Abstract
Background:
Urine is a supersaturated solution with solutes in very high concentration. Matrix gla protein (MGP) is one of the proteins involved in inhibition of extracellular calcification. Among the various polymorphisms studied, polymorphism of SNP rs4236 in the MGP gene is found to have association with nephrolithiasis. We evaluated the significance of SNP rs4236 polymorphism in nephrolithiasis among the Indian population.
Materials and Methods:
The study was conducted in 2013 and 2014 among fifty participants. Twenty-five of them were patients with documented symptomatic stone disease and the 25 controls had no history of stone disease and ultrasonogram was normal. Serum creatinine was estimated in all patients. DNA was isolated from the blood sample and amplified by polymerase chain reaction using defined primers. Single strand conformational polymorphism was done to identify abnormal bands and the same was sequenced.
Results:
The patients and controls were matched in age and sex. The serum creatinine levels were similar in both groups. The predominant change in SNP rs4236 in patients was a G to A nucleotide change, resulting in AA homozygous genotype. This was found in 60% of patients. The rest (40%) of patients and all (100%) controls had heterozygous AG genotype. This corresponds to a stone disease with an odds ratio (OR) of 75 (P < 0.003).
Conclusion:
This first study in Indian population shows that genotype AA polymorphism of SNP rs4236 of MGP gene was found to be significant risk factor for renal stone disease. Studies with larger sample size may be needed to confirm this finding and elucidate the exact OR.
Keywords: Matrix gla protein, risk factor, urolithiasis
Introduction
The lifetime prevalence of urinary stone disease is 4%–16%.[1] Most of the solutes are supersaturated in the human urine. For instance, the concentration of oxalates in urine is four times higher than that of its solubility in water.[2] Still, most of the normal individuals do not form stones. Various factors present in the urine increase the solute concentration required for crystallization and aggregation of salts. Pyrophosphates, citrates, osteopontin, and Tamm Horsfall protein are the few predominant inhibitors of crystallization and aggregation.[2]
Vascular calcification, particularly in atherosclerosis, has been studied extensively. Matrix gla protein (MGP) is an inhibitor of vascular calcification, and specific allelic modification have been found to increase the incidence of vascular calcification.[3] This protein has been found to have a role in extracellular calcification in the kidneys too, in animal studies.[4] Various single nucleotide polymorphisms (SNPs) have been studied in the MGP gene, for their role in stone formation.[5] Among them, SNP rs4236 has been found to be a significant risk factor for stone disease.[6] With this background, we studied the significance of single nucleotide polymorphisms of SNP rs4236 in MGP gene in renal calculus disease patients in local Indian population.
Materials and Methods
The study was conducted in 2013 and 2014. Institutional ethical committee clearance was obtained. Fifty subjects were selected for inclusion in the study, and informed consent was obtained. Twenty-five of them had documented stone disease, either renal or ureteric calculus and presented to the urology outpatient department. The control group was age- and sex-matched individuals, with normal ultrasonogram of the kidneys, and without any history of stone disease. Patients with preexisting deranged renal function (serum creatinine more than 1.6 mg%) were excluded from the study. In the study, 54 participants were enlisted and four of them (three in the stone group and one in the control group) had to be excluded due to high serum creatinine levels. The baseline clinical examination and ultrasound findings were documented. Serum creatinine was estimated in all patients.
DNA isolation
Genomic DNA was isolated from the blood samples. Whole blood and red blood cell (RBC) lysis buffer (0.16M NH4 Cl, 0.17M Tris-HCl) in the ratio 1:3 was vortexed for 1 min and centrifuged at 4000rpm for 5 min. The excess red supernatant was discarded, retaining 1 ml of fluid with pellet. 3 ml of RBC lysis buffer was added to this residue, and the cycles were repeated till the supernatant is clear.
The pellet which has been obtained is further processed. 400 μL of nuclei lysis buffer (10 mM Tris-HCl, 400 mM sodium chloride, 2 mM Na2 ethylenediaminetetraacetic acid (EDTA), 1% SDS and 1 mg protease K) was added, contents vortexed gently and incubated at 65°C in water bath for 150 min. At the end of incubation, contents were transferred to 2 mL microcentrifuge tube and 175 μL of 5.3% NaCl was added and centrifuged at 10,000 rpm for 5 min. To the supernatant, 1 mL of 100% ethanol was added and inverted several times to precipitate DNA and centrifuged at 1500 rpm for 10 min. Pellet was washed with 1mL of ice-cold 75% ethanol and centrifuged at 15,000 rpm for 3 min. Supernatant was removed and the pellet was air-dried to vapourise ethanol. Tris EDTA buffer (100–150 μL) was added to the air-dried pellet, and the DNA was stored at −20°C.
