Abstract
Mounting evidence has shown that miR-23b-3p, which is associated with cell proliferation, invasion, and apoptosis, acts as a biomarker for diagnosis and outcomes in numerous cancers. However, the clinicopathological implication of miR-23b-3p in hepatocellular carcinoma (HCC) remains unclear. Our study evaluated the role of miR-23b-3p in HCC and investigated its potential application as a marker for preliminary diagnosis and therapy in HCC. High-throughput data from the NCBI Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) were collected and analyzed. One hundred and one tissue sections of HCC were paired with adjacent non-cancerous HCC as further supplements. miR-23b-3p expression was detected using quantitative real-time PCR. Additionally, the relationship between miR-23b-3p expression and HCC progression and Time-to-recurrence (months) was explored. Ten algorithms were applied to predict the prospective target genes of miR-23b-3p. Next, we conducted bioinformatics analysis for further study. miR-23b-3p expression was pronouncedly decreased in HCC tissues in contrast with their paired adjacent non-cancerous HCC (P<0.001) with RT-qPCR. In total, 405 targets, acquired with consistent prediction from at least five databases, were used for the bioinformatics analysis. According to the Gene Ontology (GO) analysis, all targets were classified into biological processes, cellular components and molecular functions. In the pathway analysis, targets of miR-23b-3p were primarily enriched in the signaling pathways of renal cell carcinoma, hepatitis B and pancreatic cancer (corrected P-value <0.05). In the protein-protein interaction (PPI) network for miR-23b-3p, a total of 8 targets, including SRC, AKT1, EGFR, CTNNB1, BCL2, SMAD3, PTEN and KDM6A, were located in the key nodes with high degree (>35). In conclusion, this study provides impressive illumination of the potential role of miR-23b-3p in HCC tumorigenesis and progression. Furthermore, miR-23b-3p may act as a predictor of HCC and could be a new treatment target.
Keywords: miR-23b-3p, hepatocellular carcinoma, progression, target, pathway
Introduction
Hepatocellular carcinoma (HCC) is a malignant tumor that has one of the highest morbidity and mortality rates, and its tumorigenesis is closely related to HBV and HCV infection (1). There were estimated 782,500 hepatic carcinoma cases and 745,500 related deaths globally in 2012, according to Torre et al (2). Moreover, estimated 70–90% of primary liver cancers are HCC. Despite advances in chemotherapy, radiotherapy and surgery, the low 5-year survival and quality of life observed in HCC patients remain intractable issues. Many etiological factors, such as alcohol consumption and aflatoxin B1 exposure, are widely accepted to facilitate HCC occurrence (3,4); however, the precise molecular mechanisms remain unknown. Thus, there is an urgent need for a reliable biomarker that can be used to predict HCC progression and prognosis.
MicroRNAs (miRNAs), which are limited to a length of 19–22 nucleotides, are small non-coding RNAs that post-transcriptionally influence gene expression (5). A vast array of oncogenes and anti-oncogenes are potentially regulated by miRNAs, which act as gene regulators by conjugating the 3′-untranslated region (3′-UTR) of their mRNAs. In regulating genes, miRNAs play a crucial biological role in tumor development, especially during initiation, proliferation, differentiation, invasion and metastasis (6). Thus, miRNAs may serve as promising diagnostic and prognostic indexes.
Validating a potential target of miRNA is arduous and time-consuming due to the tremendous amounts of target sites in miRNA. However, predicting the targets of miRNA, which is a vital step in performing miRNA-target interactions, contributes to narrowing down prospective target sites and promoting experimental verification. Following the principle of sequence complementarity, many algorithms were manipulated to figure out the predicted miRNA targets.
miR-23b-3p was identified as a tumor suppressor that showed a tendency toward downregulated expression in different classes of human malignant tumors such as prostate cancer, renal cell carcinoma, acute myeloid leukemia and osteosarcoma (7–10). Nevertheless, increasing evidence has indicated that the upregulation of miR-23b-3p promoted cell proliferation and invasion in glioma, gastric cancer and breast cancer (11–13). miR-23b-3p was also found to be involved in liver stem cell differentiation, and its reduced expression may contribute to liver regeneration after partial hepatectomy (14,15). However, to our knowledge, only one study (16) has explored the relationship between miR-23b-3p expression and HCC but did not comment on the clinicopathological significance and prognosis. Therefore, further inquiry is urgently needed.
Our objectives were as follows: to study the pathophysiologic expression quantity of miR-23b-3p in HCC tissue through comparison with matching adjacent tissues as well as to elucidate the correlation between miR-23b-3p expression and HCC clinicopathological parameters. Additionally, we aimed to identify the probable target genes of miR-23b-3p and determine the potential role of miR-23b-3p in HCC development and progression.
Materials and methods
miR-23b-3p expression in HCC based on GEO datasets
Data acquisition and exclusion criteria
Twenty-one micro-array datasets were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/), and 11 were eliminated after screening. The search strategy was formulated as follows: (malignan* OR cancer OR tumor OR tumour OR neoplas* OR carcinoma) AND (hepatocellular OR liver OR hepatic OR HCC). The last dataset search was on April 2016. The exclusion criteria were as follows: i) datasets without information on miR-23b-3p; ii) datasets without complete data for analysis; iii) samples based on cell lines; iv) not all subjects of the included studies were human; or v) miR-23b-3p was determined in the HCC patients without a comparison. To identify the clinical relevance of miR-23b-3p in HCC, we collected data on miR-23b-3p expression from the 6 datasets and analyzed their association with the clinicopathological characteristics of HCC. The clinical features, which could be obtained from more than two microarray datasets, were used for the meta-analysis.
Statistical analysis
The meta-analysis was carried out with Stata 12.0 (StataCorp LP, College Station, TX, USA). A standard mean difference (SMD) and a 95% confidence interval (CI) were utilized to measure continuous outcomes, including age, sex, HBV infection, invasion and metastasis. Fixed or random effects models were applied to pool the effect sizes. Cochrane's Q test (Chi-square test; Chi2) (17) and inconsistency (I2) (18) were conducted to assess heterogeneity. A P<0.05 or I2 >50% indicates significant heterogeneity (19), and a random effects model (20) was applied. Otherwise, the fixed effects model would be adopted. Begg's funnel plot for asymmetry and Egger's funnel plot for quantitation were generated to evaluate publication bias. In addition, sensitivity analysis was performed to evaluate the reliability of results by elimination of a study each time.
miR-23b-3p expression in HCC based on TCGA dataset
Retrieval of public data
Altogether, 377 anonymized HCC tissues and 50 normal tissues were retrieved from the TCGA database (http://cancergenome.nih.gov/publications/publica-tionguidelines). The non-HCC samples or samples with data deficiency were excluded; 361 HCC patients were finally included in this study. Additionally, a total of 50 normal liver tissues were retrieved for comparison. The clinicopathological features of HCC patients in TCGA are available in Table II.
