Abstract
Oxysterol-binding protein (OSBP) and OSBP-related proteins (ORPs) constitute a large family of proteins that mainly function in lipid transport and sensing. ORP5 is an endoplasmic reticulum (ER)-anchored protein implicated in lipid transfer at the contact sites between the ER and other membranes. Recent studies indicate that ORP5 is also involved in cancer cell invasion and tumor progression. However, the molecular mechanism underlying ORP5's involvement in cancer is unclear. Here, we report that ORP5 promotes cell proliferation and motility of HeLa cells, an effect that depends on its functional OSBP-related domain (ORD). We also found that ORP5 depletion or substitutions of key residues located within ORP5–ORD and responsible for interactions with lipids interfered with cell proliferation, migration, and invasion. ORP5 interacted with the protein mechanistic target of rapamycin (mTOR), and this interaction also required ORP5–ORD. Of note, whereas ORP5 overexpression induced mTOR complex 1 (mTORC1) activity, ORP5 down-regulation had the opposite effect. Finally, ORP5-depleted cells exhibited impaired mTOR localization to lysosomes, which may have accounted for the blunted mTORC1 activation. Together, our results suggest that ORP5 expression is positively correlated with mTORC1 signaling and that ORP5 stimulates cell proliferation, at least in part, by activating mTORC1.
Keywords: mammalian target of rapamycin (mTOR), mTOR complex (mTORC), cell proliferation, phosphoinositide, cancer, cell signaling, cancer, lipids, ORP5, ORP8, OSBP
Introduction
Alterations in cell-signaling pathways that control normal cell growth are very common in cancer. In particular, deregulation of the mechanistic target of rapamycin (mTOR)3 signaling is known to play an important part in cancer development (1, 2). mTOR is an evolutionarily conserved protein kinase that is present in two distinct complexes, each containing several other proteins, termed mTOR complex 1 (mTORC1) and mTORC2. mTORC1 is a central regulator of cell proliferation. It senses nutrients, especially amino acids, as well as energy levels and stress, and regulates cell proliferation in response to these signals. In this regard, activated mTORC1 phosphorylates and activates its major downstream effector ribosomal protein S6 kinase (S6K). Once activated, S6K phosphorylates the ribosomal S6 protein to control fundamental cellular processes, including protein synthesis, lipid metabolism, and cell proliferation (3, 4).
Oxysterol-binding protein (OSBP)-related protein 5 (ORP5) belongs to a large family of lipid transfer proteins. In mammalian cells, the OSBP/ORPs protein family consists of 12 members with variant sequence identities, giving rise to six subfamily groups (5, 6). OSBP/ORPs share a characteristic feature of a conserved C-terminal OSBP-related domain (ORD) or ligand-binding domain. The N termini of these proteins often possess an FFAT motif (diphenylalanine in an acidic tract) and a pleckstrin homology (PH) domain. The FFAT motif and the PH domain are responsible for targeting OSBP/ORPs to endoplasmic reticulum (ER) membranes and non-ER organelle membranes, respectively (7–9). These two membrane-targeting determinants enable OSBP and some ORPs to function at ER-associated membrane contact sites, where they contribute to the intracellular exchange of lipids (10). For example, OSBP associates with the Golgi through its PH domain and the ER through FFAT–VAPs interaction (9). Tethering the ER and Golgi, OSBP has been shown to transport cholesterol from the ER to the Golgi by the ORD, where it exchanges cholesterol with phosphatidylinositol 4-phosphate (PI(4)P) and transfers it back to the ER (11).
Within the OSBP/ORPs family, ORP5 and ORP8, which lack the FFAT motif, are the only members that have a transmembrane domain anchoring them to the ER (12, 13). ER-anchored ORP5 has been identified as a phosphatidylserine (PS) transporter through its ORD (14, 15). In a counter-transport process occurring at the membrane contact sites, ORP5 and ORP8 were shown to transfer PS from the ER to the plasma membrane and PI(4)P and phosphatidylinositol 4,5-bisphosphate from the plasma membrane to the ER (15, 16). Interestingly, ORP5 and ORP8 were also reported to transport PS from the ER to mitochondria and maintain proper mitochondrial function (17). Despite seemingly working together to transport lipids at the membrane contact sites, ORP5 and ORP8 appear to have different functions in the regulation of cellular homeostasis (18). Notably, ORP8 expression reportedly inhibits cell proliferation and tumor growth while enhancing apoptosis (19). In contrast, ORP5 expression has been linked to increased cancer cell invasion and metastasis. For example, a previous report showed that the invasion rate of both hamster and human pancreatic cancer cells is enhanced by ORP5 overexpression and reduced by ORP5 depletion (20). Importantly, analysis of clinical samples suggested that poor prognosis in human pancreatic cancer is associated with high expression levels of ORP5 (20). In a recent study, tissue microarray analysis revealed that ORP5 is highly expressed in lung tumor tissues, especially in the lung tissues of metastasis-positive cases (21).
Why overexpression of ORP5 is positively correlated with cancer cell invasion and tumor progression is unknown. Specifically, it is unclear whether this correlation involves alterations of cell-signaling pathways. In this study, we provide evidence that mTORC1 signaling is positively regulated by ORP5. Our data suggest that ORP5 facilitates cancer cell proliferation and motility at least in part through mTORC1 activation.
Results
Overexpression of ORP5 promotes cell proliferation and migration
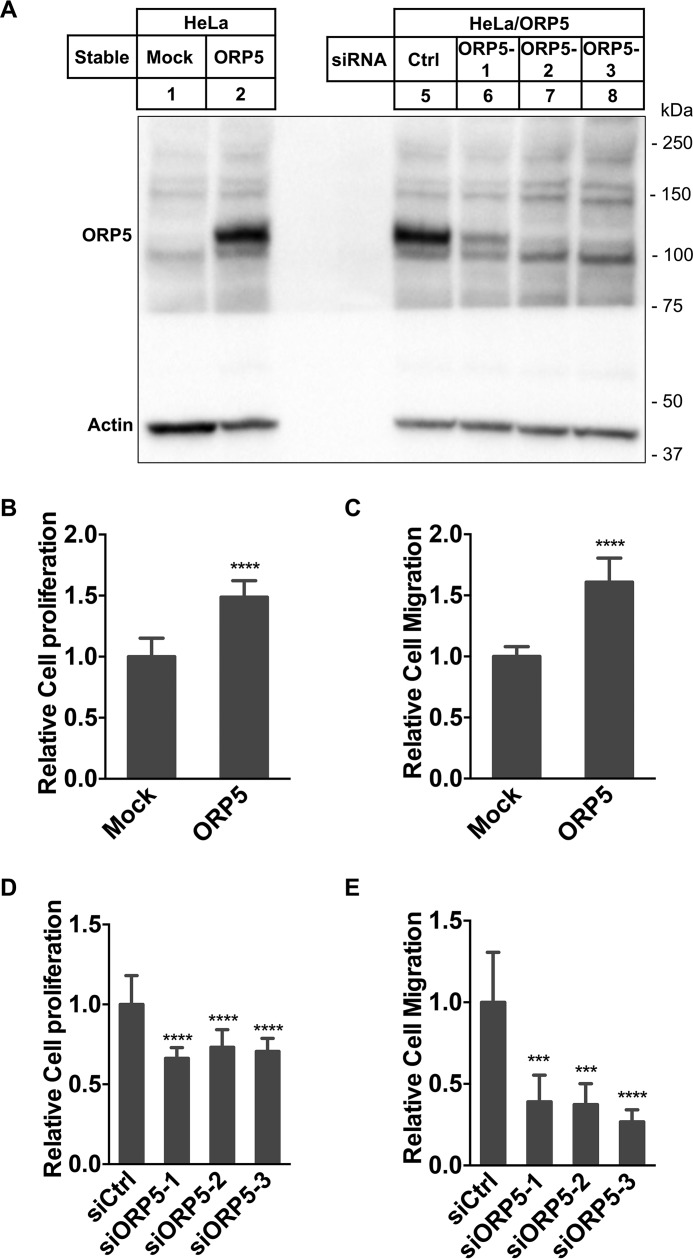
Elevated expression of ORP5 has been associated with pancreatic and lung cancers (20, 21). One of the aims of this study is to verify whether increased expression of ORP5 can drive cell proliferation. For this purpose, we chose HeLa cells because the level of endogenous ORP5 in HeLa cells is relatively low. This system not only allowed us to test wild type ORP5, but also mutant forms of ORP5 in cell proliferation. To study the effect of ORP5 overexpression, we used a retroviral vector system (Clontech pQCXIN) and generated a stable line of HeLa cells overexpressing ORP5. Using a goat polyclonal ORP5 antibody, we detected a robust band of ∼110 kDa corresponding to ORP5 in the stable cell line (HeLa/ORP5) (Fig. 1A, lane 2). It has been shown that overexpression of ORP5 induces pancreatic cancer invasion and lung tumor formation (20, 21). We predicted that overexpression of ORP5 in HeLa cells may facilitate cell proliferation and motility. To test this hypothesis, we carried out cell proliferation and migration assays in HeLa/ORP5 and Mock cells. Indeed, compared with the control, HeLa/ORP5 cells had a significantly higher proliferation rate (increased by ∼50%) (Fig. 1B). Cell migration rate was also significantly enhanced in HeLa/ORP5 cells by ∼60% compared with the Mock control (Fig. 1C). If ORP5 overexpression facilitates cell proliferation and migration, depletion of ORP5 should blunt this effect. We used a set of three specific siRNAs against ORP5 to efficiently reduce ORP5 expression in HeLa/ORP5 cells (Fig. 1A, lanes 6–8). As expected, depletion of ORP5 inhibited both cell proliferation and migration to an extent comparable with that of the mock control cells (Fig. 1, D and E).
