Preface
Bacterial ADP-ribosyltransferase toxins (bARTTs) transfer ADP-ribose to eukaryotic proteins to promote bacterial pathogenesis. In this review we use prototype bARTTs, such as diphtheria and pertussis toxins, as references for the characterization of several new bARTTs from human, insect, and plant pathogens, which were identified recently through bioinformatic analyses. Several of these toxins, including Cholix toxin from Vibrio cholerae, SpyA from Streptococcus pyogenes, HopU1 from Pseudomonas syringae, and the Tcc toxins from Photorhabdus luminescens, ADP-ribosylate novel substrates and possess unique organizations, which distinguish them from the reference toxins. The characterization of these toxins extends our appreciation for the variety of structure-function properties possessed by bARTTs and their roles in bacterial pathogenesis.
Protein toxins are potent bacterial virulence factors that disrupt host cell activity, often by modulating the activity of host proteins through covalent modifications. Bacterial ADP-ribosyltransferase toxins (bARTTs) are encoded by a range of bacterial pathogens, including human pathogens, such as Corynebacterium diphtheriae, Vibrio cholerae, Bordetella pertussis, Clostridium botulinum, and Streptococcus pyogenes, the plant pathogen Pseudomonas syringae, and the insect pathogen Photorhabdus luminescens. bARTTs confer a single post-translational modification – the addition of ADP-ribose – on a variety of eukaryotic substrates, including Rho proteins (C. botulinum C3-exoenzymes1, 2), heterotrimeric G proteins (V. cholerae cholera toxin3, 4 and B. pertussis pertussis toxin5) and actin (C. botulinum C2 toxin6). Many of these modifications inhibit normal eukaryotic protein function; however, several novel bARTTs have recently been identified that activate eukaryotic proteins to promote bacterial pathogenesis.
ADP-ribosylation is one of the most prevalent covalent modifications, with examples of ADP-ribosyltransferases in both Gram-positive and Gram-negative bacteria, as well as eukaryotic organisms7, 8. These enzymes utilize a NAD donor molecule to catalyze the transfer of the ADP-ribose moiety to a substrate acceptor residue, which can be an Arg, Asn, Thr, Cys, or a Gln, depending on the individual toxin. The ADP-ribosylation reaction is proposed to follow a SN1 strain-alleviation mechanism9–11.
The bARTT family consists of >35 members which can be divided into several discrete groups, catalogued by their structure-function properties, including AB domain organization and the eukaryotic protein target. Although bARTTs are typically organized into four AB groups, several recently characterized bARTTs have unique AB organizations (Table 1). The conservation of structure-function among bARTTs provides a basis for the identification of new bARTT family members through bioinformatics, rather than the identification of a disease state. In the past, identification of bARTTs required the detection and purification of the protein toxin and the extrapolation of a pathogenic phenotype to the purified protein, an often time-consuming process. Bioinformatics provides a mechanism to predict the presence of a candidate toxin within the genome of known pathogens, and more significantly, in bacteria not yet recognized as human or environmental pathogens (BOX 1).
Table 1. Characterization of the subfamilies of bacterial ADP-ribosyltransferase toxins (bARTTs).
Toxins initially identified through bioinformatic approaches are marked with *.
| Toxin | Bacteria | Eukaryotic substrate | Cellular receptor/delivery | Reference |
|---|---|---|---|---|
| Diphtheria-like toxins | ||||
| Diphtheria toxin | Corynebacterium diphtheriae | eukaryotic Elongation Factor 2 (eEF2) | Heparin-binding epidermal growth factor | 51 |
| Exotoxin A | Pseudomonas aeruginosa | eEF2 | Low Density lipoprotein-receptor related protein (LRP1) | 68 |
| *Cholix toxin | Vibrio cholerae | eEF2 | LRP1 | 74 |
| Cholera-like toxins | ||||
| Cholera toxin | Vibrio cholerae | Gαs | Ganglioside: GM1 | 86 |
| Heat-labile enterotoxin | E. coli (ETEC) | Gαs | Ganglioside: Various | 89 |
| Pertussis toxin | Bordetella pertussis | Gαi | Ganglioside: Various | 101 |
| C2-like binary toxins | ||||
| C2 toxin | Clostridium botulinum | G-actin | N-linked carbohydrates | 6 |
| Iota toxin | Clostridium perfringens | G-actin | Lipolysis-stimulated lipoprotein receptor (LSR) | 104 |
| CDT | Clostridium difficile | G-actin | LSR | 105 |
| CST | Clostridium spiroforme | G-actin | LSR | 106 |
| VIP | Bacillus cereus | G-actin | Unknown | 107 |
| *SpvB | Salmonella sp | G-actin | Type-III secreted | 117 |
| *AexT | Aeromonas hydrophila | G-actin | Type-III secreted | 120 |
| *Photox | Photorhabdus luminescens | G-actin | Type-VI secreted (putative) | 121 |
| C3-like toxins | ||||
| C3bot | Clostridium botulinum | RhoA,B,C | N/A | 1 |
| C3Stau (EDIN) | Staphylococcus aureus | RhoA,B,C,E | N/A | 137 |
| C3cer | Bacillus cereus | RhoA,B,C | N/A | 129 |
| ExoS | Pseudomonas aeruginosa | Ras, ERM proteins, vimentin | Type-III secreted | 125,126 |
| ExoT | Pseudomonas aeruginosa | CrkI/II | Type-III secreted | 156 |
| *HopU1 | Pseudomonas syringae | GRP7 | Type-III secreted | 154 |
| *SpyA | Streptococcus pyogenes | vimentin, actin | N/A | 142 |
| Novel toxins | ||||
| *Typhoid toxin | Salmonella Typhi | Unknown | Ganglioside: Various, GD2 preferred | 29 |
| *PTC3 | Photorhabdus luminescens | G-actin | Secreted | 30 |
| *PTC5 | Photorhabdus luminescens | RhoA,B,C | Secreted | 30 |
Box 1. Using bioinformatics to identify candidate bARTTs.
In early work, identification of the mechanism of action of a bacterial toxin required that genetic, cell biological and biochemical analyses all be used. Subsequently, classification of protein toxins into different toxin groups, including bARTTs, continued to utilize wet-bench approaches. However, the development of weighted bioinformatics analyses, like Dayhoff matrices, allowed the identification of closely related bARTT variants based on primary amino acid sequence conservation. The continued development of software algorithms, such as PSI-BLAST187, allowed deeper mining for primary amino acid sequence conservation among related proteins. An increase in the number of solved protein crystal structures and the identification of protein folds and the conservation of amino acids within these folds through fold-recognition searches18 has advanced the ability to identify new members of the bARTT family. While the number of protein crystal structures continues to increase exponentially, only a limited number of new protein folds have been identified188, indicative of a finite number of structural organizations that yield stable and functional protein folds.
A bioinformatics approach can correctly predict new bARTT members based upon strict conservation of a function, like NAD binding, but is limited in its ability to correctly predict the host protein substrate. For example, the closely related Pseudomonas aeruginosa exoenzymes ExoS and ExoT, which share ~ 76% primary amino acid homology, target disparate substrates for ADP-ribosylation126, 127, 157. In fact, the locations of the substrate recognition sites within ExoS and ExoT are different189. A more detailed characterization of how bARTTs recognize the target host protein may provide better rules to predict substrates for subgroups of bARTTs. While this approach has primarily been used to identify new bARTTS, it may be extrapolated to other groups of virulence factors with conserved properties, ultimately leading to better characterization of bacterial pathogenesis.
Other reviews have focused on in-depth descriptions of the mode of action of specific subgroups of bARTTs12–14. In this review we present several of the newest members of the bARTT family that were identified through bioinformatic searches. Bioinformatics has been used to identify virulence factors using different genomic screening approaches15–17. However, many bioinformatics approaches require a significant number of sequences or structures to generate data for a consensus protein folding domain or a conserved catalytic sequence that can then be used to predict new virulence factors. With more than 35 identified members and targets in plants, animals, and insects, the bARTT family is uniquely suited for this branch of in silico analysis18, 19. Bioinformatic analyses recently identified Cholix toxin from V. cholerae, SpyA from S. pyogenes, HopU1 from P. syringae, and the Tcc toxins from P. luminescens. In this review we discuss the novel and conserved properties of these toxins compared with the established prototype toxins.
bARTT structure and function
bARTTs can be divided into two major groups based on both toxin domain organization and conserved active site motifs: the diphtheria toxin (DT) group and the cholera toxin (CT) group; the CT group can be further subdivided into the CT-like, C2-like, and C3-like bARTTs.
Domain organization
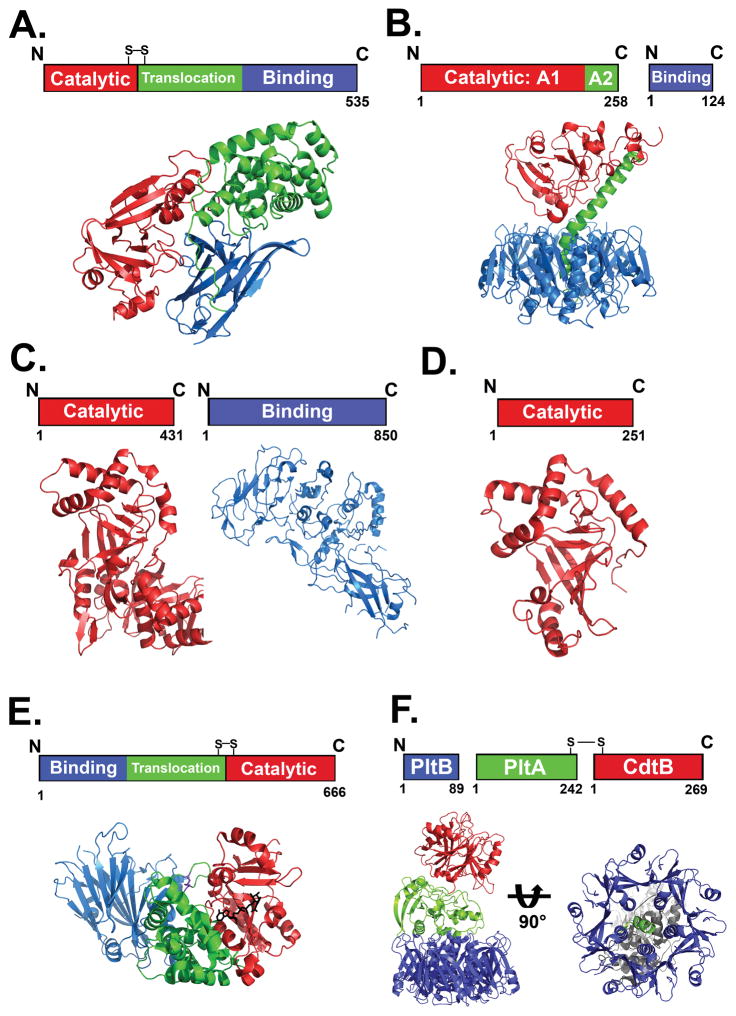
DT-like toxins are ~60 kDa single-chain AB toxins, with a catalytic (A) domain and a binding (B) domain that contains both receptor binding and translocation activities (Figure 1A)20–23. CT–like toxins are AB5 toxins with an ~28 kDa A domain, which is non-covalently bound to a B oligomer comprising five non-covalently associated ~12 kDa proteins (Figure 1B)24, 25. C2-like toxins are binary toxins, synthesized as two separate proteins comprising an ~50 kDa A component (C2I)6 and an ~ 80 kDa B component (C2II)26, 27(Figure 1C). The C3-like exoenzymes are single-chain proteins consisting solely of an ~ 25 kDa A domain28, 29. The structural organization of the C3 A domain is remarkably similar to the catalytic domain of the C2-like toxins, despite the lack of a B component (Figure 1D).
Figure 1. Structure-function organization of bARTT family members.
A. Diphtheria toxin (DT)-like toxins are single chain polypeptides possessing a catalytic A domain linked by a disulphide bond to a B domain that includes translocation and binding capacity. The figure shows the domain organization of DT (PDB:1MDT) B. Cholera toxin (CT)-like toxins are dual chain proteins, with catalytic A1 and A2 domains that insert into a pore in the pentameric binding (B) domain. The figure shows the domain organization of CT (PDB:1XTC) C. C2-like toxin catalytic A and receptor binding B domains are expressed separately. The figure shows the domain organization of C2 toxin (A domain; PDB:2J3Z) and B domain PDB:2J42). D. C3-like transferases consist solely of a catalytic A subunit. The figure shows the A subunit of C3bot-transferase (PDB:1G24) E. Domain organization and crystal structure of Cholix toxin. Cholix is organized similar to PE, with the binding domain at the N terminus and the disulfide linked translocation and catalytic domain at the C terminus. (PDB:3Q9O) F. Domain organization and crystal structure of Typhoid toxin (TT). (PDB:4K6L). TT has A2B5 organization, with two catalytic subunits (Left). CdtB shows structure-function homology to the Cytolethal distending toxins and is linked to PltA by a disulfide bond. Displaying structure-function homology to the pertussis toxin S1 and S2 subunits, respectively, PltA inserts into a PltB pentamer (Right).