Table II.
Characteristics of the HCC patients included in TCGA.
| Clinicopathological features | N | miR-23b-3p relevant expression (2−ΔCq)
|
Correlation
|
|||
|---|---|---|---|---|---|---|
| Mean ± SD | t-value | P-value | r-value | P-value | ||
| Tissue | ||||||
| Normal | 50 | 13.4039±0.51072 | −8.286 | <0.001 | 0.246 | <0.001 |
| HCC | 371 | 12.6722±0.97815 | ||||
| Age (years) | ||||||
| <60 | 166 | 12.7336±0.97231 | 0.85 | 0.396 | −0.045 | 0.396 |
| ≥60 | 194 | 12.6464±0.9000 | ||||
| Sex | ||||||
| Male | 246 | 12.6581±0.93294 | −0.613 | 0.54 | 0.032 | 0.54 |
| Female | 115 | 12.7259±1.0132 | ||||
| Neoadjuvant treatment | ||||||
| No | 359 | 12.6798±0.97924 | 0.036 | 0.972 | −0.002 | 0.972 |
| Yes | 2 | 12.6550±1.07244 | ||||
| Radiation therapy | ||||||
| No | 231 | 12.6570±0.95382 | 0.998 | 0.319 | −0.065 | 0.319 |
| Yes | 4 | 12.1762±1.03021 | ||||
| Pharmaceutical treatment | ||||||
| No | 218 | 12.6475±0.96107 | 0.041 | 0.967 | −0.003 | 0.967 |
| Yes | 12 | 12.6358±0.90350 | ||||
| Alcohol consumption | ||||||
| − | 226 | 12.7394±0.99963 | 1.374 | 0.17 | −0.074 | 0.17 |
| + | 117 | 12.5870±0.92032 | ||||
| Hepatitis B | ||||||
| − | 237 | 12.5999±0.97009 | −2.506 | 0.013 | 0.134 | 0.013 |
| + | 106 | 12.8831±0.96070 | ||||
| Hepatitis C | ||||||
| − | 289 | 12.6963±0.97544 | 0.388 | 0.698 | −0.021 | 0.698 |
| + | 54 | 12.6401±0.97811 | ||||
| Non-alcoholic fatty liver | ||||||
| − | 324 | 12.6763±0.98327 | −0.874 | 0.383 | 0.047 | 0.383 |
| + | 19 | 12.8774±0.81188 | ||||
| Smoking | ||||||
| − | 326 | 12.7127±0.96181 | 2.115 | 0.035 | −0.114 | 0.035 |
| + | 17 | 12.2025±1.11847 | ||||
| Cirrhosis | ||||||
| − | 337 | 12.6956±0.95014 | 0.564 | 0.597 | −0.063 | 0.248 |
| + | 6 | 12.2312±2.01359 | ||||
| Grade | ||||||
| GI–II | 23 | 12.6505±0.76988 | 1.493 | 0.147 | −0.214 | 0.256 |
| GIII–IV | 7 | 12.0712±1.26499 | ||||
| Vascular invasion | ||||||
| No | 199 | 12.8350±0.94514 | 2.149 | 0.032 | −0.122 | 0.032 |
| Yes | 107 | 12.5905±0.95709 | ||||
| Recurrence after treatment | ||||||
| No | 166 | 12.5805±0.98050 | −0.589 | 0.556 | 0.037 | 0.556 |
| Yes | 94 | 12.6537±0.92925 | ||||
| Survival status | ||||||
| Dead | 86 | 12.6050±1.01283 | −0.811 | 0.418 | 0.043 | 0.418 |
| Alive | 275 | 12.7031±0.96776 | ||||
| T status | ||||||
| T1 | 177 | 12.7653±0.93707 | F=1.605a | 0.202a | −0.083 | 0.117 |
| T2–T4 | 181 | 12.5926±1.01626 | ||||
| TX | 1 | 12.0451±0.00000 | ||||
| N status | ||||||
| N0 | 247 | 12.7780±0.96867 | F=5.116a | 0.006a | −0.158 | 0.003 |
| N1 | 3 | 11.7684±0.75393 | ||||
| NX | 110 | 12.4733±0.96540 | ||||
| M status | ||||||
| M0 | 262 | 12.7389±0.96998 | F=1.778a | 0.170a | −0.101 | 0.055 |
| M1 | 4 | 12.6180±0.12946 | ||||
| MX | 95 | 12.5190±1.00762 | ||||
One-way analysis of variance (ANOVA) test was utilized. t, Student's t-test; SD, standard deviation. HCC, hepatocelluar carcinoma; T, tumor; N, nodes; M, metastasis. TCGA, The Cancer Genome Atlas.
Statistical analysis
The statistics were analyzed with SPSS 22.0 software (IBM Corp., Armonk, NY, USA). The final data after calculation are shown as the mean ± standard deviation (SD). Student's t-test was conducted for a comparative analysis of two independent groups. A one-way analysis of variance (ANOVA) test was utilized to study the data, which were divided into three or four groups, such as T status, N status or M status. Spearman's method was performed to evaluate the association between miR-23b-3p expression and other clinicopathological parameters. The relationship between miR-23b-3p and recurrence was obtained with the Kaplan-Meier survival approach along with a log-rank test. To differentiate between controls and HCC tissues, a diagnostic value was identified using a receiver operator characteristic (ROC) curve. A P-value <0.05 denoted a statistically significant difference. At least 5 individual tests were applied without ambiguity.
miR-23b-3p expression in HCC hospital tissues
Patients and data collected from medical records
Regarding the cases recruited, 101 FFPE HCC tissues and their counterpart adjacent non-cancerous HCC were obtained from the First Affiliated Hospital of the Guangxi Medical University March, 2010 and December, 2011. Before curative hepatectomy, none had received radiotherapy or chemotherapy. Formalin-fixed and paraffin embedded (FFPE) tissues were assessed in both the HCC tissues and adjacent non-cancerous HCC; we retrospectively acquired the following clinicopathological parameters: age, sex, cirrhosis, tumor size, tumor nodes, differentiation, clinical TNM stages, metastasis, existence or inexistence of portal vein tumor embolus, capsular infiltration, vaso-invasion, as well as other biomarkers, such as serum AFP level, microvessel density (MVD). Two independent pathologists diagnosed all the cases. The project had already gained ethics approval from the Ethics Committee of First Affiliated Hospital of Guangxi Medical University and the patients signed informed consents and agreed their samples to be used in scientific research. The clinicopathological characteristics of the population are shown in Table III.