Figure 1.
ORP5 facilitates cell proliferation and migration. A, immunoblotting of ORP5 in HeLa/Mock and HeLa/ORP5 cells and in HeLa/ORP5 cells treated with control siRNA or three different ORP5 siRNAs. B, cell proliferation assay in HeLa/Mock and HeLa/ORP5 cells (mean ± S.D.; ****, p < 0.0001; n = 12). C, cell migration assay in HeLa/Mock and HeLa/ORP5 cells. Relative cell migration was analyzed using ImageJ (mean ± S.D.; ****, p < 0.0001; n = 6). D, cell proliferation assay in HeLa/ORP5 cells treated with control siRNA or three different ORP5 siRNAs (ORP5 siRNAs (mean ± S.D.; ****, p < 0.0001; n = 12). E, cell migration assay in HeLa/ORP5 cells treated with control siRNA or three different ORP5 siRNAs. Relative cell migration was analyzed using ImageJ (mean ± S.D.; ***, p < 0.001; n = 6). All data are representative from three to four independent experiments with similar results.
Functional ORP5–ORD is critical for facilitating cell growth and motility
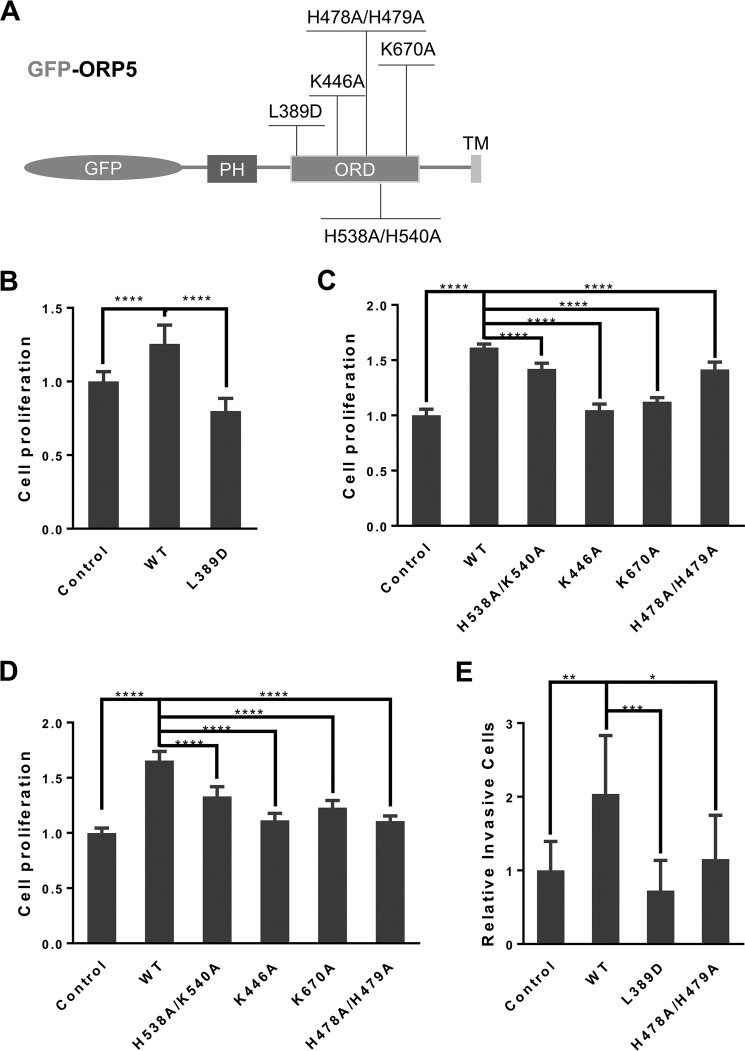
ORP5 is a lipid transfer protein, and its ORD domain is responsible for extracting, binding, and transporting lipid cargos between the ER and other organelle membranes (12, 16, 17). To investigate whether a functional ORD of ORP5 is required for facilitating cell proliferation and migration, we mutated some critical residues within ORP5–ORD conserved for binding PS (L389D) (14), PI(4)P (K446A, H478A/H479A, and K670A), or possibly cholesterol (H538A/K540A) (Fig. 2A) (16, 22). Transient transfection in HeLa cells with GFP-fused wildtype ORP5 and these mutant cDNAs displayed comparably high transfection efficiency under a fluorescent microscope (70–80%, Fig. S1). The transiently transfected HeLa cells were used for testing cell proliferation and migration. Compared with empty vector control, wildtype ORP5 transfection significantly enhanced cell proliferation and migration (Fig. 2, B–D), which is consistent with the observations in HeLa/ORP5 versus HeLa control cells (Fig. 1, B and C). A significant reduction of cell proliferation and migration rates was seen in the cells transfected with lipid-binding mutants compared with wildtype ORP5 (Fig. 2, B–D). Importantly, the PS-binding mutation (L389D) and the PI(4)P-binding mutations (H478A/H479A) also resulted in a significant reduction of cell invasiveness based on the assay using the Matrigel invasion chamber (Fig. 2E), highlighting the critical importance of a functional ORP5–ORD in facilitating cell motility. Together, these data suggest that lipid-binding/transfer function of ORP5 is essential for its role in promoting cell growth.
Figure 2.
Functional ORP5–ORD is critical for facilitating cell proliferation and motility. A, diagram of the functional mutations within the ORD of GFP-ORP5. B, cell proliferation assay in HeLa cells transfected with pGFP-ORP5 (WT) or pGFP-ORP5 L389D (mean ± S.D.; ****, p < 0.0001; n = 12). C, cell proliferation assay in HeLa cells transfected with pGFP-ORP5 (WT) or indicated pGFP-ORP5 mutants (mean ± S.D.; ****, p < 0.0001; n = 12). D, cell migration assay in HeLa cells transfected with pGFP-ORP5 (WT) or indicated pGFP-ORP5 mutants. Relative cell migration was analyzed using ImageJ (mean ± S.D.; ****, p < 0.0001; n = 6). E, cell invasion assay in HeLa cells transfected with pGFP-ORP5 WT or the two mutants, pGFP-ORP5 L389D and pGFP-ORP5 H478A/H479A. Relative invasive cells per field were counted and analyzed using ImageJ (mean ± S.D.; *, p < 0.05; **, p < 0.01; ***, p < 0.001; n = 7). All data are representative from three to five independent experiments with similar results.