While some recently described bARTTs display sequence and structural conservation to prototype toxins, like Cholix toxin and its homology to PE (Figure 1E), other recently identified bARTTs have a unique organization relative to the prototypical AB toxins. Salmonella enterica serovar Typhi expresses a novel A2B5 toxin (typhoid toxin (TT)) that comprises an A1 domain possessing DNase activity that is joined by a disulphide bond to an A2 domain with homology to the pertussis toxin S1 catalytic domain (Figure 1F). The A2 domain is non-covalently bound to a pentameric B oligomer comprising a homologue of the pertussis toxin S2 binding subunit30. P. luminescens Tc proteins display a novel ABC organization that includes one of several A subunits31, 32. Despite the divergence in overall organization, the ADP-ribosyltransferase domains of these new bARTTS are remarkably conserved33 and include an invariant Glu, a key residue among the bARTTs34.
Active site residues
DT-like toxins possess an active site HYE motif that contains a His, two Tyr, and a Glu residue. The Glu residue was originally identified as the key catalytic residue in DT by the Collier group, and was eventually shown to be invariant in all bARTTs35, 36. Mutation of this residue results in a several hundred-fold loss of ADP-ribosyltransferase activity and 10,000-fold loss in cytotoxicity34, 37. The invariant Glu positions the NAD for hydrolysis and coordination of the NAD binding pocket for ADP-ribose transfer38. The HYE motif is also conserved in mammalian poly-ADP-ribosyltransferases39, 40. The CT group of bARTTs, comprising the CT-like, C2-like and C3-like toxins, possesses a different active site motif relative to DT, termed the RSE motif29, 41, 42, which consists of an Arg that positions NAD in the active site, a Ser-Thr-Ser that stabilizes the NAD-binding pocket, and a Gln/Glu-X-Glu motif. The Gln/Glu promotes transfer of ADP-ribose to a substrate residue and, like the DT-like toxins, the second invariant Glu positions the NAD molecule to promote hydrolysis.
DT-like toxins
Diphtheria toxin, death after translation inhibition
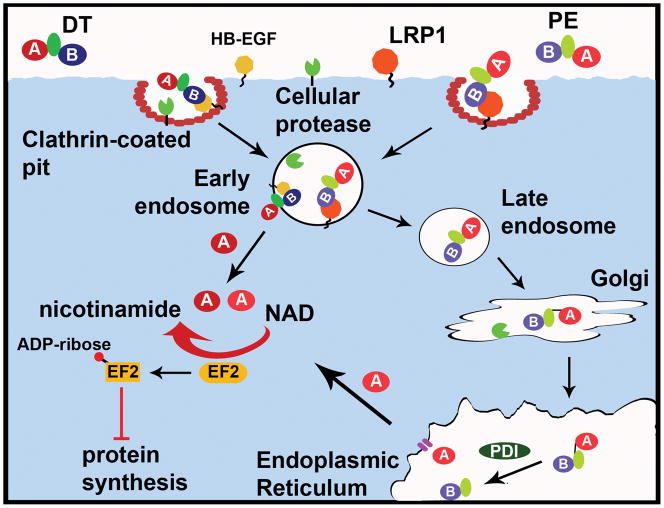
DT was discovered in 1888 through examination of sterile culture filtrates of C. diphtheriae43; injection of the filtrate into animals resulted in lesions and death characteristic of diphtheria, with subsequent studies correlating this pathology to DT44, 45. DT is encoded on a lysogenic β-phage46 and is produced by C. diphtheriae colonizing the upper respiratory tract epithelium. Lysogenized C. diphtheriae release DT, which disseminates throughout the body and targets numerous tissues via binding to a ubiquitous cellular receptor, the heparin binding-epidermal growth factor (HB-EGF)47, 48 (Figure 2A). After receptor binding, the DT-receptor complex undergoes clathrin-mediated endocytosis and traffics to an early endosome49. Once internalized, DT is processed by host furin-like proteases, resulting in a nicked di-chain linked by a disulphide bond50. Endosomal acidification promotes the insertion of two hydrophobic helices from the translocation domain of the B subunit into the membrane to deliver the A domain across the endosome membrane where disulphide reduction releases the A domain into the cytosol 51, 52.
Figure 2. Diphtheria toxin-like cellular intoxication pathways.
A. Cellular intoxication of DT-like toxins. DT-like toxins bind host membrane-bound receptors and enter via receptor-mediated endocytosis. Protease cleavage of the peptide backbone results in a single disulfide-linked chain. Upon endosomal acidification, the A domain of DT is translocated into the cytosol by insertion of the translocation domain into the endosomal membrane and reduction of the disulfide bond. Conversely, PE undergoes retrograde trafficking to the ER, where reduction of the disulfide and interactions with ER-associated chaperone proteins like PDI promote translocation of the A domain through a Sec61-like channel into the cytosol. Once in the cytosol, the A domains of DT-like toxins ADP-ribosylate EF2, inhibiting protein synthesis and killing the cell. Cholix can utilize LRP1 to enter cells and may follow the PE intoxication pathway.
DT ADP-ribosylates eukaryotic elongation factor 2 (EF2)53, 54 at residue 71555, a post-translationally modified histidine termed diphthamide, which to date has been found only in archaeal and eukaryotic EF256. ADP-ribosylation of dipthamide inhibits EF2 function and subsequently inhibits protein synthesis57. Diphthamide is located in the region of EF2 that is responsible for the interaction with the P-site of the ribosome58. ADP-ribosylated-diphthamide may have several effects on protein synthesis including steric hindrance of EF2 from binding the P-site, interference with mRNA positioning59, or decreased ribosome stability60. DT is extremely potent towards humans, with an estimated lethal dose of 100ng/kg61.
P. aeruginose PE
P. aeruginosa exotoxin A (PE) is a closely related AB toxin of 613 amino acids62. Despite limited primary amino acid homology (~18% identity between PE and DT), the A domains of PE and DT are structurally conserved, both containing the HYE motif36, 62. However, PE displays an inverted domain orientation compared to that of DT, with the B domain at the N terminus and the A domain at the C terminus63, 64. PE utilizes low-density lipoprotein receptor-related protein 1 (LRP1) as a host receptor65 (Figure 2A). Like DT, PE is cleaved by a cellular protease66 and is internalized through clathrin-mediated endocytosis67, but uniquely, PE undergoes retrograde trafficking through the Golgi to the ER via a C-terminal KDEL-like sequence. Within the ER, reduction of the disulphide bond and utilization of the cellular ER-associated degradation (ERAD) system allows the A domain to traverse the ER membrane into the cytosol68,69. Once in the cytosol, PE utilizes the same killing mechanism as DT, the ADP-ribosylation of EF270.
Cholix, a new V. cholerae virulence factor
Cholix toxin (ChxA) was identified through bioinformatic analysis of V. cholerae genomes. V. cholerae infections are typically associated with O1 or O139 strains, which produce the CT responsible for the rice water diarrhoea associated with this disease71, 72. Genome analyses of environmental73 and clinical74 strains of V. cholerae identified chxA, which has homology to the PE coding gene, toxA. Further analysis showed that roughly one-third of non-O1 and non-O139 strains encoded chxA, while O1 and O139 strains did not encode chxA, suggesting that chxA may contribute to non-CT-mediated virulence75.
chxA encodes the prototype cholix toxin (ChxA-I), a 666 residue protein with an AB organization similar to PE76, with N-terminal receptor binding and C-terminal ADP-ribosyltransferase domains (Figure 1E)77. Although Cholix shares 32% amino acid identity with PE, the GC content of chxA is 43% (the GC content of the chxA-containing V. cholerae NRT36S genome is 47%74) and the GC content of toxA is 69% (the GC content of the P. aeruginosa PAO1 genome is 67%78). This suggests that chxA was not the product of horizontal transfer between P. aeruginosa and V. cholerae. Cholix maintains the conserved HYE triad, a property of the DT-like toxins, with the catalytic Glu at position 58176. Full-length alignment with PE shows strong conservation of structure and function, and this was confirmed by ChxA-mediated ADP-ribosylation of EF276. Although ChxA can utilize the PE receptor (LRP1)65 to enter cells, ChxA also intoxicates cells deficient in LRP, indicating there may be a second toxin receptor76. Further examination into ChxA trafficking to determine whether ChxA follows the PE intoxication pathway is needed (Figure 2A). Additionally, ChxA has extended the potential of the DT-subfamily of toxins for therapeutic applications through its use as a potential cancer therapeutic agent (BOX 2). Recently, two chxA variants have been identified with substantial differences in amino acid sequence within the B domain (ChxA-II) and A domain (ChxA-III)75. In vitro, ChxA and ChxA-II are toxic to multiple cultured cell lines, whereas ChxA-III is non-toxic despite containing the catalytic Glu residue. Loss of toxicity may instead be due to replacement of the KDEL ER-targeting sequence with an HDEL sequence which could disrupt trafficking or processing steps, although this requires further testing. Injection of mice with Cholix or ChxA-II resulted in severe necrosis of the liver and death75.
Box 2. Double edged swords: utilization of bARTTs for therapy and research.
Although often considered damaging to human health, bARTTs also possess a remarkable capacity to contribute to positive health outcomes. The ability of bARTTs to kill cells at low doses, and particularly the extreme toxicity of DT-like toxins, has led to their use in a variety of novel therapeutic and basic science applications. The toxic effects of DT ADP-ribosyltransferase activity have been used for cell ablation in transgenic mice190 that do not express the DT receptor47. Expression of the DT receptor under control of a cell- or tissue-specific promoter results in loss of that cell type upon expression or injection of DT191, 192, allowing examination of cellular functions. Other DT-like toxins have been used in the development of immunotoxins. Fusion of the catalytic domain of PE to the variable domain of antibodies193 allows targeting of diseased-cell markers194. Immunotoxins targeting CD22195, which is upregulated in B-cell malignancies, have shown promise in clinical settings196. Immunotoxins engineered with the CD4 domain that binds the HIV envelope protein have been used to kill HIV infected cells197 and may be used in combination with other therapies198. Newer immunotoxins have been generated using the catalytic domain of Cholix toxin, which is non-reactive to PE-neutralizing antibodies199. bARTTs also play a role in neuron-specific applications. Fusion of DT to the binding domain of tetanus toxin demonstrated delivery of cargoes to neuronal cell lines, suggesting a mechanism for administering neuron-specific therapies200. Additionally, application of C3bot enhances growth and regeneration of neurons in cells201–203 and in vivo204, 205. Increased neuron growth and motility in a model of spinal contusion205 indicates that this toxin could serve as a therapy to promote regrowth of neurons in patients with damaged nervous systems. With the wide variety of receptors, entry mechanisms, eukaryotic substrates, and phenotypes, bARTTs will continue to be utilized to advance basic scientific knowledge and medical therapies.
CT-like toxins
CT, cAMP signalling and fluid secretion
In 1886, Koch predicted the existence of a secreted CT79, 80 that was eventually identified in 1959, after injection of a sterile spent culture filtrate of V. cholerae into rabbit intestines yielded a “rice water” fluid accumulation similar to the pathology elicited by V. cholerae infection81. CT is the prototype AB5 toxin. The CT B subunit binds up to 5 molecules of ganglioside GM182 on the membrane of intestinal epithelial cells83, which may result in receptor clustering and stimulation of endocytosis84. Upon delivery to the cytosol, A1 is activated by binding the small GTPase ADP-ribosylation factor(ARF)85–87 and ADP-ribosylates Gα of the heterotrimeric G-protein Gs88, 89. ADP-ribosylation locks Gα in a GTP-bound state to constitutively stimulate host adenylate cyclase3, 88. This leads to elevation of cAMP, which activates protein kinase A and in turn stimulates the action of a chloride ion efflux channel, leading to secretion of ions and water into the intestinal lumen, resulting in the watery diarrhoea associated with cholera. The closely related heat-labile enterotoxin from Escherichia coli also ADP-ribosylates the Gα subunit of the heterotrimeric Gs, causing activation of adenylate cyclase90–92.
Pertussis toxin, cAMP accumulation through Gi signalling
Bordetella pertussis produces pertussis toxin (PT), another member of the AB5 family of bARTTs, and of current relevance given the resurgence of pertussis in the developed world and worldwide infection of more than 16 million people annually93. PT consists of an ADP-ribosyltransferase domain (S1) noncovalently bound to a heteropentamer composed of 4 distinct subunits (S2–S5)94, 95. The B5 component of PT consists of two molecules of S4 and one molecule each of S2, S3, and S5. PT utilizes oligosaccharide receptors present on a variety of eukaryotic proteins, with separate carbohydrate binding sites within S2 and S396–98. PT is delivered to the ER99, where a disulphide bond that locks S1 in an inactive conformation is reduced100, 101 allowing delivery into the cytosol. S1 ADP-ribosylates Cys3475 on Gα of the heterotrimeric Gi-protein102, 103, uncoupling signalling from the G-protein coupled receptor104. This uncoupling prevents Gαi from regulating adenylate cyclase stimulation, resulting in amplification of cAMP signalling upon G-protein coupled receptor activation. Thus, although both stimulate cAMP production, CT and PT use disparate signalling pathways to achieve this end result, with unique physiological and clinical outcomes.
