Abstract
Background. Cryptosporidium is a protozoan parasite and a major cause of diarrhea in children and immunocompromised patients. Current diagnostic methods for cryptosporidiosis such as microscopy have low sensitivity while techniques such as PCR indicate higher sensitivity levels but are seldom used in developing countries due to their associated cost. A loop-mediated isothermal amplification (LAMP) technique, a method with shorter time to result and with equal or higher sensitivity compared to PCR, has been developed and applied in the detection of Cryptosporidium species. The test has a detection limit of 10 pg/µl (~100 oocysts/ml) indicating a need for more sensitive diagnostic tools. This study developed a more sensitive lateral flow dipstick (LFD) LAMP test based on SAM-1 gene and with the addition of a second set of reaction accelerating primers (stem primers). Results. The stem LFD LAMP test showed analytical sensitivity of 10 oocysts/ml compared to 100 oocysts/ml (10 pg/ul) for each of the SAM-1 LAMP test and nested PCR. The stem LFD LAMP and nested PCR detected 29/39 and 25/39 positive samples of previously identified C. parvum and C. hominis DNA, respectively. The SAM-1 LAMP detected 27/39. On detection of Cryptosporidium DNA in 67 clinical samples, the stem LFD LAMP detected 16 samples and SAM-2 LAMP 14 and nested PCR identified 11. Preheating the templates increased detection by stem LFD LAMP to 19 samples. Time to results from master mix preparation step took ~80 minutes. The test was specific, and no cross-amplification was recorded with nontarget DNA. Conclusion. The developed stem LFD LAMP test is an appropriate method for the detection of C. hominis, C. parvum, and C. meleagridis DNA in human stool samples. It can be used in algorithm with other diagnostic tests and may offer promise as an effective diagnostic tool in the control of cryptosporidiosis.
1. Background
Cryptosporidiosis is caused by a group of phenotypically, and genotypically diverse Cryptosporidium species [1] and transmission of infection occurs when an individual comes in contact with infective oocyst(s) via contaminated food, water, person-to-person contact [2–4], and contact with animals. Transmission is common in developing countries due to poor sanitation and limited access to safe drinking water. The disease affects enterocytes of the small intestines and is a major cause of diarrhea, hospitalization, malnutrition in children [5–7] and can be fatal among immune-compromised persons [3, 4]. The ability of low doses of oocysts to cause infection following exposure and the absence of sensitive and effective diagnostic tools and treatment regime make cryptosporidiosis a major public health concern [5, 8].
In Sub-Saharan Africa and South Asia where most diarrheal disease deaths occur, there is limited data on the burden of Cryptosporidium diarrheal cases [9]. Nevertheless, the prevalence of the infection is thought to range from 10 to 33% among Human Immunodeficiency Virus (HIV) infected adult persons [10] and can even be higher in children. Indeed, findings from an Ethiopian based study indicated the presence of Cryptosporidium species in 26.9% of HIV-positive patients [11], while a prevalence of 73.6% was recorded among HIV-positive children in Uganda presenting with persistent diarrhea [12]. Kenyan based studies involving children have revealed a higher cryptosporidiosis prevalence of up to 30.5% among HIV-infected participants compared to noninfected participants in the same cohort [5, 13]. The enormous Cryptosporidium disease burden among immune-compromised children warrants development of sensitive diagnostics tools among other control tools.
Routine diagnosis of cryptosporidiosis in Kenya and in most of the developing world is based on microscopic detection of Cryptosporidium oocysts in stool samples despite the method's low sensitivity and inability to distinguish between pathogenic and nonpathogenic Cryptosporidium species [14]. There are over 20 species and numerous genotypes of Cryptosporidium of which C. hominis and C. parvum are the most common species infecting humans [9, 15]. Several tests have been developed to detect pathogenic strains and among them are ELISA and immune chromatography [16]. Others include pathogen DNA-based tests such as real-time PCR [17], single strand conformation polymorphism [18], and restriction fragment length polymorphism [19]. PCR tests have indicated superior sensitivity and specificity compared to antigen-based tests. However, the method has limited use in the routine diagnosis of cryptosporidiosis in Kenya due to its associated costs.
In the last decade, loop-mediated isothermal amplification (LAMP) has emerged as a powerful technique for the diagnosis of parasites of both medical and veterinary importance [20]. The method is robust and can amplify DNA from partially processed samples [21]. It has a shorter time to results compared to PCR, and the large amounts of reaction products formed allow the use of various detection formats [22, 23] making it ideal for application under field conditions. LAMP has been used in the diagnosis of protozoan parasitic pathogens such as Plasmodium falciparum [24], human infective Trypanosoma rhodesiense [21] and pathogens found in stool such as Ascaris lumbricoides [25] and Necator americanus [26]. Various methods have been used to detect LAMP products [27] in particular; the use of lateral flow dipstick (LFD) method offers high test specificity and visual inspection of results. This is because the detection probe targets a specific complementary sequence in the product and the results appear as presence or absence of a line on LFD stick [28, 29]. This makes LFD ideal for field application. Previously, LAMP tests for Cryptosporidium species have been designed and evaluated based on the heat shock protein 70 (HSP 70) gene, glycoprotein 60 (GP60), and the S-adenosyl-methionine synthetase-1 (SAM-1) gene [1, 15]. Among the three tests, the SAM-1 LAMP test indicated a higher sensitivity level by detecting 10 pg/μl compared to 1 ng/μl for the LAMP targeting GP60 gene [15]. These detection levels are inadequate for deployment of LAMP tests for routine diagnosis of cryptosporidiosis. Therefore, considering the advantages of LAMP and its potential as a diagnostic tool in rural endemic setups, there is a need to explore ways of improving the diagnostic capacity of cryptosporidiosis LAMP tests.