ORP5 interacts with mTOR
We hypothesized that the role of ORP5 in promoting cell proliferation may involve other ORP5-interacting proteins. To this end, we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis to identify potential ORP5-interacting proteins. The goat polyclonal ORP5 antibody was incubated with the cell lysates prepared from HeLa/ORP5, and immunoprecipitation was performed with normal goat IgG as a control. After immunoprecipitation, the efficiency of ORP5 pulldown was verified by Coomassie Blue staining and anti-ORP5 Western blot analysis (Fig. S2, A and B). LC-MS/MS analysis of ORP5 immunoprecipitates identified a list of proteins (Fig. S2C), among which we selected mTOR (MASCOT ID score, 32.5–49.6; peptide hits = 26) for further investigation because this serine/threonine-protein kinase is a master regulator of cell proliferation and survival (1).
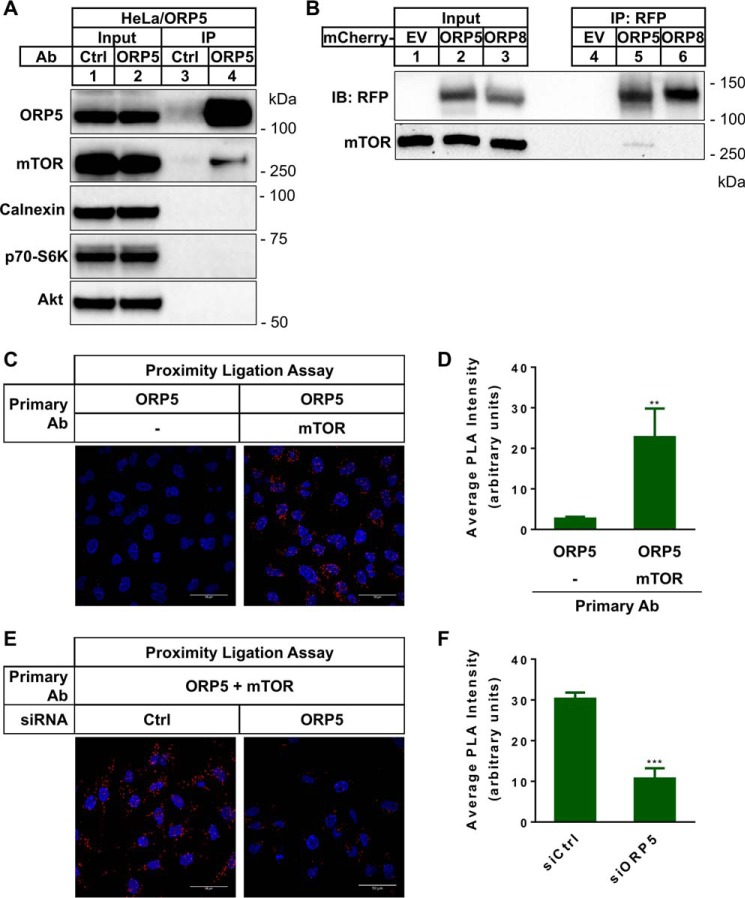
We performed immunoprecipitation experiments to confirm ORP5–mTOR interaction. First, we analyzed samples pulled down by the control or ORP5 antibody from the HeLa/ORP5 cell lysates. Western blot analysis using antibodies to ORP5, mTOR, calnexin (an ER integral-membrane protein), Akt (an upstream effector of mTOR), and p70S6K (a downstream of mTOR signaling) showed that only mTOR was detected along with ORP5 (Fig. 3A, lane 4). Second, we transiently transfected HeLa cells with cDNAs for mCherry empty vector (EV), mCherry-fused ORP5, or ORP8 and used anti-RFP antibody to carry out immunoprecipitation. Anti-mTOR Western blot analysis demonstrated that mCherry-ORP5, and not mCherry-ORP8, interacted with mTOR (Fig. 3B, lane 5). We next used the in situ proximity ligation assay (PLA) to further verify ORP5–mTOR interaction. PLA allows direct visualization and quantitation of the protein–protein interaction. When HeLa/ORP5 cells were incubated with ORP5 primary antibody alone, only negligible PLA signal was observed and was hardly detectable. However, when the cells were treated with both ORP5 and mTOR primary antibodies, there was a robust, significantly amplified PLA signal that could be clearly detected in every single cell (Fig. 3, C and D). The PLA signal observed in these experiments was specific, as the signal was significantly weakened in cells depleted of ORP5 by siRNAs (Fig. 3, E and F).
Figure 3.
ORP5 interacts with mTOR. A, immunoprecipitation of cell lysates extracted from HeLa/ORP5 cells using control IgG or ORP5 antibody (Ab). Immunoblotting (IB) analyses of indicated proteins are shown. B, RFP immunoprecipitation of cell lysates extracted from HeLa cells transfected with mCherry-EV or mCherry-tagged ORP5 or ORP8. C, PLA using goat ORP5 antibody together with or without rabbit mTOR antibody in HeLa/ORP5 cells. D, average PLA intensity in C was quantitated using ImageJ (mean ± S.D.; **, p < 0.01; n > 50 cells). E, proximity ligation assay in HeLa/ORP5 cells treated with control siRNA or ORP5 siRNA. F, average PLA intensity in E was quantitated using ImageJ (mean ± S.D.; ***, p < 0.001; n > 30 cells).
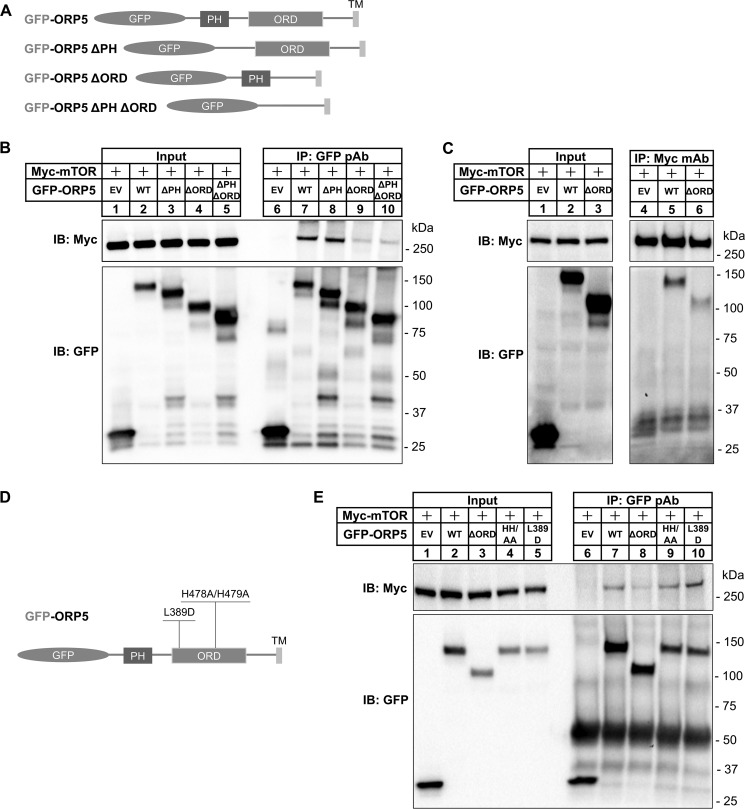
Next, we wanted to examine which domain of ORP5 is required for its interaction with mTOR. We constructed GFP-fused ORP5 deletion mutants that lack the N-terminal PH domain (GFP-ORP5ΔPH), ORD (GFP-ORP5ΔORD), as well as both the PH domain and ORD (GFP-ORP5ΔPHΔORD) (Fig. 4A). GFP EV, GFP-fused wildtype ORP5, or the deletion mutants were co-transfected into HEK-293 cells together with Myc-mTOR cDNA. Anti-GFP immunoprecipitation analysis revealed that the PH domain of ORP5 is dispensable for its interaction with mTOR, as GFP-ORP5ΔPH is similarly associated with Myc-mTOR when compared with wildtype GFP-ORP5 (Fig. 4B, lane 8). In stark contrast, deletion of ORD severely diminished the physical association between ORP5 and mTOR (Fig. 4B, lanes 9 and 10), suggesting that ORP5–ORD is required for ORP5–mTOR interaction. Consistent with this observation, the reverse anti-Myc immunoprecipitation also demonstrated that mTOR strongly associates with wildtype GFP-ORP5 but not GFP-ORP5ΔORD (Fig. 4C, lane 5 versus lane 6). The dependence on ORP5–ORD appears to be separate from its lipid transfer activity, as the point mutations within ORP5–ORD (L389D and H478A/H479A) abolishing PS or PI(4)P binding did not affect ORP5–mTOR interaction (Fig. 4, D and E). This may be due to the observation that mutations abolishing lipid transfer capability do not alter the overall conformation of ORP5–ORD (14, 22). Taken together, these results demonstrate that ORP5 interacts with mTOR and that ORP5–ORD is required for this interaction.
