Abstract
Context
Simulation models are increasingly used in both theoretical and applied studies to explore system responses to natural and anthropogenic forcing functions, develop defensible predictions of future conditions, challenge simplifying assumptions that facilitated past research, and to train students in scientific concepts and technology. Researcher’s increased use of simulation models has created a demand for new platforms that balance performance, utility, and flexibility.
Objectives
We describe HexSim, a powerful new spatially-explicit, individual-based modeling framework that will have applications spanning diverse landscape settings, species, stressors, and disciplines (e.g. ecology, conservation, genetics, epidemiology). We begin with a model overview and follow-up with a discussion of key formative studies that influenced HexSim’s development. We then describe specific model applications of relevance to readers of Landscape Ecology. Our goal is to introduce readers to this new modeling platform, and to provide examples characterizing its novelty and utility.
Conclusions
With this publication, we conclude a >10 year development effort, and assert that our HexSim model is mature, robust, extremely well tested, and ready for adoption by the research community. The HexSim model, documentation, worked examples, and other materials can be freely obtained from the website www.hexsim.net.
Keywords: HexSim, Simulation model, Individual-based model, Spatially-explicit model, Mechanistic model, Population viability analysis, Forecasting
Introduction
HexSim is a spatially-explicit, individual-based, multi-population, eco-evolutionary modeling environment in which users develop simulations of wildlife or plant population dynamics and interactions. We designed HexSim to complement and/or improve upon existing models in several key areas. HexSim is fully parameterizable through its graphical user interface (GUI); can simulate both demographic and genetic processes, and their interactions; will work with a great variety of landscapes, species, and stressors; is appropriate for simple, complex, abstract, applied, data-poor and data-rich studies; can generate a large set of model outputs such as maps, tables, and animations; and includes robust batch-file and command-line execution utilities.
As described here, HexSim is a 2-dimensional grid-based model, with landscapes being composed of a regular array of hexagonal cells. These individual hexagonal cells (hereafter “atomic hexagons”) constitute the smallest unit of space that can be resolved by simulated individuals. A complementary suite of network-based tools (e.g. allowing users to add a fractal-dimensioned river network to the hexagonal grid) has also been developed for HexSim, and these features can be easily activated by editing a single line in the model’s preferences file. The present manuscript does not describe these network-based utilities.
Model structure, complexity, and data needs are fully user-defined. HexSim features are accessed through an extensive graphical user interface (GUI), making model development visual and intuitive. HexSim uses spatial data to capture landscape structure, habitat quality, stressor distributions, and other information. Spatial data may be static or dynamic, and can be constructed by a simulation as a model is running. Simulations may include one or more populations, each built up from individuals who possess any number of life history traits. Life history traits make HexSim individuals unique, and can be used to track current or past access to resources, exposure to stress, fitness, disease status, genotype, phenotype, and myriad other quantities that may be of interest.
Simulations are executed over a sequence of time steps of arbitrary length. Each time step consists of a single pass through a sequence of life history events. Users assemble the event sequence from a suite of life history events without having to write any source code. HexSim will benefit researchers developing theory and applications within ecology, conservation, genetics, epidemiology, and other disciplines, and the model’s visualization features will make it a valuable resource for educators. Readers who are new to HexSim may find that the analysis of Lurgi et al. (2014) provides a useful framework for comparing HexSim to many of the other ecologically-oriented simulation models in use today.
In developing HexSim, our goal was the creation of a mechanistic, spatially-explicit, eco-evolutionary, IBM (individual-based model) that could be employed across a wide array of ecological systems, landscape structures, and disturbance regimes. HexSim model development began in 2006, culminating in 2017 with version 4.0. HexSim grew from an existing spatial IBM called PATCH, developed by author NHS during the period from 2000–2006. HexSim’s utility and scientific defensibility have been documented in a number of published studies examining a wide range of species, landscapes, and stressors. These studies, plus other works in progress, have addressed wildlife responses to anthropogenic disturbance, species interactions, eco-evolutionary population dynamics, disease spread, pesticide impacts, and more. We have now completed extensive testing of the HexSim platform, and consider it to be mature, robust, and ready for use. HexSim is available, free of charge, at www.hexsim.net.
While a limited numbers of ecological simulation models are available, choosing a platform for a specific study can still be difficult (Lurgi et al. 2014, White et al. 2017). Key attributes that discriminate between modeling systems include whether they are spatially-explicit, individual-based, multi-species and multi-stressor, couple demography and genetics, have rigid data requirements, are easy to use, run quickly on large data sets, and are well-established in the literature. Parsimony should also be a useful deciding factor, with for example, population models often being more appropriate for highly data-limited systems. Demographic simulators frequently ignore genetics, and genetic models tend to incorporate minimal demographic machinery. Readers interested in HexSim might compare alternatives such as Vortex + Spatial (www.vortex10.org), NetLogo (https://ccl.northwestern.edu/netlogo/), CDPOP (http://hs.umt.edu/dbs/labs/cel/software.php), Range Shifter (Bocedi et al. 2014), and Aladyn (Schiffers and Travis 2014). Other widely used models include RAMAS GIS (www.ramas.com) a highly flexible pseudo-spatial population model; Circuitscape (www.circuitscape.org) a tool for evaluating landscape connectivity and dispersal flux; and Landis (www.landis-ii.org), a sophisticated platform for simulating landscape change.
Our intent here is to introduce readers to HexSim and illustrate how it can be used to translate concepts and ecological knowledge into mechanistic models. To achieve this goal, we begin with a model overview, we review a number of published studies that were influential in HexSim’s development, and we provide illustrations showing how features available in HexSim can be used to simulate processes of interest to students and researchers. This material should help new users get started with the model and develop a feel for its unique features and powerful analytic capabilities.
Model Overview
HexSim is a Microsoft Windows application distributed as a zipped folder containing 21 separate files, primarily of type exe (executable) and dll (dynamically-linked library). Users typically interact with HexSim through its graphical user interface (GUI), by running the program HexSim.exe. The model’s source code is written in C++ (engine) and C# (interface). The model engine can be easily compiled for Linux computers. While HexSim has not been designed for parallel processing, HexSim has several features that facilitate running multiple simulations (often replicates of a single model) at the same time. Most of our work with HexSim, including running large and complex simulations, has been performed on laptop computers with a modern CPU and 16 gigabytes of RAM. Users are more likely to encounter memory limitations than processor bottlenecks. Memory use varies primarily with population size, and is less severely impacted by landscape size.