C2-like toxins
Actin ADP-ribosylating binary toxins
The reference C2-like toxins, including C. botulinum C2 toxin6, C. perfringens iota toxin105, C. difficile toxin106, C. spiroforme toxin (CST)107, and Bacillus cereus vegetative insecticidal protein (VIP)108, are binary toxins that ADP-ribosylate actin. C2-like toxins consist of A and B proteins secreted independently that associate on the surface of host cells. The B components are structurally and functionally related to protective antigen (PA), the binding component of anthrax toxin, and form PA-like heptamers109 which translocate A proteins into target cell cytosol. Iota toxin, C. difficile toxin, and CST B proteins bind the same cell membrane receptor, lipolysis-stimulated lipoprotein receptor (LSR)110, thereby comprising a subfamily of iota-like toxins13. The A component comprises two ADP-ribosyltransferase domains, likely resulting from gene duplication, one of which is inactive (unable to bind NAD) and used as an adaptor for interaction with the binding component111. bARTTs from this family ADP-ribosylate monomeric G-actin at Arg177, thereby inhibiting actin polymerization, inducing F-actin depolymerization and destruction of the actin cytoskeleton112–114.
Interestingly, destruction of the actin cytoskeleton may not be the only pathogenic role of these binary toxins. C. difficile toxin, which is frequently found in hypervirulent C. difficile strains115, causes unexpected effects at moderate toxin concentrations. C. difficile toxin-induced ADP-ribosylation of actin is accompanied by formation of microtubule-based cell protrusions116, which form a network of tentacle-like processes on the cell surface. This network embeds C. difficile toxin-producing bacteria and increases their adherence and colonization. Additionally, C. difficile toxin-induced ADP-ribosylation of actin results in re-routing of Rab11-positive vesicles containing fibronectin from the basolateral to the apical side of epithelial cells, where fibronectin is released at microtubule protrusions to generate a binding site for C. difficile117. Similar effects on microtubules were obtained with the other binary actin-ADP-ribosylating toxins116, suggesting more sophisticated consequences of actin ADP-ribosylation for host-pathogen interaction than mere destruction of the cytoskeleton.
Actin ADP-ribosylating toxins with novel organization
In addition to the binary actin-ADP-ribosylating toxins, several single-chain secreted effectors including Salmonella SpvB118, 119, Aeromonas hydrophila exoenzyme T (AexT)120, 121, and P. luminescens Photox122 possess ADP-ribosyltransferase domains that modify eukaryotic actin with similar functional consequences for the actin cytoskeleton. These effectors were identified through bioinformatics, with this strategy exemplified by the discovery of SpvB. spvB was a gene of unknown function until it was identified through PSI-BLAST analyses of known mono-ADP-ribosyltransferases119. SpvB is secreted by the Salmonella Type III secretion system and is required for full virulence in mice and human macrophages118. The C terminus of SpvB has only 19% sequence identity to the ADP-ribosyltransferase domain of VIP2, the closest bARTT relative, but the typical RSE motif of C2-like ADP-ribosyltransferases was easily identified. Function was confirmed by demonstrating ADP-ribosylation of actin at Arg177123. AexT was identified by bioinformatics through similarities to P. aeruginosa ExoS, as it is secreted through a Type III secretion system and possesses an N-terminal RhoGAP domain and a C-terminal ARTT domain120, 124, 125. However, unlike ExoS126, 127, AexT only displays ADP-ribosyltransferase activity towards actin, modifying actin at Arg177121. AexT also shows potent RhoGAP activity towards Rho, Rac, and CDC42, resulting in synergistic enhancement of actin cytoskeleton depolymerization128. Photox, a two-domain protein from P. luminescens, was identified through BLAST analysis, which revealed 39% overall sequence identity with SpvB122. The C-terminal domain shares 61% identity with the ARTT domain of SpvB and, similarly, Photox modifies Arg177 of actin. Photox secretion is through an as-yet-unidentified mechanism; however, Photox contains a unique N-terminal domain of unknown structure and function which may contribute to secretion.
C3-like toxins
C3bot, uncoupling Rho signaling
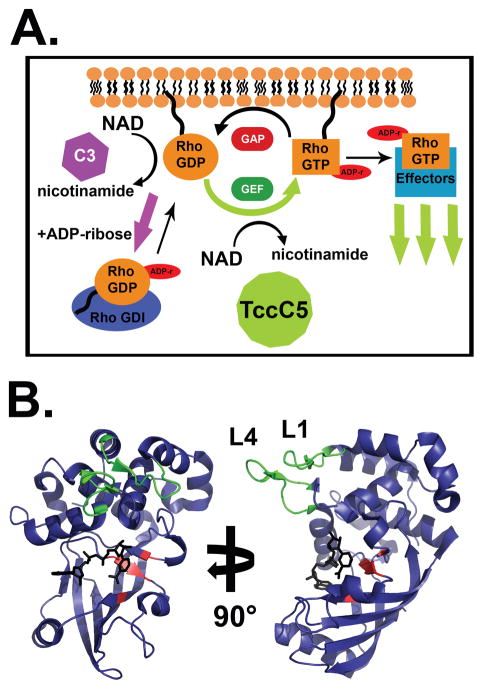
C3 exoenzymes have a single domain organization, consisting solely of an ADP-ribosyltransferase A domain. C3-like toxins ADP-ribosylate RhoGTPases, primarily RhoA, RhoB, and RhoC1, 129, 130. Rho GTPases are small-molecular weight G-proteins, which modulate the state of actin polymerization by cycling between GTP-bound active and GDP-bound inactive states though interactions with guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs) (Figure 3A). GTP binding promotes association with downstream effector proteins, primarily resulting in CDC42 modulation of cellular polarity, Rac modulation of cell motility, and Rho modulation of actin stress fibres131. However, Rho proteins also affect changes in adhesion, morphogenesis, vesicular trafficking, microtubule dynamics, and gene expression132. C3bot ADP-ribosylates Rho on Asn41 at the edge of the Switch-I region133 which typically undergoes conformational change with GDP/GTP nucleotide cycling. Unlike other bARTTs that inhibit interactions with effector proteins, ADP-ribosylated Rho can still interact with effectors134, but has an increased affinity for RhoGDI135, which sequesters Rho in the cytoplasm and prevents activation by RhoGEFs136 (Figure 3A). High affinity binding to RhoGDI blocks actin-stress fiber polymerization and collapses the cytoskeleton. C3bot also interacts with the Ras-related GTPase Ral. C3bot does not ADP-ribosylate Ral, but through high-affinity binding prevents nucleotide exchange and activation of Ral in a GDI-like manner137. The functional consequences of the C3-Ral complex formation are unclear.
Figure 3. Modulation of Rho signalling by bARTTs.
A. Rho proteins cycle between active (GTP-bound) and inactive (GDP-bound) states at the plasma membrane, or can be bound by RhoGDI in the cytoplasm. GTP-bound Rho interacts with downstream proteins to effect signal transduction and actin stress fiber polymerization. C3 ADP-ribosylation of Rho results in an increased affinity for RhoGDI and sequestration of Rho in the cytoplasm, resulting in cytoskeletal collapse. Conversely, TccC5 ADP-ribosylation of Rho inhibits GTP hydrolysis and promotes constitutive Rho activation and stress fiber polymerization. B. Crystal structure of HopU1. HopU1 shows a conserved C3-like ADP-ribosyltransferase core structure (see Figure 1D) with unique regions responsible for substrate recognition. The “RSE” motif residues (red) and the novel substrate recognition loops L1 and L4 (green) are denoted (PDB:3U0J).
As C3 exoenzymes do not have a B domain, entry into host cells occurs through other pathways. For example, C3Stau (EDIN)138 from Staphyloccus aureus is proposed to be secreted into the host cytoplasm following bacterial internalization by HeLa cells139, while other C3 homologues are taken up by macrophage-like cells140. The entry of each C3 may represent a directed mechanism to inhibit phagocytic activity of specific cells. Intriguingly, C3bot also appears to play a role in stimulating neuron growth and regeneration, suggesting it may have therapeutic potential (BOX 2). Recent bioinformatics surveys have discovered new C3-like toxins with novel substrate specificities and pathogenic mechanisms.
SpyA, a membrane-anchored bARTT
S. pyogenes is a Gram-positive human pathogen and the primary agent of Group A streptococcal infections, which cause a spectrum of diseases encompassing mild pharyngitis (“strep throat”) to streptococcal toxic shock syndrome, which if untreated can be fatal within hours. Recognized S. pyogenes virulence factors include adhesion-like molecules, a hyaluronic-acid-containing capsule, and superantigen toxins141. Genome sequencing has identified >40 putative virulence-associated genes, including a gene encoding a candidate ADP-ribosyltransferase, termed spyA142. Genomes of multiple clinical isolates of S. pyogenes encode spyA, suggesting that SpyA may have a role in pathogenesis in humans143.
Sequence analysis predicted that SpyA would exhibit C3-transferase activity; spyA encodes a 25-kDa protein consisting of 250 amino acids and a putative 30-residue leader sequence. Unexpectedly, SpyA was not detected in S. pyogenes culture supernatants; instead the predicted leader sequence formed a transmembrane helix and was inserted into the bacterium outer membrane144. SpyA contains a C3-like RSE motif with the predicted catalytic Glu at position 187143. Despite bioinformatics data suggesting that SpyA was a C3-like toxin, recombinant SpyA did not ADP-ribosylate RhoGTPases, but rather ADP-ribosylated vimentin, with actin a minor substrate143. These substrates are also targeted by other bARTTs; C2-like toxins ADP-ribosylate actin6 and ExoS of P. aeruginosa ADP-ribosylates vimentin126. Vimentin is a highly conserved multifunctional intermediate filament protein involved in promoting cell adhesion and integrating signalling pathways, as well as being key in the organization of cellular architecture145. Vimentin consists of a central coil domain, flanked by head and tail domains, and forms anti-parallel dimers and tetramers dependent on the head-tail phosphorylation state. Vimentin also plays a role in migration of immune cells, and loss of vimentin function results in impaired wound healing146.
Stoichiometric analysis showed that SpyA ADP-ribosylated vimentin at two arginine residues, Arg44 and Arg49147. These residues are located in the head domain of vimentin, and modification of residues in the head domain prevents vimentin from forming higher order structures148. ADP-ribosylation likely blocks vimentin oligomerization into intermediate filaments, resulting in disruption of vimentin filaments and cytoskeletal collapse147. SpyA contributes to S. pyogenes pathogenesis in vivo, as a SpyA-deficient mutant is internalized more readily by HeLa cells and causes smaller bacterial lesions than those caused by SpyA-containing bacteria in a mouse cutaneous infection model149.
SpyA is an excellent example of both the strengths and limitations of bioinformatics. Bioinformatics correctly predicted that SpyA was a bARTT but its initial classification as a C3-like transferase was not entirely correct. SpyA ADP-ribosylates a different subset of eukaryotic proteins to promote bacterial survival and virulence and, unlike other C3-like toxins, is anchored in the bacterial membrane to promote bacterial pathogenesis.
HopU1, modulator of plant immunity
bARTTs are not limited to human pathogens. Pseudomonas syringae infects >50 species of plants, utilizing >30 type-III secreted effector proteins150–152, most of which are of unknown function. Bioinformatic analysis of the P. syringae genome, which was sequenced in the early 2000s153, 154, identified a putative bARTT, denoted HopU1, which was predicted to be a 264 amino acid protein with a conserved RSE motif155 (Figure 3B). Although the domain fold of HopU1 displays high overall similarity to C3bot, the two proteins share only 19% amino acid identity. Additionally, HopU1 contains two unique loops, L1 and L4 (Figure 3B), which are not present in other bARTTs and have been implicated in substrate recognition and binding156. Mutation of key residues in L1 or L4 results in loss of substrate binding and ADP-ribosylation, suggesting that unique loops outside the core NAD-binding pocket may define substrate specificity in these toxins156. This is similar to Crk recognition by another C3-like toxin, ExoT of P. aeruginosa157, where an α-helix adjacent to the NAD binding pocket encodes the site of substrate recognition158.