Figure 4.
ORP5–ORD is required for ORP5–mTOR interaction. A, diagram of GFP-ORP5 and its deletion mutants lacking the domains of PH, ORD, or both PH and ORD. B, GFP immunoprecipitation (IP) of cell lysates extracted from HEK-293 cells transfected with cDNAs for Myc-mTOR together with EV or GFP-ORP5 variants as shown A. C, c-Myc immunoprecipitation of cell lysates extracted from HEK-293 cells transfected cDNAs for Myc-mTOR together with EV control or GFP-ORP5 or GFP-ORP5ΔORD. D, diagram of the functional mutations (L389D and H478A/H479A) within the ORD of GFP-ORP5. E, GFP immunoprecipitation of cell lysates extracted from HEK-293 cells transfected with cDNAs for Myc-mTOR together with EV or GFP-ORP5 variants (WT, ΔORD, H478A/H479A, and L389D). IB, immunoblot.
ORP5 is involved in mTORC1 signaling
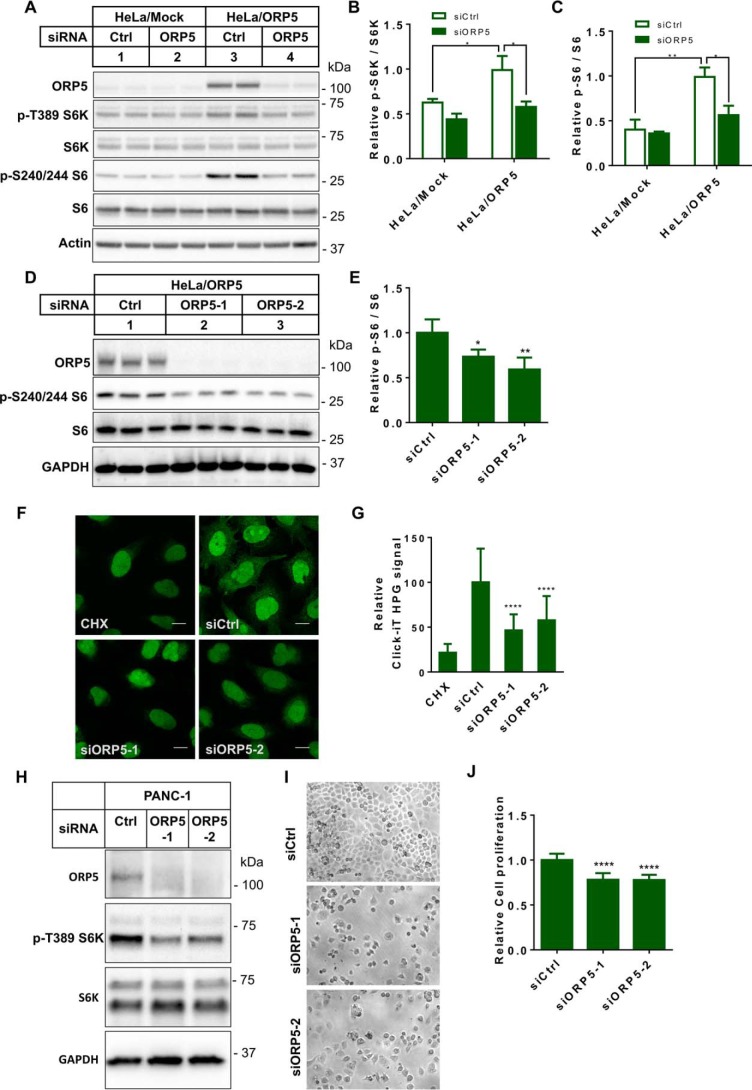
In mammalian cells, mTOR forms two functionally distinct complexes, namely mTORC1 and mTORC2. mTORC1 contributes to cell growth and proliferation by directly phosphorylating S6 kinase (S6K), which activates ribosomal S6 protein, ultimately promoting protein synthesis (3). The role of ORP5 in facilitating cell proliferation and the association between ORP5 and mTOR prompted us to ask whether ORP5 plays any role in mTORC1 signaling. We treated HeLa/Mock and HeLa/ORP5 cells with control or ORP5 siRNAs and analyzed the phosphorylation of S6K and S6. When treated with control siRNA, a significant increase of S6K and S6 phosphorylation was seen in HeLa/ORP5 cells (Fig. 5, A, lane 3 versus lane 1, B and C), indicating that overexpression of ORP5 in HeLa cells induces mTORC1 activation. ORP5 depletion resulted in down-regulation of the phosphorylation of S6K and S6, and this effect was more drastic in HeLa/ORP5 cells (Fig. 5, A, lane 4, B and C). Treatment of HeLa/ORP5 cells with two different ORP5 siRNAs showed similar results in terms of S6 phosphorylation (Fig. 5, D and E). The down-regulation of S6 phosphorylation by ORP5 knockdown appears to be mTORC1-dependent, because in our system a complete inhibition of S6K activation by rapamycin treatment also shut down S6 phosphorylation (Fig. S3). Nascent protein synthesis assay in HeLa/ORP5 cells treated with control or ORP5 siRNAs indicated that ORP5 knockdown significantly decreased protein synthesis (Fig. 5, F and G), a phenotype that reflected the down-regulation of S6K and S6 phosphorylation downstream of mTORC1. These data indicate that, at least when overexpressed, ORP5 is positively associated with mTORC1 activation.
Figure 5.
ORP5 depletion down-regulates mTORC1 signaling. A, HeLa/Mock and HeLa/ORP5 cells were treated with control or ORP5 siRNAs. Cells were starved in serum-free medium overnight prior to harvest for immunoblotting analysis. B, densitometry of activated p70 S6 kinase (p-S6K) in A (mean ± S.D.; *, p < 0.05; n = 4). C, densitometry of activated S6 ribosomal protein (p-S6) in A (mean ± S.D.; *, p < 0.05; **, p < 0.01; n = 4). D, HeLa/ORP5 cells were treated with control and two different ORP5 siRNAs. Cells were starved in serum-free medium overnight prior to harvest for immunoblotting analysis. E, densitometry of p-S6 in D (mean ± S.D.; *, p < 0.05; **, p < 0.01; n = 3). F, protein synthesis assay in HeLa/ORP5 cells treated with control or ORP5 siRNAs. Newly synthesized proteins in the cytosol were detected by the Click-iT® HPG Alexa Fluor® 488 protein synthesis assay kit. G, Click-iT HPG signals in F were quantified using ImageJ and plotted relative to the values of control siRNA-treated cells (mean ± S.D.; ****, p < 0.0001; n = 16–24 cells). H, PANC-1 cells were treated with control or ORP5 siRNAs for 72 h followed by harvest for immunoblotting analysis. I, PANC-1 cells were treated with control or ORP5 siRNAs for 72 h. Cells were imaged under a wide-field microscope. J, cell proliferation assay in PANC-1 cells treated with control siRNA or three different ORP5 siRNAs (mean ± S.D.; ****, p < 0.0001; n = 12).
Knockdown of endogenous ORP5 impairs S6K activation and cell proliferation
Next, we investigated whether endogenous ORP5 regulates mTORC1 signaling. The expression of ORP5 has been linked to the induction of pancreatic cancer cell invasion (20). This prompted us to examine the effect of ORP5 depletion on mTORC1 signaling and cell proliferation in PANC-1 cells, a pancreas ductal adenocarcinoma cell line. The two specific ORP5 siRNAs efficiently reduced endogenous ORP5 protein levels in PANC-1 cells (Fig. 5H). Anti-phosphorylated S6K immunoblotting analysis clearly revealed that S6K phosphorylation was down-regulated upon the depletion of ORP5 (Fig. 5H). Accordingly, ORP5 depletion severely impaired normal cell growth and cell number expansion (Fig. 5I). This observation was also in accordance with the results from cell proliferation assay, which indicated a significant impairment of cell proliferation when ORP5 was depleted in PANC-1 cells (Fig. 5J). We observed similar results in another pancreatic cancer cell line, Capan-1 cells. In Capan-1 cells, there was a clear decrease of mTORC1 activation and significant reduction of cell growth when ORP5 was depleted (Fig. S4). Importantly, the growth defect in ORP5-depleted Capan-1 cells was rescued when TSC1, an inhibitory factor of mTORC1 activation (23), was silenced (Fig. S4), highlighting the link between ORP5 and mTORC1.