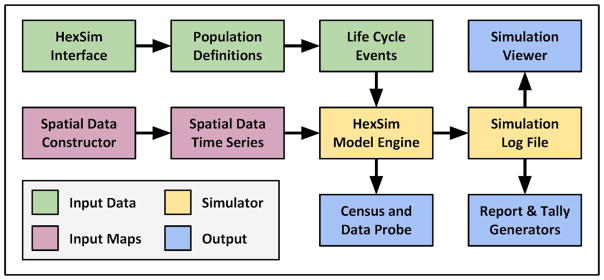
HexSim’s architecture mirrors the typical user’s workflow (Figure 1). Users launch the model interface and use it to conduct tasks such as assembling a workspace, creating spatial data, defining populations, developing a life cycle, running simulations, and analyzing simulation results. HexSim workspaces consist of a collection of nested folders containing all model inputs and outputs -- a portable design that facilitates collaboration. Below, we briefly describe key HexSim features.
Figure 1.
A high-level schematic of several key elements making up the HexSim model. Each box represents a collection of tools, all of which are accessible from the model GUI.
Workspaces
HexSim landscapes are rectangular arrays of hexagonal cells. All HexSim workspaces include a “grid file” (with extension.grid) that specifies a fixed landscape grain (hexagon size) and extent (number of rows and columns). Users normally visualize workspace grids by displaying spatial data on-screen with hexagon edges included as an overlay (the default). But HexSim also has a utility that tabulates grid characteristics in a popup window, documenting the number of hexagons, hexagon area and width, the number of rows and columns, grid geometry (narrow or wide), plus coordinates of the workspace edges in meters. Spatial data, models, and model results are all stored within separate workspace folders. Workspace construction is performed by a HexSim utility. Workspace size is defined by the atomic hexagon area, and by the extent of the map in rows and columns. HexSim workspaces can be quite large, for example >35M 22ha hexagonal cells used in a recent study of the Brazilian maned wolf. Simulation speeds tend to decline as population size increases, but are not significantly affected by workspace size.
Spatial data
HexSim spatial data fall into two categories, space-filling arrays of hexagonal cells (hereafter “hex-maps”), and collections of hexagon edges used to represent movement barriers such as roads (hereafter “barrier-maps”). HexSim spatial data may be static (a single image) or dynamic (a sequence of images). Dynamic spatial data, frequently used to simulate landscape change, need not include separate maps for every time step, as all maps remain in-use until superseded. HexSim includes a number of tools that simplify the process of creating and modifying spatial data. HexSim provides utilities for generating spatial data from raster imagery, importing spatial data from ESRI Shapefiles, and manually creating or editing spatial data. When Shapefiles are imported as hex-maps, only the database (.dbf) file is used, thus HexSim expects each polygon will represent a hexagon. The typical use-case involves exporting a blank hex-map as a Shapefile, importing and attributing this map in a GIS, then exporting the result and bringing it back into HexSim. When Shapefiles are imported as barrier-maps, HexSim snaps individual polyline segments to the nearest hexagon edge.
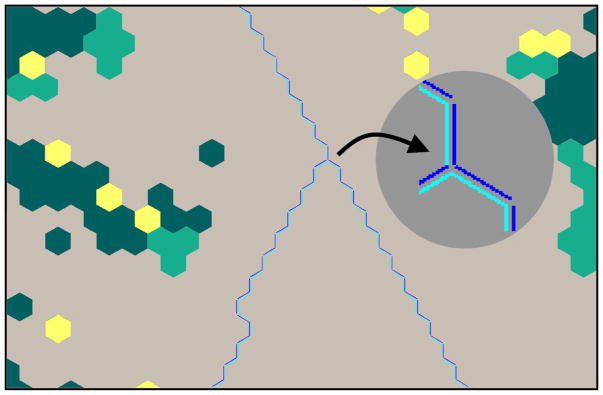
Individual hex-maps assign a single floating-point number (e.g. habitat quality or amount of stress) to each hexagon, while barrier-maps are built up from pairs of adjacent hexagon edges (Figure 2). HexSim simulations typically take advantage of multiple spatial data sets, for example using separate maps to capture food versus nesting resources, dispersal habitat, spatially-distributed stressors, and other quantities. Each barrier-map may contain multiple barrier categories, for example distinguishing between dirt roads, paved roads, and highways. Because barriers in HexSim are two-sided, the consequences of encountering a barrier can differ depending on the direction of movement (e.g. users may impose a penalty when an urban area is entered, but none if it is exited). Barriers are assigned separate probabilities for mortality, deflection, and transmission.
Figure 2.
A close-up view of a HexSim barrier-map (blue and cyan lines) overlaid on a hex-map (colored hexagonal cells). The inset illustrates how barrier maps are built-up from pairs of hexagon edge segments.
Hex-maps may also be constructed during a simulation (hereafter a “generated hex-map”), using map algebra to manipulate existing maps (hereafter “workspace spatial data”), information possessed by individuals, other model state, and global variables. For example, a population of predators capable of prey-taxis might utilize this feature to create smoothed maps of prey locations prior to moving, and then walk up-gradient in search of food items.
Populations
HexSim populations are collections of individuals that are assigned a common set of dynamic properties, and that can form groups and reproduce together. HexSim populations may include one or both sexes, though males and females may also be represented as separate populations. Specific classes of information defined on a per-population basis include initial population size and placement; map locations that are off-limits to a population (e.g. an ocean or urban area); use of space criteria and resource needs; traits, accumulators, affinities, and genetics (described below).
All populations have access to workspace spatial data, generated hex-maps, and to a user-defined set of static global variables. Some HexSim life history events (e.g. reproduction) operate on a single population, while others (e.g. species interactions or hex-map construction) may affect multiple populations.
Individuals
HexSim individuals inherit the data structures assigned to their population, including state variables, resource needs, and their genotype. Given that certain compatibility constraints are met, it is possible to switch an individual from one population to another during a simulation -- for example, aquatic-dependent members of a tadpole population might be shifted into a frog population that inhabits uplands. The use of population switching can simplify model structure, but it is usually straightforward to simulate highly distinct life stages within a single population. Individuals interact with their landscape via workspace spatial data, barrier-maps, and generated hex-maps, and can do so at multiple spatial scales. Individuals may maintain any number of variables such as traits and accumulators (see below) that in-turn affect their behavior, fitness, or other attributes. These variables may be assigned properties based on map values (e.g. resource quality or exposure to stress), interactions with other individuals, probabilistically, and so on. Interactions between individuals may also result in diverse outcomes, for example prey death coupled with predator resource acquisition, pair formation, and reproduction.