Like SpyA, HopU1 does not ADP-ribosylate Rho proteins. Instead, HopU1 ADP-ribosylates glycine-rich RNA-binding protein-7 (GRP7)155. GRP7 is involved in processing mRNA and promoting mRNA stability within plant cells159, as well as interacting with components of the translational machinery160. Loss of GRP7 function results in increased susceptibility to pathogens155, 156, suggesting a role in mediating the immune response to infection. HopU1 ADP-ribosylates GRP7 at Arg49155, which is located in conserved ribonucleoprotein consensus sequence 1 (RNP1), which has been implicated in RNA binding in other proteins161, 162. Arg49 forms a salt-bridge with the RNA phosphate backbone, promoting transcript stability163, 164, and a point mutation at Arg49 results in a decrease in GRP7 mRNA binding affinity165. In vivo, GRP7 binds various mRNA transcripts, including mRNA encoding the pattern recognition receptors (PRRs) FLS2 and EFR160. PRRs are a part of the plant innate immune system166, 167, responsible for recognizing pathogen-associated molecular patterns (PAMPs) such as LPS, peptidoglycan, and bacterial proteins like flagellin and bacterial elongation factor-Tu. Bacterial activation of PRRs results in a PAMP-mediated immune response that upregulates production of reactive oxygen species and expression of innate immunity genes168. HopU1, therefore, inhibits GRP7 binding and stabilization of the PRR transcripts, resulting in downregulation of PRR protein expression and suppression of plant immunity160. The ability to limit inhibition of translation to proteins of the innate immune system contrasts with the total inhibition of protein synthesis and cell death caused by the DT-like bARTTs. In this way, P. syringae, a biotrophic pathogen, suppresses the host immune response while sparing the life of the host cell. As with SpyA, while bioinformatics analyses correctly identified HopU1 as a novel bARTT, the substrate and mechanism of HopU1 activity were solved by conventional cell biology and biochemical approaches.
Novel bARTTs
Typhoid toxin, a novel chimera with dual activities
Salmonella enterica serovar Typhi is the causative agent of typhoid fever which kills ~ 200,000 people annually169. Until recently, the virulence factors that contribute to systemic infection with S. Typhi were not well understood. Bioinformatic analysis of the S. Typhi genome identified a gene (cdtB) with similarity to genes encoding cytolethal-distending toxins (CDTs) expressed by several other bacterial pathogens170. However, although the putative A component CdtB possessed DNase activity when transiently expressed within cultured cells, a corresponding B component was not apparent. Deeper sequence analysis of the pathogenicity islet containing cdtB identified two additional genes exhibiting similarity to the sequences of the pertussis toxin S1 (pltA) and S2 (pltB) subunit genes171. Unexpectedly, all three proteins were produced during S. Typhi intracellular infection of mammalian cells, and deletion of pltA or pltB eliminated cdtB-mediated pathology, linking the three gene products. The holotoxin was termed “typhoid toxin” (TT), after the disease caused by S. Typhi. Crystallization of TT showed a novel 2AB5 domain organization, with 2A comprising one unit of CdtB and one unit of PltA and B5 comprising 5 PltB subunits30 (Figure 1F, left). CdtB and PltA are joined by a disulfide bond created by two unique cysteine residues not present in PT or other CDTs; PltA and PltB interact through the insertion of a short PltA α-helix into the PltB B5 pentamer (Figure 1F, right). PltB and CdtB do not appear to have direct contact30. Purified TT binds a broad range of gangliosides as cellular receptors, with a preference for the disialic acid moiety on GD2.
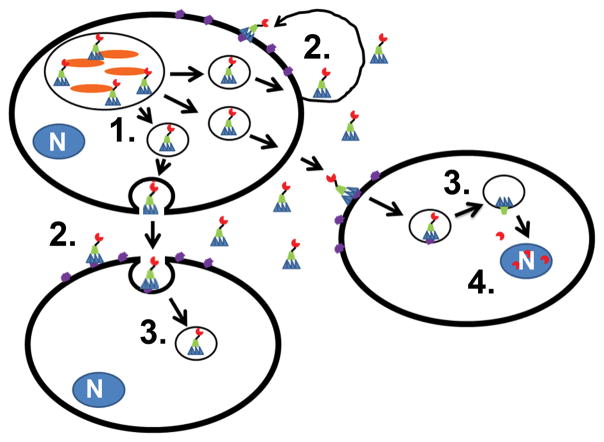
Intoxication of mice with TT elicited symptoms similar to typhoid fever, including weight loss, depletion of neutrophils, and death30. While PltA possessed significant structural homology to the pertussis S1 subunit, PltA ADP-ribosyltransferase activity targets an as-yet-unidentified protein. Each of the three components of TT are expressed by intracellular S. Typhi, but not by S. Typhi cultured alone in liquid media171. Interestingly, growth media from S. Typhi infected cells contained CDT-toxicity, which indicates that full-length TT is secreted into the extracellular media171. Subcellular studies showed that each of the three components of TT colocalized in membrane-bound vesicles, which presumably contribute to the delivery of the toxin to the extracellular media171. Addition of a TT-neutralizing antibody blocked toxic effects on neighboring cells and toxicity within S. Typhi infected cells, indicating that TT may utilize both autocrine and paracrine intoxication pathways (Figure 4). A recent report suggests that TT is packaged into LPS-positive vesicles, which are then released into the extracellular medium; the TT-containing vesicles are then taken up by neighboring cells through dynamin-dependent endocytosis and TT is delivered via a Golgi-mediated pathway 172.
Figure 4. Typhoid toxin utilizes a unique intoxication mechanism.
Salmonella Typhi (orange) produce TT within the Salmonella Containing Vacuole (SCV) and package TT into outer-membrane vesicles [1]. TT is delivered into the extracellular space and binds to ganglioside-containing proteins on infected or uninfected cells [2]. TT is taken up by endocytic pathway and is delivered into the cytoplasm via the Golgi [3]. CdtB enters the nucleus and cleaves DNA to promote cytopathology [4].
While the bioinformatics prediction that TT was similar to the CDTs and pertussis toxin was confirmed by crystallographic analyses, the novel structural organization and toxin delivery mechanism were not predicted. Additionally, the cellular target of PltA ADP-ribosyltransferase activity has not been identified, despite homology to pertussis toxin S1, again illustrating the limits of current bioinformatics analysis.
TccC proteins
bARTTs also exist in insect pathogens. Large (>1.5 MDa) tripartite toxin complexes, called Tc or ABC toxins, are produced by entomopathogenic P. luminescens, which lives in symbiosis with nematodes (e.g., Heterorhabditis species)173–175. Nematodes harbouring the bacteria in their intestine invade insect larvae and release the bacteria by regurgitation. Once released into the insect haemocoel, the bacteria produce an array of toxins to kill the insect, generating an enormous source of nutrients for proliferation of bacteria and nematodes. When the larvae have been depleted, the nematodes take up the bacteria then invade new insect prey175.
Tc toxin, which consists of three components, TcA, TcB and TcC, is one of the most potent toxins produced by P. luminescens176, 177. TcA (285 kDa) is the cell binding and translocation B component and forms B pentamers178. TcC (~105 kDa) is the biologically catalytic A component. Like TcA and TcB, TcC occurs in several isoforms179 and consists of a highly conserved N-terminal region of 660–680 amino acids and a C-terminal variable region of about 300 residues. TcB (170 kDa) is a linker between TcA and TcC.
Using bioinformatics, the variable regions of TcC were identified as bARTTs with typical RSE motifs31. In the presence of TcA and TcB, TccC3 induces clustering of F-actin in insect and mammalian cells. TccC3 ADP-ribosylates actin at Thr148, with the ADP-ribose attachment located in the binding site of thymosin-β4, which normally sequesters monomeric actin180, 181. Thus, ADP-ribosylation blocks thymosin-β4 binding and polymerization of actin is induced. Component TccC5 also induces strong stress fiber formation in the presence of TcA and TcB, although through a different mechanism31 (Figure 3A). In contrast to the C3 toxins, which inactivate Rho signaling through ADP-ribosylation of Asn41 in the Switch-I region of Rho proteins, TccC5 ADP-ribosylates RhoGTPases at Gln61/63, located in the Switch-II region31. Switch-II conformational changes are crucial for GTP hydrolysis182 and TccC5 ADP-ribosylation locks Rho proteins into a constitutively active form, resulting in activation of Rho kinase and/or formins to induce stress fiber formation31. The Tcc proteins, therefore, act antagonistically to the other bARTTs that act on the Rho proteins or actin, which inhibit protein function and block actin polymerization. TccC3- and TccC5-induced actin polymerization also inhibits functions of immune cells (e.g., blockade of phagocytosis) present in the hemocoel of insects.
Recently, the crystal structures of the components of the Tc toxin have been solved183, showing that TcA forms a pentameric prepore structure consisting of a central channel shaped like a flared funnel, with an outer shell compartment, resulting in a bell-like structure. The crystal structure of the TcdB2-TccC3 complex shows a huge β-barrel-shaped cage, with the ADP-ribosyltransferase domain in an unfolded form within the cage. The top of the barrel is closed by highly conserved rearrangement hot spot (RHS)-repeat associated domain. An aspartate protease domain is located in this region, which likely cleaves the already unfolded C-terminal ADP-ribosyltransferase domain of the TcC component and is released into the β-barrel-shaped cage. Once released, TcC can be injected into host cells by the TcA B pentamer syringe.
CONCLUSIONS
This research area has changed dramatically over the past decade with the advent of bioinformatics. bARTTs can be identified from genome sequences and can be experimentally tested without any prior association with disease. However, although bioinformatics provides a starting platform for the identification of putative virulence factors, Falkow’s molecular Koch’s postulates should still be considered for definitive identification184, 185. Ensuring that the putative virulence factor exists only in a pathogenic genus or strain and that deletion or mutation of the gene encoding the proposed virulence factor results in loss of pathogenicity must be demonstrated to show that the bioinformatics-identified determinant has a role during infection. Replacement or restoration of the wild-type gene should then be shown to restore the pathogenic phenotype. These guidelines should be utilized to triage the increasing number of putative toxins and other virulence factors that are being identified through bioinformatic methods. Following these guidelines will ensure that the most important virulence determinants are investigated and therapies or vaccines targeted against the most important molecules.
With the large number of identified members allowing rigorous formulation of conserved structural and sequence determinants to identify new toxins from sequenced genomes, the bARTT family demonstrates the power of bioinformatics to identify new toxins based on in silico analyses. While bioinformatics has been used to identify other putative virulence factors16, the number of available data sets for each group of virulence factors is limited when compared to the number of identified bARTT sequences and available crystal structures186. However, although the ability to predict a bARTT is strongly supported, this predictive capability wanes when he aim is to identify the substrate. In the near future, the “discovery” of new members of the family of bARTTs through fold-recognition searches based upon the conserved NAD binding domain and key amino acid residues will play an increasingly important role in advancing the field. The increasing number of bARTT family members will provide greater power to predict and study the properties of these toxins and may establish more defined rules for these determinations and better prediction of toxin substrates. Eventually, these rules may be extrapolated to other less-well characterized families of protein virulence determinants and increase our understanding of bacterial pathogenesis.
Online Summary.
The advent of bioinformatics has changed how the study of microbial pathogenesis is performed, allowing for development of “reverse proteomics” strategies. No longer do researchers need to identify a pathogen clinically before identifying putative virulence factors.
These analyses can be used to identify new bARTTs based on conserved function, even with novel structural organizations. This strategy has been proven through identification of putative bARTTs followed by experimental confirmation of ADP-ribosyltransferase activity. Multiple novel protein toxins have been identified and characterized in this manner.
New toxins such as Cholix, SpyA, HopU1, SpvB, and the TcC proteins have been identified through this strategy of “reverse proteomics” and shown experimentally to display the predicted ADP-ribosyltransferase enzymatic activity.
However, this strategy still has limitations. Many of the novel toxins display unique substrate, structure, or delivery properties not predicted by bioinformatic methods. Experimental confirmation of the significance of a putative virulence factor according to Falkow’s molecular postulates also needs to be considered.
Continuing growth of the family of bacterial ADP-ribosylating toxins should allow greater ability to create “rules” to properly identify substrates and structural organization of novel toxins. Extrapolation of this technique to other toxins or families of proteins may greatly enhance the field of microbiology.
Acknowledgments
NCS and JTB are supported by a grant for the NIH AI30162.
Glossary
- SN1 strain-alleviation mechanism
This proposed electrophilic reaction mechanism starts with the hydrolytic release of nicotinamide from the NAD donor, followed by formation of an oxocarbenium cation intermediate in a “strained” conformation. Rotation around the phosphodiester bond forms a second oxocarbenium cation, resulting in strain relief and moving the ribose moiety near the acceptor residue to complete the ADP-ribose transfer.
- P (peptidyl) site
Site on the small ribosomal subunit that holds the tRNA molecule linked to the growing end of the polypeptide chain.
- Retrograde trafficking
Trafficking of vesicles in a direction that starts from the host cell surface and ends in the endoplasmic reticulum (ER); for example, trafficking from the Golgi complex to the ER.
- ER-associated degradation (ERAD)
A cellular pathway that targets misfolded proteins in the endoplasmic reticulum for ubiquitylation and subsequent degradation by the proteasome.
- Intermediate filaments
Filaments formed by coiled-coil-rich cytoskeletal proteins, such as keratin.
- Pattern-recognition receptor
Soluble or membrane-associated receptor displayed by the metazoan host that can recognize complex molecular patterns on the surface of microorganisms.
