Abstract
Huntingtin N-terminal fragments (Htt-NTFs) with expanded polyglutamine tracts form a range of neurotoxic aggregates that are associated with Huntington's disease. Here, we show that aggregation of Htt-NTFs, irrespective of polyglutamine length, yields at least three phases (designated M, S, and F) that are delineated by sharp concentration thresholds and distinct aggregate sizes and morphologies. We found that monomers and oligomers make up the soluble M phase, ∼25-nm spheres dominate in the soluble S phase, and long, linear fibrils make up the insoluble F phase. Previous studies showed that profilin, an abundant cellular protein, reduces Htt-NTF aggregation and toxicity in cells. We confirm that profilin achieves its cellular effects through direct binding to the C-terminal proline-rich region of Htt-NTFs. We show that profilin preferentially binds to Htt-NTF M-phase species and destabilizes aggregation and phase separation by shifting the concentration boundaries for phase separation to higher values through a process known as polyphasic linkage. Our experiments, aided by coarse-grained computer simulations and theoretical analysis, suggest that preferential binding of profilin to the M-phase species of Htt-NTFs is enhanced through a combination of specific interactions between profilin and polyproline segments and auxiliary interactions between profilin and polyglutamine tracts. Polyphasic linkage may be a general strategy that cells utilize to regulate phase behavior of aggregation-prone proteins. Accordingly, detailed knowledge of phase behavior and an understanding of how ligands modulate phase boundaries may pave the way for developing new therapeutics against a variety of aggregation-prone proteins.
Keywords: biophysics, Huntington disease, neurodegenerative disease, polyglutamine, protein aggregation, phase transitions
Introduction
Many diseases are associated with protein misfolding and aggregation (1, 2). The aggregation process is often characterized by the presence of one or more threshold concentrations at which a sharp, discontinuous change to some aspect of the assembly state (e.g. size, conformational characteristics, material properties) occurs (3–6). Such a change can be described using the concepts of phase transitions. Phase separation, a subcategory of phase transitions, has recently received considerable attention due to increasing recognition of its importance in cell biology (7–13). Phase separation refers to aggregation-related changes in molecular density that give rise to the coexistence of dilute macromolecule-deficient phases and dense macromolecule-rich phases (3, 14, 15). Examples of multiple coexisting phases have been observed in biological contexts (15–18), and these phases can be liquid, solid, or semisolid (e.g. a gel) (10, 19–25). Driving forces for phase separation are quantified in terms of saturation concentrations (14, 19, 26). For a given two-phase system, the saturation concentration is the bulk concentration of the protein, beyond which the solution separates into two coexisting phases. The lower the saturation concentration, the stronger the driving force for aggregation and phase separation (14, 19, 26–29).
Ligand binding can alter saturation concentrations and shift phase boundaries to either stabilize or destabilize specific phases (30). Wyman and Gill (31) introduced this concept, known as polyphasic linkage, to interpret the impact of ligands on saturation concentrations. In the current work, we report results from our studies of ligand binding to N-terminal fragments of the huntingtin protein (Htt-NTFs).2
Huntington's disease (HD) is a devastating neurological disorder that affects medium spiny and cerebellar neurons (32). The age of onset as well as severity of HD are inversely correlated with the length of the polyglutamine (polyQ) tract within the N-terminal region of huntingtin (Htt) (32). The disease is characterized by deposition of insoluble intranuclear deposits of Htt-NTFs with expanded polyQ tracts in afflicted neurons (33, 34). Htt-NTFs consist of a 17-residue N-terminal stretch (MATLEKLMKAFESLKSF) designated as N17, a polyQ tract of length n (Qn), and a 38-residue C-terminal stretch (C38) that includes two polyproline (polyP) modules, P11 and P10, connected by a 17-residue linker denoted as L17 (QLPQPPPQAQPLLPQPQ) (Fig. 1a). In transgenic mouse models, Htt-NTFs produce robust degeneration of the relevant neurons (35).
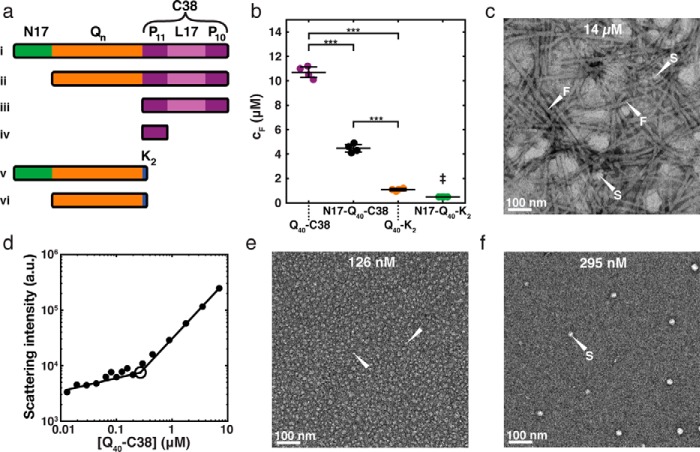
Figure 1.
Experimental evidence for Htt-NTF phase behavior with multiple phase boundaries. a, Htt-NTF sequence architectures used in this study. i, full Htt exon 1, N17-Qn-C38; ii, Qn-C38; iii, C38; iv, P11; v, N17-Qn-K2; vi, Qn-K2. Here, n = 40 in all experiments except for the in-cell FRET aggregation assay (see Fig. S1), where n = 72, and some experiments where n = 30 was compared with n = 40 (see Fig. 5e). Additionally, K2 corresponds to two lysines. b, previously reported (19) saturation concentrations (denoted here as cF) determined by measuring the concentration of soluble protein remaining in the supernatant (colorimetric micro-BCA assay) following quiescent incubation at 30 °C and centrifugation of the indicated Htt-NTF constructs. Consistent with the presence of a saturation concentration, the same concentration was arrived at for a given construct regardless of the starting concentration, so long as the starting concentration was above the saturation concentration indicated. The saturation concentration is modulated by the N17 and C38 sequence modules that flank polyQ in Htt-NTFs. Black bars, mean ± S.D. (error bars) of four independent experiments. The N17-Q40-K2 cF was below the detection limit of the assay for all trials, so the quantity plotted is the detection limit and represents an upper bound for the N17-Q40-K2 cF. Statistical significance was determined by one-way ANOVA with Tukey's range test for post hoc analysis. ***, p < 0.001. ‡, the N17-Q40-K2 cF data were not included in the statistical significance analysis for the reason cited above. c, TEM image of Q40-C38 fibrils observed at a concentration supersaturated with respect to cF (14 μm). Two of the fibrils in the image are indicated with white arrows labeled F, and two examples of spherical aggregates are indicated with white arrows labeled S. See Fig. 4c for an additional example of fibrils and spheres. d, right-angle static light scattering of Q40-C38 in 20 mm Tris, 5 mm EDTA, 1 mm DTT, pH 7.4, as a function of Q40-C38 concentration. The discontinuity at ∼290 nm is indicative of a phase boundary. e, TEM image of Q40-C38 monomers and small oligomers observed at a concentration subsaturated with respect to cS (126 nm). This particular field of view contains monomers/small oligomers, two of which are indicated with white arrows. f, TEM image of Q40-C38 spherical aggregates observed at a concentration supersaturated with respect to cS but subsaturated with respect to cF (295 nm). One of the spherical aggregates is indicated with a white arrow labeled S. All TEM images (c, e, and f) are at the same scale. Scale bar, 100 nm.
Although HD is characterized by Htt-NTF inclusions, the role of these inclusions as agents of neurotoxicity has been the topic of considerable debate (34–44). In a recent study, Ramdzan et al. (45) showed that both soluble species and insoluble deposits engender neurotoxicity, albeit through very different mechanisms. Soluble forms of Htt-NTFs promote cell death via apoptosis due to mitochondrial dysfunction. In contrast, insoluble inclusions weaken apoptosis and induce slow cell death via necrosis by impacting cellular metabolism and promoting co-aggregation and phase separation of other proteins with Htt-NTFs (45). In light of this recent study, it is reasonable to postulate that the entire aggregation process and the full spectrum of Htt-NTF aggregates might have deleterious roles to play in neurons. This postulate argues against the notion that a single species of aggregate is toxic and suggests that attempts to target a single toxic species are likely to be unfruitful. It may be more advantageous to instead consider toxicity in terms of supersaturation with respect to a phase boundary (or boundaries) and the alleviation of toxicity in terms of ligands that effectively reduce supersaturation by shifting phase boundaries via thermodynamic linkage.
A ligand of interest that has been shown to reduce toxicity of Htt-NTFs in multiple HD models is the ubiquitously expressed protein profilin-1, hereafter referred to as profilin for brevity. Profilin is a 15-kDa protein that binds to G-actin, polyP, and phosphatidylinositol lipids through three distinct binding sites (46–49) and is essential for cell division and survival during embryogenesis (50). Shao et al. (51) showed that profilin suppresses Htt-NTF aggregation through direct interactions with Htt-NTFs. Increased expression of profilin reduces intracellular aggregation of Htt-NTFs and diminishes the toxicity of Htt-NTFs in cell lines, primary neurons, model organisms, and transgenic mice (52). Therefore, a direct therapeutic approach to HD could involve the design of ligands that mimic the effects of profilin on Htt-NTF aggregation and alleviation of toxicity. To realize such a therapeutic, we need a mechanistic understanding of how profilin suppresses Htt-NTF aggregation. This is the focus of the current work.
Here, we show that Htt-NTFs form at least three distinct phases that are preferred across distinct concentration regimes, and we identify the concentration thresholds that demarcate these concentration regimes. By quantifying the driving forces for aggregation and phase separation in the presence and absence of profilin, we show that the binding of profilin to the proline-rich C38 region of Htt-NTFs decreases the driving forces for aggregation and phase separation through polyphasic linkage. Specifically, systematic measurements of profilin binding aided by coarse-grained simulations reveal that profilin preferentially binds low-molecular weight species (monomers/oligomers). This leads to the stabilization of monomers and oligomers and a suppression of large-scale aggregation and phase separation. In addition to direct interactions between profilin and polyP, our analysis points to hitherto unknown auxiliary interactions between profilin and the polyQ domain that engender an enhancement in profilin binding to Htt-NTF monomers/oligomers.