ORP5 depletion impairs the localization of mTOR to lysosomes
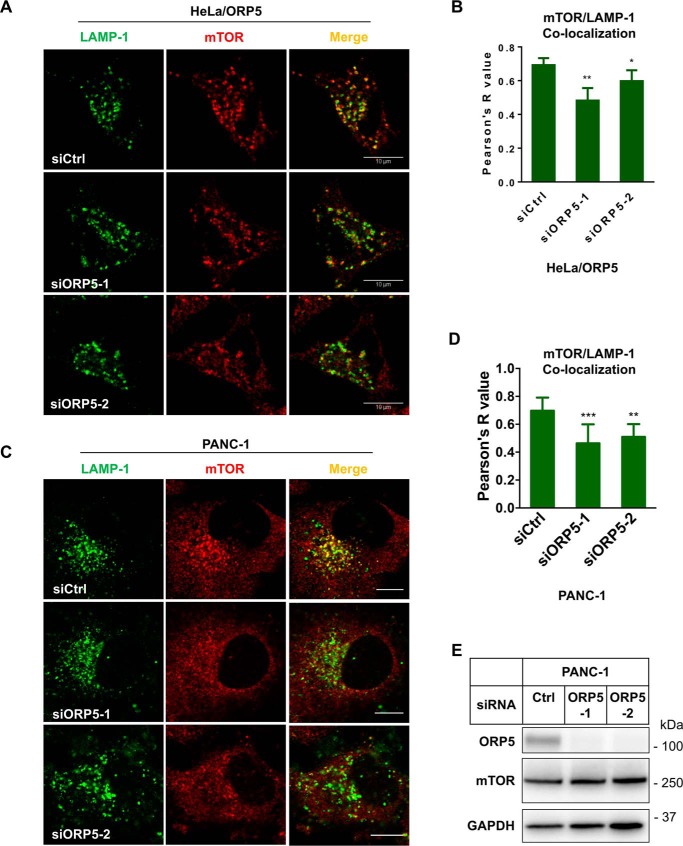
Targeting of mTORC1 to the lysosomal surface is essential for its activation (24). We hypothesized that depletion of ORP5 may interfere with the translocation of mTORC1 to lysosomes, thereby impairing mTORC1 signaling. To test this hypothesis, we examined the co-localization of endogenous mTOR with LAMP-1, a lysosomal membrane marker, by immunofluorescence analysis in HeLa/ORP5 cells. In control siRNA-treated cells and in agreement with the previous study (23), the majority of endogenous mTOR localized to LAMP-1-positive lysosomal structures (Fig. 6A, top panel). Quantification of mTOR/LAMP-1 co-localization revealed a significant decrease of the association of mTOR with lysosomes (Fig. 6, A and B) in ORP5 siRNAs-treated cells. Similar results were obtained from the immunofluorescence experiments performed in PANC-1 cells. Knockdown of endogenous ORP5 in PANC-1 cells also significantly decreased mTOR/LAMP-1 co-localization (Fig. 6, C and D), without affecting overall mTOR protein levels in the cells examined (Fig. 6E).
Figure 6.
ORP5 knockdown impairs the localization of mTOR to lysosomes. A, HeLa/ORP5 cells were treated with control siRNAs or siORP5 for 72 h followed by immunofluorescence of LAMP-1 and mTOR. B, co-localization of mTOR and LAMP-1 in A was analyzed using ImageJ. Pearson's R value is shown (mean ± S.D.; *, p < 0.05; **, p < 0.01; n = 15–20 cells). C, PANC-1 cells were treated with control siRNAs or siORP5 for 72 h followed by immunofluorescence of LAMP-1 and mTOR. D, co-localization of mTOR and LAMP-1 in C was analyzed using ImageJ. Pearson's R value is shown (mean ± S.D.; **, p < 0.01; ***, p < 0.001; n = 10 cells). E, PANC-1 cells were treated with control or ORP5 siRNAs for 72 h followed by harvest for immunoblotting analysis.
Discussion
OSBP/ORPs are unified by their characteristic ORDs that render them capable of sensing, binding, and transporting lipids between intracellular membranes. Previous studies have implicated some members of the OSBP/ORPs family in the regulation of cell-signaling pathways, such as OSBP in ERK1/2 activation (25) and ORP4L in Ca2+ signaling (26). Here, we uncover another connection between OSBP/ORPs and cellular signaling events. We show that overexpression of ORP5 promotes cell proliferation and motility, and that OPR5 expression is positively correlated with mTORC1 signaling. In this context, we present evidence that ORP5 interacts with mTOR and contributes to the targeting of mTORC1 to lysosomal surface for activation.
As a lipid transfer protein, ORP5 has been shown to play a role in intracellular lipid transport at membrane contact sites associated with the ER (12, 15–17). Membrane lipid homeostasis regulated by ORP5 and other lipid transfer proteins has an impact on mTORC1, which itself is a key regulator of lipid metabolism, as well as protein synthesis and cell proliferation (4). Notably, ORP5 appears to be a phosphatidylserine transporter (14) and has been shown to deliver phosphatidylserine from the ER to the plasma membrane and mitochondria (15–17). Phosphatidylserine is critical for the organized distribution of cholesterol in the cytosolic leaflet of the plasma membrane and possibly other organelles (27). Hence, phosphatidylserine transport mediated by ORP5 may be coupled with cholesterol movements between intracellular membranes. Because proper intracellular cholesterol trafficking is essential for mTOR signaling (28), it is possible that phosphatidylserine transport by ORP5 is favored for mTORC1 activation in certain cancer cells, contributing to increased cell proliferation. It is worth noting that membrane phosphatidylserine binds to specific residues in the PH and regulatory domain of Akt, an upstream effector of mTORC1 (29). The phosphatidylserine–Akt interaction is required for Akt activation and downstream signaling (29). This may represent another possible explanation of why ORP5 expression is positively correlated with mTORC1 signaling. In agreement with this possibility, we found that in HepG2 cells ORP5 depletion impaired insulin-induced Akt activation and mTORC1 signaling.4 Therefore, the ability of ORP5 to transport phosphatidylserine seems to be critical for the regulation of mTORC1. In line with this, mutation of a key residue in ORP5–ORD (L389D) responsible for phosphatidylserine binding and transfer failed to promote cell proliferation, migration, and invasion. Similar effects obtained from ORP5–ORD mutants incapable of transporting phosphoinositides or other lipids could be secondary to the impairment of phosphatidylserine transport. However, we could not rule out the possibility that these mutants have a direct impact on the homeostasis of membrane phosphoinositides, resulting in the down-regulation of the Akt/mTORC1 pathway (30). It should also be noted that whereas ORP5 overexpression in HeLa cells was needed for assessing the gain-of-function effects of ORP5 and its mutants, it remains possible that the use of ORP5 overexpression in HeLa cells yields non-physiological results.
An important finding of this study is that proteomics analysis identified mTOR as one of the possible ORP5-interacting proteins. The results from the co-immunoprecipitation and proximity ligation assays confirmed that ORP5 is closely associated with mTOR, suggesting that ORP5 might be able to directly regulate mTORC1. Interestingly, the association between ORP5 and mTOR depends on ORP5–ORD, and this dependence does not seem to rely on the lipid transfer activity of ORP5. Future studies are needed to pinpoint critical regions or residues responsible for ORP5–mTOR interaction. It is also possible that deletion of ORD influences the localization of ORP5 and hence the interaction between ORP5 and mTOR. For example, loss of ORD has been shown to dissociate ORP5 from the mitochondrial surface (17). Given the observation that mTORC1 can be found on mitochondria and regulates mitochondrial functions (31), the dissociation of ORP5 from mitochondria due to ORD deletion could justify the loss of the interaction between ORP5 and mTOR. Future work is necessary to investigate whether a possible ORP5/mTORC1 cooperation at the mitochondrial membrane controls the proper function of mitochondria. Finally, it should be noted that the interaction between ORP5 and mTOR was detected only when ORP5 was overexpressed. Thus, although there is a strong functional link between the ORP5 and mTORC1 pathway, the physical interaction between endogenous proteins remains to be demonstrated and warrants further analyses in the future.
