Abstract
A genetic variant in the SAND domain of autoimmune regulator (AIRE), R247C, was identified in a patient with type I diabetes mellitus (T1DM) and his mother with rheumatoid arthritis. In vitro, the variant dominantly inhibited AIRE; however, typical features of Autoimmune Polyendocrinopathy Candidiasis and Ectodermal Dysplasia (APECED) were not seen in the subjects. Rather, early manifestation of autoimmunity appeared to be dependent on additional genetic factors. On a population level, diverse variants were identified in this region. Surprisingly, many likely pathogenic variants were seen disproportionately in Africans when compared to Europeans, reinforcing the importance of these variants in altering the immune repertoire. In light of these findings, we propose that R247C and other variants within the SAND-domain alter protein function in a dominant fashion and hold potential as drivers of autoimmunity.
Keywords: Autoimmune regulator, Type I Diabetes Mellitus, Next generation sequencing, Dominant-negative mutation, Immune tolerance
1. Introduction
The loss of immune tolerance can result in autoimmunity, the phenotypic expression of the immune system aberrantly responding to self-antigen. AIRE participates in the proper generation of immune tolerance by allowing expression of tissue-restricted autoantigens in the medullary thymus where developing T cells with receptors specific to autoantigens are deleted or programmed to enforce tolerance of their cognate autoantigen. The essential role of AIRE in this immune tolerance process is known from studying patients with complete loss of function of AIRE, a situation that results in APECED, an autoimmune syndrome with overlapping but somewhat variable autoimmune manifestations including hypoparathyroidism, adrenal failure, candidiasis, gonadal failure, thyroiditis, T1DM, alopecia, vitiligo, and hepatitis.
In contrast to causing the widespread autoimmunity seen in APECED, genetic variation in AIRE has also been associated with various forms of sporadic autoimmunity. Cetani et al first identified an extended pedigree in which the mono-allelic AIRE G228W mutation segregated with either APECED or organ-specific autoimmunity, mainly autoimmune thyroiditis [1]. Subsequent studies of AIRE polymorphisms identified ethnicity-specific associations with alopecia areata [2] and rheumatoid arthritis [3,4,5]. More recently, Oftedal et al identified dominant-negative mutations in AIRE’s PHD1 domain that segregated with varied organ-specific immune diseases [6]. As a result of these studies, the role of impairment of AIRE function in the development of autoimmunity in the absence of APECED features has been re-evaluated [7]. In support of the hypothesis that functional impairment of AIRE associates with sporadic autoimmunity, we characterize a novel dominant-negative AIRE variant in a mother and son who suffer from distinct autoimmune manifestations but not APECED. We demonstrate that the identified variant impairs AIRE function in a dominant-negative manner. We then examine the SAND domain of AIRE for similar variants and demonstrate evidence of genetic selection of the region in general. These findings expand the list of variants in AIRE that can dominantly influence protein function and provide further evidence that a variety of variants in AIRE influence genetic susceptibility to autoimmunity.
2. Materials and Methods
2.1 Research Subjects
Subjects enrolled in an IRB-approved protocol at National Jewish Health and informed consent was documented.
2.2 Gene sequencing
A panel of 345 genes was designed for detection of variant in immune-related genes. Genomic DNA was isolated from whole blood and enriched for targets using a custom RNA capture kit (Sure Select QXT custom library kit, Agilent, Santa Clara, CA) prior to sequencing (Illumina MiSeq, San Diego, CA). Sequence data was mapped and variants called utilizing NextGENe (softgenetics.com). The -23HphI polymorphism was sequenced. HLA Class II typing utilized sequence specific oligonucleotides microspheres on the xMAP platform (Luminex, Austin, TX).