Results
C38 is necessary for profilin to bind to Htt-NTFs
Shao et al. (51) previously identified unphosphorylated profilin as an intracellular ligand that reduces Htt-NTF aggregation and toxicity. In that study, a direct interaction was inferred from pulldowns between non-mutant Htt-NTF (Q25) and either wildtype profilin or profilin variants with a mutated polyP binding pocket. The results indicated that the polyP binding pocket of profilin is essential for binding Htt-NTFs. We carried out follow-up experiments to assess, directly, whether the Htt-NTF C38 stretch is essential to elicit profilin-dependent reduction in aggregation. We used an intracellular aggregation assay (51, 53–55) to quantify the impact of profilin overexpression on the aggregation of constructs with and without the C38 stretch. Htt-NTFs with expanded polyQ tracts (Q72) were fused to cyan (CFP) or yellow fluorescent proteins (YFP). CFP and YFP are used as FRET pairs, and when fused to Htt-NTFs they serve as intracellular reporters of aggregation. We expressed N17-Q72-C38-CFP/YFP or N17-Q72-CFP/YFP in HEK293 cells cultured in a 96-well format (51), with or without profilin. We incubated the cells for 48 h before fixing the plates in 2% paraformaldehyde and measuring intracellular, intermolecular FRET in a fluorescence plate reader. As described previously (51), wildtype profilin reduced N17-Q72-C38-CFP/YFP aggregation by 30%. However, when the polyP-containing C38 stretch was removed, the aggregation of N17-Q72-CFP/YFP was unaffected by profilin overexpression (see first and second green bars in Fig. S1). The ability to suppress intracellular aggregation by profilin was blocked by the phosphomimic mutation S137D and was preserved by the S137A mutation (second and third gray bars, respectively, in Fig. S1), whereas these mutations had no effects on constructs lacking C38 (green bars in Fig. S1). These data confirm that direct interactions between profilin and polyP modules in C38 play a central role in the modulation of Htt-NTF aggregation via profilin binding.
Constructs for in vitro experiments
Recent in vivo studies from Yang and co-workers (56, 57) showed that a ΔN17 variant of huntingtin, which lacks the N17 module, leads to faithful reproduction of HD phenotypes in transgenic mice. In light of these observations, and given the relative ease of synthesis and purification of Htt-NTFs that lack the N17 module (19), we used constructs of the form Qn-C38 for a majority of our in vitro experiments. Here, n refers to the length of the polyQ tract. For most of the experiments, a polyQ length of n = 40 was used for the feasibility of synthesis and handling while still being long enough to be of a mutant or pathological length (58). In the interest of completeness, we also used a set of distinct Htt-NTF sequences for some of the experiments. Fig. 1a shows a detailed inventory of all of the sequence constructs used in one or more of our studies.
Htt-NTFs form at least three distinct phases
Previous studies (19) showed that Htt-NTFs separate into soluble and insoluble species at bulk concentrations that are above construct-specific saturation concentrations. Accordingly, one set of saturation concentrations can be defined in terms of the solubility limit, which we shall designate as cF for fibrillar-phase saturation concentration. It is important to clarify that cF was designated as cS in the work of Crick et al. (19). For bulk concentrations that are greater than cF, the concentration of soluble protein in the supernatant will be equal to cF, provided that equilibrium has been established between the soluble and insoluble phases. The value of cF is quantifiable using previously published colorimetric assays (19). In this approach, solutions with varying concentrations of fully disaggregated Htt-NTFs (see “Experimental procedures” and supporting Methods), were equilibrated via incubation in a temperature-controlled water bath without shaking for up to 2 weeks. The solutions were separated, via centrifugation, into soluble supernatants that coexist with insoluble pellet fractions. For bulk concentrations above a construct-specific threshold value of cF, the concentration of protein remaining in solution was found to be constant and independent of starting concentration. This is a prerequisite for the existence of a saturation concentration (59). Values of cF for different Htt-NTF constructs with polyQ lengths of n = 40 at 37 °C are shown in Fig. 1b. The values of cF varied with Htt-NTF constructs, where each construct either has a different sequence module that flanks the polyQ segment or, for a given construct, the polyQ length is different (19).
At concentrations above cF, the insoluble material in the pellet stains positively with amyloid-sensitive dyes, such as thioflavin T (ThT) (19). Transmission electron microscopy (TEM) images confirmed that the insoluble phase is predominantly composed of fibrillar aggregates (Fig. 1c). We also observed coexisting small spherical aggregates in TEM images. These observations suggest that spherical aggregates might represent either a separate stable phase that is in equilibrium with the fibrils and the unaggregated protein in the bulk solution, or they are metastable phases that form as precursors to fibrils. Given their destabilization with increased supersaturation with respect to cF, we investigated the possibility that spherical aggregates form as a distinct phase with saturation concentrations that are below cF. On average, the spherical species appeared to be too small for sedimentation by centrifugation. Therefore, we turned to right-angle static light scattering to investigate the presence of a second phase boundary at concentrations that were subsaturated with respect to cF (i.e. for concentrations less than cF).
The average size and the concentration of scattering species determine the intensity of scattered light in static light-scattering experiments. The appearance of a sharp discontinuity in scattering intensity versus concentration is indicative of the presence of a saturation concentration in a phase-separating system (60). To test for the possible presence of a distinct saturation concentration that lies below cF, we measured right-angle static light scattering of Htt-NTF solutions as a function of peptide concentration (Fig. 1d). We observed a discontinuity in the concentration dependence of the light-scattering intensity at ∼290 nm for Q40-C38. This is consistent with the presence of a phase boundary or saturation concentration, which we term cS for spherical-phase saturation concentration. TEM images indicate that species considerably smaller than the 10–20-nm size, which presumably include monomers and oligomers, are the predominant morphological species at concentrations below cS (Fig. 1e). Conversely, spherical aggregates with a diameter of ∼25 nm are the preferred morphologies for concentrations between cS and cF (Fig. 1f). We analyzed multiple independent TEM images to quantify the distribution of aggregate sizes. This analysis confirmed that a single size distribution was present at concentrations below cS, whereas two distinct size distributions were present at concentrations that lie between cS and cF (Fig. S2).
Our results, derived from a joint analysis of solubility, light scattering, and morphological measurements, lead to a phase diagram that delineates the presence of at least three distinct phases separated by two phase boundaries (Fig. 2a). For total concentrations ct that are lower than cS, Htt-NTFs populate monomers and oligomers (M phase). For concentrations between cS and cF, the concentration of monomers/oligomers saturates, and these species are in equilibrium with large spherical aggregates (S phase). Soluble, spherical aggregates that are ∼25 nm in diameter are the predominant morphologies in the S phase. For ct > cF, the concentration of soluble species saturates, and equilibrium is established between soluble species and insoluble, large, β-sheet–rich fibrils with a bottlebrush architecture that constitute the F phase (61). We use the term “aggregates” for molecules engaged in intermolecular interactions that can range from dimers to fibrils, whereas “oligomers” refers to aggregates smaller than the spherical aggregates that characterize the S phase.
Figure 2.

Schematic representation of Htt-NTF phase behavior and polyphasic linkage. a, Htt-NTF phase behavior is described by three phases (M, S, and F phase), color-coded as red, blue, and green, respectively, and two phase boundaries (cS and cF) indicated by gray lines between the phases. The total monomer-equivalent Htt-NTF concentration (horizontal axis) with respect to these boundaries will determine whether monomers and oligomers, spherical aggregates, or fibrils form. b, the influence of a binding partner (profilin) on the Htt-NTF cS phase boundary. The phase boundary is depicted as a solid gray line in the absence of profilin (i) and for three different profilin binding scenarios (ii–iv). The relative number of profilin (black circles) associated with each phase (M or S) indicates which phase is preferentially bound. The dotted gray line in ii and iii indicates the cS in the absence of profilin, and the red and blue arrows indicate the direction of the shift in the cS if the M or S phase is preferentially bound, respectively. In scenario iv, profilin binds both phases equally well, and thus no shift in cS is observed. The orientation of b is rotated 90° with respect to a for formatting purposes. See “Results” for further description of polyphasic linkage.
Profilin preferentially binds M-phase species
The quantitative phase diagrams for Htt-NTFs become useful when considering the impact of binding partners on phase behavior. Polyphasic linkage provides a framework for understanding how ligand binding modulates the phase equilibria of molecules that undergo aggregation and phase separation (30, 31) (Fig. 2b). Polyphasic linkage can be explained by considering the simple example of a macromolecular solution that separates into two distinct phases labeled as A and B, respectively. Here, A could be the dilute phase and B the dense phase. In the absence of ligand, we denote cA as the saturation concentration of the macromolecule in the A phase. The establishment of phase equilibrium in the presence of ligand, which refers to the equalization of chemical potentials of the macromolecule across the two phases, leads to the following scenarios. Assuming the ligand binds to the macromolecule in both phases, then the saturation concentration for phase separation of the macromolecule in the presence of the ligand becomes cA(L) = cA(pA/pB) (30, 31). Here, pA and pB are the binding polynomials for the binding of ligand to species in the A and B phases, respectively. For reversible macromolecular associations, a binding polynomial quantifies the sum of the activities (concentrations) of all bound and unbound species involving the macromolecule relative to the activity (concentration) of the unbound macromolecule (62). The negative logarithm of the binding polynomial is proportional to the free energy of binding. Accordingly, if pA is greater than pB, then species in the A phase will have higher affinity for the ligand when compared with species in the B phase. In this scenario, ligand binding will help retain a higher concentration of the macromolecule in the A phase, and hence cA(L) will be greater than cA. The converse is true if pA is less than pB, which would engender increased partitioning of the ligated macromolecule into the dense phase, B. Finally, ligand binding will not impact the phase equilibrium if pA = pB.
To understand the basic tenets of polyphasic linkage, we consider the impact that profilin binding has on the phase boundary between the M and S phases. In accordance with polyphasic linkage, if profilin were to bind preferentially to the M phase of Htt-NTFs, then the value of cS would shift to higher concentrations (Fig. 2b, ii). In contrast, preferential binding to the S phase would shift cS to lower concentrations (Fig. 2b, iii). Equivalent binding to species in the two phases would have no effect on the phase boundary (Fig. 2b, iv). These scenarios are depicted qualitatively in Fig. 2b.