Traits and accumulators
Accumulators are variables that store floating-point numbers. Traits are categorical variables used to assign properties to individuals. HexSim has four types of traits, called “accumulated”, “probabilistic”, “genetic”, and “genetic accumulated”. When an accumulator changes, any accumulated traits linked to it are automatically reevaluated. The assignment of values to accumulators is performed by an “accumulate event”, which in turn make use of a diverse collection of “updater functions” that can execute algebraic expressions, query maps, inspect individual properties, or evaluate other elements of model state. In addition to altering trait values, accumulators can also directly influence individuals (e.g. store a survival or reproductive rate), and are often used to record information for post-simulation analysis (e.g. record individual fitness metrics). Probabilistic trait values are modified by a “transition event”, which houses the probabilities used when transitioning individuals between trait values.
A simple example of an accumulator/accumulated trait pair is a stage class trait (e.g. specifying juvenile, subadult, adult stages) linked to an age accumulator. Each year, an accumulate event might increase every individual’s age (the accumulator), which would in some cases increment their stage class (the accumulated trait), until the terminal class was reached. The linkage between the accumulator and accumulated trait involves user-defined threshold; for example, the transition from juvenile to subadult might take place at age 2, and the transition from subadult to adult at age 5. SEIR disease models -- in which individuals are categorized as susceptible, exposed, infected, or recovered -- serve as an illustration of a probabilistic trait. The transitions between these four disease classes would typically be governed by a HexSim transition event, which could account for the influence of multiple state variables simultaneously, such as an individual’s degree of exposure, fitness, and disease status. For example, transitions to the exposed state might be allowed if a low-fitness susceptible individual experienced any exposure, or when a high-fitness susceptible individual experienced repeated exposure events.
Simulating genetics is optional in HexSim. When this feature is used, each population may be assigned a genome composed of one or more loci, with each locus having a user-defined number of alleles. Users have control over the spatial distribution of the initial population’s genotypes, including those of individuals added while a simulation is in progress. At birth, genotypes are derived automatically from a simulated recombination of the parents’ genetic data. Genetic and genetic accumulated trait values are evaluated once at birth, so while an individual’s genotype may be altered through mutation, any genetic consequences of mutation are experienced strictly by offspring.
Loci, alleles, genetic traits, mutation
Individuals in HexSim are diploid, with one allele typically being drawn from each parent, though other inheritance rules are available. Haploid systems can be simulated by forcing loci to be initialized in a homozygous state, and then restricting offspring to inherit alleles from a single parent. HexSim also permits linkage distances (the chromosomal distances separating specific loci) to influence allele selection. Mutation rates can be stratified by any combination of trait values, making it easy to impose a genetic cost of exposure to mutagens, while also (optionally) accounting for genetic predispositions to mutation. HexSim mutation events utilize probabilistic allele transition matrices that allow users to readily develop flexible mutation regimes. Mutation can also be used to initialize a population’s genetic data after a model has reached steady state, or to simulate gene drives.
Genetic traits are used to assign phenotypes to specific allele combinations at a single locus. Probabilistic traits can be used to assemble a whole-organism phenotype from multiple per-locus phenotypic elements. Genetic accumulated trait values can be linked to allele data across multiple loci, and are particularly useful for quantifying homozygosity.
The event sequence
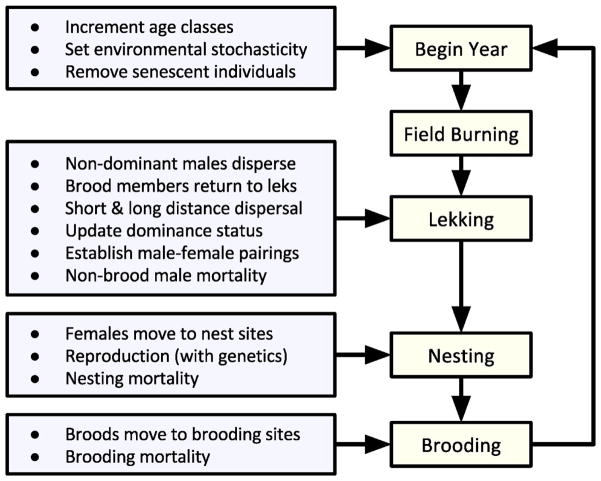
A HexSim model’s event sequence is a collection of life history events that users select (with replacement) from a list, and assemble in arbitrary order (Figure 3). The event sequence provides the model engine with a list of instructions to be executed as a simulation runs. Users may also create “event triggers” to specify the time steps at which an event will execute. A simple example might include increasing each individual’s age, performing reproduction, dispersal, resource acquisition, and finally survival. The duration of a single time step, defined as one pass through the entire event sequence, is inferred from the actions that are initiated therein. While time steps will typically represent an hour, a day, or one year, event triggers may be used to impose nested temporal scales. For example, individuals in a population may extract resources hourly, disperse monthly, and reproduce yearly. Then, a single pass through the event sequence would correspond to an hour if dispersal triggered approximately once every 720 cycles, or correspond to a year if dispersal ran 12 separate times per cycle. HexSim models have a single top-level event sequence but one of the available life history events, called an “event group”, is in itself an event sequence. Event groups may be nested and top-level event groups may be triggered, allowing users to readily create complex but well-organized simulations with specific functions collected into thematic modules.
Figure 3.
An example event sequence, in schematic form, taken from an unpublished HexSim model of the greater prairie chicken (Tympanuchus cupido). The boxes on the right correspond to event groups, each containing collections of life history events that accomplish the tasks indicated at the left.
HexSim has 24 different life history event types (Table 1) from which users may assemble the event sequence. Many of these are actually a collection of tools; for example, “accumulate” events are containers that house operators called “updater functions”, of which HexSim has 25 distinct types (Table 2). Life history traits can be used to identify discrete subsets of a population that will be acted upon by events when they run. For example, trait stratification can allow one movement events to act on juveniles while a second operates only on adults, thus allowing the two stage classes to exhibit unique dispersal behaviors.
Table 1.