2.3 Cell culture and transfection
HEK 293T cells at ~90% confluence were transiently transfected with indicated plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The plasmids pCDNA3.1 hAIRE-myc-His and pEGFPN1-mAIRE-FLAG were generous gifts from the Benoist-Mathis lab (Harvard Medical School, Boston, MA). Point mutations within AIRE plasmids were generated by using KAPA HiFi Hot start DNA polymerase (Kapa Biosystems, Wilmington, MA). The same mutagenesis strategy was used to introduce new epitope tags to make pCDNA3.1 hAIRE-HA and hAIRE-FLAG.
2.4 Immunofluorescence experiments
HEK 293T cells were seeded onto poly-Lys-coated cover slips. Cells at ~65% confluence were transiently transfected with indicated plasmids. After twenty-four hours, cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 in phosphate buffered saline (PBS). Cells were blocked with 1% BSA and probed with rabbit anti-HA (Cell Signaling Technology, Danvers, MA) and mouse anti-FLAG (M2, Sigma-Aldrich, St. Louis, MO), followed by Cy5-conjugated anti-mouse and FITC-conjugated anti-rabbit IgG secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA), and counterstained with DAPI. Fluoromount-G (SouthernBiotech, Birmingham, AL) mounted coverslips were imaged with a Zeiss Axio Imager M1 microscope.
2.5 Gene Expression Measurement
HEK 293T cells were harvested 48 hours post-transfection. Total RNA was isolated using Direct-zol RNA mini prep kit (Zymo Research, Irvine, CA) and reverse-transcribed using SuperScript II (Life Technologies) with oligo (dT)18. qPCR was performed using Power SYBR Green PCR Master Mix (Invitrogen, Carlsbad, CA) on a CFX-Connect detection system (Bio-Rad, Hercules, CA). The following primer sequences were used:
GAPDH For: TGATGACATCAAGAAGGTGGTGAAG
GAPDH Rev: TCCTTGGAGGCCATGTGGGCCAT
HPRT1 For: TGAAGAGCTATTGTAATGACCAGTCAAC
HPRT1 Rev: AGCAAGCTTGCGACCTTGACCA
IGFL1 For: CTCCCCGAGGCTGCATCGTA
IGFL1 Rev: TGGCACAGCATCAGGTAAGGA
KRT14 For: CAGTCCCTACTTCAAGACCATTGA
KRT14 Rev: ACTGTGGCTGTGAGAATCTTGTTC
2.6 Immunoprecipitation
Empty vector or hAIRE WT-FLAG was co-transfected with myc-His-tagged hAIRE WT or R247C. Forty-hours post-transfection, HEK 293T cells were harvested and incubated with hypotonic buffer (0.05% Nonidet P-40, 20mM Hepes pH 7.5, 1.5mM MgCl2, 10mM KCl, 5mM EDTA) with 1X mammalian protease inhibitor cocktail (G-Biosciences, St. Louis, MO) on ice for 15 minutes. Lysates were spun at 500g for 5 minutes and the supernatant (cytosolic fraction) was removed. The remaining nuclei were incubated in nuclear extraction buffer (50mM Bis-Tris pH 7.5, 750mM 6-aminocaproic acid, 3mM CaCl2, 10% glycerol, 1X mammalian protease inhibitor cocktail and micrococcal nuclease (Thermo Scientific, Waltham, MA) for 1 hour on ice and cleared at 18000g for 10 minutes. The resulting supernatant was nutated with anti-FLAG M2 affinity agarose beads (Sigma Aldrich, St. Louis, MO) at 4°C for 16 hours. Beads were washed and boiled in Laemmli sample buffer to elute captured proteins. Immunoblots were developed with anti-FLAG-HRP (Sigma Aldrich) and 3anti-myc followed by ant-irabbit-HRP (Cell Signaling Technology, Danvers, Massachusetts).
