S. X. (2013) Detection of protein palmitoylation in cultured hippocampal neurons by immunoprecipitation and acyl-biotin exchange (ABE). J. Vis. Exp. 18, e50031 10.3791/50031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang L., Gal J., Chen J., and Zhu H. (2014) Self-assembled FUS binds active chromatin and regulates gene transcription. Proc. Natl. Acad. Sci. U.S.A. 111, 17809–17814 10.1073/pnas.1414004111 [DOI] [PMC free article] [PubMed] [Google Scholar]