HexSim life history events. The use cases listed are just examples, and do not characterize the range of a life history event’s applications.
| Name | Purpose | Example Use Cases |
|---|---|---|
| Accumulate | Assigns values to accumulators and reevaluates accumulated traits | Update individual ages and corresponding stage classes. Measure exposure to a spatially-distributed stressor. |
| Adjust Range Parameters | Alters population resource needs and use-of-space criteria | Force individuals to alternate between summer and winter range areas. |
| Census | Records population counts stratified by trait values | Census a population by both stage and fitness classes. |
| Clone Individual | Creates exact copies of select individuals | Use reproduction to create a single meta-individual representing thousands of frog eggs. After substantial mortality, clone the meta-individual to produce a small number of tadpoles. |
| Data Lookup | Assigns accumulator values based on entries in a lookup table | Assign hourly water temperature data to individuals distributed across a large river system without having to first convert the data into a hex-map time series. |
| Data Probe | Records individual accumulator and affinity values | Track individual resource allocation data at various points during a simulation. Record how frequently individuals are moving towards specific landscape features. |
| Event Group | Houses a collection of events and adds iterative functionality | Develop a collection of events that together pair males with females. Ensure the process iterates until all adults have found mates. |
| Floater Creation | Removes individuals from groups thus allowing them to move | Remove newly created juvenile individuals from their parents group in preparation for dispersal. |
| Generated Hexmap | Constructs spatial data while a simulation is running | Let predators construct a map of prey locations to guide foraging. Create maps showing the location of diseased individuals for use in a video. |
| Interaction | Allows individuals from one or more populations to interact with each other | Let predators probabilistically consume co-located prey items. Predators acquire resources and prey die. Allow diseased individuals to infect susceptibles. |
| Introduction | Adds new individuals to a population | Simulate immigration from outside the study area. Customize a model’s starting conditions. |
| Movement | Performs dispersal and assigns space or resources to individuals and groups | Force juveniles to disperse and then prospect for a territory. Allow large seabirds to migrate between distant colonies. |
| Mutation | Alters an individual’s allelic composition | Impose random mutations that allow predators to evolve an enhanced ability to capture members of a prey species. |
| Patch Introduction | Adds a single individual to each patch detected in a map | Distribute special populations across a landscape for data gathering purposes. |
| Plant Dynamics | Simulates all aspects of plant reproduction and propagation | Model pollen and seed formation and distribution, and fertilization. |
| Propagation | Performs reproduction for both group members and floaters | Simulate reproduction by non-territorial organisms. Draw clutch sizes from a normal distribution. |
| Range Dynamics | Allows groups to modify their range size | Shrink a group’s territory size if it experiences significant mortality. |
| Reanimation | Allows users to start one simulation where another simulation ended | Record internal population data after a simulation has reached steady-state. Use this data to initialize future simulations. |
| Reproduction | Performs reproduction exclusively for group members | Simulate reproduction by territorial organisms. Enumerate the likelihood of observing each possible clutch size. |
| Set Group Affinity | Facilitates the collective movement of groups of individuals | Move a pack of wolves, or a flock of birds, without losing the group structure. |
| Set Spatial Affinity | Assigns movement targets based on spatial proximity | Select a lekking site from the collection of leks that are <1km from an individual’s current location. |
| Survival | Imposes mortality on individuals | Impose yearly survival probabilities based on stage class. |
| Switch Population | Moves individuals between populations | Complement metamorphosis by switching members of an aquatic frog population to a terrestrial population. |
| Transition | Modifies probabilistic trait values | Transition individuals between susceptible, exposed, infected, and recovered classes in a disease model. |
Table 2.
HexSim updater functions. These operators are used within accumulate events to modify accumulator values. Suggestions regarding usage illustrate just one of many possible applications.
| Name | Utility |
|---|---|
| Accumulator Expression | Sets an accumulator equal to an algebraic expression. Standard math operators, accumulators, and global variables may be included. |
| Accumulator Transfer | Transfesr a quantity from one accumulator into another. |
| Allocated Hexagons | Returns the number of hexagons in each individual’s allocated area. Useful for quantifying territory sizes. |
| Births | Returns the number of offspring produced in the most recent reproduction or propagation event. |
| Clear | Sets an accumulator to zero. |
| Explored Hexagons | Returns the number of hexagons in each individual’s explored area. Useful for quantifying home range sizes. |
| Group Size | Returns the number of members present in each individual’s group. Returns zero for floaters. |
| Group Sum | Sets a target accumulator equal to the sum, taken across all group members, of a specified target accumulator. Useful for ascertaining group composition (e.g. is an alpha-male present). |
| Hexagon Presence | Detects the presence of nonzero values in a generated hex-map. Useful for triggering premature model termination (e.g. end a disease simulation if the the pathogen disappears). |
| Increment | Adds a fixed quantity to an accumulator. |
| Individual ID | Returns each individual’s ID. Useful for simulating DNA recombination in two-sex models. |
| Individual Locations | Returns the integer ID of the patch each individual is located within, relative to a user-supplied patch-map. |
| Mate Verification | Sets a mate accumulators to zero if the corresponding mate has died. Assumes the accumulator in question stores mate IDs. |
| Quantify Environment | Returns the sum or mean of the hexagons in each individual’s explored or allocated area, relative to a specified hex-map. Useful for quantifying resource availability, exposure, population density, and many other quantities of interest. |
| Quantify Extremes | Similar to the quantify environment updater function, except the minimum or maximum value is returned. |
| Quantify Location | Returns the floating point score of the hexagon each individual is located within, relative to a user-supplied hex-map. |
| Resources (Allocated) | Returns the percentage of each individual’s resource target that is presently being met by the hexagons in its allocated area. |
| Resources (Explored) | Returns the percentage of each individual’s resource target that is presently being met by the hexagons in its explored area. Is also used to simulate resource competition between populations. |
| Stochastic Increment | Increments an accumulator by an amount drawn from a uniform distribution with user-supplied bounds. |
| Stochastic Trigger | Returns a 1 with probability p (specified by the user) and a 0 with probability 1-p. A single evaluation is made and then applies to every individual. Used to randomly trigger life history events. |
| Subpopulation Assign | Randomly selects a user-specified number of individuals from a population, and assigns them a specific accumulator value. Useful for operating on a subset of a population. |
| Subpopulation Selector | Non-randomly selects a user-specified number of individuals from each group, and sets their target accumulator to 1. Useful for maintaining alpha status in wolf packs after mortality events. |
| Time Step | Returns the current time step, but includes a modulus operator. Useful as input to data lookup life history events. |
| Trait Value Index | Returns the index value of probabilistic, genetic, or genetic accumulated traits. Provides a way for individuals to incorporate this trait information into accumulator mathematics or generated hex-map construction. |
| Uptake | Transfers a quantity from a hex-map into an accumulator. Individual allocated areas define the source region. Useful for modeling the transfer of pathogens from the environment into organisms. |
Simulation outputs
A large number of model outputs can be easily generated from HexSim simulations, providing users with a wide range of analysis options. Census events record user-defined measures of model state. Data probe events record accumulator and affinity values. Generated hex-map events construct maps that can be saved in the model results. And all simulations produce “log files” that can be used to generate reports, tallies, and to drive the simulation viewer (discussed below). HexSim includes 21 different reports ranging from simple (e.g. information about births and deaths) to complex (e.g. projection matrix analogs of the full IBM). HexSim also includes 10 different tallies, which generate maps from simulation results by compiling model output on a per-hexagon basis. HexSim’s simulation viewer animates key simulation outputs including emergent occupancy patterns and movement processes.