Given knowledge of the phase diagram for Htt-NTFs, we hypothesized that profilin must reduce aggregation/phase separation through a polyphasic linkage mechanism, whereby it binds preferentially to M-phase species to destabilize the S and F phases. This hypothesis is directly testable by measuring saturation concentrations in the absence (e.g. Fig. 1d) and presence of profilin. A lack of change in the saturation concentration in the presence of profilin may indicate no binding or equivalent binding to the two phases. Alternatively, if the saturation concentration changes in the presence of profilin, then the direction of the change will point to the phase that is stabilized or destabilized by ligand binding.
We performed static right-angle light scattering measurements as a function of Q40-C38 concentration in the presence of fixed concentrations of profilin (Fig. 3a). The discontinuity in the concentration-dependent scattering profile, which quantifies the cS of Q40-C38, increases by more than an order of magnitude in the presence of profilin. The inferred value of cS increases from ∼290 nm in the absence of profilin to ∼3 μm in the presence of 1 μm profilin (Fig. 3b and Table S1). The value of cS increases with increasing profilin concentration and appears to plateau to a value of ∼6 μm in the presence of 20 μm profilin. The increase of cS in the presence of profilin suggests that binding to profilin destabilizes the S phase. Our reasoning, based on thermodynamic linkage relations, suggests that profilin must bind preferentially to either the M or F phases, whereby ligand-induced changes to one phase boundary in a closed system will necessarily lead to changes in all phase boundaries.
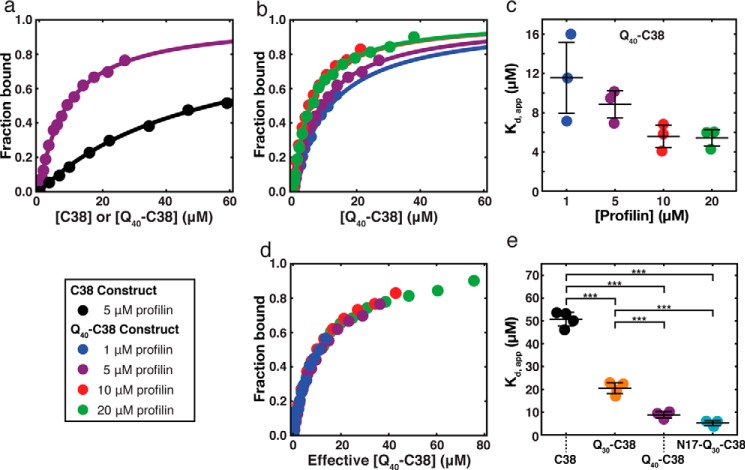
Figure 3.

Profilin shifts the cS of Q40-C38 through polyphasic linkage. a, right-angle static light scattering of Q40-C38 solution as a function of Q40-C38 concentration, in the absence of profilin (black, reproduced from Fig. 1d) and in the presence of 1, 5, 10, or 20 μm profilin (blue, purple, red, and green, respectively) in 20 mm Tris, 5 mm EDTA, 1 mm DTT, pH 7.4. One representative trial at each profilin concentration is shown, and the plots are arbitrarily offset in the vertical direction for the sake of clarity. The values of cS for Q40-C38 in the presence of each of these concentrations of profilin are indicated by the discontinuity in slope, and each discontinuity is marked using an open circle. We note that the slope of the low-concentration arm of the 10 μm profilin data set differs from the other data sets and suspect that this is the result of an experimental artifact (see “Experimental procedures”). Regardless, there was good agreement between trials concerning the location of the discontinuity, as is evident in b. b, Q40-C38 cS measured in a plotted as a function of the profilin concentration. The intersections of “best-fit” lines from jackknife sampling for at least three trials at each profilin concentration are plotted (see supporting Methods). Black bars, mean ± S.D. (error bars). Statistical significance was determined by Welch's ANOVA with Games–Howell post hoc analysis. **, p < 0.01; ***, p < 0.001. Colors are the same as in a. Even the lowest concentration of profilin tested (1 μm) resulted in more than an order of magnitude increase in the cS of Q40-C38.
To infer the impact of profilin binding on the stability of the F phase, we quantified the impact of the presence of profilin on the kinetics of fibril formation and the morphologies of fibrils that are formed in the presence of profilin. We monitored the kinetics of fibril formation by following the rate of change of ThT fluorescence. In these measurements, we fixed the total concentration (ct) of Q40-C38 to be above its intrinsic cF such that the degree of supersaturation with respect to cF, quantified as ln(ct/cF), was 0.75. A series of independent measurements were performed, each in the presence of a different concentration of profilin (Fig. 4a). As the profilin concentration increased, fewer fibrillar aggregates formed, as indicated by the maximum ThT intensity reached, and the rate of fibril formation was slowed, as indicated by the time required to reach half-maximum intensity (Fig. 4b). These results are consistent with ligand binding shifting the intrinsic cF of Q40-C38 to higher values. Negative stain TEM images of samples from the end point of aggregation assays confirms that in the presence of profilin, fibrils appear to be reduced in both length and number (Fig. 4, c and d).
Figure 4.

The influence of profilin on Q40-C38 fibril formation. a, ThT assay of Q40-C38 aggregation kinetics at a fixed concentration of Q40-C38 in the presence of various concentrations of profilin. ThT fluoresces upon binding to amyloid-like fibrils, thus providing a readout of fibril formation in the form of fluorescence intensity. b, the plateau ThT fluorescence intensity (ThT Imax) and the time to reach 50% Imax (t50) (red and blue, respectively), are plotted as a function of profilin concentration. c, representative TEM image of a sample taken from an aggregation assay in the absence of profilin showing an abundance of long fibrils coexisting with spherical aggregates. d, representative TEM image of a sample taken from an aggregation assay in the presence of 5 μm profilin shows short fibrils few in number as well as spherical aggregates and oligomers or possibly monomers. His-tagged profilin was marked on the carbon-coated copper grid with 5-nm nickel-nitrilotriacetic acid-gold nanoparticles, which appear as black dots. Examples of gold nanoparticles are indicated with black arrows. Examples of fibrils and spheres are indicated with white arrows labeled F and S, respectively, in c and d.
To determine whether profilin was bound to fibrils or other aggregates, we tagged profilin on TEM sample grids with 5-nm gold nanoparticles functionalized with a nickel moiety that binds histidine tags. Imidazole buffer washes were used to prevent nonspecific binding. The gold nanoparticles were found predominantly near oligomers and less frequently near larger spherical aggregates or fibrils (Fig. 4d). In total, 257 gold nanoparticles in three TEM images were counted and classified according to their association with M-, S-, or F-phase species. Of the 257 gold nanoparticles, 133 (51.7%) were associated with M-phase species, whereas only 49 (19.1%) were associated with S-phase species, and 75 (29.2%) were associated with F-phase species (see supporting Methods for details). Taken together, the results from Figs. 3 and 4 suggest that profilin binding destabilizes the S and F phases in favor of the M phase via polyphasic linkage. This is attributable to the preferential binding of profilin to species of the M phase. In light of this suggestion, we investigated the mode of binding of profilin to species of the M phase to uncover a mechanistic understanding of how the M-phase species are stabilized by profilin binding.
Profilin binds to polyP and binding to Htt-NTFs is aided by the presence of the polyQ region
Profilin binds to polyP through a distinct binding site (47–49, 63, 64). The C38 region of Htt-NTFs encompasses two polyP modules (P11 and P10) connected by a 17-residue linker (L17) that is rich in Pro, Gln, and Leu residues. As described above, pulldowns performed by Shao et al. (51) indicated that the polyP binding pocket of profilin is responsible for binding Htt-NTFs, and our follow-up experiments confirmed that the Htt-NTF C38 stretch is essential to elicit profilin-dependent reduction in aggregation (Fig. S1). Thus, the main mode of interaction between profilin and Htt-NTFs is through direct binding to polyP modules in the C38 region.
To place the interactions of profilin with Htt-NTFs on a quantitative footing, we first measured the binding affinity of profilin to polyP in isolation and in the context of C38 (Fig. S3a). We used a fluorescence-based binding assay developed by the Pollard laboratory (49). The binding of polyP ligands to profilin leads to increased fluorescence of Trp-3 and Trp-31 from profilin and a blue shift in the maximal wavelength for Trp fluorescence. We used the fluorescence intensity at 320 nm to quantify the binding of profilin to C38 and compared this with the binding of profilin to an 11-mer of polyP (P11). The binding data for P11 and C38 were fit to a model with a single binding site (Fig. S3a). Here, profilin is the macromolecule, and P11 and C38 are the ligands. The dissociation constant Kd,P11 that we obtain for the binding of P11 to profilin (106.5 μm) matches published estimates (49, 65).
In contrast to P11, C38 is a bivalent ligand with two polyP modules. The dissociation constant Kd,C38 for the binding of profilin to C38 is 50.7 μm, which is essentially half the value of Kd,P11. When we used the number of polyP tracts to normalize the binding isotherms, the isotherms overlay, indicating that the difference in dissociation constants results from C38 having two binding sites for profilin (Fig. S3b). The fact that Kd,P11 ≈ 2Kd,C38 suggests that profilin binds to the two sites on C38 independently. This was confirmed using Hill and Scatchard analyses as shown in Fig. S4. We obtained an average value of 1.0 as our estimate for the Hill coefficient, which quantifies the size of the cooperative unit for binding. Therefore, we conclude that profilin molecules bind independently to the individual polyP modules in C38. Accordingly, the lowering of Kd,C38 derives exclusively from doubling the concentration of polyP by doubling the polyP valency in C38. Additionally, in three separate sets of fluorescence experiments, we fixed the concentration of profilin and titrated the concentrations of N17, polyQ (Q30), and L17. We were unable to detect any measurable changes in Trp fluorescence of profilin for a range of concentrations of each of N17, polyQ, and L17 as ligands. We conclude that these modules have undetectably weak interactions with profilin at or near the polyP-binding site, although interactions with other distal sites cannot be ruled out.