The isolated DNA was run in 0.8% agarose gel with ethidium bromide as a staining dye. Electrophoresis was carried out at 50 V. The gel was viewed using ChemiDoc gel documentation system, (BioRad Inc. Berkeley, CA) and photographed. The A260/A280 values were also checked to assess DNA purity.
Polymerase chain reaction amplification
The DNA was subjected to polymerase chain reaction (PCR) gene amplification using Eppendorf Thermal Cycler, Germany. The details of PCR are given in Table 1. Primers for the SNP region rs4236 were designed using NCBI Primer BLAST tool as given in Table 2. Five microliter of master mix along with 1 μL each of forward primer, 1μL reverse primer and 1 μL DNA sample and 2 μL of nuclease-free water were used for PCR. The primers were also confirmed by UCSC in-silico PCR which resulted in a product of 261 bp. The samples were run on 1.5% agarose gel to check whether the DNA has been amplified. The amplified samples were taken for single-strand conformational polymorphism (SSCP).
Table 1.
Polymerase chain reaction amplification details

Table 2.
NCBI primer BLAST tool

Single strand conformational polymorphism analysis
To 7 μL PCR product, 15 μL SSCP dye (0.1% bromophenol blue, 0.1% xylene cyanol, 90% formamide, and 20 mM EDTA) was added. From this preparation, a 10 μL aliquot was denatured at 95°C for 6 min in PCR. The sample was then loaded on 10% polyacrylamide gel electrophoresis gel and electrophoresis was performed followed by silver staining. The gel was then viewed using ChemiDoc gel documentation system, (BioRad Inc. Berkeley, CA) and photographed. The abnormal bands in SSCP gel were sequenced using Automated DNA sequencer at Scigenome Biotech Pvt. Ltd.
Serum creatinine analysis
Serum creatinine analysis was performed from the blood samples. O-cresolphthalein complexone-based calorimetric method was used for serum calcium estimation, and the values were recorded.
Statistical analysis
The difference in the serum creatinine levels in both groups was compared using Student's t-test. Odds ratio (OR) of difference in the incidence of stone disease in different allele groups were calculated by 2 × 2 cross-tabulation using SPSS® Statistics for Windows, version 10.0 (SPSS Inc., Chicago, Ill., USA) software.
Results
A total of 50 individuals were included in the study. Twenty-five were in the control group, and 25 were in the stone group. The mean age of the patients was 47.68 years in the stone group and 47.76 years in the control group. There were 18 males and 7 females in both groups (matched pairs). The mean serum creatinine was 1.01mg% in the stone group and 0.93 mg% in the control group. The difference was not statistically significant.
The predominant change in patients was G to A nucleotide change in chromosome 12 at position 15,035,081. This results in a change from GA heterozygous genotype to AA homozygous genotype [Table 3 and Figures 1, 2]. 60% (15/25) of the patients had such change. The rest of the patients and all those in control group had persistent AG heterozygosity. The OR was 75 (P = 0.003). This shows very high risk of stone disease with AA genotype. Regarding the allelic polymorphism, the OR for stone disease with adenine nucleotide to that of guanine nucleotide was 4 (P < 0.002).
Table 3.
Genotypes observed in the present study with respect to single nucleotide polymorphism rs4236 of matrix gla protein gene

Figure 1.
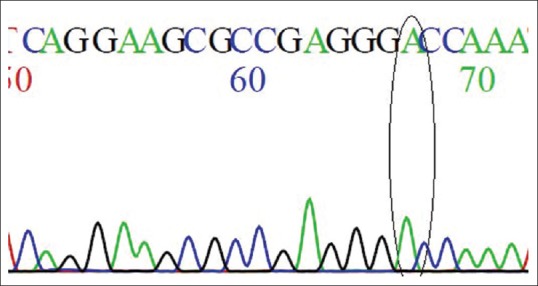
Homozygous A genotype, the predominant change in patients with respect to rs4236 as present in chromatogram
Figure 2.
