Abstract
The successful use of CRISPR/Cas9 based gene editing for therapeutics requires efficient in vivo delivery of the CRISPR components. There are, however, major challenges on the delivery front. In this Topical Review, we will highlight recent developments in CRISPR delivery, and we will present hurdles that still need to be overcome to achieve effective in vivo editing.
Graphical Abstract
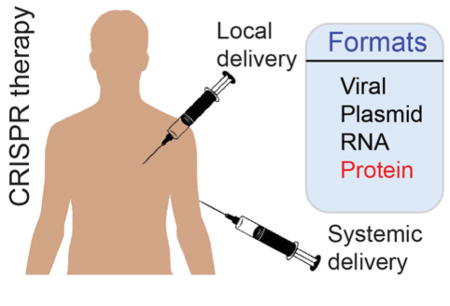
Different formats of CRISPR/Cas9 delivery through local or systemic injections for in vivo therapy.
INTRODUCTION
The use of bacterially-derived CRISPR/Cas9 systems (Clustered Regularly Interspaced Short Palindromic Repeat) to manipulate mammalian genomes presents enormous opportunities for curing human diseases.1–3 According to a report by National Institute of Health (NIH), of thousands of human diseases, only ~500 have any treatment.4 Many thousands of these diseases are caused by genetic alterations in the human genome. CRISPR technology enables correcting such genetic alterations, making a large number of these diseases therapeutic targets.
Bacterial CRISPR/Cas9 system is composed of two elements: a nuclease protein Cas9 that cuts double-stranded DNA, and a guide RNA molecule (sgRNA) that guides the Cas9 protein to a specific DNA sequence.5–9 This system (Cas9 protein and the gRNA, together called CRISPR/Cas9) has been harnessed in mammalian cells to specifically cut target genes, followed by repairing of the target gene via host cell repair machinery. The repair can occur through two basic mechanisms.10 (1) Non-Homologous End Joining (NHEJ): this mechanism allows the cell to randomly insert or delete nucleotides at the CRISPR-mediated double-stranded DNA break site, resulting in gene coding sequence disruption. (2) Homology-Directed Repair (HDR): this mechanism provides insertion of a template DNA to correct mutations at the DNA break site.
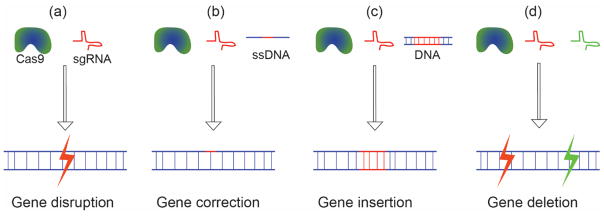
For therapeutic use the CRISPR components need to be delivered into mammalian cells to enable gene modification in the host cell. Once delivered, the CRISPR/Cas9 system can manipulate host cell genome in numerous ways. Depending on the desired genetic manipulation, various components of CRISPR/Cas9 are delivered: (a) a minimal Cas9/gRNA pair for gene disruption/mutation, (b) Cas9/gRNA, and a ‘spare’ template DNA for gene correction, (c) Cas9/gRNA, and a desired gene for gene insertion, and (d) Cas9 and two gRNAs for the complete deletion of a gene (or a portion of a gene) (Figure 1). In its simplest implementation the Cas9/gRNA pair is sufficient for gene disruption (i.e. knockout), however, the delivery of an additional piece of DNA is required for advanced functions such as gene repair or insertion (knock-in).
Figure 1.
Multiple components of CRISPR/Cas9 system are delivered into cells to achieve a specific function. (a) Cas9 and sgRNA for gene disruption (knock-out), (b) Cas9, sgRNA, and a template ssDNA for mutation correction, (c) Cas9, sgRNA, and a template DNA for gene insertion (knock-in), and (d) Cas9 and two sgRNAs for gene deletion.