2.7 In silico analysis
To determine conserved residues, the complete amino acid sequence of human AIRE (NP_000374.1) was aligned with AIRE from representative species using T-Coffee Psi-Coffee algorithm (http://tcoffee.crg.cat/apps/tcoffee/index.html) [8]. Homologous sequences were downloaded from OrthoDB (http://www.orthodb.org) [9]. Population level variant information for the AIRE gene was downloaded from the gnomAD browser (http://gnomad.broadinstitute.org/) [10]. Pathogenicity prediction scores were downloaded from the CADD server (http://cadd.gs.washington.edu/) [11]. Pathogenicity levels were assigned based on CADD PHRED score with greater than 15 being considered pathogenic and less than 15 being considered benign. Variants were further subdivided based on minor allele frequency as reported in the GnomAD database.
2.8 Statistical analysis
The Simpson Index of Diversity was used to measure diversity of variants within AIRE domains. Chi-squared test was performed to assess differences in domain allele frequency between populations.
3. Results
3.1 A rare AIRE variant identified in a patient with autoimmunity
A 17-year-old Caucasian young man presented to the immunology clinic for evaluation with a medical history remarkable for type I diabetes (T1DM) onset at age 3 and rheumatoid arthritis in his mother. Because of a previously identified association between rheumatoid arthritis and T1DM [12], the proband and his mother were enrolled in a study searching for genetic causes of immune dysregulation. DNA was isolated from the patient’s whole blood, and a panel of 345 genes with known immunologic function was sequenced. After filtering genetic variants for low allele frequency, 6 candidate genetic variants remained, of which the c.739C>T variant, rs147846074, p.Arg247Cys (R247C) in AIRE was investigated. No features of APECED were identified in the patient. Anti-cytokine antibodies typically seen in APECED were absent, as were other autoantibodies with the exception of GADA, IA-2, and MIAA (Table S1, Fig. S1). The variant was also identified in the proband’s mother who has rheumatoid arthritis. Genetic screening of additional family members was unable to be performed as they chose not to enroll in the study.
3.2 In silico analysis of AIRE R247C
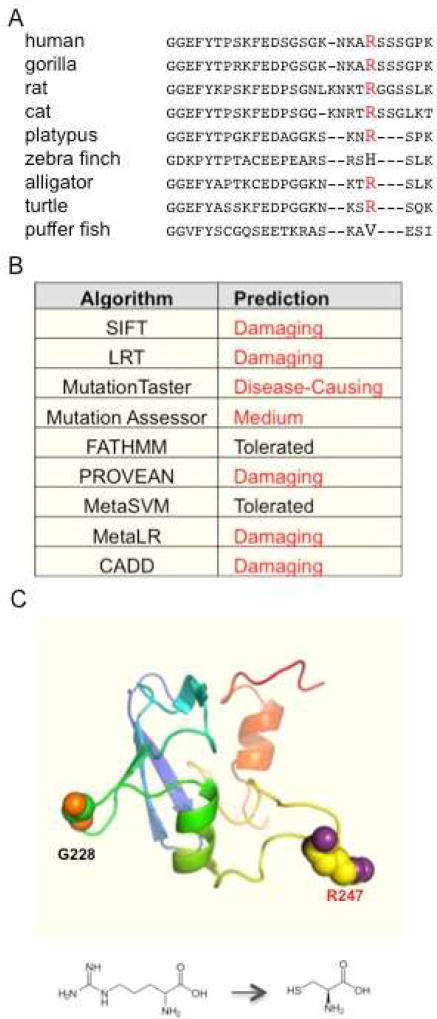
We first assessed the likelihood that R247C was capable of altering protein function utilizing various in silico methods. R247C is located within the SAND domain of AIRE, a protein subunit of unclear function that previously was shown to lack conservation in distant chordate AIRE homologues [13]. Nonetheless, the dominant-negative variant identified by Cetani et al [1], G228W, is in the SAND domain, confirming that at least one variant within this domain can cause disease. R247C occurs at a relatively conserved position among mammalian orthologues, but the position is only partially conserved in non-mammalian tetrapods (Fig. 1). Despite a lack of conservation beyond mammals, multiple variant analysis algorithms predicted the variant to have a functional impact on the gene (Fig. 1).