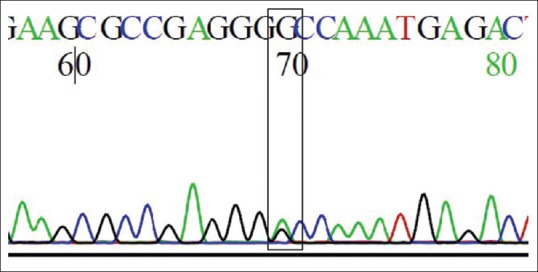
Heterozygous AG genotype, the predominant change in controls with respect to rs4236 as present in chromatogram
Discussion
Renal stone formation is the result of loss of equilibrium between the stone promoting factors and stone inhibiting factors. The mechanisms underlying extracellular calcification such as vascular calcification have been studied, and various proteins have been hypothesized to be the causative factor.[7] MGP gene is one of such genes encoding calcification inhibitory proteins.[3] Luo et al. showed that homozygous MGP-deficient mice were observed to die within 8 weeks as a result of arterial calcification that led to blood vessel rupture.[8] MGP was initially isolated from the matrix of bone but is now known to be synthesized in all tissues tested, with the highest levels of synthesis in heart, kidney, lung, and cartilage.[9] It is a vitamin K-dependent extracellular matrix protein. It acts as a molecular in determinant regulating the extracellular matrix calcification. It is an 84-amino acid gamma-carboxylated protein that binds and inhibits bone morphogenetic protein (BMP-2 and BMP-4). It has a high affinity for calcium and phosphate ions and hydroxyapatite crystals.[10]
Cancela et al. cloned a human genomic library using an MGP cDNA probe. They found that the human MGP gene spans 3.9 kb and consists of 4 exons separated by 3 large intervening sequences which account for more than 80% of the gene. The human genome contains a single copy of the gene in chromosome 12. Three of the exons of MGP correspond to the sequences of vitamin K dependent vertebrate proteins.[10]
In vitro studies suggest that SNPs in MGP are associated with altered promoter activity.[11,12,13] The SNP rs4236 is located in exon 4 of the human MGP gene, and the corresponding amino acid is located at the COOH terminus (Arg-Lys-Arg-Arg-Gly-Thr-Lys) of MGP.[6] The alteration of allele A to G leads to an amino acid change from Thr to Ala. The amino acid substitution in the COOH terminus may alter posttranslational modification of MGP, and affect MGP function due to charge or steric changes in the MGP molecule.
In the present study, the MGP gene polymorphism rs4236 was mainly investigated in 25 kidney stone patients and 25 healthy normal controls. The AA genotype was higher in kidney stone patients with A allele frequency of 0.8 and G allele frequency of 0.2 with respect to rs4236. The controls showed higher heterozygous genotype AG with an equal frequency of alleles A and G. AA genotype was present in 15 stone patients and none of the controls. Ten patients with the stone and all controls had AG genotype. This gives OR of 75 with P = 0.003, suggesting increased risk of stone disease with AA genotype. The OR value can be skewed due to one of the columns (AA genotype in controls) being null. This is mainly due to the small sample size. Regarding the allelic frequency OR, the presence of adenine increases the risk of stone disease with OR of 4.0 (P < 0.002).
Gao et al. studied MGP gene polymorphism among patients and controls. Out of 19 SNPs, they could identify, SNP rs4236 alone was significantly associated with renal stone disease. G allele to A allele translocation had a 2-fold risk in kidney stone patients.[5] Lu et al. published their study regarding the analysis of SNPs of MGP in patients with renal calculus and control group. They identified 18 polymorphisms. Among them, the Tag SNP rs4236 was associated with kidney stones. The G allele carrier had a 1.373-fold reduced kidney stone risk compared with A allele carriers in SNP rs4236.[6]
Although the increased risk and the very high OR may be due to the small sample size as described earlier, this study has provided some vital preliminary information linking the SNP rs4236 with kidney stone formation in this population for the first time. Further studies with larger sample size may be needed to know the exact OR and find a link between the SNP and the disease risk. This would help to counsel the patient better regarding the prognosis and possible gene therapy in the future.