Recent studies have indicated that it is possible to improve the detection limits of some LAMP tests through the addition of a second set of reaction accelerating primers (stem primers) that target the stem section of the LAMP amplicon [30]. The advantage of stem primers is that they can be used in addition to loop primers without affecting the LAMP test reproducibility [30]. Here we report an improved LAMP test for the detection of human infective Cryptosporidium species that uses stem primers and detection of the LAMP products using a lateral flow dipstick format.
2. Methods
2.1. Clinical Samples and DNA Extraction
This study used a total of 39 archived DNA samples determined as C. parvum and C. hominis through sequencing of Cryptosporidium GP60 gene [6]. The DNA were extracted from stool samples obtained from children aged 5 years and below presenting with diarrhea at the pediatric ward of Mbagathi district hospital and from three participating clinics (Lea Toto, Medical Missionaries of Mary, and Reuben Center) within Mukuru slums, Nairobi County [5]. To evaluate the developed LAMP test, 67 DNA samples were prepared from stool samples collected from children who presented with diarrhea in the same study area. The stool samples were preserved in 2.5% potassium dichromate and stored at −80°C. Genomic DNA was processed from the preserved specimen using the QiAmp® DNA Stool Mini Kit (Qiagen, West Sussex, United Kingdom) as per the manufacturer instructions with slight modifications. Briefly, 200 μl of fecal suspension was washed five times with triple-distilled water by centrifugation at 14,000 rpm for 5 minutes per cycle. To this suspension 1.4 ml of ASL buffer was added and subjected to five times' thawing at 80°C and freezing at −80°C to rupture the oocysts. A 100 μl suspension was used for DNA extraction, and the resulting genomic DNA was then eluted in 50 μl of nuclease-free water and stored at −20°C until use.
2.2. Design of LAMP Primers and Probe
The sequence section of SAM-1 gene for three common species of Cryptosporidia affecting humans in Kenya C. parvum (AB119646.1), C. hominis (X662396.1), and C. meleagridis (AB119648.1) were obtained from Genbank. The gene target was selected based on the findings of a recent study that indicated that LAMP test developed based on the SAM-1 gene achieved the best sensitivity levels [15]. The sequences were aligned using Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/) and the most conserved sequence section determined and used to design LAMP primers. Briefly, the forward and backward outer primers (F3/B3) and the forward and backward inner primers (FIP/BIP) were designed using the primer explorer version 3 software (https://primerexplorer.jp/e/). The loop forward and backward primers (LF/LB), stem forward and backward primers (SF/SB), and the probe were designed manually following published conditions [29, 30]. All the primers were checked for target specificity using the nucleotide basic local alignment search tool (BLASTn) https://blast.ncbi.nlm.nih.gov/Blast.cgi.
2.3. Optimization of LAMP Reactions
The designed LAMP primers were first analyzed for sensitivity using a 10-fold serial dilution of C. hominis and C. parvum reference DNA initially prepared from 107 oocysts suspended in 10 ml of fecal materials and using standard LAMP conditions [32] to select the most sensitive primer set (Table 1). This was followed by the optimization of reaction components (reagents) using the Taguchi method to select the optimum concentration of the four reagents determined to have the greatest effect on LAMP reaction. Briefly, the reaction components concentration was varied at three levels and ranged within 30–60 pmoles for inner primers, 10–30 pmoles for loop primers, 10–30 pmoles for stem primers, and 1–3 mM for dNTPs. The concentrations variables were then arranged in an orthogonal array and used to determine the amount of LAMP product formed [33]. This was followed by regression analysis to determine the concentration optima for each selected reaction component [33]. To select the optimal reaction temperature, LAMP reactions were performed at 61, 63, and 65°C, respectively. The tests were run for 30–60 minutes to obtain products. The LAMP products were analyzed using electrophoresis in 2% agarose gel.
Table 1.