Our data indicate that overexpression of ORP5 up-regulates and depletion of ORP5 down-regulates mTORC1 activation. How are these effects linked to ORP5–mTOR interaction? In response to amino acids or other stimuli, mTORC1 is translocated to the lysosomal surface for activation (24). We found significant impairment of mTOR localization to lysosomes in cells depleted with ORP5. At least in part, this could account for the down-regulation of mTORC1. How the impairment of mTORC1 translocation to lysosomes is caused by ORP5 depletion remains to be elucidated. ORP5 interacts with lysosomal membrane protein Niemann Pick C1, and ORP5 without the ER-anchoring transmembrane domain has been shown to be enriched in late endosome and lysosomes (12), suggesting an association between ORP5 and lysosomes. In this regard, ORP5 may recruit mTORC1 to lysosomes through the interaction with mTOR. This would explain why mTORC1 translocation to the lysosomal surface is impaired when ORP5 is deficient. However, it is also possible that the regulation of mTORC1 activation by ORP5 could be independent of the interaction between ORP5 and mTOR. As mentioned earlier, lipid transfer or targeting activity by ORP5 may be the key to regulate mTORC1 activation. For instance, ORP5 may regulate the homeostasis of phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2), which is concentrated in the endolysosomal membrane (32). Of note, PI(3,5)P2 has been shown to bind to the mTORC1 component raptor and could contribute to lysosomal targeting of mTORC1 (33). Future studies involving co-localization between PI(3,5)P2 and mTORC1, as well as in vitro lipid transfer assay and lipidomics analysis, are required to test this hypothesis.
In summary, we show that the lipid transfer function of ORP5 is critical for ORP5 to promote cell proliferation. Importantly, we identify a novel link between ORP5 and mTORC1 signaling, which provides another explanation for the involvement of ORP5 in cancer cell proliferation. Our work suggests that ORP5 may be a therapeutic target, alongside mTORC1 inhibition, to treat certain cancers.
Experimental procedures
Cell culture and transfection
HeLa, PANC-1 and Capan-1 cells were obtained from ATCC. Monolayers of cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS (for Capan-1 cells 20% FBS was used), 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate in 5% CO2 at 37 °C. DNA transfection was performed using LipofectamineTM LTX and Plus Reagent (Life Technologies, Inc.) according to the manufacturer's instruction. siRNA transfection was carried out in cells grown in full serum medium according to standard methods using LipofectamineTM RNAiMAX transfection reagent (Life Technologies, Inc.).
RNAi and cDNA constructs
siRNA against ORP5 was obtained from Sigma (product no. SASI_Hs02_00365256, SASI_Hs01_00039221, and SASI_Hs02_00365257). ORP5 cDNA was cloned between the NotI and PacI sites of the pQCXIN retroviral vector (Clontech) using the standard subcloning procedure. GFP-ORP5 and pmCherry-ORP8 were described previously (11). GFP-ORP5 point or deletion mutations (L389D, K446A, H478A/H479A, H538A/K540A, K670A, ΔPH, ΔORD and ΔPHΔORD) were generated by site-directed mutagenesis using the following oligonucleotides: L389D, F5′-ATGGACCTGTCCCGCGTGGTGGACCCCACGTTCGTACTGGAGCCG-3′, and R5′-CGGCTCCAGTACGAACGTGGGGTCCACCACGCGGGACAGGTCCAT-3′; K446A, F5′-GGCCCCAAAGGTGAGAGCGTGGGCTCCATCACACAGCCCCTGCCC-3′, and R5′-CTCACCTTTGGGGCCCTTCAGGCTGTCAGCCATGATGACCACGCT-3′; H478A/H479A, F5′-GGCCCCAAAGGTGAGAGCGTGGGCTCCATCACACAGCCCCTGCCC-3′, and R5′-CTCACCTTTGGGGCCCTTCAGGCTGTCAGCCATGATGACCACGCT-3′; H538A/K540A, F5′-CCCTTACCATGCCCTACGCCGCCTGCGCAGGAATCCTGTATGGCACGAT, and R5′-ATCGTGCCATACAGGATTCCTGCGCAGGCGGCGTAGGGCATGGTAAGGG; ΔPH, F5′-GATTCCCTTGGGCTTATCCTCCTCCACCGCAGCCCTGGAGAGCAG-3′, and R5′-AAGCCCAAGGGAATCAAGAAGCCGTACAACCCCATCCTGGGGGAG-3′; ΔORD, F5′-GAGGACCACAGCCCCTGGGACCCCCTGAAGGACATCGCCCAGTTT-3′, and R5′-GGGGCTGTGGTCCTCTAGCACCACGCGGGACAGGTCCATGCCTGG-3′. Myc-mTOR was a gift from David Sabatini (Addgene plasmid 1861).
Generation of HeLa/ORP5 stable cell line
Retrovirus particles were produced in the packaging cell line 293T Phoenix-AMPHO expressing pQCXIN-ORP5. HeLa cells were transduced with the retrovirus encoding ORP5 or mock control for 48 h. Transduced cells were selected with growth medium containing 500 μg/ml G418 (Sigma) for 10 days. During the selection, cells were passaged every 2 days until a stable line of cells expressing ORP5 was established.
Antibodies
Antibodies used were rabbit polyclonal to mTOR (Cell Signaling Technology (CST), catalog no. 2983, clone 7C10); S6K (CST, catalog no. 9202); p-T389 S6K (CST, catalog no. 9205); S6 (CST, catalog no. 2217, clone 5G10); p-S240/244 S6 (CST, catalog no. 5364, clone D68F8); β-actin (CST, catalog no. 4970, clone 13E5); calnexin (CST, catalog no. 2433); GAPDH (CST, catalog no. 2118, clone 14C10); Akt (CST, catalog no. 9272); RFP (Abcam, ab167453); and ORP5 (Sigma, HPA038335); antibodies used were goat polyclonal to ORP5 (Abcam, ab59016); mouse monoclonal to LAMP1 (Santa Cruz Biotechnology, sc-20011, clone H4A3); RFP (Santa Cruz Biotechnology, sc-390909, clone E-8); actin (Abcam, ab8226); and GFP (Santa Cruz Biotechnology, sc-9996, clone B-2). For immunoblotting, we obtained horseradish peroxidase-conjugated secondary antibodies from Jackson ImmunoResearch. For immunofluorescence, we obtained Alexa-Fluor–conjugated secondary antibodies from Molecular Probes/Life Technologies, Inc.
Immunoblot analysis
Samples were mixed with 2× Laemmli buffer, boiled for 5 min at 95 °C or incubated for 10 min at 70 °C, and then subjected to 7.5 or 10% SDS-PAGE. After electrophoresis, the proteins were transferred to Hybond-C nitrocellulose filters (GE Healthcare). Incubations with primary antibodies were performed at 4 °C overnight. Secondary antibodies were peroxidase-conjugated AffiniPure donkey anti-rabbit or donkey anti-mouse IgG (H+L; Jackson ImmunoResearch Laboratories) used at a 1:5000 dilution. The bound antibodies were detected by ECL Western blotting detection reagent (GE Healthcare or Merck Millipore) and visualized with Molecular Imager® ChemiDocTM XRS+ (Bio-Rad).