In addition to the identity of the delivered components, the CRISPR/Cas9 constituents can be delivered into cells in different formats: plasmids or viral vectors that carrying Cas9 and sgRNA genes in gene-based delivery; Cas9 mRNA and a synthetic sgRNA in RNA-based delivery; Cas9 protein and a synthetic sgRNA in protein-based delivery (Table 1).11 All methods have their strengths, however Cas9 protein delivery has certain advantages over gene or RNA delivery methods. Protein delivery is transient and therefore is less immunogenic. Moreover, unlike gene delivery, protein delivery does not have the potential issue of permanently integrating CRISPR genes into the host genome (Table 1). Although a number of CRISPR delivery platforms have been created so far, the effective delivery of multiple CRISPR components in vivo into host cells still remains a major challenge. In this Topical Review, we will highlight the current in vivo therapeutic CRISPR delivery platforms, and address some of the challenges of CRISPR delivery.
Table 1.
Comparison, and pros/cons of CRISPR/Cas9 delivery in different formats. Compared to other format of delivery, Cas9-RNP delivery has the most advantage as it offers no insertional mutagenesis, high editing efficiency, low off-target, and low immunogenicity.
| Cas9 delivery in different formats | ||||
|---|---|---|---|---|
| Viral | Plasmid | RNA | Protein | |
| Insertional mutagenesis | High | Moderate | No | No |
| Editing efficiency | High | Moderate | Moderate | High |
| Off-target | Low | Moderate | Moderate | High |
| Immunogenicity | High | Moderate | Moderate | Low |
CURRENT IN VIVO CRISPR/CAS9 DELIVERY STRATEGIES
Both viral and non-viral approaches have been adopted for in vivo delivery of CRISPR/Cas9.11 Viral vectors have achieved success in delivery,12 with the most prominent being adenovirus and adeno-associated virus. Non-viral delivery of CRISPR/Cas9 in the form of plasmid DNA, Cas9 mRNA or Cas9 protein along with in vitro transcribed sgRNA has also emerged as a promising delivery strategy, with inherent strengths and challenges.
Viral delivery of CRISPR/Cas9
Adenovirus (AV) is an efficient transducing agent used for CRISPR/Cas9 mediated genome editing. This system has been used to demonstrate Mcc1 gene13 and Pcsk9 gene14 editing in adult mouse liver. Retro-orbital injection of CRISPR/Cas9 AVs resulted in disruption of Pcsk9 gene with ~50% of insertion and deletion mutation (indel) and 35–40% reduction of blood cholesterol level in mice. AV can be transduced in both dividing and non-dividing cells, and does not generally integrate into the host genome, however this vector can elicit a significant immune response in the host.15
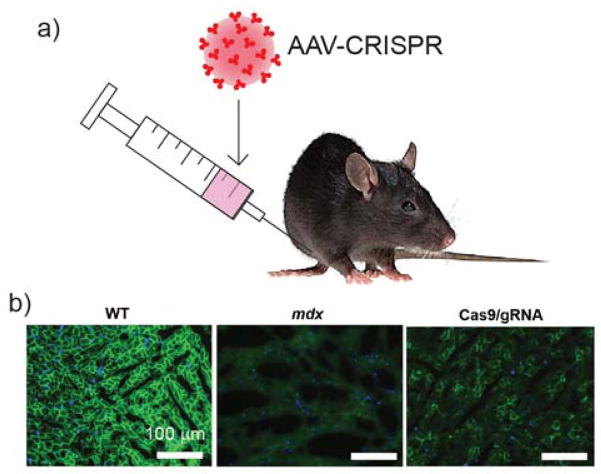
Adeno-associated virus (AAV) is the most widely used among the viral vectors due to its non-immunogenicity and its specificity towards a vast range of serotypes.16 Studies have been done to correct mutated dystrophin gene in Duchenne muscular dystrophy (Dmd) disease by CRISPR/Cas9 mediated NHEJ in muscle tissue after delivery of CRISPR components using AAV17 (Figure 2). In these studies, SpCas9 (Cas9 isolated from Streptococcus pyogenes bacteria) and sgRNA were delivered separately using two different AAV-vectors into postnatal mdx mice, a model of Dmd, via intramuscular, retro-orbital and intraperitoneal injections. The authors have reported excision of the defective dystrophin exon 23. This resulted in skipping of the premature stop codon and restoring the reading frame. The protein expressed was a shorter but more active form of dystrophin that enhanced skeletal muscle function in mice. Similar gene-editing studies for correcting Dmd in mdx mouse model using AAV vector has also been reported.18,19 However, in these studies, SpCas9 has been replaced by SaCas9 (Streptococcus aureus) due to its smaller size. SaCas9 and related sgRNAs were packed in AAV vector and injected via intramuscular or intraperitoneal injections. The results exhibited restoration of dystrophin gene reading frame in myofibers, cardiomyocytes and muscle stem cells with ~3% indel (insertion/deletion).19
Figure 2.