To conclude, the genotype AA of SNP rs4236 of MGP gene was found to be higher in the kidney stone patients compared to the controls with increased frequency of allele A. The presence of this allele is a significant risk factor for urolithiasis. However, studies with larger sample sizes are needed to confirm this finding and to understand the effect of SNP rs4236 of MGP gene in kidney stone formation.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Trinchieri A. Epidemiology of urolithiasis. Arch Ital Urol Androl. 1996;68:203–49. [PubMed] [Google Scholar]
- 2.Margaret SP, lotan Y. Urinary lithiasis: Etiology, epidemiology, and pathogenesis. In: Wein AJ, Kavoussi LR, Novick AC, Artin AW, Peters CA, editors. Campbell-Walsh Urology. 10th ed. Philadelphia: Saunders; 2012. pp. 1257–86. [Google Scholar]
- 3.Jono S, Ikari Y, Vermeer C, Dissel P, Hasegawa K, Shioi A, et al. Matrix Gla protein is associated with coronary artery calcification as assessed by electron-beam computed tomography. Thromb Haemost. 2004;91:790–4. doi: 10.1160/TH03-08-0572. [DOI] [PubMed] [Google Scholar]
- 4.Yasui T, Fujita K, Sasaki S, Sato M, Sugimoto M, Hirota S, et al. Expression of bone matrix proteins in urolithiasis model rats. Urol Res. 1999;27:255–61. doi: 10.1007/s002400050119. [DOI] [PubMed] [Google Scholar]
- 5.Gao B, Yasui T, Itoh Y, Tozawa K, Hayashi Y, Kohri K, et al. A polymorphism of matrix Gla protein gene is associated with kidney stones. J Urol. 2007;177:2361–5. doi: 10.1016/j.juro.2007.01.118. [DOI] [PubMed] [Google Scholar]
- 6.Lu X, Gao B, Liu Z, Tian X, Mao X, Emmanuel N, et al. A polymorphism of matrix Gla protein gene is associated with kidney stone in the Chinese Han population. Gene. 2012;511:127–30. doi: 10.1016/j.gene.2012.09.112. [DOI] [PubMed] [Google Scholar]
- 7.El Asmar MS, Naoum JJ, Arbid EJ. Vitamin k dependent proteins and the role of vitamin k2 in the modulation of vascular calcification: A review. Oman Med J. 2014;29:172–7. doi: 10.5001/omj.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo G, Ducy P, McKee MD, Pinero GJ, Loyer E, Behringer RR, et al. Spontaneous calcification of arteries and cartilage in mice lacking matrix GLA protein. Nature. 1997;386:78–81. doi: 10.1038/386078a0. [DOI] [PubMed] [Google Scholar]
- 9.Price PA, Faus SA, Williamson MK. Warfarin-induced artery calcification is accelerated by growth and vitamin D. Arterioscler Thromb Vasc Biol. 2000;20:317–27. doi: 10.1161/01.atv.20.2.317. [DOI] [PubMed] [Google Scholar]
- 10.Cancela L, Hsieh CL, Francke U, Price PA. Molecular structure, chromosome assignment, and promoter organization of the human matrix Gla protein gene. J Biol Chem. 1990;265:15040–8. [PubMed] [Google Scholar]
- 11.Farzaneh-Far A, Davies JD, Braam LA, Spronk HM, Proudfoot D, Chan SW, et al. A polymorphism of the human matrix gamma-carboxyglutamic acid protein promoter alters binding of an activating protein-1 complex and is associated with altered transcription and serum levels. J Biol Chem. 2001;276:32466–73. doi: 10.1074/jbc.M104909200. [DOI] [PubMed] [Google Scholar]
- 12.Herrmann SM, Whatling C, Brand E, Nicaud V, Gariepy J, Simon A, et al. Polymorphisms of the human matrix Gla protein (MGP) gene, vascular calcification, and myocardial infarction. Arterioscler Thromb Vasc Biol. 2000;20:2386–93. doi: 10.1161/01.atv.20.11.2386. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi N, Kitazawa R, Maeda S, Schurgers L, Kitazawa S. T-138C polymorphism of matrix Gla protein promoter alters its expression but is not directly associated with atherosclerotic vascular calcification. Kobe J Med Sci. 2004;50:69–81. [PubMed] [Google Scholar]