Additional HexSim utilities
HexSim’s GUI also gives users access to an array of utilities that conduct critical operations such as workspace construction, map import, export, and editing, post-simulation analysis, and so on. HexSim simulations and report-generation tasks can also be run from batch files, and a suite of command-line options are available to help users automate repetitive tasks outside the confines of the GUI.
Formative Studies
To create a complex modeling framework like HexSim, developers must take part in investigations that can reveal application errors and shortcomings, and improve usability. Here, we briefly describe a number of published studies, conducted while HexSim was under development, that helped shape the model and establish its utility as a research tool.
The first published HexSim-based study (Heinrichs et al. 2010) produced a forecasting model for Ord’s kangaroo rats (Dipodomys ordii) in western Canada. The model was used to evaluate the risk of local extinction under alternative habitat removal scenarios. Fulford et al. (2011) used HexSim to develop one of the first mechanistic spatially-explicit individual-based models (SIBM) for an estuarine system, and explored how habitat variability and dispersal behavior could account for observed distributions of fish species. A.V. Stronen and co-authors (2012) used HexSim to examine the joint impacts of roads, natural area boundaries, and human-induced mortality on wolves (Canis lupus) in western Canada. Marcot et al. (2013) used HexSim to explore the consequences of habitat patch size and spacing on northern spotted owl (Strixoccidentalis caurina) occupancy rates and population dynamics.
Fordham et al. (2014) developed a dispersal-only HexSim model for a large population of northern snake-necked turtles (Chelodina rugosa) occupying two neighboring catchments in northern Australia. Pomara et al. (2014) developed a large multi-state HexSim simulation for the eastern massasauga rattlesnake (Sistrurus catenatus) that examined the species’ vulnerability to climate change. Huber et al. (2014) conducted the first HexSim-based reintroduction study, in this case addressing tule elk (Cervus elaphus nannodes) in the Central Valley of California, USA. Zang and Gorelick (2014) used HexSim to forecast the impacts of sea level rise on the clapper rail (Rallus longirostris obsoletus), an endangered shorebird that inhabits salt marshes. Wilsey and co-authors (2014) used a HexSim model to evaluate the degree to which endangered black-capped vireos (Vireo atricapilla) on the Fort Hood military installation (Texas, USA) depend upon brown-headed cowbird (Molothrus ater) management to avoid future population declines. Schumaker et al. (2014) developed a SIBM for the northern spotted owl, and used it to illustrate HexSim’s utility for identifying emergent source-sink dynamics, and for quantifying landscape connectivity.
Nogeire et al. (2015) developed a HexSim model for the endangered San Joaquin kit fox (Vulpes macrotis mutica) and evaluated the impacts on foxes of second-generation anticoagulant rodenticides over a large area across which application rates varied significantly. Marcot et al. (2015) used a HexSim simulation of the northern spotted owl to illustrate how researchers might develop sensitivity analyses for SIBMs. Heinrichs et al. (2015a) used HexSim to illustrate why a landscape’s value to a population might not be reliably inferred from an evaluation of its properties at any single spatial scale. Heinrichs et al. (2015b) used simulated removal experiments to examine the costs and benefits of sink habitats for three federally-listed declining species: black-capped vireos in Texas, USA, Ord’s kangaroo rats in Alberta, CA, and northern spotted owls in the Pacific Northwest, USA.
Tuma et al. (2016) used HexSim to examine the impacts of a diverse array of disturbances on threatened Agassiz’s desert tortoise (Gopherus agassizii) populations occupying two distinct study areas in the western U.S. Bancroft and co-authors (2016) developed a simulation model for red-cockaded woodpeckers (Picoides borealis) that was driven by both anticipated climate change and land-use changes associated with ongoing military activities at the Fort Benning military base in Georgia, USA. Heinrichs et al. (2016a) used computer-generated landscapes to explore the effects of patch size, quality, and spatial dispersion on simulated population growth rate and source-sink dynamics. Heinrichs et al. (2016b) explored how habitat degradation, loss, and fragmentation can independently affect population performance metrics. Wiens et al. (2017) used HexSim to construct a SIBM for a population of golden eagles (Aquila chrysaetos) exposed to risks associated with rapid increases in renewable energy development in southern California, USA. Heinrichs et al. (2017) used a HexSim model for sage grouse (Centrocercusurophasianus) in Wyoming, USA to explore the efficacy of possible conservation strategies. Pacioni et al. (2018) used HexSim to examine the efficacy of landscape-scale exclusion fencing coupled with lethal culling techniques to control wild dogs in Western Australia.
Future Investigations
HexSim’s unique attributes make it useful for ecology, conservation, genetics, epidemiology, and other fields. Below, we illustrate how HexSim features might be used to address future challenges of interest within these disciplines. It is impractical to provide more than a cursory overview here; we refer readers to the model’s website for additional resources and support. For each example below we identify an existing analytic challenge, outline a HexSim-based solution, and discuss modeling methodology that researchers might employ. We have either reported on, or at least developed detailed proof-of-concept models that address each topic discussed below. It is our hope that this material will inspire graduate students to become familiar with HexSim, and use it to push further into these research areas.
Quantifying source-sink dynamics
The challenge involves making demographic source-sink metrics purely emergent properties of complex species-landscape interactions. Source-sink properties are frequently supplied to models as inputs, not observed as outputs. We believe that more defensible source-sink analyses will play an increasingly larger role in informing conservation decisions.
The HexSim solution is to evaluate source-sink properties from simulation results. This is done by tabulating either births - deaths or emigration - immigration, or both, for every object in a spatial tessellation of the study landscape. For simulations at steady-state, these two quantities will be roughly equal. When model output is collected from a simulation that is far from steady-state, source-sink properties can be assessed by measuring births + immigration -deaths - emigration, or BIDE (Pulliam 1988).