Nucleotide sequences for Cryptosporidium hominis and parvum for stem SAM-1 LAMP test. Degenerate primers have been underlined and made bold.
| Target | Primer name | Sequence (5′-3′) | Bases | Final amplicon size |
|---|---|---|---|---|
| SAM-1 | F3 | GAGGATGGGGTGCTCATGG | 19 | 220 bp |
| B3 | CCTTATTAACTATCTCCAGYAG | 22 | ||
| FIP | GACTTTGCAACAAGYCTTGCCAGCATTTAGCGGGAAAGATG | 41 | ||
| BIP | ATTGGAATAGCAARGCCTTTATCGTCATTATACCCATCTTTCGC | 44 | ||
| LF | CRCCTGAYCTATCTACTTTAG | 21 | ||
| LB | CTRTATATTAATACATTTGGCAC | 23 | ||
| SF | TACACAAKCCAGAAAAGACG | 20 | ||
| SB | TGTTTGGTRCAGGTTTCATATG | 20 | ||
| Probe | CTTGTGTAGCAGATGTTTGGTACAGG |
26 |
2.4. Lateral Flow LAMP Reactions
The selected LAMP primers (Table 1) and determined reaction concentrations were used with the forward inner primer being labeled with biotin in the 5′-end and the probe for detecting biotinylated LAMP product labeled with fluorescein isothiocyanate (FITC). Briefly, the Taguchi method determined the concentrations for inner primers at 44 pmoles, with both stem and loop primers 20 pmoles and dNTPs at 2 mM. The concentration of other reagents was as reported previously [20]. The template was 2 μl for both C. hominis and C. parvum DNA. The reactions were done at 63°C for 60 minutes followed by reaction inactivation at 80°C for 5 minutes. After the LAMP reaction, the LFD hybridization was performed by incubating LAMP products with 20 pmol of FITC-labeled probe at 63°C for 5 min in a final volume of 20 μl followed by the addition of 8 μl of the reaction mixture and 150 μl of the reaction assay buffer. The LFD strips (Millennia® HybriDetect, Millennia Biotec, Germany) were then dipped into the mixture for 5 min at room temperature. The test was considered positive when both the control and test lines appeared. The size of the obtained amplicon was 220 bp. The experimental test was labeled stem LFD-LAMP. The reactions were duplicated using an opened heating block that maintained the temperature at ~62-63°C.
2.5. Stem LFD-LAMP and Nested PCR Analytical Sensitivity and Analysis of Clinical Samples
The analytical sensitivity of the stem LFD-LAMP test was determined using tenfold serial dilution of C. hominis and C. parvum DNA. A sequenced C. parvum and C. hominis DNA [6] were used as positive controls. The C. meleagridis DNA was not used in sensitivity analysis because the concentration was very low. Two formats of stem LFD-LAMP tests (i.e., with outer F3/B3 primers and without outer primers) were compared with the SAM-1 LAMP test [15] and nested PCR targeting Cryptosporidium species small subunit rRNA [31] (Table 2). All clinical samples were analyzed in duplicate and repeated once after two weeks. To check whether the stem LFD-LAMP format analytical sensitivity could be improved further, preheated templates were used. Briefly, the LAMP master mix was divided into 25 μl reaction tubes and placed in the incubation chamber at ~63°C. After approximately 3 minutes, 2 μl of preheated template (genomic DNA) was added to each respective tube and reactions were left to run for 60 minutes. To check time to results for different LAMP formats, a dilution of 10−4 (~1000 oocysts/ml) of reference C. hominis DNA was used, and reactions ran for 25, 30, 35, 40, and 45 minutes. For each time schedule, the reaction tubes were transferred to a thermal block set at 80°C to stop the reaction. For SAM-1 LAMP and nested PCR, the expected products were analyzed using 2% agarose gel. The stem LFD-LAMP test specificity was checked using Toxoplasma gondii, Giardia duodenalis, Entamoeba histolytica, Ascaris lumbricoides, Cyclospora species, and human DNA.
Table 2.
The analytical sensitivity of stem LFD LAMP test formats, SAM-1 LAMP, and PCR tests using a 10-fold serial dilution of C. hominis DNA.
| Test | Primer combination | Probe | 10-fold serial dilution | Result (Min)e | Remarks | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 10−1 to 3 | 10−4d | 10−5 | 10−6 | 10−7 | 10−8 | |||||
| Stem LFD LAMPa | F3/B3, FIP/BIP, LF/LB, SF/SB | FITC | + | + | + | + | ±† | − | 30 | This study |
| Stem LFD LAMPb | FIP/BIP, LF/LB, SF/SB | FITC | + | ± | − | − | − | − | 40 | This study |
| LAMPc | F3/B3, LF/LB, FIP/BIP | N/A | + | ± | − | − | − | − | 40 | This study |
| SAM-1 LAMP | F3/B3, LF/LB, FIP/BIP | N/A | + | + | + | − | − | − | 35 | [15] |
| Nested PCR test | F1/R1; F2/R2 | N/A | + | + | + | − | − | − | N/A | [31] |
aLAMP test with outer F3/B3 primers; bLAMP test without outer F3/B3 primers. cPrimers designed in this study. dThe reaction done using 10−4 (1000 oocysts/ml). eTime to recording a positive reaction. †Approximately 30% of the replicates were consistently positive with preheated template and sequencing; N/A: not applicable.
