ABSTRACT
The covalent histone modifications were associated with plant development. However, the function of histone modification factors involved in gibberellins (GAs) signaling pathway remains unclear. In recent study, we reported that histone modification factors MUT9p-LIKE KINASE1 (MLK1) and MLK2 coordinate GA and circadian clock signaling in hypocotyl elongation. MLK1 and MLK2 interact with the DELLA protein REPRESSOR OF ga1-3 (RGA), and antagonize the function of RGA to interact with CIRCADIAN CLOCK ASSOCIATED1 (CCA1), resulting in promoting hypocotyl elongation. In this addendum to the report, we presented and discussed the results related to the function of MLK1 and MLK2 in GA pathway. MLK1 and MLK2 interact with RGA, which is independent on 17-amino acid DELLA, TVHYNP, or Poly S/T/V motif, suggesting that MLK1 and MLK2 might have novel functions beyond the protein degradation.
KEYWORDS: MLK1, MLK2, RGA, gibberenllins
Casein Kinase I, the serine/threonine protein kinase, plays important roles in vesicular trafficking, DNA repair, circadian rhythm, and morphogenesis in mammalian cells.1 In the alga Chlamydomonas reinhardtii, MUT9p is related to casein kinase I and phosphorylates histone H3 at threonine 3.2 The Arabidopsis MUT9p-LIKE KINASE1 (MLK1), MLK2, MLK3, and MLK4 are closely related to MUT9p(3, 4). MLK1 and MLK2 were first identified as kinases for phosphorylation of H3 at threonine 3 in vivo and are associated with the osmotic stress response,3 while MLK4 was characterized as a kinase for phosphorylation of histone H2A at serine 95 and promoted flowering time under long-day conditions,4 suggesting that MLK4 might evolved the divergent function from MLK1 and MLK2.
The GA signaling pathway is controlled by the DELLA repressors, which have a characteristic DELLA domain. The Arabidopsis thaliana genome encodes five DELLA proteins and REPRESSOR OF ga1-3 (RGA) is one of the most important DELLA proteins in elongation growth.5 In the absence of gibberellins (GAs), DELLA proteins interact with transcription factors to inhibit the transcription of GA-responsive genes.6,7 In response to GAs, the DELLA proteins were inactivated and degradated by the 26S proteasome system.6-8 The roles of GA-induced phosphorylation of DELLA proteins in degradation were controversial.9-11
Our recent study suggested that histone modification factors MLK1 and MLK2 coordinate GAs and circadian clock signaling in hypocotyl elongation.12 MLK1 and MLK2 interact with the DELLA protein RGA, and antagonize the interaction between RGA and CIRCADIAN CLOCK ASSOCIATED1 (CCA1), thus to promote hypocotyl elongation. We therefore examined which domain is critical for this interaction. We deleted the N-terminus of RGA (N-terminal DELLA regulatory domain) containing the DELLA, TVHYNP, and Poly S/T/V motifs, or C-terminal GRAS domain. Both N-terminal DELLA regulatory domain and C-terminal GRAS domain were necessary for this interaction (Fig. 1). We then deleted the motif one by one. In C-terminal GRAS domain, each motif was indispensible for this interaction. In N-terminal DELLA regulatory domain, 17-amino acid DELLA, TVHYNP, or Poly S/T/V alone is not essential for this interaction. However, deletion of TVHYNP and Poly S/T/V motifs lost interaction between MLK1/2 and RGA (Fig. 1). These results suggested that MLK1 and MLK2 might be not directly involved in RGA degradation.
Figure 1.
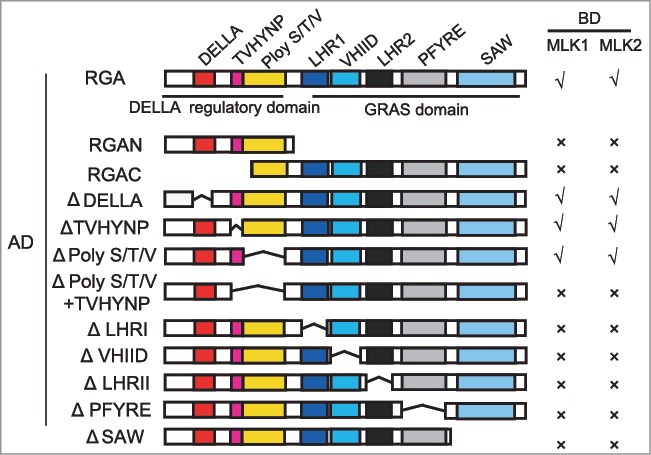
MLK1 and MLK2 interact with different motifs of RGA. Yeast two-hybrid analysis revealed an interaction between MLK1/2 and the different motifs of RGA. Different regions of RGA were indicated. The positive interactions were indicated with √, and the negative interactions were indicated with ×. Δindicated the deletion domain.