Next, we measured the binding of profilin to Q40-C38, a construct possessing both a polyQ and C38 tract. The binding data were fit to a 1:1 binding model, and the inferred apparent dissociation constant (Kd,app) in the presence of 5 μm profilin was found to be 8.8 μm for Q40-C38 (Fig. 5a). This value for Kd,app is ∼6-fold lower than the intrinsic value of Kd,C38 = 50.7 μm for C38. We also measured the binding of Q40-C38 to profilin at several different fixed concentrations of profilin. These measurements yield a modest decrease in the inferred Kd,app as profilin concentration increases (Fig. 5 (b and c) and Table S1) and suggest one of two possibilities; either profilin undergoes a concentration-dependent change, such as dimerization, that influences its binding to Q40-C38, or alternatively, the effect can be explained in terms of a shift in the concentration of Q40-C38 within the M phase that is caused by profilin binding leading to a modulation of the phase boundary. Rennella et al. (66) have shown that profilin only forms sparsely populated oligomers at millimolar concentrations. In our studies, we never exceeded 20 μm profilin. Therefore, it appears safe to conclude that profilin oligomerization is not relevant.
Figure 5.
The effect of polyQ on profilin binding to Htt-NTFs. a, binding isotherms of 5 μm profilin with C38 (black) versus 5 μm profilin with Q40-C38 (purple). The presence of the polyQ tract enhances binding at a given profilin concentration. b, binding isotherms of 1, 5, 10, and 20 μm profilin with Q40-C38. c, apparent Kd values extracted from fits to the isotherms in a. A slight dependence of the Kd,app on profilin concentration is apparent, although a one-way ANOVA test of the data revealed that the difference is at the threshold of significance, with a p value of 0.0507. d, when the concentration of each binding isotherm is adjusted by the magnitude of the profilin-dependent shift in cS (cS,Pfn/cS,intrinsic), then the binding isotherms collapse to a single curve, confirming that the observed dependence of Kd,app on the profilin concentration is due to profilin-dependent changes in partitioning of Q40-C38 molecules between the M- and S-phases (see “Results” for details). e, comparison of apparent Kd values of C38 (black) and Q40-C38 (purple) with Q30-C38 (orange) and N17-Q30-C38 (teal), all measured at 5 μm profilin. Statistical significance was determined by one-way ANOVA with Tukey's range test for post hoc analysis. ***, p < 0.001. Binding of profilin appears to correlate with aggregation propensity, which is dictated by polyQ length (Q30 versus Q40) and flanking sequence context (absence or presence of N17). Black bars in c and e indicate the mean ± S.D. (error bars) of three or four independent experiments.
We know now that profilin binding increases the value of cS. Because the cS dictates the relative amounts of molecules in the M and S phases, which in turn influences Kd,app, the observed dependence of Kd,app on the profilin concentration could be due to changes in the partitioning of Q40-C38 molecules between the M and S phases resulting from profilin-dependent changes in the cS. In support of this hypothesis, we find that adjusting the concentration of each binding isotherm by the magnitude of the profilin-dependent shift in cS (defined here as cS,Pfn/cS,intrinsic, where cS,Pfn is the cS measured in the presence of a given concentration of profilin, and cS,intrinsic is the cS measured in the absence of profilin (see Fig. 3)), leads to binding isotherms that collapse onto a single master curve (Fig. 5d). Therefore, lowering of Kd,app at higher profilin concentrations is thermodynamically linked to the profilin-dependent increase in cS, which increases the M-phase concentration of Q40-C38 molecules.
Our measurements also show that the binding of profilin to Htt-NTF constructs depends on the length of the polyQ segment and the presence of the N17 module (Fig. 5e and Table S1). Specifically, the value of Kd,app, measured in the presence of a fixed profilin concentration, is lowered upon increasing the polyQ length in Qn-C38 constructs. The value of Kd,app is further lowered upon the addition of the N17 module, which is shown by comparing the values for Kd,app obtained for N17-Q30-C38 with those obtained for Q30-C38 and Q40-C38. The decrease in Kd,app in going from Q30-C38 to N17-Q30-C38 is equivalent to the decrease we observe in going from Q30-C38 to Q40-C38. Given that increasing the polyQ length and the presence of N17 both promote Htt-NTF oligomerization/aggregation (19, 68, 69), these results are consistent with the idea that the binding of profilin to Htt-NTFs is enhanced due to polyQ- and N17-dependent assembly. These results further suggest that oligomerization can engender positive cooperativity in binding, which results in a reduced Kd,app. Consistent with this interpretation, Scatchard analysis of profilin binding to Q40-C38 indicated positive cooperativity, which was absent in the binding of C38 to profilin (Fig. S4). Additionally, binding of profilin to Q30-C38 and N17-Q30-C38 also exhibited positive cooperativity (Fig. S5), offering a clear correlation between the observed cooperativity and the presence of polyQ or N17-polyQ. However, the evidence for cooperativity in binding does not illuminate the specific mechanism through which profilin binding is enhanced in the presence of polyQ and N17. To explore possible origins for this behavior, we used coarse-grained simulations to explicitly test distinct models.
Modeling suggests that profilin modulates Htt-NTF phase behavior through a combination of specific interactions with polyP and auxiliary interactions with polyQ
To understand the source of the enhanced profilin binding in constructs with flanking N-terminal polyQ regions, we turned to coarse-grained simulations to identify the most plausible mechanistic models for our observations. Our modeling probes how the apparent binding affinity is influenced by increased local concentration and auxiliary interactions that involve regions on profilin that do not overlap with its polyP-binding site. Here, increased local concentration refers to the clustering of polyP segments due to Q40-C38 oligomerization. Auxiliary interactions can also contribute to the apparent increase in binding affinity through multivalency of both Q40-C38 oligomers and profilin (70–72).
Using a phenomenological coarse-grained model, in which we represent profilin and Q40-C38 as multiple spheres or “beads” connected by “springs” that mimic their atomistic architectures, we performed simulations in which binding was quantified between profilin and C38 or Q40-C38 as a function of Q40-C38 oligomer size. The general architecture of the coarse-grained model is shown in Fig. 6a. Each profilin molecule has a polyP-binding site that can interact with a polyP segment in either C38 or Q40-C38. We refer to this interaction as the primary interaction between profilin and C38 or Q40-C38, given that this is the interaction that is measured in the fluorescence experiments. If an increased local concentration of polyP segments leads to the observed experimental results, then the inclusion of this primary interaction between profilin and C38 or Q40-C38 should be sufficient to reproduce trends in binding that are consistent with the experimental results. In this scenario, Q40-C38 oligomerization should increase the local concentration of C38 modules. To be consistent with experimentally measured data, the simulation results would need to reproduce two key findings: 1) binding to C38 alone should be weaker than binding to Q40-C38 (Fig. 5a), and 2) binding to M-phase oligomers should be stronger than binding to S-phase aggregates (Fig. 3). Our results show that a model restricted to inclusion of primary interactions alone will not reproduce either of the experimental observations listed above. Instead, in a model that only accounts for primary interactions, we found that binding to C38 alone was stronger than binding to Q40-C38, and the fraction of profilin molecules bound decreased from that of the Q40-C38 monomer with increasing Q40-C38 oligomer size (Fig. 6c). This decrease originates from steric-effects, whereby bound profilin molecules, as well as other C38 stretches, get in the way of unbound profilin, occluding the C38-binding sites and reducing the effective concentration of free C38. This result suggests that an increased local concentration of polyP segments alone cannot account for the experimentally observed trends.
Figure 6.

Coarse-grained model identifies an auxiliary interaction necessary for the experimentally observed profilin effect. a, visual representation of the architecture used for the increased local concentration model. Q40-C38 and profilin are represented by a set of spheres or “beads” as defined in the top legend (see supporting Methods for model details). The primary interaction between profilin (Pfn) and the polyP segment of C38 is denoted by a green arrow. b, visual representation of the architecture used for the polyQ-Pfn auxiliary interaction model. This model is identical to the increased local concentration model, except an auxiliary interaction is added between the polyQ bead and a nonspecific region of the profilin molecule that does not overlap with the polyP interaction site (red arrow). c and d, fraction of profilin molecules bound to polyP segments observed in coarse-grained simulations of the increased local concentration model and the polyQ-Pfn auxiliary interaction model, respectively. Each simulation contained 210 Pfn molecules and 630 C38 stretches. Additionally, each simulation had a homogeneous distribution of cluster sizes, X (X = 1, 3, 6, 9, 15, 35, 70, 210), such that the number of clusters multiplied by X equaled 630. Each bar denotes results from simulations performed with a distinct cluster size as indicated by the bottom legend. A cluster size of zero denotes C38 without the polyQ domain. Gray beads in the legend denote beads that interact with profilin via the primary Pfn-PolyP interaction. Error bars, S.E. from five independent simulations.
Next, we tested three distinct models for auxiliary interactions that engender multivalency and might explain the observed experimental results. The distinct auxiliary interactions are denoted as polyQ-Pfn interactions, linker-Pfn interactions, and Pfn-Pfn interactions. Here, Pfn refers to profilin. In each of these models, the coarse-grained profilin molecule can engage in an auxiliary interaction, in addition to the primary polyP interaction. In the polyQ-Pfn interaction model, the auxiliary interaction is between the polyQ bead of a Q40-C38 molecule and a nonspecific region on the profilin molecule that is distinct from the polyP-binding site (Fig. 6b). This model accounts for experimental results, which suggest that long polyQ tracts and polyQ aggregates can interact nonspecifically with other molecules, given the uniformly “sticky” surface of the polyQ domain (73–76). This model allows for the possibility of a heterotypic multivalent complex in which a single profilin molecule is engaged in two different types of interactions with a Q40-C38 oligomer. In the linker-Pfn interaction model, the auxiliary interaction is between the non-proline residues of L17 and the face opposing the polyP-binding site on profilin, which is enriched in polar residues (Fig. S6b) (64). This model queries the possibility of a heterotypic complex in which a single profilin molecule interacts with the glutamine and leucine residues of L17 from one Q40-C38 molecule and a polyP region from another Q40-C38 molecule. Finally, in the Pfn-Pfn interaction model, the auxiliary interaction is a stable dimerization of two profilin molecules (Fig. S6c). This model is intended to address published conjectures suggesting that profilin can undergo dimerization (66, 77) and allows for the possibility of a homotypic complex in which a profilin dimer engages in interactions with two distinct polyP sites.