2.6. Detection and Confirmation of Stem LAMP and Nested PCR Product
The formation of LAMP product was first monitored through gel electrophoresis in 2.0% agarose gel and through addition of 1/10 dilution of SYBR® Green 1 dye. Later the product detection was exclusively done using the lateral flow dipstick format. Two approaches were used to confirm that the LAMP test amplified the predicted product, namely, the restriction enzyme digestion and sequencing. The restriction enzyme Ndel (New England BioLabs, MA, USA) which has a single cut site within the selected product sequence was used to digest LAMP product at 37°C for 3 h, followed by electrophoresis in 3% agarose gel. The predicated two bands were sized with molecular markers. Secondly, the uppermost LAMP amplicon were excised from the agarose gel, cloned, and transformed and inserts sequenced using an automated DNA 3730 analyzer. The resulting sequences were aligned with the target sequences using the DNAman computer software (Lynnon, USA). In nested PCR, the two-step restriction digestion of the secondary PCR products was carried out using endonucleases SspI and VspI and Cryptosporidium species and genotypes determined analyzed as described previously [34].
3. Results
3.1. Detection and Confirmation of LAMP Products
The positive stem LAMP products showed ladder-like pattern on the agarose gel indicating the formation of stem-loops with inverted repeats (Figure 1(a)) and the expected test line on the LFD strip (Figure 1(b)). Occasional nonspecific products were noted in the agarose gels if the LAMP reactions were run for over 70 minutes and which turned green (false positive) on the addition of SYBR Green 1 dye (Figures 2(a) and 2(b)). These bands varied in patterns from reaction to reaction while consistent patterns were recorded for true positive samples (Figure 1(a)). The false positive samples were not detectable with the LFD strips (Figures 2(a) and 2(b)). The restriction enzyme digestion of the stem LAMP amplicons indicated the predicted amplicons of approximately 117 bp and 103 bp, respectively (Figure 3(a)). The sequence from the uppermost amplicon of randomly selected stem LAMP positive samples indicated high sequence homology with SAM gene sequences from C. hominis and C. parvum (Figure 3(b)). No restriction enzyme digestion was recorded from samples with inconsistent banding patterns. The LAMP test was reproducible using the open heat block, and no cross-reactivity was recorded with nontarget DNA from Toxoplasma gondii, Giardia duodenalis, Entamoeba histolytica, Ascaris lumbricoides, Cyclospora spp., or human DNA.
Figure 1.
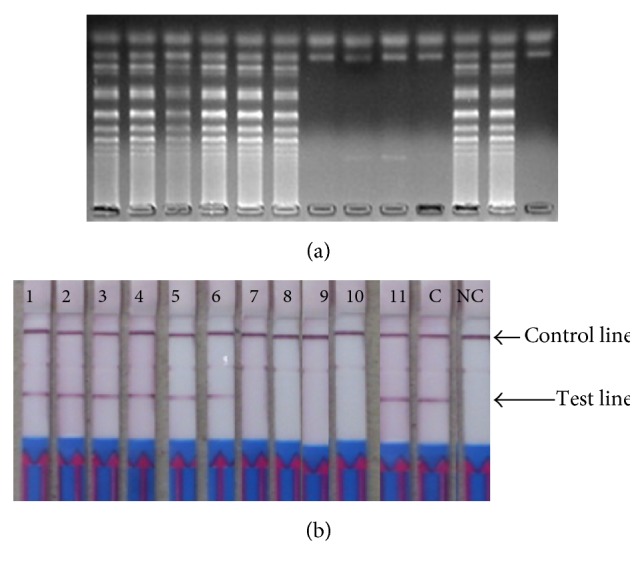
The detection Cryptosporidium spp. using the stem LAMP amplification product (220 bp) using 2.0% agarose gel stained with ethidium bromide and LFD format. The genomic DNA was prepared from specimen collected from children presenting with diarrhea. The faint line between the test line and the positive control line is nonspecific binding at DIG test line because the strips were done to detect two products. 1 = MB407, 2 = MB419, 3 = MB491, 4 = MB501, 5 = MB502, 6 = M1492, 7 = M1599, 8 = M009, 9 = M016, 10 = M044, 11 = M074, C = C. hominis DNA, and NC = PCR water.
Figure 2.