Figure 1. In silico analysis of R247C predicts an important functional role.
Panel A shows a multiple sequence alignment of the AIRE gene from various chordates demonstrates conservation of R247 among mammals and a representative reptile and amphibian. R247 is highlighted in red. Panel B shows the results of various variant effect prediction algorithms. Data was retrieved using dbNSFP3. (https://sites.google.com/site/jpopgen/dbNSFP) Panel C is a homology model of the SAND domain of human AIRE. R247 is labeled in red. For reference, G228 is labeled in black.
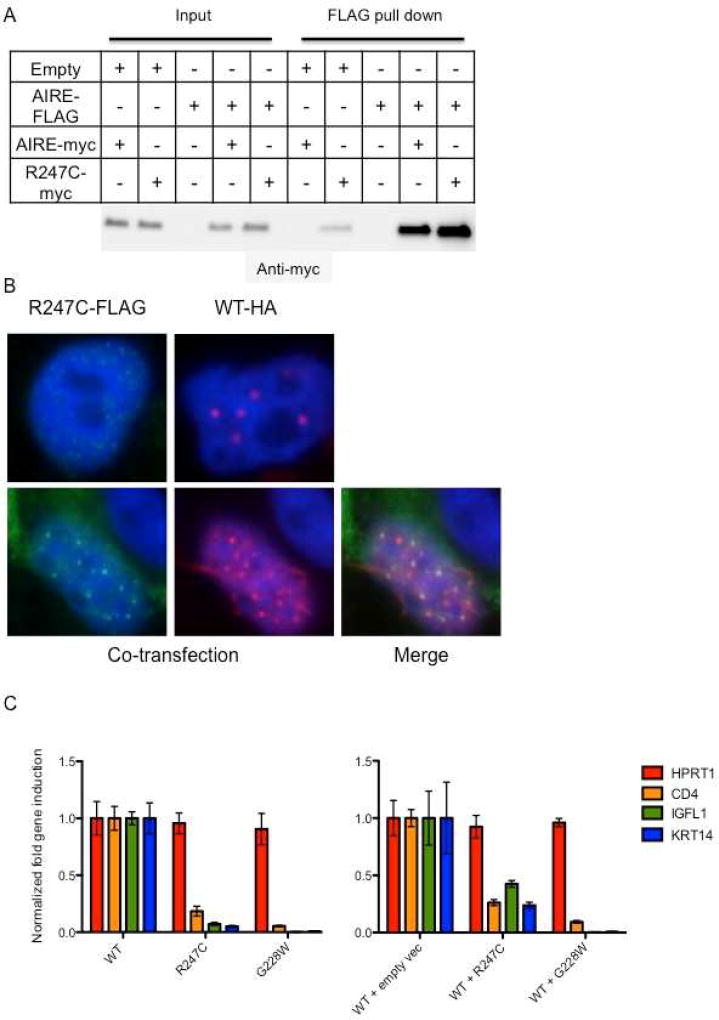
3.3 R247C forms complexes with WT AIRE and localizes to the nucleus
Experiments were performed to confirm expression, binding, and localization of R247C. Co-immunoprecipitation showed that WT AIRE binds to R247C within the nucleus of transfected cells (Fig. 2A). Transfection of HEK293T cells with R247C-FLAG resulted in clearly visible nuclear punctae, similar to what was observed in WT-transfected cells (Fig. 2B). Co-transfection of R247C and WT resulted in nuclear punctae appearing to contain both R247C-FLAG and WT-HA (Fig. 2B). These findings suggested that R247C retains the ability to complex with WT AIRE, and these complexes localize to nuclear punctae in a manner similar to WT alone.
Figure 2. R247C impairs transcriptional regulation of WT AIRE.