Table III.
The clinicopathological characteristics related to miR-23b-3p expression in patients.
| Clinicopathological features | N | miR-23b-3p relevant expression (2−ΔCq)
|
Correlation
|
|||
|---|---|---|---|---|---|---|
| Mean ± SD | t-value | P-value | r-value | P-value | ||
| Tissue | ||||||
| Adjacent non-cancerous | 101 | 4.4694±1.99328 | −6.847 | <0.001 | 0.46 | <0.001 |
| HCC | 101 | 2.7376±1.57739 | ||||
| Age (years) | ||||||
| <50 | 50 | 2.7160±1.53973 | 0.136 | 0.892 | −0.019 | 0.85 |
| ≥50 | 51 | 2.7588±1.62852 | ||||
| Sex | ||||||
| Male | 80 | 2.7162±1.55445 | −0.265 | 0.792 | 0.057 | 0.568 |
| Female | 21 | 2.8190±1.69930 | ||||
| Cirrhosis | ||||||
| − | 54 | 2.8426±1.53274 | −0.715 | 0.476 | 0.112 | 0.256 |
| + | 47 | 2.6170±1.63539 | ||||
| Tumor size | ||||||
| <5 cm | 21 | 2.5905±1.59371 | −0.478 | 0.633 | 0.067 | 0.505 |
| ≥5 cm | 80 | 2.7762±1.58092 | ||||
| Tumor nodes | ||||||
| Single | 57 | 3.0158±1.45979 | 2.049 | 0.043 | −0.255 | 0.01 |
| Multiple | 44 | 2.3773±1.66606 | ||||
| Differentiation | ||||||
| High | 7 | 2.9857±1.65673 | F=0.394a | 0.675a | −0.044 | 0.66 |
| Moderate | 64 | 2.8062±1.71722 | ||||
| Low | 30 | 2.5333±1.23995 | ||||
| Clinical TNM stage | ||||||
| I–II | 25 | 3.0520±1.9796 | 1.151 | 0.253 | −0.192 | 0.055 |
| III–IV | 76 | 2.6342±1.67766 | ||||
| Metastasis | ||||||
| − | 49 | 3.3959±1.57506 | 4.435 | <0.001 | −0.447 | <0.001 |
| + | 52 | 2.1173±1.31762 | ||||
| Portal vein tumor embolus | ||||||
| − | 69 | 3.1029±1.60797 | 3.618 | <0.001 | −0.34 | 0.001 |
| + | 32 | 1.9500±1.19055 | ||||
| Tumor capsular infiltration | ||||||
| With complete capsule | 49 | 2.7449±1.60663 | 0.045 | 0.964 | 0.001 | 0.989 |
| Infiltration or no capsule | 52 | 2.7308±1.56500 | ||||
| Vaso-invasion | ||||||
| − | 63 | 2.9635±1.63917 | 1.876 | 0.064 | −0.169 | 0.092 |
| + | 38 | 2.3632±1.41123 | ||||
One-way analysis of variance (ANOVA) test was utilized; t, Student's t-test; SD, standard deviation; HCC, hepatocellular carcinoma; TNM, tumor node metastasis; ANOVA, analysis of variance.
RT-qPCR
The total RNAs were isolated from clinical tissue samples with a miRNeasy FFPE kit (Qiagen, Limburg, The Netherlands) according to the manufacturer's instructions. To assess the miRNA purity and concentration, a quantitative real-time PCR method was utilized in 101 HCC tissues and their counterpart adjacent non-cancerous HCC.
Statistical analysis
The statistical software and methods that were used to analyze the data from clinical patient files downloaded from TCGA were also used to identify the relationship between miR-23b-3p expression and HCC based on qPCR data.
Meta-analysis for GEO datasets, TCGA datasets and PCR verification in-house
Data acquisition
We next added the data from TCGA datasets and PCR verification in-house into meta-analysis to expand the sample size and enhance the credibility. The expression of hsa-miR-23b-3p in HCC tissues and adjacent non-cancerous tissues were extracted in meta-analysis.
Statistical analysis
As for statistical analysis of meta-analysis of GEO datasets, the same statistical methods and software were used to perform the meta-analysis of GEO datasets, TCGA datasets and qPCR data.
Biological information analysis
Prediction targets of miR-23b-3p collection
Ten algorithms, including those from TargetScan (http://www.targetscan.org), microRNA.org (http://www.microrna.org), RNA22 (https://cm.jefferson.edu/rna22/Precomputed), PicTar-vert (http://pictar.mdc-berlin.de/cgi-bin/PicTar_vertebrate.cgi), miRDB (http://mirdb.org/miRDB), PolymiRTS Database (http://compbio.uthsc.edu/miRSNP/), PITA (http://genie.weiz-mann.ac.il/pubs/mir07/mir07_dyn_data.html#), TargetMiner (http://www.isical.ac.in/~bioinfo_miu/targetminer20.htm), TarBase (http://diana.imis.athenainnovation.gr/DianaTools/index.php?r=tarbase), and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw), were applied. Only target genes appearing more than or equal to five times amongst all 10 algorithms were finally applied for investigation of the next step.
Validation targets of miR-23b-3p collection
We searched the PubMed database using the following search strategy: (MIRN23 OR microRNA23 OR microRNA-23 OR miR-23 OR hsa-mir-23 OR miR-23b OR microRNA-23b OR miRNA-23b OR 'miR23b' OR 'miRNA23b' OR 'microRNA23b' OR miR-23b-3p OR miRNA-23b-3p OR microRNA-23b-3p) and target*. The validation targets of hsa-miR-23b-3p were recorded from the identified articles by two authors (P-R Wu and X-L Xiang), and the third author resolved controversies. These targets were integrated with prediction targets after combination.
Bioinformatics analysis of miR-23b-3p
Gene Ontology (GO) analysis. GO analysis was conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://David.abcc.ncifcrf.gov/). The targets of miR-23b-3p were divided into three primary classes: biological processes, cellular components and molecular functions.
Pathway analysis
For pathway analysis, we downloaded pathway data from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.ad.jp/KEGG) to investigate gene interactions. miR-23b-3p targets were mapped to the KEGG pathway database using KOBAS 2.0 (http://www.genome.jp/kegg/pathway.html), and P-values [and a corresponding false discovery rate (FDR)] were applied to estimate each enriched pathway.
Network analysis
The Search Tool for the Retrieval of Interacting Genes (STRING), which provides information for experimental and predicted interactions, is an online database. STRING was applied to search and to determine an interaction network of targets of miR-23b-3p via confidence score calculation.