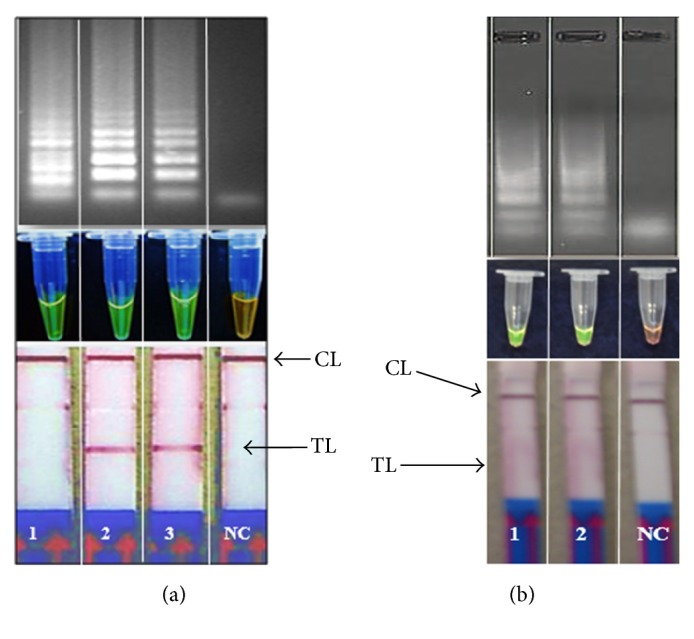
The detection of stem LAMP product from some selected reactions done for over 60 minutes using 2.0% agarose gels stained with ethidium bromide, SYBR Green 1 dye, and LFD dipstick format. 1 = (false positive), 2 = C. hominis, 3 = C. parvum, and NC = PCR water. (b) The appearance of nonspecific products at 75 minutes' reaction cut-off time. 1, M044 (false positive), 2, M099 (false positive), and NC = PCR water. TL = test line; CL = control line. The nonspecific products show different patterns agarose gel and turn green on addition of SYBR Green 1. However, none was positive using the LFD format.
Figure 3.
3.2. Analytical Sensitivity of LAMP and Nested PCR Tests
The stem LFD-LAMP indicated unequivocal detection limit of 10 oocysts/ml using the 10-fold serial dilution of the C. hominis and C. parvum reference DNA. However, an average of 2 of 6 replicates in every run, or approximately 30% of the replicates consistently showed detection limit of ~1 oocyst/ml when the template was preheated (Tables 2 and 3). The SAM-1 LAMP test and the nested PCR indicated detection limit of 100 oocysts/ml (Table 2). Time to results from master mix preparation was 80 minutes for the stem LFD-LAMP format and 120 minutes for SAM-1 LAMP test using gel electrophoresis (Table 3).
Table 3.
The comparative analysis of stem LFD LAMP, SAM-1 LAMP, and nested PCR test in the detection of previously confirmed and archived C. hominis and C. parvum DNA samples and clinical samples.
| Indices | Types of test | |||
|---|---|---|---|---|
| Stem LFD-LAMP | SAM-1 LAMPa | Nested PCRb | ||
| Cryptosporidium spp. DNA (N = 39) | Number of positive samples | 29 (74.4%) | 27 (69.2%) | 25 (64.1%) |
|
| ||||
| Clinical samples (N = 67) | Number of positive samples | 16 (23.9%); 19 (28.4%)c | 14 (20.8%)† | 11 (16.4%)† |
| Time to results (Min)f | 80 | 120d | 320d | |
| Accelerating primers | Loop and stem | Loop | nd | |
3.3. Clinical Samples Result
The stem LFD-LAMP and nested PCR detected 29/39 and 25/39 positive samples of previously identified C. parvum and C. hominis DNA, respectively. The SAM-1 LAMP detected 27/39 (Table 3). On detection of Cryptosporidium DNA in 67 clinical samples, the stem LFD-LAMP detected 16 samples and SAM-2 LAMP 14 and nested PCR identifies 11. Preheating the templates increased detection by stem LFD-LAMP to 19 samples.
4. Discussion
Loop-mediated isothermal amplification of DNA is a method that has gained momentum in the diagnosis of different microorganisms due to its inherent advantages of high sensitivity and specificity and its potential applicability in resource-poor endemic areas [20]. In this study, we successfully used a second set of reaction accelerating primers (stem primers) combined with a lateral flow dipstick format to design a sensitive LAMP test capable of detecting C. hominis, C. parvum, and C. meleagridis based on the SAM-1 gene. The stem LFD-LAMP test indicated a shorter time to results, higher analytical sensitivity, and better comparative analysis, a characteristic that translated to superior detection of pathogen DNA in clinical specimen (Table 3) when compared to SAM-1 LAMP and nested PCR. The higher detection levels of stem LFD-LAMP test may be attributed to the use of two reaction accelerating primers (loop and stem primers) which lead to formation of larger amounts of product compared to the SAM-1 LAMP format which relies only on loop primers. The loop primers accelerate the reaction by priming the sequence loops [35] while the stem primers accelerate the reaction by targeting the stem section of the sequence [30]. The use of preheated template marginally improved the stem LFD-LAMP test sensitivity by 10-fold and detection of pathogen DNA by ~4.6% from clinical specimen compared to SAM-1 LAMP test (Tables 2 and 3). It can be assumed that preheating of the template unwinds target DNA and hence provides more target for priming. Moreover, heating accelerates betaine destabilization of the target DNA bonds, hence easier displacement by the outer primers. The omission of outer F3 and B3 primers in this stem LAMP format indicated poor test performance (Table 2) confirming that outer primers have varied effects on different stem LAMP tests [30]. The higher detection of rates of Cryptosporidium DNA from clinical samples compared to SSU rRNA nested PCR agrees with previous results [15, 36].