- Autocrine
Activation of cellular receptors on the same cell that produces the ligand.
- Paracrine
Activation of cellular receptors on cells adjacent to the cell producing the ligand.
Biographies
Nathan Simon is a graduate student with Joseph Barbieri in the Department of Microbiology and Molecular Genetics at the Medical College of Wisconsin. He received his undergraduate training in biology and chemistry at Lawrence University in Appleton, Wisconsin. His research characterizes the biochemistry and cell biology of bacterial ADP-ribosylating toxins.
Klaus Aktories is Professor of Pharmacology and Toxicology at the University of Freiburg in Freiburg, Germany. He received a M.D. from the University Frankfurt in Germany and a Ph.D. in pharmacology from University Heidelberg in Germany. His laboratory has pioneered studies on the pharmacology, toxicology and cell biology of bacterial toxins, focusing toxins that modulate small GTPases.
Joseph Barbieri is Professor of Microbiology and Molecular Genetics at the Medical College of Wisconsin. He received a Ph.D. in microbiology from the University of Massachusetts- Amherst. His laboratory has pioneered studies on the mechanisms of protein toxin action, focusing on the properties of ADP-ribosylating toxins and the clostridial neurotoxins.
References
- 1.Aktories K, Braun U, Rösener S, Just I, Hall A. The rho gene product expressed in E. Coli is a substrate of botulinum ADP-ribosyltransferase C3. Biochemical and Biophysical Research Communications. 1989;158:209–213. doi: 10.1016/s0006-291x(89)80199-8. [DOI] [PubMed] [Google Scholar]
- 2.Braun U, Habermann B, Just I, Aktories K, Vandekerckhove J. Purification of the 22 kDa protein substrate of botulinum ADP-ribosyltransferase C3 from porcine brain cytosol and its characterization as a GTP-binding protein highly homologous to the rho gene product. FEBS Letters. 1989;243:70–76. doi: 10.1016/0014-5793(89)81220-7. [DOI] [PubMed] [Google Scholar]
- 3.Cassel D, Selinger Z. Mechanism of adenylate cyclase activation by cholera toxin: Inhibition of GTP hydrolysis at the regulatory site. Proc Natl Acad Sci U S A. 1977;74:3307–3311. doi: 10.1073/pnas.74.8.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gill DMaRM. ADP-ribosylation of membrane proteins catalyzed by cholera toxin: basis of the activation of adenylate cyclase. Proc Natl Acad Sci. 1978;75:3050–3054. doi: 10.1073/pnas.75.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West RE, Moss J, Vaughan M, Liu T, Liu TY. Pertussis toxin-catalyzed ADP-ribosylation of transducin. Cysteine 347 is the ADP-ribose acceptor site. Journal of Biological Chemistry. 1985;260:14428–14430. [PubMed] [Google Scholar]
- 6.Aktories K, et al. Botulinum C2 toxin ADP-ribosylates actin. Nature. 1986;322:390–392. doi: 10.1038/322390a0. [DOI] [PubMed] [Google Scholar]
- 7.Holbourn KP, Shone CC, Acharya KR. A family of killer toxins. FEBS Journal. 2006;273:4579–4593. doi: 10.1111/j.1742-4658.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 8.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends in Biochemical Sciences. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Tsuge H, et al. Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens ι-toxin. Proceedings of the National Academy of Sciences. 2008;105:7399–7404. doi: 10.1073/pnas.0801215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsurumura T, et al. Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex. Proceedings of the National Academy of Sciences. 2013;110:4267–4272. doi: 10.1073/pnas.1217227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jank T, Aktories K. Strain-alleviation model of ADP-ribosylation. PNAS. 2013;110:4163–4164. doi: 10.1073/pnas.1302537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barth H, Aktories K. New insights into the mode of action of the actin ADP-ribosylating virulence factors Salmonella enterica SpvB and Clostridium botulinum C2 toxin. European Journal of Cell Biology. 2011;90:944–950. doi: 10.1016/j.ejcb.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Barth H, Aktories K, Popoff MR, Stiles BG. Binary Bacterial Toxins: Biochemistry, Biology, and Applications of Common Clostridium and Bacillus Proteins. Microbiol Mol Biol Rev. 2004;68:373–402. doi: 10.1128/MMBR.68.3.373-402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogelsgesang M, Pautsch A, Aktories K. C3 exoenzymes, novel insights into structure and action of Rho-ADP-ribosylating toxins. Naunyn-Schmiedeberg’s Archives of Pharmacology. 2007;374:347–360. doi: 10.1007/s00210-006-0113-y. [DOI] [PubMed] [Google Scholar]
- 15.Dueholm MS, Albertsen M, Otzen D, Nielsen PH. Curli Functional Amyloid Systems Are Phylogenetically Widespread and Display Large Diversity in Operon and Protein Structure. PLoS ONE. 2012;7:e51274. doi: 10.1371/journal.pone.0051274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doxey AC, McConkey BJ. Prediction of molecular mimicry candidates in human pathogenic bacteria. Virulence. 2013;4:453–466. doi: 10.4161/viru.25180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Priest NK, et al. From genotype to phenotype: can systems biology be used to predict Staphylococcus aureus virulence? Nat Rev Micro. 2012;10:791–797. doi: 10.1038/nrmicro2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masignani V, et al. In silico identification of novel bacterial ADP-ribosyltransferases. International Journal of Medical Microbiology. 2004;293:471–478. doi: 10.1078/1438-4221-00296. [DOI] [PubMed] [Google Scholar]
- 19.Fieldhouse RJ, Turgeon Z, White D, Merrill AR. Cholera- and Anthrax-Like Toxins Are among Several New ADP-Ribosyltransferases. PLoS Comput Biol. 2010;6:e1001029. doi: 10.1371/journal.pcbi.1001029. Demonstrated that bioinformatics could “discover” new toxins through the use of protein fold homology and amino acid sequence conservation searches. Characterized several toxins in silico, several of which were later confirmed to possess ADP-ribosyltransferase activity experimentally. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choe S, et al. The crystal structure of diphtheria toxin. Nature. 1992;357:216–222. doi: 10.1038/357216a0. [DOI] [PubMed] [Google Scholar]
- 21.Gill DM, Dinius LL. Observations on the structure of diphtheria toxin. Journal of Biological Chemistry. 1971;246:1485–1491. [PubMed] [Google Scholar]
- 22.Collier RJ, Kandel J. Structure and activity of diphtheria toxin. I. Thiol-dependent dissociation of a fraction of toxin into enzymically active and inactive fragments. Journal of Biological Chemistry. 1971;246:1496–1503. [PubMed] [Google Scholar]
- 23.Gill DM, Pappenheimer AM., Jr Structure-activity relationships in diphtheria toxin. Journal of Biological Chemistry. 1971;246:1492–1495. [PubMed] [Google Scholar]
- 24.Zhang RG, et al. The Three-dimensional Crystal Structure of Cholera Toxin. Journal of Molecular Biology. 1995;251:563–573. doi: 10.1006/jmbi.1995.0456. [DOI] [PubMed] [Google Scholar]
- 25.Sixma TK, et al. Refined Structure of Escherichia coli Heat-labile Enterotoxin, a Close Relative of Cholera Toxin. Journal of Molecular Biology. 1993;230:890–918. doi: 10.1006/jmbi.1993.1209. [DOI] [PubMed] [Google Scholar]
- 26.Blöcker D, et al. The C Terminus of Component C2II of Clostridium botulinum C2 Toxin Is Essential for Receptor Binding. Infection and Immunity. 2000;68:4566–4573. doi: 10.1128/iai.68.8.4566-4573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schleberger C, Hochmann H, Barth H, Aktories K, Schulz GE. Structure and Action of the Binary C2 Toxin from Clostridium botulinum. Journal of Molecular Biology. 2006;364:705–715. doi: 10.1016/j.jmb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Aktories K, Weller U, Chhatwal GS. Clostridium botulinum type C produces a novel ADP-ribosyltransferase distinct from botulinum C2 toxin. FEBS Letters. 1987;212:109–113. doi: 10.1016/0014-5793(87)81566-1. [DOI] [PubMed] [Google Scholar]
- 29.Han S, Arvai AS, Clancy SB, Tainer JA. Crystal structure and novel recognition motif of Rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: structural insights for recognition specificity and catalysis. Journal of Molecular Biology. 2001;305:95–107. doi: 10.1006/jmbi.2000.4292. [DOI] [PubMed] [Google Scholar]
- 30.Song J, Gao X, Galan JE. Structure and function of the Salmonella Typhi chimaeric A2B5 typhoid toxin. Nature. 2013;499:350–354. doi: 10.1038/nature12377. Identified the crystal structure of Typhoid toxin with novel A2B5 arrangement and demonstrated its similarity to pertussis toxin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang AE, et al. Photorhabdus luminescens Toxins ADP-Ribosylate Actin and RhoA to Force Actin Clustering. Science. 2010;327:1139–1142. doi: 10.1126/science.1184557. Identified the substrates of the TcC components of Photorhabdus toxin complex proteins. Demonstrated that these toxins promote actin polymerization. [DOI] [PubMed] [Google Scholar]
- 32.Blackburn M, Golubeva E, Bowen D, Ffrench-Constant RH. A Novel Insecticidal Toxin from Photorhabdus luminescens, Toxin Complex a (Tca), and Its Histopathological Effects on the Midgut of Manduca sexta. Applied and Environmental Microbiology. 1998;64:3036–3041. doi: 10.1128/aem.64.8.3036-3041.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han S, Tainer J. The ARTT motif and a unified structural understanding of substrate recognition in ADP-ribosylating bacterial toxins and eukaryotic ADP-ribosyltransferases. Int J Med Microbiol. 2002;291:523–529. doi: 10.1078/1438-4221-00162. Proposed that toxins could be identified by core ARTT motif and suggested that substrate specificity was dependent on external protein structure. [DOI] [PubMed] [Google Scholar]
- 34.Wilson BA, Reich KA, Weinstein BR, Collier RJ. Active-site mutations of diphtheria toxin: Effects of replacing glutamic acid-148 with aspartic acid, glutamine, or serine. Biochemistry. 1990;29:8643–8651. doi: 10.1021/bi00489a021. [DOI] [PubMed] [Google Scholar]
- 35.Carroll SF, Collier RJ. NAD binding site of diphtheria toxin: identification of a residue within the nicotinamide subsite by photochemical modification with NAD. Proceedings of the National Academy of Sciences. 1984;81:3307–3311. doi: 10.1073/pnas.81.11.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll SF, Collier RJ. Active site of Pseudomonas aeruginosa exotoxin A. Glutamic acid 553 is photolabeled by NAD and shows functional homology with glutamic acid 148 of diphtheria toxin. Journal of Biological Chemistry. 1987;262:8707–8711. [PubMed] [Google Scholar]
- 37.Douglas D, Collier R. Exotoxin A of Pseudomonas aeruginosa: Substitution of Glutamic Acid 553 with Aspartic Acid Drastically Reduces Toxicity and Enzymatic Activity. Journal of Bacteriology. 1987;169:4967–4971. doi: 10.1128/jb.169.11.4967-4971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell CE, Eisenberg D. Crystal Structure of Diphtheria Toxin Bound to Nicotinamide Adenine Dinucleotide†. Biochemistry. 1996;35:1137–1149. doi: 10.1021/bi9520848. [DOI] [PubMed] [Google Scholar]
- 39.Oliver AW, et al. Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Research. 2004;32:456–464. doi: 10.1093/nar/gkh215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruf A, Mennissier de Murcia J, de Murcia G, Schulz GE. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proceedings of the National Academy of Sciences. 1996;93:7481–7485. doi: 10.1073/pnas.93.15.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuge H, et al. Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin. Journal of Molecular Biology. 2003;325:471–483. doi: 10.1016/s0022-2836(02)01247-0. [DOI] [PubMed] [Google Scholar]
- 42.Domenighini M, Rappuoli R. Three conserved consensus sequences identify the NAD-binding site of ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Molecular Microbiology. 1996;21:667–674. doi: 10.1046/j.1365-2958.1996.321396.x. [DOI] [PubMed] [Google Scholar]
- 43.Roux E, Jr, Yersin A. Contribution a l’etude de la diphtherie. Ann Inst Pasteur. 1888;2:620–629. [Google Scholar]
- 44.Eaton MD. The purification and concentration of diphtheria toxin. J Bact. 1936;31:347. doi: 10.1128/jb.31.4.347-366.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pappenheimer AM., Jr Diphtheria toxin - I: Isolation and characterization of a toxic protein from C. diphtheriae filtrates. J Biol Chem. 1937;125:543–553. [Google Scholar]