a) Strategy to deliver CRISPR/Cas9 components through viral delivery. b) AAV8-CRISPR delivery into Duchenne muscular dystrophy mice (mdx) to correct mutated dystrophin gene resulted restoration of dystrophin protein. Reprinted with permission from ref 19. Copyright 2016 Nature Publishing Group.
Non-viral delivery of CRISPR/Cas9
More recently, delivery of DNA components via hydrodynamic injection (HDI) has been used to bypass the challenges of viral delivery. Delivery of CRISPR components in plasmid format and a single stranded-DNA by tail vein HDI resulted in correction of Fah mutation in hepatocytes in a mouse model of tyrosinemia with a gene correction efficiency of 1 in 250 cells.20 Furthermore, this technique has also been used to inhibit hepatitis B virus (HBV) replication and expression in mice. CRISPR/Cas9 targeted to the surface antigen (HBsAg)-encoding region of HBV created a mutation in HBV DNA with 60–65% indel formation, disabling the virus in mice.21 There are also reports on sub-retinal injection of CRISPR components in plasmid format in combination with electroporation to disrupt the S334ter mutation in rhodopsin gene (RhoS334) in a rat model of severe autosomal dominant retinitis pigmentosa.22 The authors have reported cleavage efficiencies of 33–36% that resulted in improved retinal function and no retinal degeneration. In another report, CRISPR/Cas9 technology has been used to target P23H mutation in Rho gene for the same retinal defect in its mouse model.23 Sub-retinal electroporation of plasmid carrying Cas9 and sgRNAs demonstrated a significant reduction of mutated Rho protein.
As mentioned earlier, Cas9 protein delivery is a very suitable format for in vivo therapeutic applications. However, only a few Cas9-RNP delivery methods have been reported so far. Recently, Liu et al.24 have demonstrated topical delivery of Cas9/gRNA in the mouse inner ear using a cationic lipid-based nucleic acid transfection reagent (RNAiMAX). The authors reported 13% loss in GFP fluorescence at the injection site when Cas9/gRNA-RNAiMAX complexes (targeting GFP were injected in mouse cochlea. In another study, Gu et al. 25 have evaluated GFP disruption in U2OS-GFP bearing tumor mouse model by delivering Cas9/gRNA using DNA nanoclews (NCs). DNA NCs are DNA nanoparticles that are designed to be partially complimentary to the gRNA used. Additionally, the authors have coated the DNA NCs with cationic polymer polyethylenimine for endosomal escape. This study reports intratumoral injection of DNA NCs loaded with Cas9-RNPs that resulted in 25% loss of GFP fluorescence at the injection site. However, potential immunogenicity of the DNA NCs requires further validation before clinical translation can be attempted.
CHALLENGES
While a few delivery systems have achieved some level of in vivo therapeutic gene editing, efficient editing remains a challenge. Different challenges associated with efficient in vivo therapeutic gene editing are discussed below.
Packaging challenges
Packaging of CRISPR components into a single vector is a major challenge for therapeutic applications. As stated above, multiple components of CRISPR system are required to utilize the system. The packaging challenge is present in all the formats of delivery strategy i.e. gene, RNA, or protein-based delivery. For gene-based delivery through AAV, the size limit of a cargo gene is ~4.7-kilo base pair (kbp).26–28 However, the size of SpCas9 gene alone is ~4.3 kbp. Thus, inserting additional CRISPR components such as sgRNA, spare oligonucleotide or extra genes is challenging for single AAV vector-based CRISPR gene delivery.28 To overcome this problem, splitting Cas9 into two AAV vectors,29 or a smaller sized Cas9 has been demonstrated (SaCas9),30 however, their versatility for genome engineering applications remains to be investigated.