There was a general agreement in the detection of the stem LAMP products using gel electrophoresis and LFD (Figures 1(a) and 1(b)). However, SYBR Green 1 dye could not differentiate some false positive products limiting its use as detection format in this assay. This is because intercalating dyes bind to any double stranded DNA including the primer dimers (Figures 2(a) and 2(b)) leading to erroneous results interpretation. The appearance of false positive products was further confirmed by failure of digestion using restriction enzyme Ndel compared to the positive products that gave the predicted amplicons of 117 bp and 103 bp, respectively (Figure 3(a)). Theoretically, LAMP test should not amplify nonspecific products since amplification specificity is supposedly enhanced by using several primers. Nevertheless, spurious products are formed if the test is not optimized or left to run for too long (Figures 2(a) and 2(b)). Determination of nonspecific products is valuable since their presence reduces the amplification efficiency and ultimately the accuracy of the test. Since most LAMP product detection formats are developed for visual inspection of results, a product confirmation step ought to be built into the test development protocol and/or a specific detection product format is recommended [29]. In this regard, the designed LFD format in this study showed superior specificity to the intercalating dyes. The dipstick format relies on a specific DNA sequence probe that binds to a specific complementary sequence in LAMP product. The lateral flow strips used in this study have dual detection ability for FITC- and DIG-labeled products, but only the FITC was used (Figure 1, Table 2). There is a nonspecific faint line at DIG section (Figure 1) that does not affect the results interpretation. Nonetheless, it indicates the need of using specific FITC-labeled strips only.
All samples that were positive with nested PCR and SAM-1 LAMP test were also positive with stem LFD-LAMP test (Table 3) indicating that the tests were detecting the same thing. Moreover, specificity of the stem LAMP test was confirmed through sequencing of product from four samples (Figure 3(a)). The stem LFD-LAMP assay described here can further be improved by using a dipstick cartridge which allows insertion of the sample followed by a locking mechanism that cuts and pours the product directly into the LFD strip. The development of such technologies will eliminate the need to open the tube and potentially reduces contamination. Since stem LFD-LAMP test is faster to perform, the technique could form part of diagnostic algorithms for Cryptosporidium species detection where it can be used to select cases for further analysis.
5. Conclusion
This work reports the use of stem primers and lateral flow dipstick format to improve the detection of Cryptosporidium oocyst DNA from stool samples. The LFD format showed superior specificity in detection of the target DNA compared to DNA intercalating dyes and without compromising the test sensitivity. To advance the LFD format, a novel single-step reaction that will allow direct detection of product with the LFD strips without necessarily opening the tube needs to be considered. Such integration of key technologies will contribute towards making stem LFD-LAMP a suitable complementary test to the current tests (microscopy and PCR) used in the detection of cryptosporidiosis, especially in resource-poor countries.
Acknowledgments
The authors would like to acknowledge the provision of materials through Meru University of Science and Technology, Kenya, and Murdoch University, Australia. The views expressed in this article do not reflect the views of their respective institutions.
Abbreviations
- LAMP:
Loop-mediated isothermal amplification
- FITC:
Fluorescein isothiocyanate
- LFD:
Lateral flow dipstick
- SAM-1:
S-Adenosyl-methionine -1
- HIV:
Human Immunodeficiency Virus
- FIP/BIP:
Inner primer
- LF/LB:
Loop primers
- SF/SB:
Stem primers
- GP 60:
Glycoprotein 60
- HSP 70:
Heat shock proteins 70v.
Data Availability
The datasets supporting the conclusions of this article are included within the article.
Ethical Approval
The study was approved by the Scientific Ethics Review Unit of the Kenya Medical Research Institute (KEMRI) (SSC no. 2891).
Consent
All parents and/or guardians of participating children were informed of the study objectives and voluntary written consent was sought and obtained before inclusion into the study. A copy of the signed consent was filed and stored in a password protected cabinet at KEMRI and was only accessible to the principal investigator. The samples were analyzed anonymously.
Disclosure
Meru University of Science and Technology, Kenya, and Murdoch University, Australia, had no role in study design, data collection and analysis, or preparation of the manuscript. The views expressed in this article do not reflect the views of these institutions. This article is published with permission from the Director, KEMRI.
Conflicts of Interest
The authors declare that they have no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Zablon K. Njiru, Gitonga Nkanata Mburugu, and Timothy S. Mamba conceived of and designed the study protocol. Timothy S. Mamba, Erastus Mulinge, and Cecilia K. Mbae performed data collection and analysis. Zablon K. Njiru performed data presentation. Timothy S. Mamba drafted the manuscript. Zablon K. Njiru, Cecilia K. Mbae, Johnson Kinyua, Erastus Mulinge, and Gitonga Nkanata Mburugu critically reviewed the manuscript. Coordination and supervision of the project were conducted by Cecilia K. Mbae, Johnson Kinyua, and Gitonga Nkanata Mburugu. All authors read and approved the final manuscript.