Fig. 6d and Fig. S6 (d–f) show the results obtained from simulations based on each of the three auxiliary interaction models. Only the polyQ-Pfn interaction model (Fig. S7) was able to reproduce both experimental findings. Specifically, our calculations based on this model show that binding to C38 alone was weaker than binding to Q40-C38, and binding to small oligomers was stronger than binding to large aggregates (Fig. 6d and Fig. S6d). Neither the linker-Pfn interaction model nor the Pfn-Pfn interaction model was able to reproduce both experimental results (Fig. S6, e and f). Taken together, the results from coarse-grained modeling suggest that an auxiliary interaction between profilin and the polyQ domain within Htt-NTFs probably plays an important role in modulating Q40-C38 phase behavior.
Discussion
We showed that aggregation and phase separation of Htt-NTF constructs is characterized by at least three distinct phases. We also showed that the values of cS shift to higher concentrations, and fibril formation is destabilized in the presence of profilin. This implies that profilin binds preferentially to M-phase species of Htt-NTFs. In most situations that involve binding between a protein and a heterogeneous mixture of oligomers or aggregates, it would be impossible to determine which species in the heterogeneous mixture is/are preferentially bound. However, with a quantitative description of the phase boundaries that govern the distribution of species, we can apply the concepts of polyphasic linkage to identify the phases that are preferentially bound. This is important because if one or more types of aggregate species are toxic to cells, it is essential that one knows which phase is preferentially bound by a ligand or small molecule to understand its effect on the equilibrium and therefore toxicity. This is essential, given that preferential binding to any one species will influence the equilibrium populations of all aggregate species.
We complemented experiments that probe the impact of profilin binding on phase boundaries with measurements of binding isotherms. Our results from experiments and modeling suggest that increasing the local concentration of C38 domains through polyQ-dependent oligomerization cannot explain the results of the binding experiments. Instead, the experimental results are consistent with a model that invokes a combination of primary interactions between polyP and profilin and weak, auxiliary interactions between the polyQ domain and profilin.
Previous studies have shown that increases in apparent affinity can result from a combination of specific and nonspecific interactions and that the magnitude of the increase depends on the size of the nonspecific interaction surface (78). This result is consistent with the decreasing Kd,app between Qn-C38 and profilin observed as the length of the polyQ domain was increased. Based on our coarse-grained simulations, we propose that these auxiliary interactions will involve sites on the surface of profilin that are non-overlapping with the polyP-binding site.
Given that overexpression of profilin in cellular models reduces Htt-NTF dependent toxicity (51) and that multivalency appears to be important for the relevant interactions needed between profilin and Htt-NTFs, it appears that exploiting multivalent interactions might be an important strategy for designing therapeutics targeting Htt-NTFs. Such strategies have been developed for the design of antimicrobial peptides as a means to treat bacterial infections (79) and for selective targeting of different amyloid-β species as a means to alleviate neurotoxicity (80). Our results suggest that the design of multivalent peptides can increase binding affinity, thus allowing these peptides to compete more effectively with other ligands in the cell.
Understanding the effects of Htt-NTF binding partners in terms of polyphasic linkage
Htt-NTFs engage in a wide variety of protein-protein interactions (75, 81–90). These “interactomes” are tissue-specific, and within a tissue type, the nodes and edges in an interaction network are known to be different for Htt-NTFs with wildtype versus mutant polyQ lengths (90). The polyphasic linkage formalism of Wyman and Gill (30, 31) provides a route for modeling the phase behavior of Htt-NTFs and quantifying the impact of protein-protein interaction nodes on this phase behavior.
From a phenotypic standpoint, gain-of-function toxicity resulting from the accumulation of S- and F-phase species appears to be weakened by interactions with profilin (91). It is also conceivable that other heterotypic interactions with Htt-NTFs modulate phase boundaries in very different ways. For example, preferential binding to S-phase species will, via linkage effects, lead to a lowering of cS and a widening of the gap between cS and cF. Conversely, preferential binding to F-phase fibrils will lower cF, destabilize the M and S phases, and promote the formation of long, stable fibrils. Indeed, there appear to be proteins in interactomes that preferentially interact with high-molecular weight S- and F-phase species. For example, SH3GL3, a protein involved in endocytosis, binds the linker region of C38 and has been shown to bind preferentially to mutant Htt-NTF and accelerate mutant Htt-NTF aggregation (92, 93). Recent studies showed that SH3GL3 binds monomeric wildtype and mutant Htt-NTF similarly (92). However, upon introduction of aggregated mutant Htt-NTF, retention of mutant Htt-NTF on SH3GL3 pull-downs was increased in a manner that correlated with increased aggregation of mutant Htt-NTF. Together, these results suggest that the binding of SH3GL3 increases the stability of the F phase of mutant as opposed to wildtype Htt-NTFs due to the increased aggregation propensity of mutant Htt-NTFs. This points to preferential binding of SH3GL3 to larger fibrillar aggregates. This should shift cF to lower values. Our suggestion is consistent with findings of increased fibril formation when SH3GL3 is co-expressed with Htt-NTFs in cellular models (93).
Profilin is one of the most abundant cellular proteins, with a copy number of around 107/cell (94). We speculate that the high natural abundance coupled to its intrinsic ability to bind polyP tracts leads to the serendipitous consequence of being able to influence cellular aggregation and phase separation of Htt-NTFs. This raises the possibility that there may be other naturally occurring proteins that show even greater potency but are not readily identified in cellular screens due to their low cellular concentrations. Indeed, given the wide prevalence of polyP-binding modules, such as WW, SH3, and EVH1 domains (95), a plausible approach would be to identify other, potentially more powerful modifiers of intracellular aggregation and phase separation. This could be achieved by driving overexpression of putative modifiers based on the combined criteria of polyP-binding domains and low molecular weight. Similarly, profilin itself may represent a useful scaffold upon which protein engineering (96) could be used to tune its specificity and affinity for Htt-NTFs. Regardless of the “physiological relevance” of interactions between profilin and Htt-NTFs, it is clear that profilin is capable of reducing Htt-NTF aggregation and toxicity, and therefore it serves as a useful model for the design of therapeutics for Huntington's disease. The same logic applies to any protein or molecule shown to bind a pathologically aggregating protein and reduce its toxicity, irrespective of the “physiological relevance” of that interaction.
Experimental procedures
Synthesis of peptide constructs
Peptides were purchased from Watsonbio Sciences (Houston, TX) in crude form and stored at −20 °C until use.
Peptide purification
Crude peptides were disaggregated in undiluted formic acid, diluted to 30% formic acid with deionized water, and purified using reverse-phase HPLC on a semipreparative 300SB-C3 reverse-phase column (Agilent, Santa Clara, CA). Elution fractions containing pure peptide were pooled, lyophilized, and stored at −20 °C until use.
Peptide disaggregation
Peptides were disaggregated using the method reported previously (19). Further details are given in the supporting Methods.
Recombinant expression and purification of profilin
The genetic sequence of human profilin-1 (hereafter referred to as profilin) in the pRK174 vector, which includes a T7 promoter, a His6 tag, and an ampicillin resistance gene, was verified by DNA sequencing (GENEWIZ, South Plainfield, NJ). The pRK174-His6-profilin plasmid was transformed into Escherichia coli BL21 (DE3) cells. Cells were grown at 37 °C in Luria broth with 100 μg/ml ampicillin to an A600 of ∼0.8 with shaking at 220 rpm. Protein expression was induced by the addition of 1.0 mm isopropyl 1-thio-β-d-galactopyranoside and harvested by centrifugation at 4,500 rpm for 30 min at 4 °C, ∼4 h after induction. The resulting cell pellets were resuspended in nickel column buffer (20 mm sodium phosphate, 500 mm NaCl, 30 mm imidazole, pH 7.5). Protease inhibitors (Roche Applied Science) were added to the suspension, and cells were lysed on ice by sonication using four 30-s pulses with 2-min rests between each pulse. Lysates were clarified by centrifugation at 20,000 rpm for 30 min at 4 °C in a Sorvall SS34 rotor. Protein was isolated from the resulting supernatant by affinity chromatography using a HisTrap HP column on an AKTA pure FPLC system (GE Healthcare). Fractions containing His-tagged proteins were pooled and dialyzed against 20 mm Tris, 100 mm NaCl, 5 mm EDTA, 0.5 mm tris(2-carboxyethyl)phosphine, pH 7.7, using Spectra/Por dialysis membrane (Spectrum Laboratories, Rancho Dominguez, CA) at 4 °C for 48 h with multiple changes to fresh buffer. Sample purity was typically greater than 95%, as determined by Coomassie G250-stained SDS-PAGE. Protein concentrations were determined by UV spectroscopy using an extinction coefficient of 18,450 m−1 cm−1. The protein stock was aliquoted, flash-frozen in liquid nitrogen, and stored at −80 °C until use. Protein yield was typically >160 mg/liter of cell culture.
Steady-state tryptophan fluorescence
Steady-state fluorescence emission spectra of profilin were measured using a PTI QuantaMaster fluorometer (Horiba Scientific, Edison, NJ). Disaggregated peptide was reconstituted in Tris buffer as described, and profilin was added from a concentrated stock solution. Profilin was maintained at a constant concentration while the concentration of the Htt-NTF construct was titrated by dilution using a profilin solution that was identical to the Htt-NTF + profilin solution, except that it contained no Htt-NTF peptide. This solution was then used for stepwise dilution of the Htt-NTF + profilin solution. All experiments were repeated at least three times. The reverse titration was also performed (by adding concentrated Htt-NTF stock to increase the total sample concentration) with C38 and profilin, and the same result was obtained regardless of the direction of the titration. Binding experiments were carried out in a 3 × 3-mm fluorometer cell (Starna, Atascadero, CA) at 22 °C. Emission spectra were recorded from 310 to 380 nm, with excitation at 295 nm (5-nm slit width). All spectra were corrected for the contribution of buffer. Fluorescence emission intensities at 320 nm were recorded as a function of Htt-NTF concentration. Binding isotherms were fit using nonlinear least-squares analysis as described in the supporting Methods.
Right-angle static light scattering
The intensity of static right-angle light scattering at 350 nm (I) was measured at each titration point immediately before each fluorescence measurement using the same instrument. The intersection of linear fits to I versus [Q40-C38] (log-log plot) identified discontinuity points in the concentration dependence of the scattering intensity, which is indicative of a phase transition. Due to the sensitive nature of the measurement, it was somewhat prone to experimental artifact (such as persistent aggregates resulting from incomplete peptide disaggregation or the introduction of dust or other foreign material to the sample). Even minute contaminants can introduce noise or otherwise obscure the signal of interest, and it was not always possible to completely exclude minor contaminants from the sample. Because of this, none of the trials carried out were completely devoid of artifact. However, despite these challenges, our method yielded good agreement across trials with regard to the location of the discontinuity. In contrast, a physical interpretation of the slope on either side of a discontinuity was not pursued further because of sensitivity to the potential artifacts discussed above. The method of determining optimal linear fits is given in the supporting Methods.