Although attractive, protein-based CRISPR delivery poses other challenges: while SpCas9 protein is a large protein (160 kDa, ~7.5 nm hydrodynamic diameter) with a net positive surface charge, sgRNA (~31 kDa, 5.5 nm hydrodynamic diameter) is negatively charged (~100 PO3− groups).31 Thus, packaging these elements through supramolecular chemistry may be a major limitation for designing delivery vehicles. Moreover, incorporation of additional spare DNA (of size in kbp) for multiple applications may further complicate the vector design.
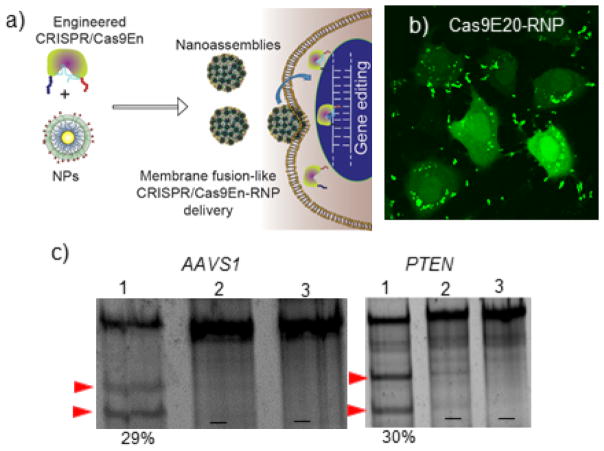
Recently, Rotello et al. have engineered Cas9 protein to carry a negative charge so that the protein electrostatically resembled sgRNA.31 In this study an oligo glutamic acid tag (E-tag) was fused to the N-terminus of Cas9 protein. The engineered Cas9En (n= number of glutamic acid) and the gRNA co-assembled with positively charged arginine gold nanoparticles (Arg-NPs) giving rise to a single delivery vector (Figure 3).31 This vector delivered Cas9En directly into cytoplasm and nucleus in ~90% of the cells grown in a culture dish. However, systemic in vivo applicability of this systems remains to be seen.
Figure 3.
Co-engineering of Cas9 nuclease and carrier nanoparticles facilitates the packaging of CRISPR components and thus delivery efficiency. a) Glutamic acid tagged (E-tagged) Cas9 protein (Cas9En) self-assembled with arginine functionalized gold nanoparticles (NPs) to form large nanoassemblies. The sgRNA was also packaged into these nanoassemblies. b) The resultant assemblies delivered FITC labelled Cas9En-RNP directly into cell cytoplasm/nucleus through a membrane fusion-like mechanism with concomitant gene editing. c) Indel gene editing of AAVS1 and PTEN genes in HeLa cells. [1—NP:Cas9E20-RNP; 2—Cas9E20-RNP only; 3—cells only. Indel percentage is given in parenthesis.] Reprinted with permission from ref 31. Copyright 2017 American Chemical Society.
Systemic delivery
Although systemic in vivo delivery of CRISPR/Cas9 has been achieved through viral vectors, no Cas9-RNP systemic delivery via synthetic vehicles has been reported so far. A few reports on local Cas9-RNP delivery have been published. Local delivery such as transdermal delivery offers advantage for certain applications, however, many therapeutic applications require systemic delivery.
Targeted delivery
Viral vectors can be used for targeted CRISPR/Cas9 delivery, as these vectors provide tissue tropism.32 However, non-viral delivery of CRISPR components will require targeting moieties such as peptides or antibodies.33 This targeting is particularly difficult to achieve, as incorporation of additional biomolecules to a delivery vector alongside the CRISPR components complicates the packaging.