- 46.Freeman VJ. Studies on the virulence of bacteriophage infected strains of C. diphtheriae. J Bact. 1951;61 doi: 10.1128/jb.61.6.675-688.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naglich JG, Metherall JE, Russell DW, Eidels L. Expression cloning of a Diphtheria toxin receptor: Identity with a heparin-binding EGF-like growth factor precursor. Cell. 1992;69:1051–1061. doi: 10.1016/0092-8674(92)90623-k. [DOI] [PubMed] [Google Scholar]
- 48.Louie GV, Yang W, Bowman ME, Choe S. Crystal structure of the complex of diphtheria toxin with an extracellular fragment of its receptor. Molecular Cell. 1997;1:67–78. doi: 10.1016/s1097-2765(00)80008-8. [DOI] [PubMed] [Google Scholar]
- 49.Lemichez E, et al. Membrane translocation of diphtheria toxin fragment A exploits early to late endosome trafficking machinery. Molecular Microbiology. 1997;23:445–457. doi: 10.1111/j.1365-2958.1997.tb02669.x. [DOI] [PubMed] [Google Scholar]
- 50.Gordon VM, Klimpel KR, Arora N, Henderson MA, Leppla SH. Proteolytic activation of bacterial toxins by eukaryotic cells is performed by furin and by additional cellular proteases. Infection and Immunity. 1995;63:82–87. doi: 10.1128/iai.63.1.82-87.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandvig K, Olsnes S. Diphtheria toxin entry into cells is facilitated by low pH. Journal of Cell Biology. 1980;87:828–832. doi: 10.1083/jcb.87.3.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Draper RK, Simon MI. The entry of diphtheria toxin into the mammalian cell cytoplasm: Evidence for lysosomal involvement. Journal of Cell Biology. 1980;87:849–854. doi: 10.1083/jcb.87.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collier RJ. Effect of diphtheria toxin on protein synthesis: Inactivation of one of the transfer factors. Journal of Molecular Biology. 1967;25:83–98. doi: 10.1016/0022-2836(67)90280-x. [DOI] [PubMed] [Google Scholar]
- 54.Honjo T, Nishizuka Y, Hayaishi O, Kato I. Diphtheria Toxin-dependent Adenosine Diphosphate Ribosylation of Aminoacyl Transferase II and Inhibition of Protein Synthesis. Journal of Biological Chemistry. 1968;243:3553–3555. [PubMed] [Google Scholar]
- 55.Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. Journal of Biological Chemistry. 1980;255:10717–10720. [PubMed] [Google Scholar]
- 56.Su X, Lin Z, Lin H. The biosynthesis and biological function of diphthamide. Critical Reviews in Biochemistry and Molecular Biology. 2013;48:515–521. doi: 10.3109/10409238.2013.831023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strauss N, Hendee ED. THE EFFECT OF DIPHTHERIA TOXIN ON THE METABOLISM OF HELA CELLS. The Journal of Experimental Medicine. 1959;109:145–163. doi: 10.1084/jem.109.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor D, et al. Critical Movements of a Single Diphthamide Residue of Eukaryotic Elongation Factor 2 Monitored by Cryo-EM. Microscopy and Microanalysis. 2007;13:388–389. [Google Scholar]
- 59.Liu S, et al. Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proceedings of the National Academy of Sciences. 2012;109:13817–13822. doi: 10.1073/pnas.1206933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta PK, Liu S, Batavia MP, Leppla SH. The diphthamide modification on elongation factor-2 renders mammalian cells resistant to ricin. Cellular Microbiology. 2008;10:1687–1694. doi: 10.1111/j.1462-5822.2008.01159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gill DM. Bacterial toxins: a table of lethal amounts. Microbiol Rev. 1982;46:86–94. doi: 10.1128/mr.46.1.86-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carroll SF, Collier RJ. Amino acid sequence homology between the enzymic domains of diphtheria toxin and Pseudomonas aeruginosa exotoxin A. Molecular microbiology. 1988;2:293–296. doi: 10.1111/j.1365-2958.1988.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 63.Collier RJ, McKay DB. Crystallization of exotoxin A from Pseudomonas aeruginosa. Journal of Molecular Biology. 1982;157:413–415. doi: 10.1016/0022-2836(82)90243-1. [DOI] [PubMed] [Google Scholar]
- 64.Allured VS, Collier RJ, Carroll SF, McKay DB. Structure of exotoxin A of Pseudomonas aeruginosa at 3.0-Angstrom resolution. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:1320–1324. doi: 10.1073/pnas.83.5.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kounnas MZ, et al. The alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein binds and internalizes Pseudomonas exotoxin A. Journal of Biological Chemistry. 1992;267:12420–3. [PubMed] [Google Scholar]
- 66.Ogata M, Fryling CM, Pastan I, FitzGerald DJ. Cell-mediated cleavage of Pseudomonas exotoxin between Arg279 and Gly280 generates the enzymatically active fragment which translocates to the cytosol. Journal of Biological Chemistry. 1992;267:25396–25401. [PubMed] [Google Scholar]
- 67.Fitzgerald D, Morris RE, Saelinger CB. Receptor-mediated internalization of pseudomonas toxin by mouse fibroblasts. Cell. 1980;21:867–873. doi: 10.1016/0092-8674(80)90450-x. [DOI] [PubMed] [Google Scholar]
- 68.Weldon JE, Pastan I. A guide to taming a toxin – recombinant immunotoxins constructed from Pseudomonas exotoxin A for the treatment of cancer. FEBS Journal. 2011;278:4683–4700. doi: 10.1111/j.1742-4658.2011.08182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McKee ML, FitzGerald DJ. Reduction of furin-nicked Pseudomonas exotoxin A: An unfolding story. Biochemistry. 1999;38:16507–16513. doi: 10.1021/bi991308+. [DOI] [PubMed] [Google Scholar]
- 70.Iglewski BH, Liu PV, Kabat D. Mechanism of action of Pseudomonas aeruginosa exotoxin A: adenosine diphosphate ribosylation of mammalian elongation factor 2 in vitro and in vivo. Infection and Immunity. 1977;15:138–144. doi: 10.1128/iai.15.1.138-144.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Faruque SM, et al. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2123–2128. doi: 10.1073/pnas.0308485100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Colwell RR. Global Climate and Infectious Disease: The Cholera Paradigm*. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- 73.Purdy AE, et al. Diversity and distribution of cholix toxin, a novel ADP-ribosylating factor from Vibrio cholerae. Environmental Microbiology Reports. 2010;2:198–207. doi: 10.1111/j.1758-2229.2010.00139.x. [DOI] [PubMed] [Google Scholar]
- 74.Chen Y, Johnson JA, Pusch GD, Morris JG, Stine OC. The Genome of Non-O1 Vibrio cholerae NRT36S Demonstrates the Presence of Pathogenic Mechanisms That Are Distinct from Those of O1 Vibrio cholerae. Infection and Immunity. 2007;75:2645–2647. doi: 10.1128/IAI.01317-06. First identified the presence of the gene encoding Cholix toxin in non-cholera toxin producing strains of Vibrio cholerae using sequence analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Awasthi SP, et al. Novel Cholix Toxin Variants, ADP-Ribosylating Toxins in Vibrio cholerae Non-O1/Non-O139 Strains, and Their Pathogenicity. Infection and Immunity. 2013;81:531–541. doi: 10.1128/IAI.00982-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jørgensen R, et al. Cholix Toxin, a Novel ADP-ribosylating Factor from Vibrio cholerae. Journal of Biological Chemistry. 2008;283:10671–10678. doi: 10.1074/jbc.M710008200. Characterized the structure of Cholix toxin, demonstrating the similarity to other diphtheria-like toxins. Also proposed a new mechanism for diphtheria-like toxin ADP-ribosylation activity. [DOI] [PubMed] [Google Scholar]
- 77.Fieldhouse RJ, Jørgensen R, Lugo MR, Merrill AR. The 1.8 Å Cholix Toxin Crystal Structure in Complex with NAD+ and Evidence for a New Kinetic Model. Journal of Biological Chemistry. 2012;287:21176–21188. doi: 10.1074/jbc.M111.337311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 79.Koch R. Sechster Bericht der deutschen Wissenschaftlichen Commission zur Enforschung der Cholera. Dtsch Med Wochenschr. 1884;10:191–192. [Google Scholar]
- 80.Howard-Jones N. Robert Koch and the cholera vibrio: a centenary. BMJ. 1984;288:379–381. doi: 10.1136/bmj.288.6414.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De SN. Enterotoxicity of Bacteria-free Culture-filtrate of Vibrio cholerae. Nature. 1959;183:1533–1534. doi: 10.1038/1831533a0. [DOI] [PubMed] [Google Scholar]
- 82.Lencer WI, Hirst TR, Holmes RK. Membrane traffic and the cellular uptake of cholera toxin. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1999;1450:177–190. doi: 10.1016/s0167-4889(99)00070-1. [DOI] [PubMed] [Google Scholar]
- 83.Eidels L, Proia R, Hart D. Membrane receptors for bacterial toxins. Microbiol Rev. 1983;47:596–620. doi: 10.1128/mr.47.4.596-620.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ewers H, Helenius A. Lipid-Mediated Endocytosis. Cold Spring Harbor Perspectives in Biology. 2011;3 doi: 10.1101/cshperspect.a004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bobak D, et al. Mechanism of activation of cholera toxin by ADP-ribosylation factor (ARF): both low- and high-affinity interactions of ARF with guanine nucleotides promote toxin activation. Biochemistry. 1990;29:855–861. doi: 10.1021/bi00456a600. [DOI] [PubMed] [Google Scholar]
- 86.O’Neal CJ, Jobling MG, Holmes RK, Hol WGJ. Structural Basis for the Activation of Cholera Toxin by Human ARF6-GTP. Science. 2005;309:1093–1096. doi: 10.1126/science.1113398. [DOI] [PubMed] [Google Scholar]
- 87.Kahn RA, Gilman AG. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. Journal of Biological Chemistry. 1986;261:7906–7911. [PubMed] [Google Scholar]
- 88.Gill DM, Meren R. ADP-ribosylation of membrane proteins catalyzed by cholera toxin: basis of the activation of adenylate cyclase. Proceedings of the National Academy of Sciences. 1978;75:3050–3054. doi: 10.1073/pnas.75.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cassel D, Pfeuffer T. Mechanism of cholera toxin action: covalent modification of the guanyl nucleotide-binding protein of the adenylate cyclase system. Proc Natl Acad Sci U S A. 1978;75:2669–2673. doi: 10.1073/pnas.75.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moss J. Activation of Adenylate Cyclase by Heat-Labile Escherichia Coli Enterotoxin: EVIDENCE FOR ADP-RIBOSYLTRANSFERASE ACTIVITY SIMILAR TO THAT OF CHOLERAGEN. The Journal of Clinical Investigation. 1978;62:281–285. doi: 10.1172/JCI109127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chang PP, Moss J, Twiddy EM, Holmes RK. Type II heat-labile enterotoxin of Escherichia coli activates adenylate cyclase in human fibroblasts by ADP ribosylation. Infection and Immunity. 1987;55:1854–1858. doi: 10.1128/iai.55.8.1854-1858.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kantor HS, Tao P, Wisdom C. Action of Escherichia coli Enterotoxin: Adenylate Cyclase Behavior of Intestinal Epithelial Cells in Culture. Infection and Immunity. 1974;9:1003–1010. doi: 10.1128/iai.9.6.1003-1010.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.WHO. Immunization, Vaccines and Biologicals-Pertussis. 2011. [Google Scholar]
- 94.Tamura MNK, Murai S, Yajima M, Ito K, Katada T, Ui M, Ishii S. Subunit structure of islet-activating protein pertussis toxin in conformity with the A-B model. Biochemistry. 1982;21:5516–22. doi: 10.1021/bi00265a021. [DOI] [PubMed] [Google Scholar]
- 95.Stein PE, et al. The crystal structure of pertussis toxin. Structure (London, England : 1993) 1994;2:45–57. doi: 10.1016/s0969-2126(00)00007-1. [DOI] [PubMed] [Google Scholar]
- 96.Armstrong GD, Howard LA, Peppler MS. Use of glycosyltransferases to restore pertussis toxin receptor activity to asialoagalactofetuin. Journal of Biological Chemistry. 1988;263:8677–8684. [PubMed] [Google Scholar]
- 97.Stein P, et al. Structure of a pertussis toxin-sugar complex as a model for receptor binding. Nat Struct Biol. 1994;1:591–596. doi: 10.1038/nsb0994-591. [DOI] [PubMed] [Google Scholar]
- 98.Brennan MJ, David JL, Kenimer JG, Manclark CR. Lectin-like binding of pertussis toxin to a 165-kilodalton Chinese hamster ovary cell glycoprotein. Journal of Biological Chemistry. 1988;263:4895–4899. [PubMed] [Google Scholar]
- 99.el Bayâ A, Linnemann R, von Olleschik-Elbheim L, Robenek H, Schmidt M. Endocytosis and retrograde transport of pertussis toxin to the Golgi complex as a prerequisite for cellular intoxication. Eur J Cell Biol. 1997;73:40–48. [PubMed] [Google Scholar]
- 100.Burns DL, Manclark CR. Role of cysteine 41 of the A subunit of pertussis toxin. Journal of Biological Chemistry. 1989;264:564–568. [PubMed] [Google Scholar]