References
- 1.Karanis P., Thekisoe O., Kiouptsi K., Ongerth J., Igarashi I., Inoue N. Development and preliminary evaluation of a loop-mediated isothermal amplification procedure for sensitive detection of Cryptosporidium oocysts in fecal and water samples. Applied and Environmental Microbiology. 2007;73(17):5660–5662. doi: 10.1128/aem.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koloren Z., Sotiriadou I., Karanis P. Investigations and comparative detection of Cryptosporidium species by microscopy, nested PCR and LAMP in water supplies of ordu, middle Black Sea, Turkey. Annals of Tropical Medicine and Parasitology. 2011;105(8):607–615. doi: 10.1179/2047773211Y.0000000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snelling J., Xiao L., Ortega-Perres G., Lowery J., Moore E., Rao R., et al. Cryptosporidiosis in developing countries. J Infect Dev Ctries. 2007;1(3):242–256. [PubMed] [Google Scholar]
- 4.Shirley D.-A. T., Moonah S. N., Kotloff K. L. Burden of disease from cryptosporidiosis. Current Opinion in Infectious Diseases. 2012;25(5):555–563. doi: 10.1097/QCO.0b013e328357e569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mbae C. K., Nokes D. J., Mulinge E., Nyambura J., Waruru A., Kariuki S. Intestinal parasitic infections in children presenting with diarrhoea in outpatient and inpatient settings in an informal settlement of Nairobi, Kenya. BMC Infectious Diseases. 2013;13(1, article 243) doi: 10.1186/1471-2334-13-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mbae C., Mulinge E., Waruru A., Ngugi B., Wainaina J., Kariuki S. Genetic Diversity of Cryptosporidium in Children in an Urban Informal Settlement of Nairobi, Kenya. PLoS ONE. 2015;10(12) doi: 10.1371/journal.pone.0142055.e0142055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotloff K. L., Blackwelder W. C., Nasrin D., et al. The Global Enteric Multicenter Study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clinical Infectious Diseases. 2012;55(supplement 4):S232–S245. doi: 10.1093/cid/cis753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheun H.-I., Kim K., Yoon S., et al. Cryptosporidium hominis infection diagnosed by real-time PCR-RFLP. The Korean Journal of Parasitology. 2013;51(3):353–355. doi: 10.3347/kjp.2013.51.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samba O., Muhsen K., Nasrin D., Blackwelder W., Wu Y., Tamer H., et al. The burden of Cryptosporidium diarrheal disease among children < 24 months of age in moderate/high mortality rate regions of Sub-Saharan Africa and South Asia. Utilizing data from the Global enteric Multicenter Study (GEMS) Plos Negl Trop Dis. 2016;10(5) doi: 10.1371/journal.pntd.0004729.e0004729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muchiri J. M., Ascolillo L., Mugambi M., et al. Seasonality of Cryptosporidium oocyst detection in surface waters of Meru, Kenya as determined by two isolation methods followed by PCR. Journal of Water and Health. 2009;7(1):67–75. doi: 10.2166/wh.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adamu H., Petros B., Zhang G., et al. Distribution and Clinical Manifestations of Cryptosporidium Species and Subtypes in HIV/AIDS Patients in Ethiopia. PLOS Neglected Tropical Diseases. 2014;8(4) doi: 10.1371/journal.pntd.0002831.e2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatei W., Wamae C. N., Mbae C., et al. Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. The American Journal of Tropical Medicine and Hygiene. 2006;75(1):78–82. [PubMed] [Google Scholar]
- 13.Pavlinac P. B., John-Stewart G. C., Naulikha J. M., et al. High-risk enteric pathogens associated with HIV infection and HIV exposure in Kenyan children with acute diarrhea. AIDS. 2014;28(15):2287–2296. doi: 10.1097/QAD.0000000000000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeChevallier M. W., Di Giovanni G. D., Clancy J. L., et al. Comparison of method 1623 and cell culture-PCR for detection of Cryptosporidium spp. in source waters. Applied and Environmental Microbiology. 2003;69(2):971–979. doi: 10.1128/AEM.69.2.971-979.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakheit M. A., Torra D., Palomino L. A., et al. Sensitive and specific detection of Cryptosporidium species in PCR-negative samples by loop-mediated isothermal DNA amplification and confirmation of generated LAMP products by sequencing. Veterinary Parasitology. 2008;158(1-2):11–22. doi: 10.1016/j.vetpar.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Helmy Y. A., Krücken J., Nöckler K., Von Samson-Himmelstjerna G., Zessin K.-H. Comparison between two commercially available serological tests and polymerase chain reaction in the diagnosis of Cryptosporidium in animals and diarrhoeic children. Parasitology Research. 2014;113(1):211–216. doi: 10.1007/s00436-013-3645-3. [DOI] [PubMed] [Google Scholar]