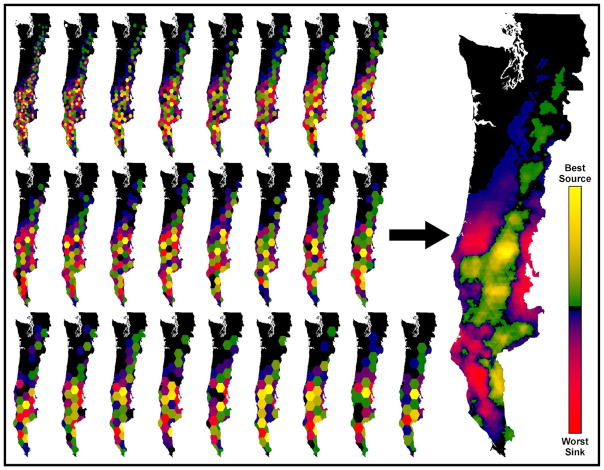
The HexSim methodology involves developing one or more “patch-maps” that can be used in a simulation to track every individual’s location each time it moves. A patch-map is simply a hex-map containing integer-valued hexagons. Once a simulation has finished running, HexSim Productivity or Projection Matrix reports can be used to obtain values for births - deaths or emigration - immigration, respectively. HexSim makes it easy for users to collect source-sink data at multiple spatial scales. We provide an illustration (Figure 4) in which a final multi-scale source-sink map was assembled using data collected simultaneously from a suite of 25 different patch-maps. These results were developed using the HexSim northern spotted owl model described in Schumaker et al. (2014). A more detailed explanation of this process can be found at HexSim’s website.
Figure 4.
Results from a HexSim model for the northern spotted owl that was sampled at 25 spatial scales simultaneously, over the course of 100 replicate simulations. The colored hexagonal patches shown in the maps on the left exhibited non-zero source-sink value, and are themselves composed of large numbers of atomic hexagons. The large image on the right is a summation of the 25 individual source-sink analyses.
Measuring landscape connectivity
The challenge involves adding biological realism to assessments of functional connectivity (Calabrese and Fagan 2004). Landscape connectivity is frequently inferred using methods from graph theory (Minor and Urban 2007) or using resistance surfaces to inform least cost path or circuit theory models (McRae 2006; McRae and Beier 2007; McClure et al. 2016; Bond et al. 2017; Marrotte et al. 2017). These standard approaches incorporate minimal life history information and cannot account for the influence on connectivity of complex demographic processes, species-landscape interactions, or interactions between species. HexSim will help researchers begin to add more biological nuance to connectivity assessments.
The HexSim solution is to develop a life history or movement-only simulation that includes key biological processes and disturbances. Users can then examine individual or aggregate movement patterns directly from simulation results, and observe how they respond when conditions change. Even relatively simple models can catalyze novel insights into the patterns and importance of landscape connectivity.
Several methods are available in HexSim for quantifying landscape connectivity based on simulation results. A HexSim Dispersal Flux tally can map dispersal patterns across a landscape. Three additional tallies will map the frequency of barrier interactions depending on outcome (transmission, deflection, mortality). Barriers can be used as integral parts of a simulation (e.g. representing high-impact roads) or as benign movement probes. A HexSim Projection Matrix report can record rates or counts of individuals moving between user-defined landscape locations. Such count-based matrices quantify landscape source-sink properties, with row and column sums representing per-patch immigration and emigration events, respectively. The rate-based matrices can be used to project population size and distribution forward in time via matrix multiplication. This allows users to convert a SIBM into a projection matrix model that can reveal the population-scale importance of local-scale landscape connections. For example, eigenanalysis can be used to identify the sensitivity of population growth rates (λ) to specific movement pathways. These model outputs also directly connect source-sink and connectivity metrics.
Simulating gene flow
The challenge involves forecasting effective rates of gene flow across complex landscapes while accounting for demographic complexity. Demo-genetic models often represent space as a resistance surface (e.g. Landguth and Cushman 2009), which necessitates simplifying both landscape structure and species’ movement behavior. By design, this class of models tends to incorporate limited demographic detail. HexSim is the first demo-genetic model to include a complex demographic submodel, and a great number of opportunities exist for its application.
The HexSim solution is to build genetics into a full spatial demographic model, possibly including features such as selection pressure or mutation. Such a model will allow rates of gene flow to become emergent properties of species-landscape interactions, and can incorporate density dependence, mate-finding, dynamic disturbance regimes, and other factors.
The methods involved are straightforward. Users select a check-box turning genetics on for a population, and add any number of loci. Each locus is assigned alleles and initialization rules, and upon reproduction HexSim will simulate recombination. Genetic traits are used to link genotypes to phenotypes, and mutation can be used to alter individual’s allelic data. Selection pressure is added by making genetic trait values influence vital rates or behaviors. Gene flow can be measured or inferred by using generated hex-maps to track the spatial distribution of alleles or phenotypes, by mapping or otherwise recording the movements of select individuals and their progeny, by mapping or tabulating amounts of homozygosity or genetic drift, and using HexSim reports to analyze population genetic structure.
Visualizing disease spread
The challenge involves estimating rates of disease spread in complex settings. The nonspatial mathematical models often used to study epidemics (e.g. Briggs et al. 2005) cannot account for the effect of landscape structure on rates of disease spread, and this can limit the utility of these models for guiding intervention efforts. Detailed HexSim disease simulators can be quicker to construct and easier to explain than traditional mathematical models, while remaining relatively parsimonious.
The HexSim solution is to let rates and patterns of disease spread emerge as the collective consequence of myriad interactions between individuals also exhibiting movement behaviors and reacting to landscape characteristics and conditions. We illustrate this example using an unpublished HexSim study of Chytridiomycosis in mountain yellow-legged frogs (Ranamuscosa), caused by the fungal pathogen Batrachochytrium dendrobatidis (Bd).
The methods involved construction of a four-stage HexSim amphibian model with a sophisticated disease submodel. We simulated stage class (tadpole-1, tadpole-2, subadult, adult) using a probabilistic trait because individuals did not advance on a strict chronological schedule. Infection load was tracked using a pair of accumulated traits that distinguished between subadults and adults. These metamorphs could be infection free, have sublethal bd loads, or could be terminally infected, with the threshold values separating the infection categories varying with developmental stage. Tadpoles cleared their infections upon metamorphosis, but while infected were effective Bd multipliers, shedding large numbers of pathogen zoospores into their ponds. Infected metamorphs shed zoospores into the upland landscape and could transmit the disease directly to conspecifics. Through careful accumulator arithmetic that accounted for both zoospore growth and death rates, which varied by media, we were able to track absolute numbers of the pathogens per-water body and per-individual. The simulation time step was a single day, but collections of life history events were also triggered on a monthly, seasonally, and a yearly basis. By swimming, infected tadpoles spread Bd throughout their ponds, and metamorphs carried the infection up into the landscape. Adults travelled to the nearest pond to lay eggs, thus functioning as a vector that made the landscape permeable to the pathogen. A multi-year HexSim video of the Bd invasion of our
Prioritizing mitigation efforts
The challenge involves prioritizing efforts aimed at mitigating human impacts on wildlife. Populations with large geographic distributions often encounter multiple distinct threats, which in turn complicate efforts to intercede on their behalf. Data limitations exacerbate these challenges. Here, we use the case of the maned wolf (Chrysocyon brachyurus) as an illustration. Our HexSim study of the maned wolf in Brazil covers an area exceeding three million km2 and includes a 200 km road network. Roadkill impacts are sufficient to threaten the population (Carvalho 2014), but road mortality data has not been collected systematically and is thus geographically biased. Limited government resources are anticipated for the development of crossing structures (e.g. wildlife overpasses), but the species’ vast range makes identifying the most efficacious locations a vexing problem.