Panel A shows that R247C forms complexes with WT AIRE. WT AIRE-FLAG was co-transfected with R247C-myc and complexes precipitated with anti-FLAG M2 beads. Eluted proteins were probed with anti-myc rabbit antibody. Panel B demonstrates the formation of nuclear punctae by R247C. WT AIRE-HA and R247C-FLAG were transfected into HEK293T cells and visualized using fluorescent secondary antibodies to either HA or FLAG. Multiple images were taken and staining patterns were compared for presence of cytoplasmic staining and nuclear punctae. Both WT AIRE and R247C demonstrated nuclear punctae with minimal cytoplasmic staining. When cotransfected, WT AIRE and R247C overlap, indicating likely complex formation. Panel C shows that HEK-293T cells transfected with either the patient mutation R247C or the known dominant negative mutant G228W produced less downstream gene transcripts than WT AIRE. Likewise, when WT AIRE and mutant AIRE were simultaneously transfected into HEK293T cells, downstream gene expression was decreased compared with cotransfection of empty vector and WT AIRE. qPCR was used to examine the expression of AIRE-regulated genes CD4, IGFL1, and KRT14 along with the AIRE-independent gene HPRT1. Expression levels were normalized against the endogenous control GAPDH. The results (mean ± SD) are shown as fold gene induction as compared to cells transfected with WT AIRE. The data is representative of two independent experiments. Cluster of differentiation 4 (CD4); IGF-life family member 1 (IGFL1); keratin 14 (KRT14); hypoxanthine phosphoribosyltransferase 1 (HPRT1); glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
3.4 R247C induces less downstream transcripts than wild type (WT) AIRE
Since R247C formed complexes and translocated to the nucleus, we transfected HEK293T cells with mutant and WT constructs and measured induced gene expression. Transfection of R247C resulted in substantially less expression of the known AIRE targets CD4, IGFL1, and KRT14, compared to WT-transfected cells (Fig. 2C). Additionally, co-transfection of WT AIRE with the mutants resulted in decreased expression of downstream genes when compared to WT co-transfected with control vector, indicating a dominant-negative effect (Fig. 2C). Overall, these data support the conclusion that despite apparently normal nuclear localization, R247C reduces downstream transcriptional activation significantly through dominant-negative inhibition.
3.5 Genetic risk factors for T1DM work in concert with the R247C variant
Prior studies of APECED have demonstrated that T1DM appears in patients with T1DM genetic risk factors and without high-affinity type-I-interferon neutralizing antibodies [14–15]. In the absence of APECED clinical features, we looked to see if these principles still applied to the proband. The combination of MHC class II DR DQ alleles accounts for up to 40% of heritability of T1DM in Europeans [16]. The proband’s alleles were DRB1*0301, DQA1*0501, DQB*0201 and DRB1*0404, DQA1*0301, DQB*0302, a combination that carries up to 5% risk of T1DM in the general population [17]. Sequencing of the INS promoter revealed that both the proband and his mother were homozygous for the “-23HphI” T1DM risk allele. This allele correlates with a truncated promoter region and an up to threefold reduction in expression of insulin in human thymic epithelial cells [18]. These findings support the theory that the development of T1DM in AIRE deficiency requires a genetically predisposed host deficient in ameliorating factors such as high-affinity type-I-interferon neutralizing antibodies.