Immunofluorescence and confocal microscopy
Cells grown on coverslips were fixed with 4% paraformaldehyde for 30 min at room temperature. Immunofluorescence was carried out as described previously (34), and the primary antibodies used were goat anti-ORP5 (Abcam catalog no. Ab59016, 1:50), rabbit anti-mTOR (CST catalog no. 2983, 1:400), and mouse anti-LAMP-1 (Santa Cruz Biotechnology, catalog no. sc-18821, 1:50). Cells were mounted in ProLong® Gold antifade reagent (Life Technologies, Inc.). Confocal images were acquired on an Olympus FV1200 laser-scanning microscope. The manufacturer's software and Fiji software (35) were used for data acquisition and analysis. For co-localization study, the Fiji Coloc 2 plugin was used.
Immunoprecipitation assay
Transfected cells grown in 100-mm dishes were harvested, washed with cold PBS, resuspended in 1 ml of cell lysis buffer (50 mm Tris-HCl, pH 7.8, 100 mm NaCl, 1% Triton X-100) containing protease inhibitor mixture (100×, Sigma) and phosphatase inhibitor mixture (100×, CST). Cell lysates were passed through a 22-gauge needle 20 times, incubated on ice for 30 min, and clarified by centrifugation at 18,000 × g for 15 min at 4 °C. Immunoprecipitation of the lysates with a polyclonal antibody against NPC1 was performed using the Dynabeads® Protein G (Life Technologies, Inc.) according to the manufacturer's instructions. The immunoprecipitated pellets were resuspended in 60 μl of 2× Laemmli buffer (Sigma) and then incubated for 10 min at 70 °C. The resultant samples were subjected to SDS-PAGE and immunoblotting.
MTS assay
Cells were seeded in 96-well plates at 2.5 × 103 cells/well and transfected with siRNA (20 nm) for 72 h. For non-transfected or plasmid cDNA-transfected cells (24 h), cells were seeded at 5 × 103 cells/well in 96-well plates. Six wells were seeded for each transfection. Cell proliferation was measured with an MTS assay kit (CellTiter 96® AQueous One Solution Cell Proliferation Assay, Promega). For each reading, 20 μl of MTS solution was added; plates were incubated for 1–2 h, and absorbance read on a Spectra MAX340 Microplate Reader at 490 nm.
Crystal violet staining
Cells were fixed with 4% paraformaldehyde for 5 min at room temperature. The fixed cells were stained with 0.05% (w/v) crystal violet diluted in distilled water for 30 min at room temperature. Cells were washed twice with water and drained to dry. The dishes were then scanned, and cell numbers were counted using Fiji software (35).
Cell migration assay
Cells were seeded in 24-well plates containing Culture-Insert 24 (Ibidi® cells in focus, Planegg, Germany). For overexpression or knockdown experiments, cells were transfected with plasmid cDNAs for 24 h or siRNAs for 48 h, respectively, prior to seeding. After Culture-Insert was removed, images were taken at 0 and 24 h using an Olympus CKX41 microscope (Olympus, Japan) fitted with an Infinity 1-2CB camera (Lumenera Corp., Ontario, Canada) connected to a computer. Percentage of the cell migration was quantitated using Fiji software (35). Data from 24 h were normalized to those from 0 h.
Proximity ligation assay
HeLa/ORP5 cells were seeded in 6-well plates containing coverslips at 4 × 105 cells/well and grown for 48 h. For knockdown experiment, cells were seeded at 2 × 105 cells/well and treated with siRNAs for 72 h. Cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 15 min and permeabilized with 0.2% Triton X-100 (Sigma) for 10 min at room temperature. The slides were then blocked with 3% BSA in PBS for 1 h and incubated with appropriate combinations of goat anti-ORP5 (Abcam catalog no. Ab59016, 1:50) and rabbit anti-mTOR (CST catalog no. 2983, 1:400) antibodies diluted in blocking solution for 1 h at room temperature. After washing, the coverslips were incubated with Duolink PLA Rabbit MINUS and PLA Mouse PLUS proximity probes (Olink Bioscience, Uppsala, Sweden), and proximity ligation was performed using the Duolink detection reagent kit (Olink Bioscience) according to the manufacturer's protocol. Fluorescence images were acquired using an Olympus Fluoview FV1200 confocal microscope with a ×60/1.35 UPlanSApo objective. Images were prepared and analyzed with Fiji software (35). All data were analyzed using GraphPad PRISM software.
Cell invasion assay
Invasion assay was performed using the Corning® BioCoatTM Matrigel® invasion chambers (Corning Life Sciences). The chambers of the 24-well cell culture inserts were rehydrated with serum-free medium at 37 °C, 5% CO2 atmosphere for 2 h and then transfer to a 24-well plate containing normal growth medium. To the upper chamber, 0.5 ml of cell suspension (2.5 × 104 cells) were added. The invasion chambers were incubated for 24 h at 37 °C in the cell culture incubator. Non-invasive cells on the upper insert membranes were removed using cotton-tipped swabs by gentle scrubbing. Invasive cells on the lower insert membranes were fixed with 100% methanol and stained with 1% toluidine blue (Sigma). The membranes from the insert housing were removed by cutting and mounted on microscope slides. The invading cells were counted under the microscope at ×100 magnification. Data are expressed relative to cell numbers migrating through control membranes.
Protein synthesis assay
HeLa/ORP5 cells were seeded in a 6-well plate containing coverslips at 2 × 105 cells/well treated control siRNA or ORP5 siRNAs for 72 h. The detection of nascent protein synthesis was carried out using the Click-iT® HPG Alexa Fluor® 488 protein synthesis assays kit (Life Technologies, Inc., catalog no. C10428) according to the manufacturer's instructions. Fluorescence images were acquired using an Olympus FluoView FV1200 confocal microscope equipped with DAPI filter and FITC filter for Alexa Fluor® 488. Nascent protein synthesis was assessed by determining the signal intensity in the fluorescent channel in the ring around the nucleus as defined by NuclearMaskTM blue stain. Images were prepared and analyzed with Fiji software (35). All data were analyzed using GraphPad PRISM software.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
To identify potential ORP5-interacting proteins, HeLa/ORP5 cell lysates were immunoprecipitated with control goat IgG or goat anti-ORP5 polyclonal antibody. The immunoprecipitates were subject to gel electrophoresis, and the gel was stained with Coomassie Blue. Stained protein bands were excised, and in-gel trypsin digestion was performed prior to LC-MS/MS analysis, which was carried out as described previously (36). All MS/MS spectra data from human proteins were searched using Mascot search engine, and the results from control and ORP5 immunoprecipitates were combined and filtered using Scaffold software (Version: Scaffold_4.0.7). Positive hits from ORP5 immunoprecipitates were identified with the following criteria: peptide thresholds ≥90.0% and protein thresholds ≥99.0% and ≥2 peptides.
Statistical analyses
Statistical analysis between groups was performed using GraphPad PRISM software (Version 6.03) with Student's unpaired t tests or one-way analysis of variance. Data are expressed as mean ± S.D. unless otherwise stated. Significant differences are indicated in the figures.
Author contributions
X. D. and H. Y. conceptualization; X. D. and A. J. B. data curation; X. D. formal analysis; X. D. and A. Z. validation; X. D. investigation; X. D. visualization; X. D., I. E. L., Y. Q., and A. J. B. methodology; X. D. writing-original draft; X. D., A. J. B., and H. Y. writing-review and editing; A. Z. and A. J. B. resources; H. Y. supervision; H. Y. funding acquisition; H. Y. project administration.
Supplementary Material
Acknowledgments
Mass spectrometric analysis was performed at the Bioanalytical Mass Spectrometry Facility, University of New South Wales, and is supported in part by infrastructure funding from the New South Wales Government. We thank Dr. Nigel Turner from the School of Medical Sciences, University of New South Wales, Sydney, Australia, for providing PANC-1 cells, and Dr. Phoebe Phillips from the Lowy Cancer Research Centre, University of New South Wales, Sydney, Australia, for providing Capan-1 cells.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
X. Du and H. Yang, unpublished data.
- mTOR
- mechanistic target of rapamycin
- OSBP
- oxysterol-binding protein
- ORP
- OSBP-related protein
- ER
- endoplasmic reticulum
- ORD
- OSBP-related domain
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- PH
- pleckstrin homology
- PI(4)P
- phosphatidylinositol 4-phosphate
- PS
- phosphatidylserine
- S6K
- S6 kinase
- PI(3,5)P2
- phosphatidylinositol 3,5-bisphosphate
- FBS
- fetal bovine serum
- CST
- Cell Signaling Technology
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- PLA
- proximity ligation assay
- RFP
- red fluorescent protein
- EV
- empty vector.