Results
miR-23b-3p expression in HCC based on GEO datasets
Characteristics of the included datasets
The characteristics of the GEO datasets included are presented in Table I. A total of 10 datasets, including GSE6857 (USA, 2013) (21), GSE10694 (China, 2008) (22), GSE21362 (Japan, 2010) (23), GSE12717 (China, 2008) (24), GSE54751 (USA, 2014) (25), GSE57555 (Japan, 2015) (26), GSE41874 (Japan, 2013) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41874), GSE67138 (USA, 2015) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67138), GSE69580 (China 2015) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE69580+), and GSE67139 (USA, 2015) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67139), were identified to meet the standards. All of the datasets were derived from tissues.
Table I.
Characteristics of hsa-miR-23b-3p gene expression profiling datasets included in meta-analysis.
| Citation | Country | Data source | HCC patients | Healthy controls | Platform |
|---|---|---|---|---|---|
| Budhu et al 2013 (21) | USA | GEO: GSE6857 | 240 | 241 | GPL4700 |
| Li et al 2008 (22) | China | GEO: GSE10694 | 75 | 87 | GPL6542 |
| Sato et al 2011 (23) | Japan | GEO: GSE21362 | 73 | 73a | GPL10312 |
| Su et al 2009 (24) | USA | GEO: GSE12717 | 5 | 3 | GPL7274 |
| Shen et al 2015 (25) | USA | GEO: GSE54751 | 10 | 10a | GPL18262 |
| Murakami et al 2015 (26) | Japan | GEO: GSE57555 | 5 | 5a+11 | GPL16699 |
| GPL18044 | |||||
| Morita 2013b | Japan | GEO: GSE41874 | 6 | 4 | GPL7722 |
| Hung 2015b | China | GEO: GSE69580 | 5 | 5a | GPL10850 |
| Barry 2015b | USA | GEO: GSE67138 | 23 | 34 | GPL8786 |
| Barry 2015b | USA | GEO: GSE67139 | 60 | 60 | GPL8786 |
Cases from adjacent tumor tissues;
no relevant references. HCC, hepatocellular carcinoma.
Value of miR-23b-3p as a biomarker for HCC
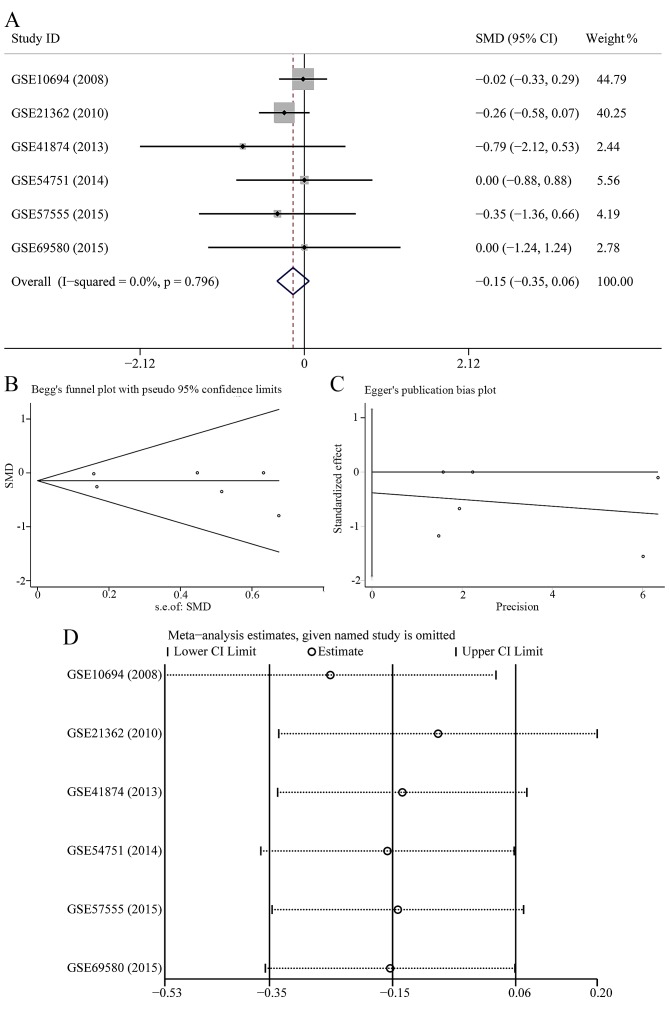
In 7 data-sets presenting miR-23b-3p expression with comparable groups, 414 patients with HCC and 343 healthy subjects were recruited. The expression of miR-23b-3p was lower in HCC patients than healthy controls (Fig. 1A); however, no statistical significance was found (SMD=−0.081; 95% CI, −0.216 to 0.054; P=0.239). A fixed effects model was selected to pool the effect variables that did not show heterogeneity (P=0.807, I2=0%). The results of Begg's and Egger's test shown in Fig. 1B and C was 0.133 and 0.251, respectively. As a consequence, no publication bias was discovered. According to sensitivity analysis shown in Fig. 1D, the result was altered when we eliminated the study of GSE6857, suggesting that this study may influence the pooled SMD and 95% CI. The forest plot after the removing of GSE6857 is shown in Fig. 2A; however, no statistical significance was found (SMD=−0.145; 95% CI, −0.352 to 0.061; P=0.168). According to Begg's and Egger's test (Fig. 2B and C), no publication bias was observed (P>0.05). The sensitivity analysis did not show any alteration when we removed a single study (Fig. 2D).
Figure 1.
Meta-analysis of miR-23b-3p expression in hepatocellular carcinoma (HCC) patients and healthy controls retrieved from Gene Expression Omnibus (GEO) datasets. (A) Forest plot. The center dot and the horizontal line represent the standard mean difference (SMD) and 95% confidence interval (CI), respectively. The pooled SMD and its corresponding 95% CI are presented below the list of studies; (B) Begg's funnel plot. Hazard ratios are presented on a logarithmic scale. (C) Egger's publication bias plot. Hazard ratios are presented on a logarithmic scale; (D) sensitivity analysis.
Figure 2.
Meta-analysis of removing a single study from Gene Expression Omnibus (GEO) datasets. (A) Forest plot. The center dot and the horizontal line represent the standard mean difference (SMD) and 95% confidence interval (CI), respectively. The pooled SMD and its corresponding 95% CI are presented below the list of studies; (B) Begg's funnel plot. Hazard ratios are presented on a logarithmic scale. (C) Egger's publication bias plot. Hazard ratios are presented on a logarithmic scale; (D) sensitivity analysis. Pooled SMDs and 95% CIs by eliminating each study.