- 17.Bouzid M., Elwin K., Nader J. L., Chalmers R. M., Hunter P. R., Tyler K. M. Novel real-time PCR assays for the specific detection of human infective Cryptosporidium species. Virulence. 2016;7(4):395–399. doi: 10.1080/21505594.2016.1149670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Power M. L., Holley M., Ryan U. M., Worden P., Gillings M. R. Identification and differentiation of Cryptosporidium species by capillary electrophoresis single-strand conformation polymorphism. FEMS Microbiology Letters. 2011;314(1):34–41. doi: 10.1111/j.1574-6968.2010.02134.x. [DOI] [PubMed] [Google Scholar]
- 19.Waldron L. S., Ferrari B. C., Gillings M. R., Power M. L. Terminal restriction fragment length polymorphism for identification of cryptosporidium species in human feces. Applied and Environmental Microbiology. 2008;75(1):108–112. doi: 10.1128/AEM.01341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notomi T., Mori Y., Tomita N., Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. Journal of Microbiology. 2015;53(1):1–5. doi: 10.1007/s12275-015-4656-9. [DOI] [PubMed] [Google Scholar]
- 21.Njiru Z. K., Mikosza A. S. J., Armstrong T., Enyaru J. C., Ndung'u J. M., Thompson A. R. C. Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLOS Neglected Tropical Diseases. 2008;2(1, article e147) doi: 10.1371/journal.pntd.0000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochemical and Biophysical Research Communications. 2001;289(1):150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 23.Parida M. M., Sannarangaiah S., Dash P. K., Rao P. V. L., Morita K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Reviews in Medical Virology. 2008;18(6):407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohon A. N., Elahi R., Khan W. A., Haque R., Sullivan D. J., Alam M. S. A new visually improved and sensitive loop mediated isothermal amplification (LAMP) for diagnosis of symptomatic falciparum malaria. Acta Tropica. 2014;134(1):52–57. doi: 10.1016/j.actatropica.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiraho E. A., Eric A. L., Mwangi I. N., et al. Development of a Loop Mediated Isothermal Amplification for Diagnosis of Ascaris lumbricoides in Fecal Samples. Journal of Parasitology Research. 2016;2016 doi: 10.1155/2016/7376207.7376207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mugambi R. M., Agola E. L., Mwangi I. N., Kinyua J., Shiraho E. A., Mkoji G. M. Development and evaluation of a Loop Mediated Isothermal Amplification (LAMP) technique for the detection of hookworm (Necator americanus) infection in fecal samples. Parasites & Vectors. 2015;8(1, article no. 1183) doi: 10.1186/s13071-015-1183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esmatabadi M. J. D., Bozorgmehr A., Zadeh H. M., et al. Techniques for evaluation of LAMP amplicons and their applications in molecular biology. Asian Pacific Journal of Cancer Prevention. 2015;16(17):7409–7414. doi: 10.7314/APJCP.2015.16.17.7409. [DOI] [PubMed] [Google Scholar]
- 28.Kiatpathomchai W., Jaroenram W., Arunrut N., Jitrapakdee S., Flegel T. W. Shrimp Taura syndrome virus detection by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. Journal of Virological Methods. 2008;153(2):214–217. doi: 10.1016/j.jviromet.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 29.Njiru Z. K. Rapid and sensitive detection of human African Trypanosomiasis by loop-mediated isothermal amplification combined with a lateral-flow dipstick. DIAGNOSTIC MICROBIOLOGY AND INFECTIOUS DISEASE. 2011;69(2):205–209. doi: 10.1016/j.diagmicrobio.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 30.Gandelman O., Jackson R., Kiddle G., Tisi L. Loop-mediated amplification accelerated by stem primers. International Journal of Molecular Sciences. 2011;12(12):9108–9124. doi: 10.3390/ijms12129108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao L., Singh A., Limor J., Graczyk T. K., Gradus S., Lal A. Molecular characterization of Cryptosporidiumoocysts in samples of raw surface water and wastewater. Applied and Environmental Microbiology. 2001;67(3):1097–1101. doi: 10.1128/aem.67.3.1097-1101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Notomi T., Okayama H., Masubuchi H., et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research. 2000;28(12, article E63) doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cobb B. D., Clarkson J. M. A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids Research. 1994;22(18):3801–3805. doi: 10.1093/nar/22.18.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao L., Bern C., Limor J., et al. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. The Journal of Infectious Diseases. 2001;183(3):492–497. doi: 10.1086/318090. [DOI] [PubMed] [Google Scholar]
- 35.Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Molecular and Cellular Probes. 2002;16(3):223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 36.Gallas-Lindemann C., Sotiriadou I., Plutzer J., Noack M. J., Mahmoudi M. R., Karanis P. Giardia and Cryptosporidium spp. dissemination during wastewater treatment and comparative detection via immunofluorescence assay (IFA), nested polymerase chain reaction (nested PCR) and loop mediated isothermal amplification (LAMP) Acta Tropica. 2016;158:43–51. doi: 10.1016/j.actatropica.2016.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.



