ABSTRACT
The CUP-SHAPED COTYLEDON (CUC) transcription factors play a fundamental role in plant morphogenesis by defining boundary domains throughout plant development. Despite their central roles in plant development, little is known about the CUC molecular network. In a recent work, we identified a role for MUR1, a protein involved in the production of GDP-L-Fucose, in this network and showed that fucose per se is required for proper boundary definition in various developmental contexts. Which pathway involving fucose is required to determine boundary is not yet known. Here, we use a previously described mutant and transgenic line with reduced fucosylated xyloglucans (XyG) to explore one such pathway. By quantitatively comparing leaf shape, we show that defects in XyG fucosylation do not impact leaf serrations development suggesting that fucose absence in XyG does not impact boundary development in mur1-1 mutant. Thus another – not yet identified – pathway or fucosylated compound contribute to boundary domain definition.
KEYWORDS: Arabidopsis, boundary, growth, fucose, leaf, xyloglucans
Boundaries act both as frontiers defining functional units and as organizing centres providing positional clues to control the fate of neighbouring cells.1,2 Both functions are important to correctly pattern developing organs. Failure to establish and maintain boundaries can result in organ fusion, meristem loss and developmental arrest.3 Boundaries often display reduced cell proliferation,4,5 while adjacent tissues or organs actively grow out via an auxin-dependent mechanism.6,7 As boundary definition regulators, CUP-SHAPED COTYLEDON transcription factors are involved in both shoot meristem formation8 and correct organ separation in various developmental contexts.3,9,10 In addition, CUC genes are key regulators of leaf shape through their roles on leaf margin development.11–13 They are expressed at the sinus of leaf margin serrations where they are thought to repress growth while allowing it in adjacent serration tips in a manner that is similar to other boundaries. While wild-type Arabidopsis leaves are serrated, cuc2 loss-of-function mutants have smooth leaves with no serrations.11,12 Our recent work uses this system and joins the decades long effort to characterize the molecular network centred around the CUC genes.14 In this work we identified a mutation that simplifies leaf dissection and affects the protein encoded by MURUS1 (MUR1),14 which participates in GDP-L-fucose production.15 Expression analyses revealed that CUC2 levels are reduced in mur1 mutant backgrounds. Our study shows that GDP-L-fucose has an important role in different developmental contexts where it contributes to organ separation in the same pathway as CUC2. Key questions remain on how fucose modifies CUC2 expression and how it contributes to boundary definition. Here we expand on our understanding of this role by showing complementary results on the role of fucosylated xyloglucans in leaf margin development.
Fucose is naturally incorporated into various cell wall glycoconjugates such as xyloglucans (XyG), rhamnogalacturonan II (RGII), and arabinogalactans.16–18 In addition, fucose participates in post-translational protein glycosylation.19 GDP-L-fucose is synthesised in three steps catalysed by two different enzymes the first of which GDP-D-mannose 4,6-dehydratase is encoded by MUR1. Fucose content of cell wall components in the mur1 mutant is strongly reduced in aerial parts but only partially in root tissues, which may be due to the presence of its homologue GMD1.15,20,21 It has been hypothesized that activity of this homolog together with the residual fucose and/or its replacement by α-L-galactosyl residues in aerial parts is responsible for the weak phenotype of mur1 mutants.20,22 Nevertheless incorporation of fucose into the different cell wall components is important for plant development as illustrated by the phenotypes of loss of GDP-L-fucose import into the Golgi lumen,23 and inhibition of fucosyltransferase activity.24,25
Among the diverse roles of GDP-L-Fucose, its impact on XyG structure and its implications in terms of growth are of especial interest. XyG is the most abundant hemicellulose in dicot cell walls and is thought to play a crucial role in cell elongation and cell wall rigidity.26 XyG molecules can be hydrolysed in muro and the resulting oligosaccharides can act as signalling molecules (termed oligosaccharins27). Fucosylated XXFG oligosaccharins in particular have been shown to antagonize at low concentration the synthetic auxin 2,4-D-stimulated elongation of pea stem segments.28 Furthermore, a role for fucosylated XyG in an auxin dependent in muro remodeling of XyG has been suggested. The overexpression of the xyloglucan fucosyl hydrolase AXY8/FUC95A29 is able to complement the short hypocotyl phenotype of dark-grown seedlings of transgenic lines impaired in auxin responses while removing XyG fucosyltransferase activity with mur2-1 mutation30 is not.31 CUC2 and auxin act together to regulate boundary domain formation, lateral organ development and leaf margin development in an intricate feed-back loop mechanism.6,7,32 Therefore we hypothesized that the decrease in fucosylated XyG and consequent reduced levels of XXFG residues in the mur1 mutant may result in modified auxin responses and subsequent developmental defects in boundary and leaf development.