Six datasets were enrolled to assess the association between miR-23b-3p expression and the clinical aspects of HCC. However, five parameters, including invasion (83 patients), age (90 patients), sex (8 patients), HBV infection (66 patients) and metastasis (27 patients) were estimated in this study. The pooled SMD were −1.302 (95% CI, −3.055 to 0.451, P=0.146) (Fig. 3A), 0.101 (95% CI, −0.203 to 0.405, P=0.515) (Fig. 3B), −0.266 (95% CI, −1.293 to 0.762, P=0.613) (Fig. 3C), −0.186 (95% CI, −1.093 to 1.465, P=0.775) (Fig. 3D), and −0.288 (95% CI, −0.787 to 0.210, P=0.257) (Fig. 3E), respectively. None of the five parameters were associated with the level of miR-23b-3p expression.
Figure 3.
Meta-analysis of the relationships between miR-23b-3p and clinicopathological parameters in hepatocellular carcinoma (HCC) patients. (A) Invasion; (B) age; (C) sex; (D) HBV infection; (E) metastasis. Pooled standard mean differences (SMDs) and 95% confidence intervals (CIs) by eliminating each study.
miR-23b-3p expression in HCC based on the TCGA dataset
The expression level of miR-23b-3p was downregulated in HCC samples
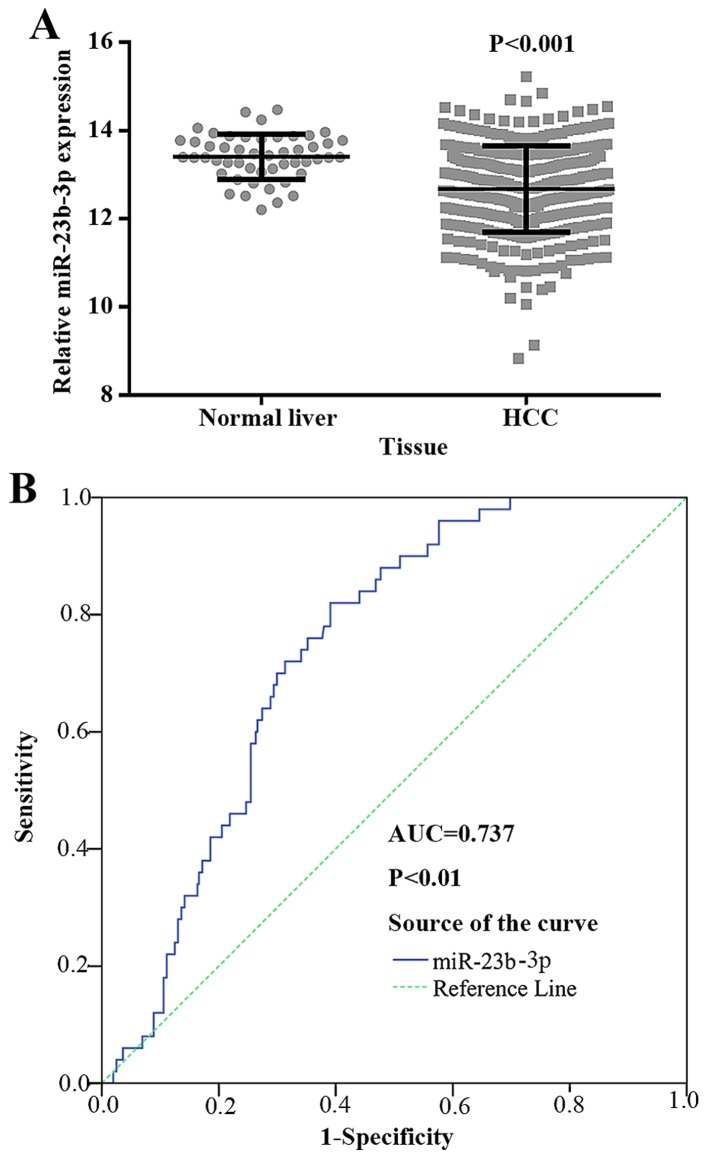
To explore the relationship between miR-23b-3p and HCC, we further obtained data for 361 HCC tissues and 50 normal liver tissues from TCGA. The clinicopathological features are presented in Table II. According to the TCGA data, the expression level of miR-23b-3p was substantially downregulated in HCC (12.6722±0.978150) being compared to normal liver tissues (13.4039±0.51072, P<0.001) (Fig. 4A).
Figure 4.
miR-23b-3p expression in hepatocellular carcinoma (HCC) tissues from The Cancer Genome Atlas (TCGA). (A) The difference in relevant miR-23b-3p expression between HCC and adjacent non-cancerous HCC; (B) ROC curve to distinguish HCC from normal tissues. Error bars represented standard deviation (SD).
Relationships between miR-23b-3p and clinicopathological features
Due to the downregulated miR-23b-3p expression observed in HCC, the association between clinicopathological features and miR-23b-3p was studied (Table II and Fig. 5). The data indicated that a higher level of miR-23b-3p was closely related with hepatitis B (12.8831±0.96070), in contrast to that without hepatitis B infection (12.5999±0.97009, P=0.013). miR-23b-3p was also downregulated in smoking patients (12.2025±1.11847) compared to those without smoking history (12.7127±0.96181, P=0.035). The results also gave a demonstration that the downregulated expression of miR-23b-3p was closely associated with vascular invasion (12.5905±0.95709) compared with non-vascular invasion (12.8530±0.94514, P=0.032). According to the universal standard pathologic neoplasm staging methods of AJCC, there was an obvious negative relationship between tumor node metastasis and miR-23b-3p. The expression level of miR-23b-3p was screened in regional lymphatic metastasis patients (11.7684±0.75393). The expression level was 12.7780±0.96867 in the patients who never had lymphatic metastasis; those with samples that remained in an undefined stage had a level of 12.4733±0.96540 (P=0.006). No other clinicopathological parameters had a significant relationship with miR-23b expression, such as HCV infection or alcohol consumption, nor did neoadjuvant therapy, radiation therapy or pharmaceutical treatment. In accordance with Table II, Spearman's method was applied when correlating miR-23b-3p with clinicopathological parameters, including hepatitis B (r=0.134, P=0.013), smoking (r=−0.014, P=0.035), vascular invasion (r=−0.122, P=0.032), and N status (r=−0.158, P=0.003). There were no other positive correlations with the remaining clinical features.
Figure 5.
The relationship between miR-23b-3p expression and clinicopathological parameters of hepatocellular carcinoma (HCC) patients from The Cancer Genome Atlas (TCGA). (A) hepatitis B; (B) smoking history; (C) vascular invasion; (D) N status. Error bars represent standard deviation (SD).