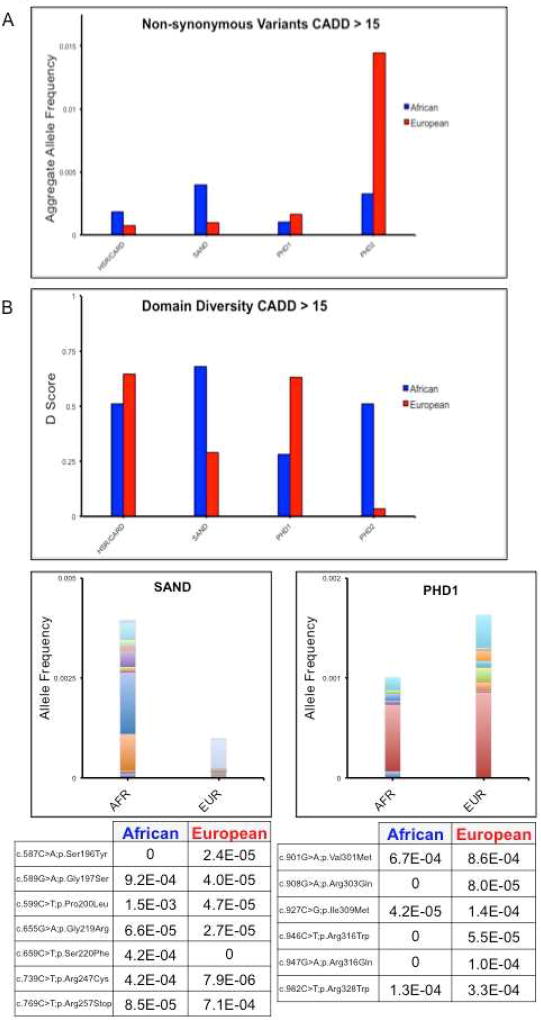
3.5 SAND-domain variant population frequency differs between Africans and Europeans
We then queried the GnomAD database [10] to understand variation in the AIRE gene on a population level. We compared the African and European population cohorts within GnomAD and found that the allele frequency of predicted-pathogenic variants within the SAND domain was greater in Africans than Europeans (Fig. 3A). We then calculated the Simpson’s diversity index for each domain within African and European samples (Fig. 3B). Interestingly, in European samples, the SAND domain was dominated by a single allele, Arg257Stop, a known loss-of-function allele that results from a bottleneck in out-of-Africa populations. Excepting Arg257Stop, the number of likely pathogenic SAND variants tolerated in Europeans was minimal. In Africans, the SAND domain carried a broad array of non-private predicted-pathogenic variants suggesting that in Africans the SAND domain is under neutral or positive selection. This ethnic disparity was not observed in the other genetic regions examined (Fig 3B). In light of these observations, we conclude that variation within AIRE has domain-specific consequences that are either being selected for in Africans or selected against in Europeans, implicating these variants as potent drivers of a clinical phenotype.
Figure 3. Population genetics of AIRE reveal domain-specific trends.
Panel A shows the allele frequency of all variants with a CADD PHRED score >15 for each domain in AIRE including the areas between defined domains. African and European populations as defined in GnomAD are plotted separately. Panel B shows the Simpson’s Diversity Index score for each domain. Only variants with CADD > 15 were included. The proportional allele frequencies of the variants in the SAND, PHD1, and PHD2 domains are plotted below. Each color represents a different variant. Actual allele counts are listed in the tables underneath each plot with ingleton and doubleton alleles removed in order to eliminate bias towards rare variation seen as a result of greater population sampling of Europeans.
4. Discussion
In this article, we demonstrate that a second variant within the SAND domain of AIRE acts in a dominant negative fashion to decrease transactivation of downstream genes. The mechanism by which R247C exerts a dominant-negative effect is unknown; however, the data indicates that R247C forms AIRE hetero-oligomers that localize to discrete locations within the nucleus. It remains unclear whether the assembled complexes arrive at different destinations than WT AIRE or if the complexes localize correctly but suffer from alteration of further downstream events. Given our findings, further studies investigating the structure and function of this domain are justified.