To test whether XyG fucosylation impacts leaf margin patterning, we studied two previously reported lines with reduced XyG fucosylation levels: the 35S:AXY8 line overexpressing the fucosidase AXY8/FUC95A29 and the mur2-1 mutant line defective for a XyG specific fucosyltransferase.30 While the near total absence of fucose in the mur1-1 mutant results in smooth leaf margins compared to the wild-type control in short-day (Fig. 1A), as previously reported in long-day conditions (Gonçalves et al. 2017), neither the mur2-1 mutation nor the overexpression of AXY8 altered mature leaf shape in short day conditions (Fig. 1A). To compare leaf serration levels between genotypes we calculated the dissection index (DI)33,34 for leaves 11, 12 and 13 of plants grown in short-day conditions. Our analysis shows that leaf serration as measured by its DI is not significantly reduced in the mur2-1 mutant or 35S::AXY8 plants when compared to the wild-type, while DI for mur1-1 is significantly reduced compared to the wild-type (ANOVA, p < 0.0001, Fig. 1B).
Figure 1.
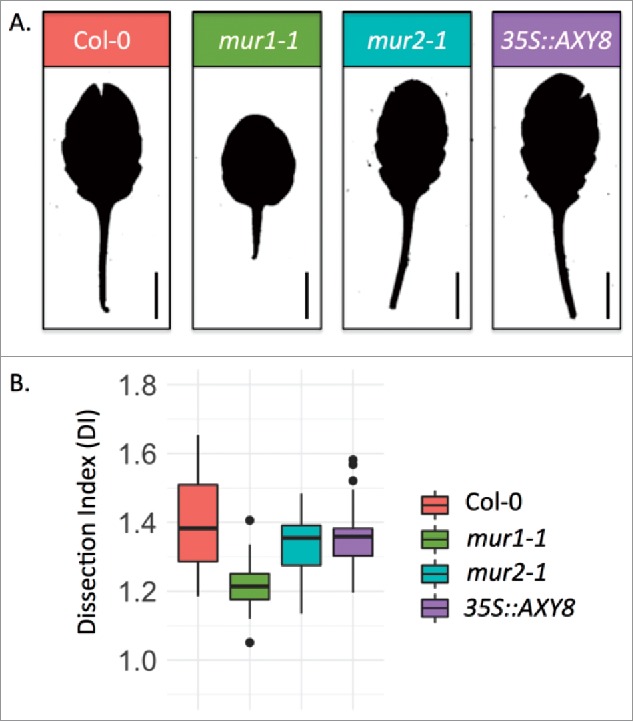
(A) Representative silhouettes of rank 13 leaves from 8 weeks-old plants grown in short-day conditions from Col-0, mur1-1, mur2-1 and 35S::AXY8 plants. (B) Quantification of leaf shape (pooled leaves of rank 11, 12 and 13) from 8 weeks-old plants grown in short-day conditions from Col-0 (n = 27), mur1-1 (n = 36), mur2-1 (n = 35) and 35S::AXY8 (n = 33) plants using the Dissection Index as a shape descriptor (DI = leaf perimeter2/(4π.leaf area).
Together these results show that correct leaf margin patterning can still occur in the absence of XyG fucosylation, suggesting that our previously reported boundary definition defects in the mur1 mutants are independent of the lack of fucose in XyG. Because GDP-L-fucose deficiency in mur1 mutants leads to defects in several glycosylation processes, it is probable that another fucosylated compound is responsible for the defects in boundary domain definition in mur1 mutants. The pectic polymer RGII is a key component of the cell wall and it has been suggested that its cross-linked dimerization has a role in cell wall expansion.17,35 Interestingly, fucosylation state of RGII has been shown to impact the stability of the borate di-ester facilitated cross-linked dimers.17 Therefore we can hypothesize that absence of fucose in mur1 mutants impacts cell wall expansion properties affecting leaf margin development through a reduction of cross-linked RGII. Alternatively it is possible that the fucosylation of a protein within the CUC2 molecular network is required for its function.
Competing interests
The authors declare no competing or financial interests.
Funding Statement
French National Research Agency (ANR), ANR-12-PDOC-0003 and ANR-11-BSV5-0007; AgreenSkillsPlus, PCOFUND-GA-2013-609398.