The diagnostic accuracy of miR-23b-3p in HCC tissues
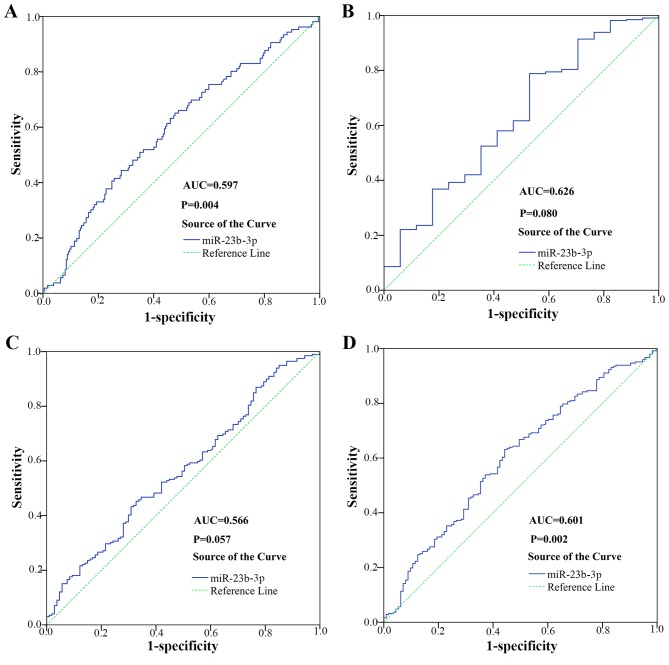
To further determine miR-23b-3p as a biomarker in diagnosing HCC, we performed ROC for verification. As shown in Fig. 4B, the AUC of miR-23b-3p in HCC and its counterpart normal tissues was 0.737 (95% CI, 0.680 to 0.794; P<0.001). The cut-off value reached 0.429; the sensitivity and specificity were 82.0 and 60.9%, respectively. The result suggested that miR-23b-3p may be treated as a reliable biomarker in diagnosing HCC. As shown in Fig. 6, the AUC to judge hepatitis B was 0.597 (95% CI, 0.532 to 0.662; P=0.004). The cut-off value for miR-23b-3p was 0.174; the sensitivity and specificity were 65.1 and 52.3% respectively. The AUC to judge smoking was 0.626 (95% CI, 0.485 to 0.767; P=0.080). The cut-off value for miR-23b-3p was 0.259; the sensitivity and specificity were 78.8 and 47.1%, respectively. The AUC to judge vascular invasion was 0.566 (95% CI, 0.499 to 0.633, P=0.057). The cut-off value for miR-23b-3p was 0.125; the sensitivity and specificity were 45.2 and 67.3%, respectively. The AUC to judge N status was 0.601 (95% CI, 0.538 to 0.664, P=0.002). The cut-off value for miR-23b-3p was 0.125; the sensitivity and specificity were 63.2 and 55.8%, respectively.
Figure 6.
Diagnostic significance between miR-23b-3p and different parameters on hepatocellular carcinoma (HCC) tissues from The Cancer Genome Atlas (TCGA). (A) Hepatitis B; (B) smoking history; (C) vascular invasion; (D) N status. Error bars represented standard deviation (SD).
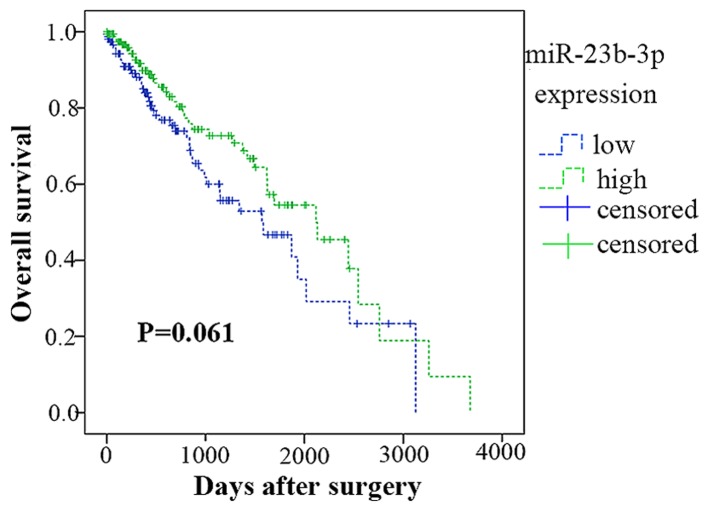
miR-23b-3p expression in surviving HCC
According to the data from TCGA, 359 of the 361 patients were followed up. The Kaplan-Meier analysis in Fig. 7 showed that 176 patients had a lower miR-23b-3p expression (lower than the mean expression level of 12.6797), whereas the remaining 183 patients had a higher level (higher than the mean expression level of 12.6797). In contrast, the lower group had an average survival time of 1,617.668±151.722 days, whereas the higher group had a survival time of 1,944.626±160.587 days. However, the survival times of the high and low miR-23b-3p expression groups were not significantly different (Chi-square =3.351, P=0.061).
Figure 7.
Kaplan-Meier curve for overall survival (months) of hepatocellular carcinoma (HCC) patients included in The Cancer Genome Atlas (TCGA) and grouped by the level of miR-23b-3p expression. There was no significant association between miR-203 expression and survival times in patients with HCC (P=0.061). Error bars represented standard deviation (SD).
miR-23b-3p expression in HCC tissues from PCR verification in-house
The expression of miR-23b-3p was downregulated in HCC
To study the expression level of miR-23b-3p, a quantitative real-time PCR method was performed in 101 HCC tissues and counterpart adjacent non-cancerous HCC. The clinicopathological features of the patients, obtained from medical records, are available in Table III. The expression of miR-23b-3p was obviously downregulated in HCC cases (2.7376±1.99328) in contrast to their counterpart adjacent non-cancerous HCC (2.7376±1.57739, P<0.001) (Fig. 8A). The TCGA and PCR data provided similar results.
Figure 8.
Expression of miR-23b-3p in hepatocellular carcinoma (HCC) and adjacent non-cancerous HCC. Quantitative real-time PCR was performed to detect the expression of miR-23b-3p. (A) The difference of relevant miR-23b-3p expression between HCC and adjacent non-cancerous HCC; (B) ROC curve of miR-23b-3p expression to distinguish HCC from comparative cancerous liver. Error bars represent standard deviation (SD).