- 101.Antoine R, Locht C. Roles of the disulfide bond and the carboxy-terminal region of the S1 subunit in the assembly and biosynthesis of pertussis toxin. Infection and Immunity. 1990;58:1518–1526. doi: 10.1128/iai.58.6.1518-1526.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bokoch GM, Katada T, Northup JK, Hewlett EL, Gilman AG. Identification of the predominant substrate for ADP-ribosylation by islet activating protein. Journal of Biological Chemistry. 1983;258:2072–5. [PubMed] [Google Scholar]
- 103.Katada T, Ui M. Direct modification of the membrane adenylate cyclase system by islet-activating protein due to ADP-ribosylation of a membrane protein. Proceedings of the National Academy of Sciences. 1982;79:3129–3133. doi: 10.1073/pnas.79.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kurose H, Katada T, Amano T, Ui M. Specific uncoupling by islet-activating protein, pertussis toxin, of negative signal transduction via alpha-adrenergic, cholinergic, and opiate receptors in neuroblastoma x glioma hybrid cells. Journal of Biological Chemistry. 1983;258:4870–5. [PubMed] [Google Scholar]
- 105.Simpson LL, Stiles BG, Zepeda HH, Wilkins TD. Molecular basis for the pathological actions of Clostridium perfringens iota toxin. Infection and Immunity. 1987;55:118–122. doi: 10.1128/iai.55.1.118-122.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perelle S, Gibert M, Bourlioux P, Corthier G, Popoff MR. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infection and Immunity. 1997;65:1402–7. doi: 10.1128/iai.65.4.1402-1407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Simpson LL, Stiles BG, Zepeda H, Wilkins TD. Production by Clostridium spiroforme of an iotalike toxin that possesses mono(ADP-ribosyl)transferase activity: identification of a novel class of ADP-ribosyltransferases. Infection and Immunity. 1989;57:255–261. doi: 10.1128/iai.57.1.255-261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aktories K, Lang AE, Schwan C, Mannherz HG. Actin as target for modification by bacterial protein toxins. FEBS Journal. 2011;278:4526–4543. doi: 10.1111/j.1742-4658.2011.08113.x. [DOI] [PubMed] [Google Scholar]
- 109.Young JAT, Collier RJ. Anthrax Toxin: Receptor Binding, Internalization, Pore Formation, and Translocation. Annual Review of Biochemistry. 2007;76:243–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- 110.Papatheodorou P, et al. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT) Proceedings of the National Academy of Sciences. 2011;108:16422–16427. doi: 10.1073/pnas.1109772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Han S, Craig JA, Putnam CD, Carozzi NB, Tainer JA. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat Struct Mol Biol. 1999;6:932–936. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- 112.Vandekerckhove J, Schering B, Bärmann M, Aktories K. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Letters. 1987;225:48–52. doi: 10.1016/0014-5793(87)81129-8. [DOI] [PubMed] [Google Scholar]
- 113.Vandekerckhove J, Schering B, Bärmann M, Aktories K. Botulinum C2 toxin ADP-ribosylates cytoplasmic beta/gamma-actin in arginine 177. Journal of Biological Chemistry. 1988;263:696–700. [PubMed] [Google Scholar]
- 114.Gülke I, et al. Characterization of the Enzymatic Component of the ADP-Ribosyltransferase Toxin CDTa from Clostridium difficile. Infection and Immunity. 2001;69:6004–6011. doi: 10.1128/IAI.69.10.6004-6011.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gerding DN, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT: Mechanism, epidemiology, and potential clinical importance. Gut Microbes. 2014;5:6–18. doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schwan C, et al. Clostridium difficile Toxin CDT Induces Formation of Microtubule-Based Protrusions and Increases Adherence of Bacteria. PLoS Pathog. 2009;5:e1000626. doi: 10.1371/journal.ppat.1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schwan C, et al. Clostridium difficile toxin CDT hijacks microtubule organization and reroutes vesicle traffic to increase pathogen adherence. Proceedings of the National Academy of Sciences. 2014 doi: 10.1073/pnas.1311589111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lesnick ML, Reiner NE, Fierer J, Guiney DG. The Salmonella spvB virulence gene encodes an enzyme that ADP-ribosylates actin and destabilizes the cytoskeleton of eukaryotic cells. Molecular Microbiology. 2001;39:1464–1470. doi: 10.1046/j.1365-2958.2001.02360.x. [DOI] [PubMed] [Google Scholar]
- 119.Otto H, et al. The spvB gene-product of the Salmonella enterica virulence plasmid is a mono(ADP-ribosyl)transferase. Molecular Microbiology. 2000;37:1106–1115. doi: 10.1046/j.1365-2958.2000.02064.x. Identified the spvB gene through PSI-BLAST protein homology searches as similar to other mono-ADP-ribosyltransferases. [DOI] [PubMed] [Google Scholar]
- 120.Braun M, et al. Characterization of an ADP-Ribosyltransferase Toxin (AexT) from Aeromonas salmonicida subsp, salmonicida. Journal of Bacteriology. 2002;184:1851–1858. doi: 10.1128/JB.184.7.1851-1858.2002. Used bioinformatics to identify AexT as similar to Pseudomonas Exoenzyme S through sequence similarity and characterized the cytotoxic activity of AexT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fehr D, et al. Aeromonas Exoenzyme T of Aeromonas salmonicida Is a Bifunctional Protein That Targets the Host Cytoskeleton. Journal of Biological Chemistry. 2007;282:28843–28852. doi: 10.1074/jbc.M704797200. Identified AexT as a bifunctional toxin and characterized its ADP-ribosyltransferase substrates. [DOI] [PubMed] [Google Scholar]
- 122.Visschedyk DD, et al. Photox, a Novel Actin-targeting Mono-ADP-ribosyltransferase from Photorhabdus luminescens. Journal of Biological Chemistry. 2010;285:13525–13534. doi: 10.1074/jbc.M109.077339. Used bioinformatics to identify Photorhabdus toxin through amino acid homology with previously identified toxin SpvB. Showed structure-function similarity between Photox and other actin-ADP-ribosylating toxins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hochmann H, Pust S, von Figura G, Aktories K, Barth H. Salmonella enterica SpvB ADP-Ribosylates Actin at Position Arginine-177Characterization of the Catalytic Domain within the SpvB Protein and a Comparison to Binary Clostridial Actin-ADP-Ribosylating Toxins†. Biochemistry. 2006;45:1271–1277. doi: 10.1021/bi051810w. Demonstrated that SpvB displayed amino acid residues conserved in bacterial actin-ADP-ribosylating toxins despite not being a binary toxin. Functionally confirmed this conservation through demonstration of actin ADP-ribosylation at Arg177. [DOI] [PubMed] [Google Scholar]
- 124.Vilches S, et al. Aeromonas hydrophila AH-3 AexT is an ADP-ribosylating toxin secreted through the type III secretion system. Microbial Pathogenesis. 2008;44:1–12. doi: 10.1016/j.micpath.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 125.Barbieri JT, Sun J. Springer; Berlin Heidelberg: 2005. pp. 79–92. [Google Scholar]
- 126.Coburn J, Dillon ST, Iglewski BH, Gill DM. Exoenzyme S of Pseudomonas aeruginosa ADP-ribosylates the intermediate filament protein vimentin. Infection and Immunity. 1989;57:996–998. doi: 10.1128/iai.57.3.996-998.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Coburn J, Wyatt RT, Iglewski BH, Gill DM. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. Journal of Biological Chemistry. 1989;264:9004–9008. [PubMed] [Google Scholar]
- 128.Litvak Y, Selinger Z. Aeromonas salmonicida Toxin AexT Has a Rho Family GTPase-Activating Protein Domain. Journal of Bacteriology. 2007;189:2558–2560. doi: 10.1128/JB.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rubin EJ, Gill DM, Boquet P, Popoff MR. Functional modification of a 21-kilodalton G protein when ADP-ribosylated by exoenzyme C3 of Clostridium botulinum. Molecular and Cellular Biology. 1988;8:418–426. doi: 10.1128/mcb.8.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wilde C, Vogelsgesang M, Aktories K. Rho-Specific Bacillus cereus ADP-Ribosyltransferase C3cer Cloning and Characterization. Biochemistry. 2003;42:9694–9702. doi: 10.1021/bi034583b. [DOI] [PubMed] [Google Scholar]
- 131.Hanna S, El-Sibai M. Signaling networks of Rho GTPases in cell motility. Cellular Signalling. 2013;25:1955–1961. doi: 10.1016/j.cellsig.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 132.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 133.Sekine A, Fujiwara M, Narumiya S. Asparagine residue in the rho gene product is the modification site for botulinum ADP-ribosyltransferase. Journal of Biological Chemistry. 1989;264:8602–8605. [PubMed] [Google Scholar]
- 134.Genth H, Schmidt M, Gerhard R, Aktories K, Just I. Activation of phospholipase D1 by ADP-ribosylated RhoA. Biochemical and Biophysical Research Communications. 2003;302:127–132. doi: 10.1016/s0006-291x(03)00112-8. [DOI] [PubMed] [Google Scholar]
- 135.Genth H, et al. Entrapment of Rho ADP-ribosylated by Clostridium botulinum C3 Exoenzyme in the Rho-Guanine Nucleotide Dissociation Inhibitor-1 Complex. Journal of Biological Chemistry. 2003;278:28523–28527. doi: 10.1074/jbc.M301915200. [DOI] [PubMed] [Google Scholar]
- 136.Barth H, et al. Neosynthesis and Activation of Rho by Escherichia coli Cytotoxic Necrotizing Factor (CNF1) Reverse Cytopathic Effects of ADP-ribosylated Rho. Journal of Biological Chemistry. 1999;274:27407–27414. doi: 10.1074/jbc.274.39.27407. [DOI] [PubMed] [Google Scholar]
- 137.Pautsch A, Vogelsgesang M, Tränkle J, Herrmann C, Aktories K. Crystal structure of the C3bot–RalA complex reveals a novel type of action of a bacterial exoenzyme. The EMBO Journal. 2005;24:3670–3680. doi: 10.1038/sj.emboj.7600813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wilde C, Chhatwal GS, Schmalzing G, Aktories K, Just I. A Novel C3-like ADP-ribosyltransferase from Staphylococcus aureus Modifying RhoE and Rnd3. Journal of Biological Chemistry. 2001;276:9537–9542. doi: 10.1074/jbc.M011035200. [DOI] [PubMed] [Google Scholar]
- 139.Molinari G, et al. Localization of the C3-Like ADP-Ribosyltransferase from Staphylococcus aureus during Bacterial Invasion of Mammalian Cells. Infection and Immunity. 2006;74:3673–3677. doi: 10.1128/IAI.02013-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fahrer J, et al. Selective and specific internalization of clostridial C3 ADP-ribosyltransferases into macrophages and monocytes. Cellular Microbiology. 2010;12:233–247. doi: 10.1111/j.1462-5822.2009.01393.x. [DOI] [PubMed] [Google Scholar]
- 141.Wollein Waldetoft K, Råberg L. To harm or not to harm? On the evolution and expression of virulence in group A streptococci. Trends in Microbiology. 2014;22:7–13. doi: 10.1016/j.tim.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 142.Ferretti JJ, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proceedings of the National Academy of Sciences. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. Proposed that the spyA gene was a C3-like ADP-ribosyltransferase based on sequence data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Coye LH, Collins CM. Identification of SpyA, a novel ADP-ribosyltransferase of Streptococcus pyogenes. Molecular Microbiology. 2004;54:89–98. doi: 10.1111/j.1365-2958.2004.04262.x. Characterized the ADP-ribosyltransferase activity of SpyA, demonstrating that despite predicted C3-like activity, SpyA displayed activity towards different eukaryotic substrates. [DOI] [PubMed] [Google Scholar]
- 144.Korotkova N, et al. SpyA is a membrane-bound ADP-ribosyltransferase of Streptococcus pyogenes which modifies a streptococcal peptide, SpyB. Molecular Microbiology. 2012;83:936–952. doi: 10.1111/j.1365-2958.2012.07979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Experimental Cell Research. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 146.Eckes B, et al. Impaired wound healing in embryonic and adult mice lacking vimentin. Journal of Cell Science. 2000;113:2455–2462. doi: 10.1242/jcs.113.13.2455. [DOI] [PubMed] [Google Scholar]
- 147.Icenogle LM, et al. Molecular and Biological Characterization of Streptococcal SpyA-mediated ADP-ribosylation of Intermediate Filament Protein Vimentin. Journal of Biological Chemistry. 2012;287:21481–21491. doi: 10.1074/jbc.M112.370791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gohara R, et al. Phosphorylation of vimentin head domain inhibits interaction with the carboxyl-terminal end of α-helical rod domain studied by surface plasmon resonance measurements. FEBS letters. 2001;489:182–186. doi: 10.1016/s0014-5793(01)02108-1. [DOI] [PubMed] [Google Scholar]