Acknowledgments
We would like to thank Grégory Mouille for critical reading of the manuscript. This work was supported by the Agence Nationale de la Recherche grant LEAFNET (ANR-12-PDOC-0003). JS is supported by the AuxiWall Project (ANR-11-BSV5-0007) and AgreenSkillsPlus (PCOFUND-GA-2013-609398). The IJPB benefits from the support of the Labex Saclay Plant Sciences - SPS (ANR-10-LABX-0040-SPS).
References
- 1.Irvine KD, Rauskolb C. Boundaries in development: formation and function. Annu Rev Cell Dev Biol. 2001;17:189–214. doi: 10.1146/annurev.cellbio.17.1.189. [DOI] [PubMed] [Google Scholar]
- 2.Dahmann C, Oates AC, Brand M. Boundary formation and maintenance in tissue development. Nat Rev Genet. 2011;12:43–55. doi: 10.1038/nrg2902. [DOI] [PubMed] [Google Scholar]
- 3.Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M, Tasaka2 M, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–57. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breuil-Broyer S, Morel P, de Almeida-Engler J, Coustham V, Negrutiu I, Trehin C. High-resolution boundary analysis during Arabidopsis thaliana flower development. Plant J. 2004;38:182–92. doi: 10.1111/j.1365-313X.2004.02026.x. [DOI] [PubMed] [Google Scholar]
- 5.Hepworth SR, Pautot VA. Beyond the Divide: Boundaries for Patterning and Stem Cell Regulation in Plants. Front Plant Sci. 2015;6:1052. doi: 10.3389/fpls.2015.01052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12:507–18. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/S0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 8.Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 1999;126:1563–70. [DOI] [PubMed] [Google Scholar]
- 9.Gonçalves B, Hasson A, Belcram K, Cortizo M, Morin H, Nikovics K, Vialette-Guiraud A, Takeda S, Aida M, Laufs P, et al.. A conserved role for CUP-SHAPED COTYLEDON genes during ovule development. Plant J. 2015;83:732–42. doi: 10.1111/tpj.12923. [DOI] [PubMed] [Google Scholar]
- 10.Burian A, Raczyńska-Szajgin M, Borowska-Wykręt D, Piatek A, Aida M, Kwiatkowska D. The CUP-SHAPED COTYLEDON2 and 3 genes have a post-meristematic effect on Arabidopsis thaliana phyllotaxis. Ann Bot. 2015;115:807–20. doi: 10.1093/aob/mcv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–45. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, Boudaoud A, Damerval C, Laufs P. Evolution and Diverse Roles of the CUP-SHAPED COTYLEDON Genes in Arabidopsis Leaf Development. Plant Cell. 2011;23:54–68. doi: 10.1105/tpc.110.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P. A conserved molecular framework for compound leaf development. Science. 2008;322:1835–9. doi: 10.1126/science.1166168. [DOI] [PubMed] [Google Scholar]
- 14.Gonçalves B, Maugarny-Calès A, Adroher B, Cortizo M, Borrega N, Blein T, Hasson A, Gineau E, Mouille G, Laufs P, et al.. GDP-L-fucose is required for boundary definition in plants. J Exp Bot. 2017;68:5801–5811. doi: 10.1093/jxb/erx402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonin CP, Potter I, Vanzin GF, Reiter W-DD. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc Natl Acad Sci U S A. 1997;94:2085–90. doi: 10.1073/pnas.94.5.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rayon C, Cabanes-Macheteau M, Loutelier-Bourhis C, Salliot-Maire I, Lemoine J, Reiter W-D, Lerouge P, Faye L. Characterization of N-glycans from Arabidopsis. Application to a fucose-deficient mutant. Plant Physiol. 1999;119:725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Neill MA, Eberhard S, Albersheim P, Darvill AG. Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science. 2001;294:846–9. doi: 10.1126/science.1062319. [DOI] [PubMed] [Google Scholar]
- 18.Van Hengel AJ Roberts K. Fucosylated arabinogalactan-proteins are required for full root cell elongation in arabidopsis. Plant J. 2002;32:105–13. doi: 10.1046/j.1365-313X.2002.01406.x. [DOI] [PubMed] [Google Scholar]
- 19.Strasser R. Plant protein glycosylation. Glycobiology. 2016;26:926–39. doi: 10.1093/glycob/cww023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiter W-DD, Chapple CCS, Somerville CR. Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science. 1993;261:1032–5. doi: 10.1126/science.261.5124.1032. [DOI] [PubMed] [Google Scholar]