The relationships between miR-23b-3p and clinicopathological features
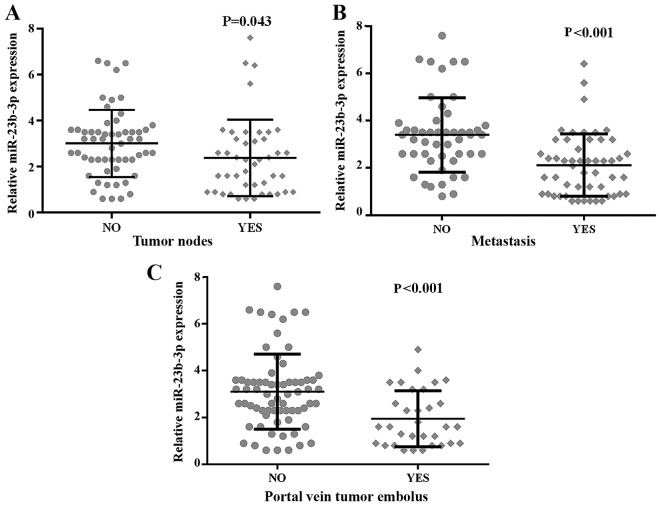
As noted above, the expression of miR-23b-3p was remarkably lower in the HCC tissues than their counterpart adjacent non-cancerous HCC, which indicated that miR-23b-3p may perform the function of a feasible tumor suppressive miRNA in HCC. We investigated the relationships between miR-23b-3p expression and clinicopathological features. The results shown in Table III and Fig. 9 validated that the down-regulated level of miR-23b-3p was positively related to patients without metastasis (3.3959±1.57506), in contrast with those with metastasis (2.1173±1.31762, P<0.001). Subsequently, the miR-23b-3p level was significantly positively associated with multiple tumor nodes (3.0158±1.45979) compared with single nodes (2.3773±1.66606, P=0.043). The presence of a portal vein tumor embolus showed a lower expression of miR-23b-3p (1.9500±1.19055) than did its absence (3.1029±1.60797, P<0.001). Furthermore, we used Spearman's method to record the correlations between the expression of miR-23b-3p and clinicopathological parameters, including tumor nodes (r=−0.255, P=0.010), metastasis (r=−0.447, P<0.001), and portal vein embolus (r=−0.340, P=0.001). Nonetheless, we failed to gain any positive correlations between miR-23b-3p expression and other clinical features.
Figure 9.
The relationship between miR-23b-3p expression and clinicopathological parameters of hepatocellular carcinoma (HCC). (A) tumor nodes, (B) metastasis; (C) portal vein tumor embolus. Error bars represent standard deviation (SD).
The diagnostic value of the miR-23b-3p level in HCC tissues
To verify the diagnostic value of the miR-23b-3p level in HCC tissues, a ROC curve was utilized. As shown in Fig. 8B, the area under the curve (AUC) of miR-23b-3p in HCC and adjacent non-cancerous HCC was 0.765 (95% CI, 0.700 to 0.830, P<0.001). At 0.396 (the cut-off value of miR-23b-3p), the sensitivity and specificity was 88.2 and 55.4%, respectively. In Fig. 10, the AUC to judge tumor nodes was 0.648 (95% CI, 0.538 to 0.758, P=0.011). The cut-off value for miR-23b-3p was 0.272; the sensitivity and specificity was 52.6 and 68.2%, respectively. The AUC to judge metastasis was 0.758 (95% CI, 0.664 to 0.851, P<0.001). The cut-off value for miR-23b-3p was 0.468; the sensitivity and specificity was 77.6 and 69.2%, respectively. The AUC to judge portal vein tumor embolus was 0.711 (95% CI, 0.601 to 0.820, P=0.001). The cut-off value for miR-23b-3p was 0.29; the sensitivity and specificity was 88.4 and 40.6%, respectively.
Figure 10.
Diagnostic significance between miR-23b-3p and various parameters for hepatocellular carcinoma (HCC). (A) tumor nodes; (B) metastasis; (C) portal vein tumor embolus. Error bars represent standard deviation (SD).
miR-23b-3p expression in HCC recurrence
Of the 101 patients recruited for the present study, 76 completed the follow-up. A Kaplan-Meier analysis is presented in Fig. 11. Among the 76 patients, 45 had a lower expression of miR-23b-3p (lower than the median level of 2.600), whereas 31 had higher miR-23b-3p expression. The lower expression group had 54.257±2.943 months of survival time without recurrence; the higher expression group had 54.509±3.962 months. Thus, there were no significant differences in survival time between high and low miR-23b-3p expression (Chi-Square =0.706, P=0.401).
Figure 11.
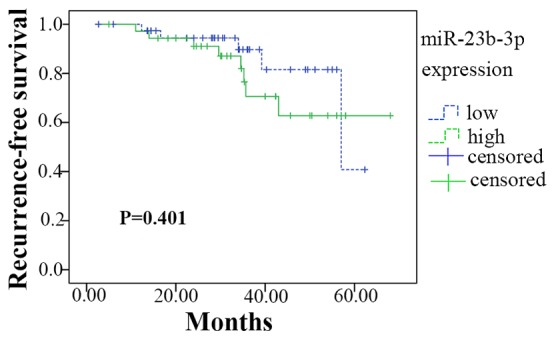
Kaplan-Meier curve for recurrence-free survival in hepatocellular carcinoma (HCC) patients grouped by the level of miR-23b-3p expression. There was no significant association between miR-203 expression and recurrence free survival in patients with HCC (P=0. 401). Error bars represented standard deviation (SD).
Meta-analysis for GEO datasets, TCGA datasets and PCR verification in-house
Characteristics of included studies
Ten GEO datasets, TCGA dataset and the data derived from medical records, as mentioned above with 886 HCC patients and 587 normal persons in total, were included in the new meta-analysis. The characteristics of the studies were previously described (which were shown in sections 'Characteristics of the included data-sets', 'The expression level of miR-23b-3p was downregulated in HCC samples' and 'The expression of miR-23b-3p was downregulated in HCC') in detail.
Value of miR-23b-3p as a biomarker for HCC
The pooled SMD and its 95% CI (Fig. 12A) indicated that people with lower expression of miR-23b-3p had a significant risk for HCC (SMD=−0.368; 95% CI, −0.689 to −0.048; P=0.024). A random effects model was selected to pool the effect variables with heterogeneity (P=0.000, I2=81.3%). The results of Begg's and Egger's test were 0.754 and 0.687, respectively (Fig. 12B and C); however, no publication bias was discovered. The sensitivity analysis indicated that our own PCR data exercised a certain influence on that result (Fig. 12D). Therefore, we removed PCR data and constructed the forest plot again (Fig. 13A). The SMD was −0.258 (95% CI, −0.529 to 0.014) and no statistical significance was found (P=0.063). The results of Begg's and Egger's test were 0.386 and 0.602, respectively (Fig. 13B and C); therefore, no publication bias was observed. The sensitivity analysis showed that the results were altered when we removed the study of TCGA dataset (Fig. 13D).
Figure 12.