- 149.Hoff JS, DeWald M, Moseley SL, Collins CM, Voyich JM. SpyA, a C3-Like ADP-Ribosyltransferase, Contributes to Virulence in a Mouse Subcutaneous Model of Streptococcus pyogenes Infection. Infection and Immunity. 2011;79:2404–2411. doi: 10.1128/IAI.01191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Guttman DS, et al. A Functional Screen for the Type III (Hrp) Secretome of the Plant Pathogen Pseudomonas syringae. Science. 2002;295:1722–1726. doi: 10.1126/science.295.5560.1722. [DOI] [PubMed] [Google Scholar]
- 151.Petnicki-Ocwieja T, et al. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proceedings of the National Academy of Sciences. 2002;99:7652–7657. doi: 10.1073/pnas.112183899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lindeberg M, et al. Closing the Circle on the Discovery of Genes Encoding Hrp Regulon Members and Type III Secretion System Effectors in the Genomes of Three Model Pseudomonas syringae Strains. Molecular Plant-Microbe Interactions. 2006;19:1151–1158. doi: 10.1094/MPMI-19-1151. [DOI] [PubMed] [Google Scholar]
- 153.Fouts DE, et al. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proceedings of the National Academy of Sciences. 2002;99:2275–2280. doi: 10.1073/pnas.032514099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Buell CR, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proceedings of the National Academy of Sciences. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fu ZQ, et al. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–288. doi: 10.1038/nature05737. Identified the substrate of effector HopU1 ADP-ribosyltransferase activity and demonstrated the unique immunosuppressive phenotype of HopU1 activity. [DOI] [PubMed] [Google Scholar]
- 156.Jeong B-r, et al. Structure Function Analysis of an ADP-ribosyltransferase Type III Effector and Its RNA-binding Target in Plant Immunity. Journal of Biological Chemistry. 2011;286:43272–43281. doi: 10.1074/jbc.M111.290122. Solved the crystal structure of HopU1 and demonstrated the necessity of unique loops framing the NAD binding pocket for substrate recognition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sun J, Barbieri JT. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 Regulator of Kinase (Crk) Proteins. Journal of Biological Chemistry. 2003;278:32794–32800. doi: 10.1074/jbc.M304290200. [DOI] [PubMed] [Google Scholar]
- 158.Sun J, Maresso AW, Kim JJ, Barbieri JT. How bacterial ADP-ribosylating toxins recognize substrates. Nature structural & molecular biology. 2004;11:868–76. doi: 10.1038/nsmb818. [DOI] [PubMed] [Google Scholar]
- 159.Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis-thaliana. Proceedings of the National Academy of Sciences. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nicaise V, et al. Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J. 2013;32:701–712. doi: 10.1038/emboj.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.THJ, CO, CHT, CP, KN Identification of molecular contacts between the U1 A small nuclear ribonucleoprotein and U1 RNA. EMBO J. 1991;10:3447–3456. doi: 10.1002/j.1460-2075.1991.tb04909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Burd C, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 163.Nagai K, Oubridge C, Jessen TH, Li J, Evans PR. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 164.Lee AL, et al. Chemical Shift Mapping of the RNA-Binding Interface of the Multiple-RBD Protein Sex-Lethal†. Biochemistry. 1997;36:14306–14317. doi: 10.1021/bi970830y. [DOI] [PubMed] [Google Scholar]
- 165.Schöning JC, et al. Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. The Plant Journal. 2007;52:1119–1130. doi: 10.1111/j.1365-313X.2007.03302.x. [DOI] [PubMed] [Google Scholar]
- 166.Boller T, He SY. Innate Immunity in Plants: An Arms Race Between Pattern Recognition Receptors in Plants and Effectors in Microbial Pathogens. Science. 2009;324:742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 168.Segonzac C, Zipfel C. Activation of plant pattern-recognition receptors by bacteria. Current Opinion in Microbiology. 2011;14:54–61. doi: 10.1016/j.mib.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 169.Maurice J. A first step in bringing typhoid fever out of the closet. The Lancet. 2012;379:699–700. doi: 10.1016/s0140-6736(12)60294-3. [DOI] [PubMed] [Google Scholar]
- 170.Haghjoo E, Galán JE. Salmonella typhiencodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4614–4619. doi: 10.1073/pnas.0400932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Spanò S, Ugalde JE, Galán JE. Delivery of a Salmonella Typhi Exotoxin from a Host Intracellular Compartment. Cell Host & Microbe. 2008;3:30–38. doi: 10.1016/j.chom.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 172.Guidi R, et al. Salmonella enterica delivers its genotoxin through outer membrane vesicles secreted from infected cells. Cellular Microbiology. 2013;15:2034–2050. doi: 10.1111/cmi.12172. [DOI] [PubMed] [Google Scholar]
- 173.Forst S, Dowds B, Boemare N, Stackebrandt E. XENORHABDUS AND PHOTORHABDUS SPP.:Bugs That Kill Bugs. Annual Review of Microbiology. 1997;51:47–72. doi: 10.1146/annurev.micro.51.1.47. [DOI] [PubMed] [Google Scholar]
- 174.Waterfield N, Hares M, ffrench-Constant R, Wren B, Hinchliffe S. In: The Genus Yersinia. Perry R, Fetherston J, editors. Springer; New York: 2007. pp. 247–257. [DOI] [PubMed] [Google Scholar]
- 175.Waterfield NR, Ciche T, Clarke D. Photorhabdus and a Host of Hosts. Annual Review of Microbiology. 2009;63:557–574. doi: 10.1146/annurev.micro.091208.073507. [DOI] [PubMed] [Google Scholar]
- 176.Waterfield NR, Bowen DJ, Fetherston JD, Perry RD, ffrench-Constant RH. The tc genes of Photorhabdus: a growing family. Trends in Microbiology. 2001;9:185–191. doi: 10.1016/s0966-842x(01)01978-3. [DOI] [PubMed] [Google Scholar]
- 177.ffrench-Constant R, Waterfield N. In: Advances in Applied Microbiology. Allen I, Laskin JWBGMG, Sima S, editors. Academic Press; 2005. pp. 169–183. [Google Scholar]
- 178.Gatsogiannis C, et al. A syringe-like injection mechanism in Photorhabdus luminescens toxins. Nature. 2013;495:520–523. doi: 10.1038/nature11987. [DOI] [PubMed] [Google Scholar]
- 179.ffrench-Constant R, et al. Photorhabdus: towards a functional genomic analysis of a symbiont and pathogen. FEMS Microbiol Rev. 2003;26:433–456. doi: 10.1111/j.1574-6976.2003.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 180.Cassimeris L, Safer D, Nachmias VT, Zigmond SH. Thymosin beta 4 sequesters the majority of G-actin in resting human polymorphonuclear leukocytes. The Journal of Cell Biology. 1992;119:1261–1270. doi: 10.1083/jcb.119.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mannherz HG, Hannappel E. The β-thymosins: Intracellular and extracellular activities of a versatile actin binding protein family. Cell Motility and the Cytoskeleton. 2009;66:839–851. doi: 10.1002/cm.20371. [DOI] [PubMed] [Google Scholar]
- 182.Vetter IR, Wittinghofer A. The Guanine Nucleotide-Binding Switch in Three Dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 183.Meusch D, et al. Mechanism of Tc toxin action revealed in molecular detail. Nature. 2014;508:61–65. doi: 10.1038/nature13015. [DOI] [PubMed] [Google Scholar]
- 184.Falkow S. Molecular Koch’s postulates applied to microbial pathogenicity. Reviews of Infectious Diseases. 1988;10 doi: 10.1093/cid/10.supplement_2.s274. Explained Falkow’s molecular application of Koch’s postulates for confirmation of the role of putative virulence factors in bacterial pathogenesis. [DOI] [PubMed] [Google Scholar]
- 185.Falkow S. Molecular Koch’s postulates applied to bacterial pathogenicity- a personal recollection 15 years later. Nat Rev Micro. 2004;2:67–72. doi: 10.1038/nrmicro799. [DOI] [PubMed] [Google Scholar]
- 186.Zheng L-L, et al. A Comparison of Computational Methods for Identifying Virulence Factors. PLoS ONE. 2012;7:e42517. doi: 10.1371/journal.pone.0042517. Demonstrated the functionality and importance of protein amino acid sequence analyses, while explaining caveats and issues with common search methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jones DT, Swindells MB. Getting the most from PSI-BLAST. Trends Biochem Sci. 2002;27:161–4. doi: 10.1016/s0968-0004(01)02039-4. [DOI] [PubMed] [Google Scholar]
- 188.Qi Y, Sadreyev RI, Wang Y, Kim BH, Grishin NV. A comprehensive system for evaluation of remote sequence similarity detection. BMC Bioinformatics. 2007;8:314. doi: 10.1186/1471-2105-8-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sun J, Maresso AW, Kim JJ, Barbieri JT. How bacterial ADP-ribosylating toxins recognize substrates. Nat Struct Mol Biol. 2004;11:868–76. doi: 10.1038/nsmb818. [DOI] [PubMed] [Google Scholar]
- 190.Palmiter RD, et al. Cell lineage ablation in transgenic mice by cell-specific expression of a toxin gene. Cell. 1987;50:435–443. doi: 10.1016/0092-8674(87)90497-1. [DOI] [PubMed] [Google Scholar]
- 191.Saito M, et al. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat Biotech. 2001;19:746–750. doi: 10.1038/90795. [DOI] [PubMed] [Google Scholar]
- 192.Buch T, et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Meth. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- 193.Chaudhary VK, et al. A recombinant immunotoxin consisting of two antibody variable domains fused to Pseudomonas exotoxin. Nature. 1989;339:394–397. doi: 10.1038/339394a0. [DOI] [PubMed] [Google Scholar]
- 194.Pastan I. Immunotoxins containing Pseudomonas exotoxin A: a short history. Cancer Immunology, Immunotherapy. 2003;52:338–341. doi: 10.1007/s00262-002-0353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kreitman RJ, Pastan I. Antibody Fusion Proteins: Anti-CD22 Recombinant Immunotoxin Moxetumomab Pasudotox. Clinical Cancer Research. 2011;17:6398–6405. doi: 10.1158/1078-0432.CCR-11-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kreitman RJ, et al. Efficacy of the Anti-CD22 Recombinant Immunotoxin BL22 in Chemotherapy-Resistant Hairy-Cell Leukemia. New England Journal of Medicine. 2001;345:241–247. doi: 10.1056/NEJM200107263450402. [DOI] [PubMed] [Google Scholar]
- 197.Chaudhary VK, et al. Selective killing of HIV-infected cells by recombinant human CD4-Pseudomonas exotoxin hybrid protein. Nature. 1988;335:369–372. doi: 10.1038/335369a0. [DOI] [PubMed] [Google Scholar]
- 198.Berger EA, Pastan I. Immunotoxin Complementation of HAART to Deplete Persisting HIV-Infected Cell Reservoirs. PLoS Pathog. 2010;6:e1000803. doi: 10.1371/journal.ppat.1000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sarnovsky R, et al. Initial characterization of an immunotoxin constructed from domains II and III of cholera exotoxin. Cancer Immunology, Immunotherapy. 2010;59:737–746. doi: 10.1007/s00262-009-0794-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Francis JW, et al. Enhancement of Diphtheria Toxin Potency by Replacement of the Receptor Binding Domain with Tetanus Toxin C-Fragment. Journal of Neurochemistry. 2000;74:2528–2536. doi: 10.1046/j.1471-4159.2000.0742528.x. [DOI] [PubMed] [Google Scholar]
- 201.Ahnert-Hilger G, et al. Differential effects of Rho GTPases on axonal and dendritic development in hippocampal neurones. Journal of Neurochemistry. 2004;90:9–18. doi: 10.1111/j.1471-4159.2004.02475.x. [DOI] [PubMed] [Google Scholar]
- 202.Bertrand J, Di Polo A, McKerracher L. Enhanced survival and regeneration of axotomized retinal neurons by repeated delivery of cell-permeable C3-like Rho antagonists. Neurobiology of Disease. 2007;25:65–72. doi: 10.1016/j.nbd.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 203.Höltje M, et al. A 29-amino acid fragment of Clostridium botulinum C3 protein enhances neuronal outgrowth, connectivity, and reinnervation. The FASEB Journal. 2009;23:1115–1126. doi: 10.1096/fj.08-116855. [DOI] [PubMed] [Google Scholar]
- 204.Bertrand J, Winton MJ, Rodriguez-Hernandez N, Campenot RB, McKerracher L. Application of Rho Antagonist to Neuronal Cell Bodies Promotes Neurite Growth in Compartmented Cultures and Regeneration of Retinal Ganglion Cell Axons in the Optic Nerve of Adult Rats. The Journal of Neuroscience. 2005;25:1113–1121. doi: 10.1523/JNEUROSCI.3931-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Boato F, et al. C3 peptide enhances recovery from spinal cord injury by improved regenerative growth of descending fiber tracts. Journal of Cell Science. 2010;123:1652–1662. doi: 10.1242/jcs.066050. [DOI] [PubMed] [Google Scholar]