- 21.Bonin CP, Freshour G, Hahn MG, Vanzin GF, Reiter WD. The GMD1 and GMD2 genes of Arabidopsis encode isoforms of GDP-D-mannose 4,6-dehydratase with cell type-specific expression patterns. Plant Physiol. 2003;132:883–92. doi: 10.1104/pp.103.022368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zablackis E, York WS, Pauly M, Hantus S, Reiter W-DD, Chapple CCS, Albersheim P, Darvill AG. Substitution of L-fucose by L-galactose in cell walls of Arabidopsis mur1. Science. 1996;272:1808–10. doi: 10.1126/science.272.5269.1808. [DOI] [PubMed] [Google Scholar]
- 23.Rautengarten C, Ebert B, Liu L, Stonebloom S, Smith-Moritz AM, Pauly M, Orellana A, Scheller HV, Heazlewood JL. The Arabidopsis Golgi-localized GDP-L-fucose transporter is required for plant development. Nat Commun. 2016;7:1–10. doi: 10.1038/ncomms12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumont M, Lehner A, Bardor M, Burel C, Vauzeilles B, Lerouxel O, Anderson CT, Mollet JC, Lerouge P. Inhibition of fucosylation of cell wall components by 2-fluoro 2-deoxy- l -fucose induces defects in root cell elongation. Plant J. 2015;84:1137–51. doi: 10.1111/tpj.13071. [DOI] [PubMed] [Google Scholar]
- 25.Villalobos JA, Yi BR, Wallace IS. 2-fluoro-L-fucose is a metabolically incorporated inhibitor of plant cell wall polysaccharide fucosylation. PLoS One. 2015;10:1–16. doi: 10.1371/journal.pone.0139091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010;61:263–89. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 27.Fry SC, Aldington S, Hetherington PR, Aitken J. Oligosaccharides as signals and substrates in the plant cell wall. Plant Physiol. 1993;103:1–5. doi: 10.1104/pp.103.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.York WS, Darvill a G, Albersheim P. Inhibition of 2,4-dichlorophenoxyacetic Acid-stimulated elongation of pea stem segments by a xyloglucan oligosaccharide. Plant Physiol. 1984;75:295–7. doi: 10.1104/pp.75.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Günl M, Neumetzler L, Kraemer F, de Souza A, Schultink A, Pena M, York WS, Pauly M. AXY8 Encodes an α-Fucosidase, Underscoring the Importance of Apoplastic Metabolism on the Fine Structure of Arabidopsis Cell Wall Polysaccharides. Plant Cell. 2011;23:4025–40. doi: 10.1105/tpc.111.089193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanzin GF, Madson M, Carpita NC, Raikhel N V, Keegstra K, Reiter W-D. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci U S A. 2002;99:3340–5. doi: 10.1073/pnas.052450699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paque S, Mouille G, Grandont L, Alabadí D, Gaertner C, Goyallon A, Muller P, Primard-Brisset C, Sormani R, Blázquez M, et al.. AUXIN BINDING PROTEIN1 links cell wall remodeling, auxin signaling, and cell expansion in arabidopsis. Plant Cell. 2014;26:280–95. doi: 10.1105/tpc.113.120048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bilsborough GGD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci. 2011;108:3424–9. doi: 10.1073/pnas.1015162108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Backhaus A, Kuwabara A, Bauch M, Monk N, Sanguinetti G, Fleming A. Leafprocessor: A new leaf phenotyping tool using contour bending energy and shape cluster analysis. New Phytol. 2010;187:251–61. doi: 10.1111/j.1469-8137.2010.03266.x. [DOI] [PubMed] [Google Scholar]
- 34.Sicard A, Thamm A, Marona C, Lee YW, Wahl V, Stinchcombe JR, Wright SI, Kappel C, Lenhard M. Repeated Evolutionary Changes of Leaf Morphology Caused by Mutations to a Homeobox Gene. Curr Biol. 2014;24:1880–6. doi: 10.1016/j.cub.2014.06.061. [DOI] [PubMed] [Google Scholar]
- 35.Fleischer A, O'Neill M, Ehwald R. The Pore Size of Non-Graminaceous Plant Cell Walls Is Rapidly Decreased by Borate Ester Cross-Linking of the Pectic Polysaccharide Rhamnogalacturonan II. Plant Physiol. 1999;121:829–38. doi: 10.1104/pp.121.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]