While the study demonstrates the dominant negative activity of R247C, we unfortunately were unable to definitively link this variant to human autoimmunity. The major pitfall of this study was an inability to investigate segregation of the variant in more family members than the proband and his mother. Despite this deficiency, the evidence suggests R247C and possibly other SAND-domain variants do associate with autoimmunity. Both subjects carrying the variant suffered from significant, early-onset autoimmunity. The proband had similar genetic predisposing factors to APECED patients with T1DM, and his mother had rheumatoid arthritis, a disease that has previously been linked to variation in the AIRE locus [3,4,5]. In addition, the population analysis suggests that variation within the SAND domain is either selected for in Africans, selected against in Europeans, or both. As Africa has greater microbial richness than Europe [19] and degree of pathogen exposure is a driving force of evolutionary selection [20], we believe that the level of immune tolerance generated by wild type AIRE could be undesirable, and subjects with reduced AIRE function could have a survival advantage that balances the risk of autoimmunity. Loss of the selective advantage in modern societies is demonstrated by the purifying selection evidenced by the lack of SAND-domain variation in Europeans, possibly attributable to an increased risk of significant autoimmunity. Despite these intriguing observations, studies designed to directly test the association between SAND variants and autoimmunity will ultimately be required to validate the association to autoimmunity.
The rarity of APECED-associated AIRE mutations and the lack of association between common AIRE SNPs and some forms of autoimmunity previously led many to discount the association to autoimmunity; however, dominant-negative AIRE variants were not part of any of the past negative-association studies. Therefore, substantial but not complete loss of AIRE function--as seen with R247C and other dominant-negative variants--remains a valid candidate for association with various types of autoimmunity. We argue that the situation of reduced or altered AIRE activity acts as a driver of autoimmunity by causing partial impairment of immune tolerance. Moreover, as seen in the proband and his mother, the specific autoimmune manifestation depends on additional genetic factors and possibly the lack of disease-ameliorating antibodies that can be seen in APECED syndrome. Given these external factors, we anticipate that AIRE variants can be identified in other autoimmune disease beyond T1DM and rheumatoid arthritis. In light of these findings, population based studies of dominant-negative AIRE variant carriers are urgently required to determine the impact of these variants on human autoimmunity.
Supplementary Material
The 3 target cytokines were transcribed and translated in the presence of [35] S methionine and radioligand binding assays were performed on the patient, normal controls, and APECED patients.
Autoantibodies evaluated during the clinical care of the proband were largely negative with the exception of insulin-specific autoantibodies
Highlights.
A novel variant in the SAND domain of AIRE is identified in a family with multiple autoimmune members.
The SAND variant dominantly inhibits transactivation of target genes.
Variants in the SAND domain are disproportionately present in populations with an African genetic background.
Acknowledgments
M.S.A. is supported by NIAID grant AI097457. S.H is supported by NIAID grant AI106912. Dr. Reddy was supported by a fellowship from Grifols USA, LLC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.K.A, E.W.G., and S.H. conceived of and designed the study; Y.S.H performed transfection, qPCR, immunofluorescence, and immunoprecipitation; P.R.R. designed and performed genetic studies; P.R.R and J.K.A. performed genetic analysis; J.K.A. performed bioinformatics analysis; M.S.A. tested for cytokine autoantibodies; Y.L and M.R. performed HLA typing and tested for insulin autoantibodies, J.K.A. wrote the manuscript; E.W.G and M.R. enrolled the subjects and provided clinical information. All authors reviewed, edited, and approved of the manuscript.
Conflict of Interest
The investigators have no competing financial interests to declare.