The HexSim solution was to construct a range-wide model for the species and estimate cumulative road impact rates under a variety of assumptions. Data limitations significantly limited the detail we could incorporate into our model; nonetheless, we were able to simulate the basic life cycle, movement behaviors, and species-landscape interactions. We examined the effect of the road network on the population under a wide range of possible road crossing, deflection, and mortality rates.
Our HexSim methodology involved varying parameterized road mortality rates between 0.1 and 1.0 and then summing observed roadkill frequency per ¼ km road segment. In most simulations, the probability of road transmission and deflection were set equal. Through this analysis we identified five distinct and relatively small stretches of roadway that consistently accounted for a hugely disproportionate share of the range-wide roadkill events, regardless of assumptions about road properties. Because we used a SIBM to conduct the study, our results were robust to possible compounding effects of roads on the large-scale population distribution, dispersal fluxes, and emergent source-sink properties.
Constructing population models
The challenge involves adding nuanced movement behavior to population models. The latter have advantages over IBMs when population sizes are large, minimal data exists, or scant time is available for model construction. But their focus on collections rather than individuals means that population models like RAMAS GIS (Akcakaya and Root 2005) must represent movement as mean flows between locations of interest. This can limit a population model’s predictive ability (Fordham et. al. 2014).
The HexSim solution is to develop a population/IBM hybrid that combines the simplicity of a population model with the movement sophistication of an IBM. At the HexSim website, we provide a template hybrid model called HexSimPLE (HexSim Populations Linked by Emigration) that readers may download and customize. While HexSimPLE models can be made large and complex, their minimum data requirements include only a habitat map, a patch-map, stage-specific estimates for maximum survival and fecundity rates, and a few other parameters that determine carrying capacity and the functional form of density dependence.
The HexSimPLE methodology involves distributing a spatial array of 3-stage Leslie matrices (Leslie 1945) across a landscape. HexSim does this automatically for the user once a patch-map is supplied. Per-matrix subpopulations are represented by stage vectors, and matrix vital rates vary as a function of local habitat quality, population density, and disturbance. When movement executes, HexSimPLE converts portions of the population vectors into emigrating individuals, simulates their independent movement decisions, and then absorbs those migrants into the population vectors associated with the landscape locations into which they arrived. While the HexSimPLE model itself is complicated, parameterizing it for novel systems is very straightforward.
Conclusions
We developed HexSim to help address contemporary problems in ecology, conservation, genetics, epidemiology, and other disciplines. HexSim users will, among other things, challenge simplifying assumptions that made earlier models tractable, explore how natural and anthropogenic forcing functions drive the spatial and temporal dynamics of simple and complex ecological systems, build scientifically defensible mechanistic forecasting models, and use the model as a visual aid for communicating ecological concepts to students. HexSim is a large and diverse modeling environment that will require dedication to master. We encourage readers to make that effort. HexSim is mature and stable, extremely well tested, and we believe it will prove to be an important contribution to the modern ecologist’s quantitative toolbox.
Acknowledgments
We dedicate this paper to the memory of Dr. Brad McRae (1966 – 2017), our colleague and treasured friend. While insignificant in comparison to his multiple contributions to ecology and conservation science, Brad is due recognition here as an original visionary force behind HexSim’s genetics toolkit.
We are indebted to Jianguo Wu and two anonymous reviewers who made significant and insightful contributions that improved the paper’s structure, style, and impact. The information in this document has been funded in part by the U.S. Environmental Protection Agency. It has been subjected to review by the National Health and Environmental Effects Research Laboratory’s Western Ecology Division and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
References
- Akcakaya HR, Root WT. RAMAS GIS: linking landscape data with population viability analysis (version 5.0) Setaukey, NY: Applied Biomathematics; 2005. [Google Scholar]
- Bancroft BA, Lawler JJ, Schumaker NH. Weighing the relative potential impacts of climate change and land-use change on an endangered bird. Ecology and Evolution. 2016;6:4468–4477. doi: 10.1002/ece3.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocedi G, Palmer S, Pe’er G, Heikkinen RK, Matsinos YG, Watts K, Travis J. RangeShifter: a platform for modelling spatial eco-evolutionary dynamics and species’ responses to environmental changes. Methods in Ecology and Evolution. 2014;5:388–396. [Google Scholar]
- Bond ML, Bradley CM, Kiffner C, Morrison TA, Lee DE. A multi-method approach to delineate and validate migratory corridors. Landscape Ecology. 2017;32:1705–1721. [Google Scholar]
- Briggs CJ, Vredenburg VT, Knapp RA, Rachowicz LJ. Investigating the population-level effects of Chytridiomycosis: an emerging infectious disease of amphibians. Ecology. 2005;86:3149–3159. [Google Scholar]
- Calabrese JM, Fagan WF. A comparison-shopper’s guide to connectivity metrics. Frontiers in Ecology and the Environment. 2004;2:529–536. [Google Scholar]
- Carvalho CF. Master thesis in Ecology and Conservation of Natural Resources – Universidade Federal de Uberlândia, Uberlândia, Minas Gerais. 2014. Atropelamento de vertebrados, hotspots de atropelamentos e parâmetros associados, BR-050, trecho Uberlândia-Uberaba; p. 57. [Google Scholar]
- Fordham DA, Shoemaker KT, Schumaker NH, Akçakaya HR, Clisby N, Brook BW. How interactions between animal movement and landscape processes modify range dynamics and extinction risk. Biology Letters. 2014;10:20140198. doi: 10.1098/rsbl.2014.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulford RS, Peterson MS, Grammer PO. An ecological model of the habitat mosaic in estuarine nursery areas: Part I — Interaction of dispersal theory and habitat variability in describing juvenile fish distributions. Ecological Modelling. 2011;222:3202–3215. [Google Scholar]