Negative-stain TEM
Samples were either recovered from fluorescence and light scattering experiments or freshly prepared in the same manner (i.e. samples were disaggregated using the TFA/HFIP method, reconstituted in buffer, and then diluted to various concentrations). Sample solutions were briefly vortexed, and 10 μl of each sample was applied to a parafilm strip. Carbon-coated copper grids (Ted Pella, catalog no. 01843, 300-mesh) were inverted on the sample droplets for 15–20 s. The grids were then washed by inverting them on 50-μl droplets of deionized water for about 5 s, after which the water was wicked away with Whatman filter paper. Finally, the grids were inverted on droplets of 0.5% uranyl acetate for 1 min, wicked dry with Whatman filter paper, and air-dried overnight. For samples in the presence of (His-tagged) profilin, the location of profilin was marked on the EM grid with 5-nm nickel-nitrilotriacetic acid-gold nanoparticles. Unbound profilin was rinsed away with buffer before applying the gold nanoparticle solution, and unbound or nonspecifically bound gold nanoparticles were washed away with imidazole buffer before imaging. TEM images were collected on an FEI Tecnai G2 Spirit transmission electron microscope operating at 120 kV and equipped with a Gatan digital camera. All images are representative of multiple distinct locations on the EM grid.
Sedimentation assay
Peptides were disaggregated using the TFA/HFIP method described in the supporting Methods and aliquoted (1.5 ml) into 1.6-ml Eppendorf tubes, which were then capped and sealed with Parafilm. The samples were incubated in a water bath at the specified temperature for 2 weeks. After 2 weeks of incubation, the samples were centrifuged for 100 min at 25,000 × g. Two aliquots of the supernatant were removed, and the peptide concentrations were determined using the micro-BCA (bicinchoninic acid) assay (97) (Pierce).
Thioflavin T fluorescence measurements
ThT assays were performed as described by Crick et al. (19). Further details are given in the supporting Methods.
Statistical significance tests
Data sets were first subjected to either Bartlett's or Levene's test for equal variance. Data sets exhibiting equal variance were subjected to a one-way ANOVA with Tukey's range test for post hoc analysis. Data sets that did not pass the equal variance test were subjected to Welch's ANOVA with Games–Howell post hoc analysis. Significance levels are indicated with one, two, or three asterisks corresponding to the following p value cutoffs: *, p < 0.05; **, p < 0.01; ***, p < 0.001. Analyses were implemented in Python.
Molecular simulations
All atom simulations used to parameterize our coarse-grained model were performed using the CAMPARI simulation package utilizing the ABSINTH implicit solvation model and force field paradigm (98, 99). Coarse-grained Langevin dynamics simulations were performed using the LAMMPS simulation package (67). Details regarding the set-up and execution of the simulations are given in the supporting Methods.
Author contributions
A.E.P., K.M.R., T.S.H., and R.V.P. conceptualization; A.E.P. K.M.R., S. L. C., and A. L. data curation; A.E.P., K.M.R., and S.L.C., A.L. formal analysis; A.E.P., K.M.R., S.L.C., and A.L. investigation; A.E.P. and K.M.R. visualization; A.E.P., K.M.R., T.S.H., S.L.C., A.L., M.I.D., and R.V.P. methodology; A.E.P., K.M.R., and R.V.P. writing-original draft; A.E.P., K.M.R., and R.V.P. writing-review and editing; K.M.R. and T.S.H. software; M.I.D. and R.V.P. funding acquisition; R.V.P. resources; R.V.P. supervision; R.V.P. project administration.
Supplementary Material
Acknowledgments
We thank Carlos Castañeda, Jeong-Mo Choi, Alex Holehouse, Ralf Langen, and J. Paul Taylor for helpful discussions.
This work was supported by National Institutes of Health Grants R01NS056114 (to R. V. P.) and R01NS089932 (to M. I. D. and R. V. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1, Figs. S1–S7, and Methods.
- Htt-NTF
- huntingtin N-terminal fragment
- HD
- Huntington's disease
- polyQ
- polyglutamine
- polyP
- polyproline
- Qn
- polyQ tract of length n
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- ThT
- thioflavin T
- TEM
- transmission electron microscopy
- ANOVA
- analysis of variance.
References
- 1. Ross C. A., and Poirier M. A. (2004) Protein aggregation and neurodegenerative disease. Nat. Med. 10, S10–S17 10.1038/nm1066 [DOI] [PubMed] [Google Scholar]
- 2. Aguzzi A., and O'Connor T. (2010) Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat. Rev. Drug Discov. 9, 237–248 10.1038/nrd3050 [DOI] [PubMed] [Google Scholar]
- 3. Pappu R. V., Wang X., Vitalis A., and Crick S. L. (2008) A polymer physics perspective on driving forces and mechanisms for protein aggregation. Arch. Biochem. Biophys. 469, 132–141 10.1016/j.abb.2007.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greer S. C. (2002) Reversible polymerizations and aggregations. Annu. Rev. Phys. Chem. 53, 173–200 10.1146/annurev.physchem.53.082301.111949 [DOI] [PubMed] [Google Scholar]
- 5. Sfatos C. D., Gutin A. M., and Shakhnovich E. I. (1994) Phase transitions in a “many-letter” random heteropolymer. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics 50, 2898–2905 10.1103/PhysRevE.50.2898 [DOI] [PubMed] [Google Scholar]
- 6. Shakhnovich E. I., and Finkelstein A. V. (1989) Theory of cooperative transitions in protein molecules. I. Why denaturation of globular protein is a first-order phase transition. Biopolymers 28, 1667–1680 10.1002/bip.360281003 [DOI] [PubMed] [Google Scholar]
- 7. Li P., Banjade S., Cheng H. C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q. X., Nixon B. T., and Rosen M. K. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., and Hyman A. A. (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- 9. Zhang H., Elbaum-Garfinkle S., Langdon E. M., Taylor N., Occhipinti P., Bridges A. A., Brangwynne C. P., and Gladfelter A. S. (2015) RNA controls PolyQ protein phase transitions. Mol. Cell 60, 220–230 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A. P., Kim H. J., Mittag T., and Taylor J. P. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Su X., Ditlev J. A., Hui E., Xing W., Banjade S., Okrut J., King D. S., Taunton J., Rosen M. K., and Vale R. D. (2016) Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 10.1126/science.aad9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Banani S. F., Lee H. O., Hyman A. A., and Rosen M. K. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hyman A. A., Weber C. A., and Jülicher F. (2014) Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- 14. Brangwynne C. P., Tompa P., and Pappu R. V. (2015) Polymer physics of intracellular phase transitions. Nat. Phys. 11, 899–904 10.1038/nphys3532 [DOI] [Google Scholar]
- 15. Harmon T. S., Holehouse A. S., Rosen M. K., and Pappu R. V. (2017) Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife 6, e30294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feric M., Vaidya N., Harmon T. S., Mitrea D. M., Zhu L., Richardson T. M., Kriwacki R. W., Pappu R. V., and Brangwynne C. P. (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacobs W. M., and Frenkel D. (2017) Phase transitions in biological systems with many components. Biophys. J. 112, 683–691 10.1016/j.bpj.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jain S., Wheeler J. R., Walters R. W., Agrawal A., Barsic A., and Parker R. (2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crick S. L., Ruff K. M., Garai K., Frieden C., and Pappu R. V. (2013) Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. U.S.A. 110, 20075–20080 10.1073/pnas.1320626110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Riback J. A., Katanski C. D., Kear-Scott J. L., Pilipenko E. V., Rojek A. E., Sosnick T. R., and Drummond D. A. (2017) Stress-triggered phase separation is an adaptive, Evolutionarily tuned response. Cell 168, 1028–1040.e19 10.1016/j.cell.2017.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murakami T., Qamar S., Lin J. Q., Schierle G. S., Rees E., Miyashita A., Costa A. R., Dodd R. B., Chan F. T., Michel C. H., Kronenberg-Versteeg D., Li Y., Yang S. P., Wakutani Y., Meadows W., et al. (2015) ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88, 678–690 10.1016/j.neuron.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Audas T. E., Audas D. E., Jacob M. D., Ho J. J., Khacho M., Wang M., Perera J. K., Gardiner C., Bennett C. A., Head T., Kryvenko O. N., Jorda M., Daunert S., Malhotra A., Trinkle-Mulcahy L., et al. (2016) Adaptation to stressors by systemic protein amyloidogenesis. Dev. Cell 39, 155–168 10.1016/j.devcel.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weber S. C., and Brangwynne C. P. (2012) Getting RNA and protein in phase. Cell 149, 1188–1191 10.1016/j.cell.2012.05.022 [DOI] [PubMed] [Google Scholar]
- 24. Patel A., Lee H. O., Jawerth L., Maharana S., Jahnel M., Hein M. Y., Stoynov S., Mahamid J., Saha S., Franzmann T. M., Pozniakovski A., Poser I., Maghelli N., Royer L. A., Weigert M., et al. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- 25. Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., et al. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ciryam P., Kundra R., Morimoto R. I., Dobson C. M., and Vendruscolo M. (2015) Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol. Sci. 36, 72–77 10.1016/j.tips.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ciryam P., Tartaglia G. G., Morimoto R. I., Dobson C. M., and Vendruscolo M. (2013) Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 5, 781–790 10.1016/j.celrep.2013.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ciryam P., Lambert-Smith I. A., Bean D. M., Freer R., Cid F., Tartaglia G. G., Saunders D. N., Wilson M. R., Oliver S. G., Morimoto R. I., Dobson C. M., Vendruscolo M., Favrin G., and Yerbury J. J. (2017) Spinal motor neuron protein supersaturation patterns are associated with inclusion body formation in ALS. Proc. Natl. Acad. Sci. U.S.A. 114, E3935–E3943 10.1073/pnas.1613854114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. So M., Hall D., and Goto Y. (2016) Revisiting supersaturation as a factor determining amyloid fibrillation. Curr. Opin. Struct. Biol. 36, 32–39 10.1016/j.sbi.2015.11.009 [DOI] [PubMed] [Google Scholar]