In the absence of GAs, DELLA proteins interact with transcriptional factors and this interaction helped DELLA proteins bind to transcriptional factors and reduced their transcriptional activity.6,7 However, how did these transcriptional factors get rid of DELLA proteins repression is still unclear. Our study shown RGA interacts with CCA1 and suppressed the ability of CCA1 to bind to the promoter of DWARF4 (DWF4), whereas this repression were reversed in the presence of MLK1/2.12
MLK1 and MLK2 were nuclear proteins (Fig. 2A) and expressed in vascular tissues of the cotyledons, hypocotyls, and roots (Fig. 2B), suggesting MLK1/2 might be involved in transcription regulation. Loss of MLK1 and MLK2 function resulted in late flowering phenotype, similar to that of mlk4.4-12 The transcripts of CO and FT were downregulated in mlk1 mlk2 at night (Fig. 2C), suggested MLK1 and MLK2 might be involved in photoperiod pathway like MLK4. mlk1 mlk2 mlk4 triple mutant displayed the flowering time later than mlk4 and mlk1 mlk2 double mutant, suggesting that MLK1/2 and MLK4 are redundant in flowering time.13 The similar expression pattern and interaction with CCA1 might contribute to the redundancy in flowering time. MLK1 and MLK2, but not MLK4, interact with RGA, suggesting that the functions of MLK1 and MLK2 in hypocotyl elongation were different from those of MLK4. Together, our study demonstrated MLK1 and MLK2 integrate the GAs and the circadian clock signaling to modulate plant development.
Figure 2.
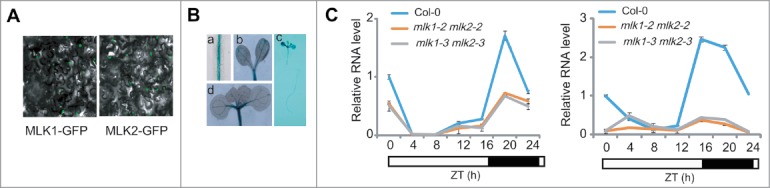
The subcellular localization of MLK2 and expression pattern of MLK2. A. MLK1 and MLK2 are located in nucleus. B. The expression pattern of MLK2 revealed that MLK2 expressed in vascular tissues of the roots (a, c), cotyledons (b, d), and hypocotyls (c). C. The relative transcripts of CO and FT in wild-type mlk1 mlk2 plants. RNA was isolated from the leaves of 3-week-old plants and was used to test the transcription levels of CO (left panel) and FT (right panel). The black bars indicate the dark period, and the white bars indicate the light period. Experiments were repeated at least three times, and each data point indicates the mean ± SE, n = 3 replicates. ZT, Zeitgeber time.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31371306 31571315 and 91435101).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 31371306, 31571315 and 91435101 to Y.D.), and the Strategic Priority Research Program “Molecular Mechanism of Plant Growth and Development” of CAS (grant XDPB04).
References
- 1.McKay RM, Peters JM, Graff JM. The casein kinase I family: Roles in morphogenesis. Dev biol. 2001;235(2):378–87. doi: 10.1006/dbio.2001.0307. [DOI] [PubMed] [Google Scholar]
- 2.Casas-Mollano JA, B-r Jeong, Xu J, Moriyama H, Cerutti H. The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc Natl Acad Sci U S A. 2008;105(17):6486–91. doi: 10.1073/pnas.0711310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Casas-Mollano JA, Xu J, Riethoven J-JM, Zhang C, Cerutti H. Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2015;112(27):8487–92. doi: 10.1073/pnas.1423325112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su Y, Wang S, Zhang F, Zheng H, Liu Y, Huang T, Ding Y. Phosphorylation of histone H2A at serine 95: A plant-specific mark involved in flowering time regulation and H2A. Z deposition. Plant Cell. 2017;29(9):2197–213. doi: 10.1105/tpc.17.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dill A, Sun T-p. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;159(2):777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.T-p Sun. The molecular mechanism and evolution of the GA–GID1–DELLA signaling module in plants. Curr Biol. 2011;21(9):R338–R45. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 7.T-p Sun, F Gubler. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- 8.Dill A, Thomas SG, Hu J, Steber CM, Sun T-p. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell. 2004;16(6):1392–405. doi: 10.1105/tpc.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299(5614):1896–8. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- 10.Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP. The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell. 2004;16(6):1406–18. doi: 10.1105/tpc.021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson SK, Steber CM. Gibberellin hormone signal perception: down‐regulating DELLA repressors of plant growth and development. Annual Plant Reviews, Volume 49: Gibberellins. 2016:153–88. doi: 10.1002/9781119210436.ch6. [DOI] [Google Scholar]
- 12.Zheng H, Zhang F, Wang S, Su Y, Jiang P, Cheng R, Ji X, Hou S, Ding Y. MLK1 and MLK2 coordinate RGA and CCA1 activity to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell. 2017:tpc. 00830.2017. doi: 10.1105/tpc.17.00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H, Alvarez S, Bindbeutel R, Shen Z, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, Nusinow DA. Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol Cell Proteomics. 2016;15(1):201–17. doi: 10.1074/mcp.M115.054064. [DOI] [PMC free article] [PubMed] [Google Scholar]