Delivery and editing efficiency
CRISPR/Cas9 in vivo editing efficiency is significantly lower compared to in vitro editing. Anderson et al. found that hydrodynamic injection of CRISPR components resulted only 1 in 250 edited cells.20 In another example, local delivery of Cas9-RNP into mouse inner ear resulted in 20% GFP fluorescence loss. Such low editing percentage may be enough for alleviating certain diseases (e.g. muscular dystrophy, liver tyrosinemia), however, other diseases such as cancer require essentially 100% editing efficiency. Unfortunately, editing efficiency is also determined by delivery efficiency. In majority of the recently published CRISPR/Cas9 delivery research papers they did not mention the delivery efficiency. We have recently achieved Cas9-RNP delivery up to ~95% in cultured cells, however, in vivo delivery efficiency of this system has not yet been investigated.31
Off-target effect
One of the major limitations of CRISPR-based genome editing is its off-target effects.34,35 Even though the sgRNA is designed to target a specific gene of interest, often a significant number of non-specific genes are targeted by the same Cas9/sgRNA. In gene-based CRISPR delivery, the long-term constitutive expression of Cas9/sgRNA further makes the problem worse: repeated exposure of Cas9/sgRNA to non-specific genes can lead to large off-target effects. Different methods have been designed to reduce off-target effect including engineering high specificity Cas9 protein.36,37 However, off-target effects of these systems in vivo have not been fully explored. Protein-based CRISPR delivery, on the other hand, offers transient exposure of the host genome to the Cas9/sgRNA, that may result in reduced off-target events.38
Immunogenicity
Since Cas9 or other CRISPR-based genome editing proteins are derived from bacteria, these systems are expected to elicit host immune response. Specially, gene-based delivery of CRISPR elements can permanently integrate Cas9 gene into host cells. The constitutive expression of foreign Cas9 protein in the host cell will engage the MHC class I immune response which may result in the elimination of Cas9 expressing cells in the host.39 Indeed, Church and his colleague recently showed that AAV-based CRISPR delivery in vivo elicits a strong immune response against Cas9 protein but not against the vector itself.29 On the other hand, protein based CRISPR delivery system may offer minimal potential immunogenicity as delivered Cas9 protein will transiently present in the host cell (Table 1).
Insertional mutagenesis
Many viral vectors are incorporated into random locations in the genome causing mutagenesis of essential genes. Gene therapy trials with retroviruses to treat SCID (Severe combined immunodeficiency) led to leukemic transformation after integration of the virus into the host genome.40 Insertional mutagenesis due to the integration of genes near a protooncogene may lead to tumorigenesis, illustrating the danger of gene-based CRISPR therapy. Protein/RNA delivery based CRISPR therapy, on the other hand, avoids this problem and thus is an attractive alternative to gene therapy (Table 1).
CONCLUSIONS
Overall, CRISPR/Cas9 ribonucleoprotein delivery seems to be superior to gene delivery as it offers numerous advantages: transient delivery, no insertional mutagenesis, low immunogenicity, and low off target effect. As highlighted above, a very few Cas9-RNP in vivo delivery methods have been reported. Numerous challenges in the delivery front needed to be solved before translating this technology into clinics. These challenges, however, open up exciting opportunities in the CRISPR/Cas9 in vivo delivery front.
Acknowledgments
We acknowledge the University of Massachusetts Presidents’ Science and Technology Fund as well as the NIH (GM077173).
Footnotes
Notes: The authors declare no competing financial interest.
References
- 1.Doudna JA, Charpentier E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox DB, Platt RJ, Zhang F. Therapeutic Genome Editing: Prospects and Challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.https://ncats.nih.gov/files/NCATS-factsheet
- 5.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA Ribonucleoprotein Complex Mediates Specific DNA Cleavage for Adaptive Immunity in Bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho SW, Kim S, Kim JM, Kim JS. Targeted Genome Engineering in Human Cells with the Cas9 RNA-Guided Endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 10.Sander JD, Joung JK. CRISPR-Cas Systems for Editing, Regulating and Targeting Genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gori JL, Hsu PD, Maeder ML, Shen S, Welstead GG, Bumcrot D. Delivery and Specificity of CRISPR/Cas9 Genome Editing Technologies for Human Gene Therapy. Hum Gene Ther. 2015;26:443–451. doi: 10.1089/hum.2015.074. [DOI] [PubMed] [Google Scholar]