References
- 1.Cetani F, Barbesino G, Borsari S, et al. A novel mutation of the autoimmune regulator gene in an Italian kindred with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy, acting in a dominant fashion and strongly cosegregating with hypothyroid autoimmune thyroiditis. J Clin Endocrinol Metab. 2001;86(10):4747–52. doi: 10.1210/jcem.86.10.7884. [DOI] [PubMed] [Google Scholar]
- 2.Tazi-Ahnini R, Cork MJ, Gawkrodger DJ, et al. Role of the autoimmune regulator (AIRE) gene in alopecia areata: strong association of a potentially functional AIRE polymorphism with alopecia universalis. Tissue Antigens. 2002;60(6):489–95. doi: 10.1034/j.1399-0039.2002.600604.x. [DOI] [PubMed] [Google Scholar]
- 3.Terao C, Yamada R, Ohmura K, et al. The human AIRE gene at chromosome 21q22 is a genetic determinant for the predisposition to rheumatoid arthritis in Japanese population. Human Molecular Genetics. 2011;20(13):2680–5. doi: 10.1093/hmg/ddr161. [DOI] [PubMed] [Google Scholar]
- 4.García-Lozano J-R, Torres-Agrela B, Montes-Cano M-A, et al. Association of the AIRE gene with susceptibility to rheumatoid arthritis in a European population: a case control study. Arthritis Res Ther. 2013;15(1):R11. doi: 10.1186/ar4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao S, Li X-R, Cen H, Yin Z-S. Association of AIRE polymorphisms with genetic susceptibility to rheumatoid arthritis in a Chinese population. Inflammation. 2014;37(2):495–9. doi: 10.1007/s10753-013-9763-3. [DOI] [PubMed] [Google Scholar]
- 6.Oftedal BE, Hellesen A, Erichsen MM, et al. Dominant Mutations in the Autoimmune Regulator AIRE Are Associated with Common Organ-Specific Autoimmune Diseases. Immunity. 2015;42(6):1185–96. doi: 10.1016/j.immuni.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Anderson MS, Casanova J-L. More than Meets the Eye: Monogenic Autoimmunity Strikes Again. Immunity. 2015;42(6):986–8. doi: 10.1016/j.immuni.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Di Tommaso P, Moretti S, Xenarios I, et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–7. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zdobnov EM, Tegenfeldt F, Kuznetsov D, et al. OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 2017;45(D1):D744–749. doi: 10.1093/nar/gkw1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao KP, Gunnarsson M, Källberg H, et al. Specific association of type 1 diabetes mellitus with anti-cyclic citrullinated peptide-positive rheumatoid arthritis. Arthritis Rheum. 2009;60(3):653–60. doi: 10.1002/art.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saltis M, Criscitiello MF, Ohta Y, et al. Evolutionarily conserved and divergent regions of the autoimmune regulator (Aire) gene: a comparative analysis. Immunogenetics. 2008;60(2):105–14. doi: 10.1007/s00251-007-0268-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paquette J, Varin DSE, Hamelin CE, et al. Risk of autoimmune diabetes in APECED: association with short alleles of the 5'insulin VNTR. Genes Immun. 2010;11(7):590–7. doi: 10.1038/gene.2010.33. [DOI] [PubMed] [Google Scholar]
- 15.Meyer S, Woodward M, Hertel C, et al. AIRE-Deficient Patients Harbor Unique High-Affinity Disease-Ameliorating Autoantibodies. Cell. 2016;166(3):582–95. doi: 10.1016/j.cell.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. The American Journal of Human Genetics. 1996;59(5):1134–48. [PMC free article] [PubMed] [Google Scholar]
- 17.Aly TA, Ide A, Jahromi MM, et al. Extreme genetic risk for type 1A diabetes. Proc Natl Acad Sci USA. 2006;103(38):14074–9. doi: 10.1073/pnas.0606349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vafiadis P, Bennett ST, Todd JA, et al. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet. 1997;15(3):289–92. doi: 10.1038/ng0397-289. [DOI] [PubMed] [Google Scholar]
- 19.Henn BM, Botigué LR, Peischl S, et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci USA. 2016;113(4):E440–9. doi: 10.1073/pnas.1510805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fumagalli M, Sironi M, Pozzoli U, et al. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7(11):e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 3 target cytokines were transcribed and translated in the presence of [35] S methionine and radioligand binding assays were performed on the patient, normal controls, and APECED patients.
Autoantibodies evaluated during the clinical care of the proband were largely negative with the exception of insulin-specific autoantibodies