- 30. Wyman J., and Gill S. J. (1980) Ligand-linked phase changes in a biological system: applications to sickle cell hemoglobin. Proc. Natl. Acad. Sci. U.S.A. 77, 5239–5242 10.1073/pnas.77.9.5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wyman J., and Gill S. J. (1990) Binding and Linkage: Functional Chemistry of Biological Macromolecules, University Science Books, Mill Valley, CA [Google Scholar]
- 32. Walker F. O. (2007) Huntington's disease. Lancet 369, 218–228 10.1016/S0140-6736(07)60111-1 [DOI] [PubMed] [Google Scholar]
- 33. Crook Z. R., and Housman D. E. (2012) Dysregulation of dopamine receptor D2 as a sensitive measure for Huntington disease pathology in model mice. Proc. Natl. Acad. Sci. U.S.A. 109, 7487–7492 10.1073/pnas.1204542109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., and Finkbeiner S. (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]
- 35. Bates G. (2003) Huntingtin aggregation and toxicity in Huntington's disease. Lancet 361, 1642–1644 10.1016/S0140-6736(03)13304-1 [DOI] [PubMed] [Google Scholar]
- 36. Arrasate M., and Finkbeiner S. (2012) Protein aggregates in Huntington's disease. Exp. Neurol. 238, 1–11 10.1016/j.expneurol.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nucifora L. G., Burke K. A., Feng X., Arbez N., Zhu S., Miller J., Yang G., Ratovitski T., Delannoy M., Muchowski P. J., Finkbeiner S., Legleiter J., Ross C. A., and Poirier M. A. (2012) Identification of novel potentially toxic oligomers formed in vitro from mammalian-derived expanded huntingtin exon-1 protein. J. Biol. Chem. 287, 16017–16028 10.1074/jbc.M111.252577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Preisinger E., Jordan B. M., Kazantsev A., and Housman D. (1999) Evidence for a recruitment and sequestration mechanism in Huntington's disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354, 1029–1034 10.1098/rstb.1999.0455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Donaldson K. M., Li W., Ching K. A., Batalov S., Tsai C. C., and Joazeiro C. A. (2003) Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proc. Natl. Acad. Sci. U.S.A. 100, 8892–8897 10.1073/pnas.1530212100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim W. Y., Fayazi Z., Bao X., Higgins D., and Kazemi-Esfarjani P. (2005) Evidence for sequestration of polyglutamine inclusions by Drosophila myeloid leukemia factor. Mol. Cell. Neurosci. 29, 536–544 10.1016/j.mcn.2005.04.005 [DOI] [PubMed] [Google Scholar]
- 41. Schipper-Krom S., Juenemann K., Jansen A. H., Wiemhoefer A., van den Nieuwendijk R., Smith D. L., Hink M. A., Bates G. P., Overkleeft H., Ovaa H., and Reits E. (2014) Dynamic recruitment of active proteasomes into polyglutamine initiated inclusion bodies. FEBS Lett. 588, 151–159 10.1016/j.febslet.2013.11.023 [DOI] [PubMed] [Google Scholar]
- 42. Zeng L., Wang B., Merillat S. A., N Minakawa E., Perkins M. D., Ramani B., Tallaksen-Greene S. J., Costa M. D. C., Albin R. L., and Paulson H. L. (2015) Differential recruitment of UBQLN2 to nuclear inclusions in the polyglutamine diseases HD and SCA3. Neurobiol. Dis. 82, 281–288 10.1016/j.nbd.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. O'Nuallain B., and Wetzel R. (2002) Conformational Abs recognizing a generic amyloid fibril epitope. Proc. Natl. Acad. Sci. U.S.A. 99, 1485–1490 10.1073/pnas.022662599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang W., Dunlap J. R., Andrews R. B., and Wetzel R. (2002) Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum. Mol. Genet. 11, 2905–2917 10.1093/hmg/11.23.2905 [DOI] [PubMed] [Google Scholar]
- 45. Ramdzan Y. M., Trubetskov M. M., Ormsby A. R., Newcombe E. A., Sui X., Tobin M. J., Bongiovanni M. N., Gras S. L., Dewson G., Miller J. M. L., Finkbeiner S., Moily N. S., Niclis J., Parish C. L., Purcell A. W., et al. (2017) Huntingtin inclusions trigger cellular quiescence, deactivate apoptosis, and lead to delayed necrosis. Cell Rep. 19, 919–927 10.1016/j.celrep.2017.04.029 [DOI] [PubMed] [Google Scholar]
- 46. Jockusch B. M., Murk K., and Rothkegel M. (2007) The profile of profilins. Rev. Physiol. Biochem. Pharmacol. 159, 131–149 [DOI] [PubMed] [Google Scholar]
- 47. Goldschmidt-Clermont P. J., Machesky L. M., Baldassare J. J., and Pollard T. D. (1990) The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science 247, 1575–1578 10.1126/science.2157283 [DOI] [PubMed] [Google Scholar]
- 48. Goldschmidt-Clermont P. J., Machesky L. M., Doberstein S. K., and Pollard T. D. (1991) Mechanism of the interaction of human platelet profilin with actin. J. Cell Biol. 113, 1081–1089 10.1083/jcb.113.5.1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Petrella E. C., Machesky L. M., Kaiser D. A., and Pollard T. D. (1996) Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry 35, 16535–16543 10.1021/bi961498d [DOI] [PubMed] [Google Scholar]
- 50. Witke W., Sutherland J. D., Sharpe A., Arai M., and Kwiatkowski D. J. (2001) Profilin I is essential for cell survival and cell division in early mouse development. Proc. Natl. Acad. Sci. U.S.A. 98, 3832–3836 10.1073/pnas.051515498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shao J., Welch W. J., Diprospero N. A., and Diamond M. I. (2008) Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol. Cell. Biol. 28, 5196–5208 10.1128/MCB.00079-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li M., Huang Y., Ma A. A. K., Lin E., and Diamond M. I. (2009) Y-27632 improves rotarod performance and reduces huntingtin levels in R6/2 mice. Neurobiol. Dis. 36, 413–420 10.1016/j.nbd.2009.06.011 [DOI] [PubMed] [Google Scholar]
- 53. Fuentealba R. A., Marasa J., Diamond M. I., Piwnica-Worms D., and Weihl C. C. (2012) An aggregation sensing reporter identifies leflunomide and teriflunomide as polyglutamine aggregate inhibitors. Hum. Mol. Genet. 21, 664–680 10.1093/hmg/ddr500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fuentealba R. A., Udan M., Bell S., Wegorzewska I., Shao J., Diamond M. I., Weihl C. C., and Baloh R. H. (2010) Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J. Biol. Chem. 285, 26304–26314 10.1074/jbc.M110.125039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pollitt S. K., Pallos J., Shao J., Desai U. A., Ma A. A. K., Thompson L. M., Marsh J. L., and Diamond M. I. (2003) A rapid cellular FRET assay of polyglutamine aggregation identifies a novel inhibitor. Neuron 40, 685–694 10.1016/S0896-6273(03)00697-4 [DOI] [PubMed] [Google Scholar]
- 56. Veldman M. B., Rios-Galdamez Y., Lu X. H., Gu X., Qin W., Li S., Yang X. W., and Lin S. (2015) The N17 domain mitigates nuclear toxicity in a novel zebrafish Huntington's disease model. Mol. Neurodegener. 10, 67 10.1186/s13024-015-0063-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gu X., Cantle J. P., Greiner E. R., Lee C. Y., Barth A. M., Gao F., Park C. S., Zhang Z., Sandoval-Miller S., Zhang R. L., Diamond M., Mody I., Coppola G., and Yang X. W. (2015) N17 Modifies mutant Huntingtin nuclear pathogenesis and severity of disease in HD BAC transgenic mice. Neuron 85, 726–741 10.1016/j.neuron.2015.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Andrew S. E., Goldberg Y. P., Kremer B., Telenius H., Theilmann J., Adam S., Starr E., Squitieri F., Lin B., and Kalchman M. A. (1993) The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat. Genet. 4, 398–403 10.1038/ng0893-398 [DOI] [PubMed] [Google Scholar]
- 59. Garai K., Sahoo B., Sengupta P., and Maiti S. (2008) Quasihomogeneous nucleation of amyloid β yields numerical bounds for the critical radius, the surface tension, and the free energy barrier for nucleus formation. J. Chem. Phys. 128, 045102 10.1063/1.2822322 [DOI] [PubMed] [Google Scholar]
- 60. Sedlák M. N., and Amis E. J. (1992) Concentration and molecular weight regime diagram of salt-free polyelectrolyte solutions as studied by light scattering. J. Chem. Phys. 96, 826 10.1063/1.462468 [DOI] [Google Scholar]
- 61. Bugg C. W., Isas J. M., Fischer T., Patterson P. H., and Langen R. (2012) Structural features and domain organization of huntingtin fibrils. J. Biol. Chem. 287, 31739–31746 10.1074/jbc.M112.353839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Freire E., Schön A., and Velazquez-Campoy A. (2009) Isothermal titration calorimetry: general formalism using binding polynomials. Methods Enzymol. 455, 127–155 10.1016/S0076-6879(08)04205-5 [DOI] [PubMed] [Google Scholar]
- 63. Mahoney N. M., Janmey P. A., and Almo S. C. (1997) Structure of the profilin-poly-l-proline complex involved in morphogenesis and cytoskeletal regulation. Nat. Struct. Biol. 4, 953–960 10.1038/nsb1197-953 [DOI] [PubMed] [Google Scholar]
- 64. Mahoney N. M., Rozwarski D. A., Fedorov E., Fedorov A. A., and Almo S. C. (1999) Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat. Struct. Biol. 6, 666–671 10.1038/10722 [DOI] [PubMed] [Google Scholar]
- 65. Kaiser D. A., and Pollard T. D. (1996) Characterization of actin and poly-l-proline binding sites of Acanthamoeba profilin with monoclonal antibodies and by mutagenesis. J. Mol. Biol. 256, 89–107 10.1006/jmbi.1996.0070 [DOI] [PubMed] [Google Scholar]