References
- 1. Laplante M., and Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pópulo H., Lopes J. M., and Soares P. (2012) The mTOR signalling pathway in human cancer. Int. J. Mol. Sci. 13, 1886–1918 10.3390/ijms13021886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Magnuson B., Ekim B., and Fingar D. C. (2012) Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 441, 1–21 10.1042/BJ20110892 [DOI] [PubMed] [Google Scholar]
- 4. Ricoult S. J., and Manning B. D. (2013) The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 14, 242–251 10.1038/embor.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ngo M. H., Colbourne T. R., and Ridgway N. D. (2010) Functional implications of sterol transport by the oxysterol-binding protein gene family. Biochem. J. 429, 13–24 10.1042/BJ20100263 [DOI] [PubMed] [Google Scholar]
- 6. Du X., Turner N., and Yang H. (2017) The role of oxysterol-binding protein and its related proteins in cancer. Semin. Cell Dev. Biol. 2017, S108409521 [DOI] [PubMed] [Google Scholar]
- 7. Levine T. P., and Munro S. (1998) The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol. 8, 729–739 10.1016/S0960-9822(98)70296-9 [DOI] [PubMed] [Google Scholar]
- 8. Loewen C. J., Roy A., and Levine T. P. (2003) A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22, 2025–2035 10.1093/emboj/cdg201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wyles J. P., McMaster C. R., and Ridgway N. D. (2002) Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J. Biol. Chem. 277, 29908–29918 10.1074/jbc.M201191200 [DOI] [PubMed] [Google Scholar]
- 10. Kentala H., Weber-Boyvat M., and Olkkonen V. M. (2016) OSBP-related protein family: mediators of lipid transport and signaling at membrane contact sites. Int. Rev. Cell Mol. Biol. 321, 299–340 10.1016/bs.ircmb.2015.09.006 [DOI] [PubMed] [Google Scholar]
- 11. Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., and Antonny B. (2013) A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 155, 830–843 10.1016/j.cell.2013.09.056 [DOI] [PubMed] [Google Scholar]
- 12. Du X., Kumar J., Ferguson C., Schulz T. A., Ong Y. S., Hong W., Prinz W. A., Parton R. G., Brown A. J., and Yang H. (2011) A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J. Cell Biol. 192, 121–135 10.1083/jcb.201004142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan D., Mäyränpää M. I., Wong J., Perttilä J., Lehto M., Jauhiainen M., Kovanen P. T., Ehnholm C., Brown A. J., and Olkkonen V. M. (2008) OSBP-related protein 8 (ORP8) suppresses ABCA1 expression and cholesterol efflux from macrophages. J. Biol. Chem. 283, 332–340 10.1074/jbc.M705313200 [DOI] [PubMed] [Google Scholar]
- 14. Maeda K., Anand K., Chiapparino A., Kumar A., Poletto M., Kaksonen M., and Gavin A. C. (2013) Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature 501, 257–261 10.1038/nature12430 [DOI] [PubMed] [Google Scholar]
- 15. Ghai R., Du X., Wang H., Dong J., Ferguson C., Brown A. J., Parton R. G., Wu J. W., and Yang H. (2017) ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane. Nat. Commun. 8, 757 10.1038/s41467-017-00861-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chung J., Torta F., Masai K., Lucast L., Czapla H., Tanner L. B., Narayanaswamy P., Wenk M. R., Nakatsu F., and De Camilli P. (2015) Intracellular transport. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 349, 428–432 10.1126/science.aab1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galmes R., Houcine A., van Vliet A. R., Agostinis P., Jackson C. L., and Giordano F. (2016) ORP5/ORP8 localize to endoplasmic reticulum-mitochondria contacts and are involved in mitochondrial function. EMBO Rep. 17, 800–810 10.15252/embr.201541108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J., Zheng X., Lou N., Zhong W., and Yan D. (2016) Oxysterol binding protein-related protein 8 mediates the cytotoxicity of 25-hydroxycholesterol. J. Lipid Res. 57, 1845–1853 10.1194/jlr.M069906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo X., Zhang L., Fan Y., Zhang Y., Zhang D., Qin L., Dong S., and Li G. (2017) Oxysterol binding protein-related protein 8 inhibits gastric cancer growth through induction of ER stress, inhibition of Wnt signaling and activation of apoptosis. Oncol. Res. 25, 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koga Y., Ishikawa S., Nakamura T., Masuda T., Nagai Y., Takamori H., Hirota M., Kanemitsu K., Baba Y., and Baba H. (2008) Oxysterol binding protein-related protein-5 is related to invasion and poor prognosis in pancreatic cancer. Cancer Sci. 99, 2387–2394 10.1111/j.1349-7006.2008.00987.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagano K., Imai S., Zhao X., Yamashita T., Yoshioka Y., Abe Y., Mukai Y., Kamada H., Nakagawa S., Tsutsumi Y., and Tsunoda S. (2015) Identification and evaluation of metastasis-related proteins, oxysterol binding protein-like 5 and calumenin, in lung tumors. Int. J. Oncol. 47, 195–203 10.3892/ijo.2015.3000 [DOI] [PubMed] [Google Scholar]
- 22. de Saint-Jean M., Delfosse V., Douguet D., Chicanne G., Payrastre B., Bourguet W., Antonny B., and Drin G. (2011) Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195, 965–978 10.1083/jcb.201104062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Menon S., Dibble C. C., Talbott G., Hoxhaj G., Valvezan A. J., Takahashi H., Cantley L. C., and Manning B. D. (2014) Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785 10.1016/j.cell.2013.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., and Sabatini D. M. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang P. Y., Weng J., and Anderson R. G. (2005) OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science 307, 1472–1476 10.1126/science.1107710 [DOI] [PubMed] [Google Scholar]
- 26. Li J. W., Xiao Y. L., Lai C. F., Lou N., Ma H. L., Zhu B. Y., Zhong W. B., and Yan D. G. (2016) Oxysterol-binding protein-related protein 4L promotes cell proliferation by sustaining intracellular Ca2+ homeostasis in cervical carcinoma cell lines. Oncotarget 7, 65849–65861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maekawa M., and Fairn G. D. (2015) Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J. Cell Sci. 128, 1422–1433 10.1242/jcs.164715 [DOI] [PubMed] [Google Scholar]
- 28. Xu J., Dang Y., Ren Y. R., and Liu J. O. (2010) Cholesterol trafficking is required for mTOR activation in endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 107, 4764–4769 10.1073/pnas.0910872107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang B. X., Akbar M., Kevala K., and Kim H. Y. (2011) Phosphatidylserine is a critical modulator for Akt activation. J. Cell Biol. 192, 979–992 10.1083/jcb.201005100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dibble C. C., and Cantley L. C. (2015) Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 25, 545–555 10.1016/j.tcb.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schieke S. M., Phillips D., McCoy J. P. Jr, Aponte A. M., Shen R. F., Balaban R. S., and Finkel T. (2006) The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 281, 27643–27652 10.1074/jbc.M603536200 [DOI] [PubMed] [Google Scholar]
- 32. Hong N. H., Qi A., and Weaver A. M. (2015) PI(3,5)P2 controls endosomal branched actin dynamics by regulating cortactin-actin interactions. J. Cell Biol. 210, 753–769 10.1083/jcb.201412127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bridges D., Ma J. T., Park S., Inoki K., Weisman L. S., and Saltiel A. R. (2012) Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol. Biol. Cell 23, 2955–2962 10.1091/mbc.E11-12-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Du X., Kazim A. S., Brown A. J., and Yang H. (2012) An essential role of Hrs/Vps27 in endosomal cholesterol trafficking. Cell Rep. 1, 29–35 10.1016/j.celrep.2011.10.004 [DOI] [PubMed] [Google Scholar]
- 35. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fei W., Zhong L., Ta M. T., Shui G., Wenk M. R., and Yang H. (2011) The size and phospholipid composition of lipid droplets can influence their proteome. Biochem. Biophys. Res. Commun. 415, 455–462 10.1016/j.bbrc.2011.10.091 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.