- 13.Cheng R, Peng J, Yan Y, Cao P, Wang J, Qiu C, Tang L, Liu D, Tang L, Jin J, et al. Efficient Gene Editing in Adult Mouse Livers via Adenoviral Delivery of CRISPR/Cas9. FEBS Lett. 2014;588:3954–3958. doi: 10.1016/j.febslet.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Ding Q, Strong A, Patel KM, Ng S-L, Gosis BS, Regan SN, Rader DJ, Musunuru K. Permanent Alteration of PCSK9 with in vivo CRISPR-Cas9 Genome Editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang AY, Peng PD, Ehrhardt A, Storm TA, Kay MA. Comparison of Adenoviral and Adeno-Associated Viral Vectors for Pancreatic Gene Delivery in vivo. Hum Gene Ther. 2004;15:405–413. doi: 10.1089/104303404322959551. [DOI] [PubMed] [Google Scholar]
- 16.Nelson CE, Gersbach CA. Engineering Delivery Vehicles for Genome Editing. Annu Rev Chem Biomol Eng. 2016;7:637–662. doi: 10.1146/annurev-chembioeng-080615-034711. [DOI] [PubMed] [Google Scholar]
- 17.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal Genome Editing Partially Restores Dystrophin Expression in a Mouse Model of Muscular Dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. In vivo Gene Editing in Dystrophic Mouse Muscle and Muscle Stem Cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Rivera RMC, Madhavan S, Pan X, Ran FA, Yan WX, et al. In vivo Genome Editing Improves Muscle Function in a Mouse Model of Duchenne Muscular Dystrophy. Science. 2015;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome Editing with Cas9 in Adult Mice Corrects a Disease Mutation and Phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY, Dong LH, Song HF, Gao X. Harnessing the Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-Associated Cas9 System to Disrupt the Hepatitis B Virus. Gene Ther. 2015;22:404–412. doi: 10.1038/gt.2015.2. [DOI] [PubMed] [Google Scholar]
- 22.Bakondi B, Lv W, Lu B, Jones MK, Tsai Y, Kim KJ, Levy R, Akhtar AA, Breunig JJ, Svendsen CN, Wang S. In vivo CRISPR/Cas9 Gene Editing Corrects Retinal Dystrophy in the S334ter-3 Rat Model of Autosomal Dominant Retinitis Pigmentosa. Mol Ther. 2016;24:556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latella MC, Di Salvo MT, Cocchiarella F, Benati D, Grisendi G, Comitato A, Marigo V, Recchia A. In vivo Editing of the Human Mutant Rhodopsin Gene by Electroporation of Plasmid-based CRISPR/Cas9 in the Mouse Retina. Mol Ther Nucleic Acids. 2016;5:e389. doi: 10.1038/mtna.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. Cationic Lipid-Mediated Delivery of Proteins Enables Efficient Protein-Based Genome Editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL, Gu Z. Self-Assembled DNA Nanoclews for the Efficient Delivery of CRISPR–Cas9 for Genome Editing. Angew Chem Int Ed. 2015;54:12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Z, Yang H, Colosi P. Effect of Genome Size on AAV Vector Packaging. Mol Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong B, Nakai H, Xiao W. Characterization of Genome Integrity for Oversized Recombinant AAV Vector. Mol Ther. 2010;18:87–92. doi: 10.1038/mt.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Senís E, Fatouros C, Große S, Wiedtke E, Niopek D, Mueller AK, Börner K, Grimm D. CRISPR/Cas9-Mediated Genome Engineering: An Adeno-Associated Viral (AAV) Vector Toolbox. Biotechnol J. 2014;9:1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 29.Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM. A Multifunctional AAV-CRISPR-Cas9 and its Host Response. Nat Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo Genome Editing using Staphylococcus Aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mout R, Ray M, Tonga GY, Lee Y-W, Tay T, Sasaki K, Rotello VM. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 2017 doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV Serotypes 1–9 Mediated Gene Expression and Tropism in Mice After Systemic Injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 33.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an Emerging Platform for Cancer Therapy. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 34.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of Off-Target Effects of CRISPR/Cas-Derived RNA-Guided Endonucleases and Nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-Frequency Off-Target Mutagenesis Induced by CRISPR-Cas Nucleases in Human Cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-Fidelity CRISPR-Cas9 Nucleases with no Detectable Genome-Wide Off-Target Effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally Engineered Cas9 Nucleases with Improved Specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramakrishna S, Kwaku DAB, Beloor J, Gopalappa R, Lee SK, Kim H. Gene Disruption by Cell-Penetrating Peptide-Mediated Delivery of Cas9 Protein and Guide RNA. Genome Res. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a Systems Understanding of MHC Class I and MHC Class II Antigen Presentation. Nat Rev Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 40.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, Beldjord K, et al. Insertional Oncogenesis in 4 Patients After Retrovirus-Mediated Gene Therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]