- 66. Rennella E., Sekhar A., and Kay L. E. (2017) Self-assembly of human profilin-1 detected by Carr-Purcell-Meiboom-Gill nuclear magnetic resonance (CPMG NMR) spectroscopy. Biochemistry 56, 692–703 10.1021/acs.biochem.6b01263 [DOI] [PubMed] [Google Scholar]
- 67. Plimpton S. (1995) Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 117, 1–19 10.1006/jcph.1995.1039 [DOI] [Google Scholar]
- 68. Shen K., Calamini B., Fauerbach J. A., Ma B., Shahmoradian S. H., Serrano Lachapel I. L., Chiu W., Lo D. C., and Frydman J. (2016) Control of the structural landscape and neuronal proteotoxicity of mutant Huntingtin by domains flanking the polyQ tract. Elife 5, e18065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sahoo B., Singer D., Kodali R., Zuchner T., and Wetzel R. (2014) Aggregation behavior of chemically synthesized, full-length huntingtin exon1. Biochemistry 53, 3897–3907 10.1021/bi500300c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Krishnamurthy V. M., Estroff L. A., and Whitesides G. M. (2006) Multivalency in ligand design. In Fragment-based Approaches in Drug Discovery (Jahnke W., and Erlanson D. A., eds), pp. 11–53, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany [Google Scholar]
- 71. Mammen M., Choi S.-K., and Whitesides G. M. (1998) Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. Engl. 37, 2754–2794 10.1002/(SICI)1521-3773(19981102)37:20%3C2754::AID-ANIE2754%3E3.0.CO%3B2-3 [DOI] [PubMed] [Google Scholar]
- 72. Jencks W. P. (1981) On the attribution and additivity of binding energies. Proc. Natl. Acad. Sci. U.S.A. 78, 4046–4050 10.1073/pnas.78.7.4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kayatekin C., Matlack K. E., Hesse W. R., Guan Y., Chakrabortee S., Russ J., Wanker E. E., Shah J. V., and Lindquist S. (2014) Prion-like proteins sequester and suppress the toxicity of huntingtin exon 1. Proc. Natl. Acad. Sci. U.S.A. 111, 12085–12090 10.1073/pnas.1412504111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wear M. P., Kryndushkin D., O'Meally R., Sonnenberg J. L., Cole R. N., and Shewmaker F. P. (2015) Proteins with intrinsically disordered domains are preferentially recruited to polyglutamine aggregates. PLoS One 10, e0136362 10.1371/journal.pone.0136362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ripaud L., Chumakova V., Antonin M., Hastie A. R., Pinkert S., Körner R., Ruff K. M., Pappu R. V., Hornburg D., Mann M., Hartl F. U., and Hipp M. S. (2014) Overexpression of Q-rich prion-like proteins suppresses polyQ cytotoxicity and alters the polyQ interactome. Proc. Natl. Acad. Sci. U.S.A. 111, 18219–18224 10.1073/pnas.1421313111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Warner J. B. 4th, Ruff K. M., Tan P. S., Lemke E. A., Pappu R. V., and Lashuel H. A. (2017) Monomeric huntingtin exon 1 has similar overall structural features for wild type and pathological polyglutamine lengths. J. Am. Chem. Soc. 139, 14456–14469 10.1021/jacs.7b06659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Korupolu R. V., Achary M. S., Aneesa F., Sathish K., Wasia R., Sairam M., Nagarajaram H. A., and Singh S. S. (2009) Profilin oligomerization and its effect on poly (l-proline) binding and phosphorylation, Int. J. Biol. Macromol. 45, 265–273 10.1016/j.ijbiomac.2009.06.001 [DOI] [PubMed] [Google Scholar]
- 78. Lundgren A., Agnarsson B., Zirbs R., Zhdanov V. P., Reimhult E., and Höök F. (2016) Nonspecific colloidal-type interaction explains size-dependent specific binding of membrane-targeted nanoparticles. ACS Nano 10, 9974–9982 10.1021/acsnano.6b04160 [DOI] [PubMed] [Google Scholar]
- 79. Liu S. P., Zhou L., Lakshminarayanan R., and Beuerman R. W. (2010) Multivalent antimicrobial peptides as therapeutics: design principles and structural diversities. Int. J. Pept. Res. Ther. 16, 199–213 10.1007/s10989-010-9230-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kim Y., Lee J. H., Ryu J., and Kim D. J. (2009) Multivalent and multifunctional ligands to β-amyloid. Curr. Pharm. Des. 15, 637–658 10.2174/138161209787315648 [DOI] [PubMed] [Google Scholar]
- 81. Riechers S. P., Butland S., Deng Y., Skotte N., Ehrnhoefer D. E., Russ J., Laine J., Laroche M., Pouladi M. A., Wanker E. E., Hayden M. R., and Graham R. K. (2016) Interactome network analysis identifies multiple caspase-6 interactors involved in the pathogenesis of HD. Hum. Mol. Genet. 25, 1600–1618 10.1093/hmg/ddw036 [DOI] [PubMed] [Google Scholar]
- 82. Kim Y. E., Hosp F., Frottin F., Ge H., Mann M., Hayer-Hartl M., and Hartl F. U. (2016) Soluble oligomers of polyQ-expanded huntingtin target a multiplicity of key cellular factors. Mol. Cell 63, 951–964 10.1016/j.molcel.2016.07.022 [DOI] [PubMed] [Google Scholar]
- 83. O'Brien R., DeGiacomo F., Holcomb J., Bonner A., Ring K. L., Zhang N., Zafar K., Weiss A., Lager B., Schilling B., Gibson B. W., Chen S., Kwak S., and Ellerby L. M. (2015) Integration-independent transgenic Huntington disease fragment mouse models reveal distinct phenotypes and life span in vivo. J. Biol. Chem. 290, 19287–19306 10.1074/jbc.M114.623561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kalathur R. K., Giner-Lamia J., Machado S., Barata T., Ayasolla K. R., and Futschik M. E. (2015) The unfolded protein response and its potential role in Huntington's disease elucidated by a systems biology approach. F1000Research 4, 103 10.12688/f1000research.6358.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hosp F., Vossfeldt H., Heinig M., Vasiljevic D., Arumughan A., Wyler E., Genetic and Environmental Risk for Alzheimer's Disease GERAD1 Consortium, Landthaler M., Hubner N., Wanker E. E., Lannfelt L., Ingelsson M., Lalowski M., Voigt A., and Selbach M. (2015) Quantitative interaction proteomics of neurodegenerative disease proteins. Cell Rep. 11, 1134–1146 10.1016/j.celrep.2015.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Brehme M., Voisine C., Rolland T., Wachi S., Soper J. H., Zhu Y., Orton K., Villella A., Garza D., Vidal M., Ge H., and Morimoto R. I. (2014) A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 9, 1135–1150 10.1016/j.celrep.2014.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shirasaki D. I., Greiner E. R., Al-Ramahi I., Gray M., Boontheung P., Geschwind D. H., Botas J., Coppola G., Horvath S., Loo J. A., and Yang X. W. (2012) Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron 75, 41–57 10.1016/j.neuron.2012.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ratovitski T., Chighladze E., Arbez N., Boronina T., Herbrich S., Cole R. N., and Ross C. A. (2012) Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle 11, 2006–2021 10.4161/cc.20423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Berchtold L. A., Størling Z. M., Ortis F., Lage K., Bang-Berthelsen C., Bergholdt R., Hald J., Brorsson C. A., Eizirik D. L., Pociot F., Brunak S., and Størling J. (2011) Huntingtin-interacting protein 14 is a type 1 diabetes candidate protein regulating insulin secretion and β-cell apoptosis. Proc. Natl. Acad. Sci. U.S.A. 108, E681–E688 10.1073/pnas.1104384108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Langfelder P., Cantle J. P., Chatzopoulou D., Wang N., Gao F., Al-Ramahi I., Lu X. H., Ramos E. M., El-Zein K., Zhao Y., Deverasetty S., Tebbe A., Schaab C., Lavery D. J., Howland D., et al. (2016) Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat. Neurosci. 19, 623–633 10.1038/nn.4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li M., Yasumura D., Ma A. A. K., Matthes M. T., Yang H., Nielson G., Huang Y., Szoka F. C., Lavail M. M., and Diamond M. I. (2013) Intravitreal administration of HA-1077, a ROCK inhibitor, improves retinal function in a mouse model of Huntington disease. PLoS One 8, e56026 10.1371/journal.pone.0056026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Davranche A., Aviolat H., Zeder-Lutz G., Busso D., Altschuh D., Trottier Y., and Klein F. A. (2011) Huntingtin affinity for partners is not changed by polyglutamine length: aggregation itself triggers aberrant interactions. Hum. Mol. Genet. 20, 2795–2806 10.1093/hmg/ddr178 [DOI] [PubMed] [Google Scholar]
- 93. Sittler A., Wälter S., Wedemeyer N., Hasenbank R., Scherzinger E., Eickhoff H., Bates G. P., Lehrach H., and Wanker E. E. (1998) SH3GL3 associates with the Huntingtin exon 1 protein and promotes the formation of polygln-containing protein aggregates. Mol. Cell 2, 427–436 10.1016/S1097-2765(00)80142-2 [DOI] [PubMed] [Google Scholar]
- 94. Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., and Aebersold R. (2011) The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zarrinpar A., Bhattacharyya R. P., and Lim W. A. (2003) The structure and function of proline recognition domains. Sci. STKE 2003, RE8 [DOI] [PubMed] [Google Scholar]
- 96. Zhang W., Ben-David M., and Sidhu S. S. (2017) Engineering cell signaling modulators from native protein-protein interactions. Curr. Opin. Struct. Biol. 45, 25–35 [DOI] [PubMed] [Google Scholar]
- 97. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., and Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 10.1016/0003-2697(85)90442-7 [DOI] [PubMed] [Google Scholar]
- 98. Radhakrishnan A., Vitalis A., Mao A. H., Steffen A. T., and Pappu R. V. (2012) Improved atomistic Monte Carlo simulations demonstrate that poly-l-proline adopts heterogeneous ensembles of conformations of semi-rigid segments interrupted by kinks. J. Phys. Chem. B 116, 6862–6871 10.1021/jp212637r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Vitalis A., and Pappu R. V. (2009) ABSINTH: a new continuum solvation model for simulations of polypeptides in aqueous solutions. J. Comput. Chem. 30, 673–699 10.1002/jcc.21005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.