- Heinrichs JA, Aldridge CL, O’Donnell M, Schumaker NH. Using dynamic population simulations to extend resource selection analysis and prioritize habitat for conservation. Ecological Modelling. 2017;359:449–459. [Google Scholar]
- Heinrichs JA, Bender DJ, Gummer DL, Schumaker NH. Effects of landscape and patch-level attributes on regional population persistence. Journal for Nature Conservation. 2015a;26:56–64. [Google Scholar]
- Heinrichs JA, Bender DJ, Gummer DL, Schumaker NH. Assessing critical habitat: Evaluating the relative contribution of habitats to population persistence. Biological Conservation. 2010;143:2229–2237. [Google Scholar]
- Heinrichs JA, Bender DJ, Schumaker NH. Habitat degradation and loss as key drivers of regional population extinction. Ecological Modelling. 2016b;335:64–73. [Google Scholar]
- Heinrichs JA, Lawler JJ, Schumaker NH. Intrinsic and extrinsic drivers of source-sink dynamics. Ecology and Evolution. 2016a;6:892–904. doi: 10.1002/ece3.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs JA, Lawler JJ, Schumaker NH, Wilsey C, Bender DJ. Divergence in sink contributions to population persistence. Conservation Biology. 2015b;29:1674–1683. doi: 10.1111/cobi.12540. [DOI] [PubMed] [Google Scholar]
- Huber PR, Greco SE, Schumaker NH, Hobbs J. A priori assessment of reintroduction strategies for a native ungulate: using HexSim to guide release site selection. Landscape Ecology. 2014;29:689–701. [Google Scholar]
- Landguth EL, Cushman SA. CDPOP: A spatially explicit cost distance population genetics program. Molecular Ecology Resources. 2009;10:156–161. doi: 10.1111/j.1755-0998.2009.02719.x. [DOI] [PubMed] [Google Scholar]
- Leslie PH. On the use of matrices in certain population mathematics. Biometrika. 1945;33:183–212. doi: 10.1093/biomet/33.3.183. [DOI] [PubMed] [Google Scholar]
- Lurgi M, Brook BW, Saltré F, Fordham DA. Modelling range dynamics under global change: which framework and why? Methods in Ecology and Evolution. 2015;6:247–256. [Google Scholar]
- Marcot BG, Raphael MG, Schumaker NH, Galleher B. How big and how close? Habitat patch size and spacing to conserve a threatened species. Natural Resources Modeling. 2013;26:194–214. [Google Scholar]
- Marcot BG, Singleton PH, Schumaker NH. Analysis of sensitivity and uncertainty in an individual-based model of a threatened wildlife species. Natural Resource Modeling. 2015;28:37–58. [Google Scholar]
- Marrotte RR, Bowman J, Brown MG, Cordes C, Morris KY, Prentice MB, Wilson PJ. Multi-species genetic connectivity in a terrestrial habitat network. Movement Ecology. 2017;5:21. doi: 10.1186/s40462-017-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure ML, Hansen AJ, Inman RM. Connecting models to movements: testing connectivity model predictions against empirical migration and dispersal data. Landscape Ecology. 2016;31:1–14. [Google Scholar]
- McRae BH. Isolation by resistance. Evolution. 2006;60:1551–1561. [PubMed] [Google Scholar]
- McRae BH, Beier P. Circuit theory predicts gene flow in plant and animal populations. PNAS. 2007;104:19885–19890. doi: 10.1073/pnas.0706568104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor ES, Urban DL. Graph theory as a proxy for spatially explicit population models in conservation planning. Ecol Appl. 2007;17:1771–1782. doi: 10.1890/06-1073.1. [DOI] [PubMed] [Google Scholar]
- Nogeire TM, Lawler JJ, Schumaker NH, Cypher BL, Phillips SE. Land use as a driver of patterns of rodenticide exposure in modeled kit fox populations. PLoS ONE. 2015;10(8):e0133351. doi: 10.1371/journal.pone.0133351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacioni C, Kennedy MS, Berry O, Stephens D, Schumaker NH. Spatially-explicit model for assessing wild dog control strategies in Western Australia. Ecological Modelling. 2018;368:246–256. doi: 10.1016/j.ecolmodel.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomara LY, Ledee OE, Martin KJ, Zuckerberg B. Demographic consequences of climate change and land cover help explain a history of extirpations and range contraction in a declining snake species. Global Change Biology. 2014;20:2087–2099. doi: 10.1111/gcb.12510. [DOI] [PubMed] [Google Scholar]
- Pulliam RH. Sources, sinks, and population regulation. Am Nat. 1988;132:652–661. [Google Scholar]
- Schiffers KH, Travis J. ALADYN - a spatially explicit, allelic model for simulating adaptive dynamics. Ecography. 2014;37:1288–1291. doi: 10.1111/ecog.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumaker NH, Brookes A, Dunk JR, Woodbridge B, Heinrichs J, Lawler JJ, Carroll C, LaPlante D. Mapping sources, sinks, and connectivity using a simulation model of northern spotted owls. Landscape Ecology. 2014;29:579–592. [Google Scholar]
- Stronen AV, Schumaker NH, Forbes GJ, Paquet PC, Brook RK. Landscape resistance to dispersal: simulating long-term effects of human disturbance on a small and isolated wolf population in southwestern Manitoba, Canada. Environmental Monitoring and Assessment. 2012;184:6923–6934. doi: 10.1007/s10661-011-2469-9. [DOI] [PubMed] [Google Scholar]
- Tuma MW, Millington C, Schumaker NH, Burnett P. Modeling Agassiz’s desert tortoise population response to anthropogenic stressors. Journal of Wildlife Management. 2016;80:414–429. [Google Scholar]
- White LA, Forester JD, Craft ME. Dynamic, spatial models of parasite transmission in wildlife: Their structure, applications and remaining challenges. Journal of Animal Ecology. 2017 doi: 10.1111/1365-2656.12761. [DOI] [PubMed] [Google Scholar]
- Wiens JD, Schumaker NH, Inman RD, Esque TC, Longshore KM, Nussear KE. Spatial demographic models to inform conservation planning of Golden Eagles in renewable energy landscapes. Journal of Raptor Research. 2017;51:234–257. doi: 10.3356/JRR-16-77.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsey CB, Lawler JJ, Cimprich D, Schumaker NH. Dependence of the endangered black-capped vireo on sustained cowbird management. Conservation Biology. 2014;28:561–571. doi: 10.1111/cobi.12176. [DOI] [PubMed] [Google Scholar]
- Zang H, Gorelick SM. Coupled impacts of sea-level rise and tidal marsh restoration on endangered California clapper rail. Biological Conservation. 2014;172:89–100. [Google Scholar]