Meta-analysis of miR-23b-3p expression in hepatocellular carcinoma (HCC) patients and healthy controls retrieved from Gene Expression Omnibus (GEO) datasets, The Cancer Genome Atlas (TCGA) datasets and PCR data. (A) Forest plot. The center dot and the horizontal line represent the standard mean difference (SMD) and 95% confidence interval (CI), respectively. The pooled SMD and its corresponding 95% CI are presented below the list of studies; (B) Begg's funnel plot. Hazard ratios are presented on a logarithmic scale. (C) Egger's publication bias plot. Hazard ratios are presented on a logarithmic scale; (D) sensitivity analysis. Pooled SMDs and 95% CIs by eliminating each study.
Figure 13.
Meta-analysis of removing PCR data from all the studies. (A) Forest plot. The center dot and the horizontal line represent the standard mean difference (SMD) and 95% confidence interval (CI), respectively. The pooled SMD and its corresponding 95% CI are presented below the list of studies; (B) Begg's funnel plot. Hazard ratios are presented on a logarithmic scale. (C) Egger's publication bias plot. Hazard ratios are presented on a logarithmic scale; (D) sensitivity analysis. Pooled SMDs and 95% CIs by eliminating each study.
Bioinformatics analysis
Target genes of miR-23b-3p collection and integration
Using the 10 algorithms, 14,810 target genes were predicted to be modulated potentially by miR-23b-3p. The number of miR-23b-3p prediction targets of the 10 algorithms varied, ranging from 74 to 6,192 with an average of 2,375. Only targets that appeared more than or equal to five times among all 10 algorithms were finally applied for further analysis. In total, 357 targets from the algorithms were included in the next step.
After screening 203 articles from PubMed, we ultimately gathered 69 validation targets and found that 21 targets (ATG12, CA2, CFL2, CHUK, GLS, HMGB2, IL6R, KIAA1467, MET, NOTCH2, PLAU, PNRC2, PRDM1, PTEN, PTK2B, RRAS2, SEMA6D, SPRY2, TAB3, VHL and WBP2) were duplicated with prediction targets. By combining targets from algorithms and articles, we finally used 405 targets to conduct the bioinformatics analysis.
Bioinformatics analysis of miR-23b-3p targets
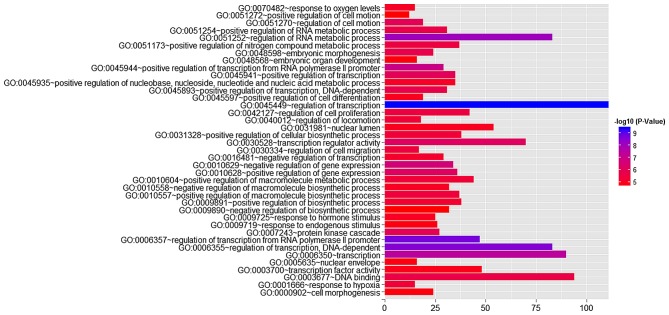
Using GO analysis, 405 targets regulated by miR-23b-3p were classified as biological processes, cellular components, and molecular functions (Table IV and Fig. 14). In the pathway analysis, targets of miR-23b-3p were primarily enriched in the signaling pathways of renal cell carcinoma, hepatitis B (shown in http://www.genome.jp/kegg-bin/show_pathway?hsa05161/hsa:6777%09red/hsa:4318%09red/hsa:8503%09red/hsa:353376%09red/hsa:596%09red/hsa:317%09red/hsa:1147%09red/hsa:5728%09red/hsa:6714%09red/hsa:4214%09red/hsa:355%09red/hsa:4088%09red/hsa:4089%09red/hsa:2185%09red/hsa:1869%09red/hsa:207%09red) and pancreatic cancer (corrected P<0.05). In the protein-protein interaction (PPI) network for miR-23b-3p (Fig. 15), 338 target genes were identified to be involved in 1,074 nodes. A total of 8 targets, including SRC, AKT1, EGFR, CTNNB1, BCL2, SMAD3, PTEN and KDM6A, were located in the key nodes with high degree (>35).
Table IV.
Four hundred five genes were categorized in GO as biological processes, cellular components and molecular functions.
| Term | Count | P-value | FDR |
| Biological process | |||
| Regulation of transcription | 111 | 3.87E-10 | 6.73E-07 |
| Regulation of transcription from RNA polymerase II promoter | 47 | 1.98E-09 | 3.44E-06 |
| Regulation of transcription, DNA-dependent | 83 | 2.93E-09 | 5.10E-06 |
| Regulation of RNA metabolic process | 83 | 8.33E-09 | 1.45E-05 |
| Transcription | 90 | 3.70E-08 | 6.44E-05 |
| Positive regulation of transcription from RNA polymerase II promoter | 29 | 1.03E-07 | 1.80E-04 |
| Negative regulation of gene expression | 34 | 2.12E-07 | 3.68E-04 |
| Positive regulation of gene expression | 36 | 6.48E-07 | 0.001128 |
| Positive regulation of transcription | 35 | 9.33E-07 | 0.001624 |
| Regulation of cell motion | 19 | 1.07E-06 | 0.001868 |
| Protein kinase cascade | 27 | 1.19E-06 | 0.002065 |
| Positive regulation of transcription, DNA-dependent | 31 | 1.79E-06 | 0.003114 |
| Positive regulation of RNA metabolic process | 31 | 2.12E-06 | 0.003697 |
| Positive regulation of nitrogen compound metabolic process | 37 | 2.57E-06 | 0.004477 |
| Regulation of cell proliferation | 42 | 2.93E-06 | 0.005101 |
| Regulation of cell migration | 17 | 3.52E-06 | 0.006119 |
| Positive regulation of macromolecule biosynthetic process | 37 | 3.65E-06 | 0.006356 |
| Positive regulation of cellular biosynthetic process | 38 | 4.10E-06 | 0.007132 |
| Positive regulation of macromolecule metabolic process | 44 | 4.31E-06 | 0.007507 |
| Regulation of locomotion | 18 | 4.36E-06 | 0.007589 |
| Response to hypoxia | 15 | 4.57E-06 | 0.007949 |
| Positive regulation of biosynthetic process | 38 | 5.68E-06 | 0.009878 |
| Negative regulation of transcription | 29 | 7.16E-06 | 0.012463 |
| Response to oxygen levels | 15 | 8.29E-06 | 0.014432 |
| Positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 35 | 8.58E-06 | 0.014931 |
| Negative regulation of macromolecule biosynthetic process | 32 | 1.01E-05 | 0.01756 |
| Response to hormone stimulus | 25 | 1.05E-05 | 0.018246 |
| Positive regulation of cell differentiation | 19 | 1.21E-05 | 0.021016 |
| Response to endogenous stimulus | 26 | 1.84E-05 | 0.032008 |
| Embryonic organ development | 16 | 1.91E-05 | 0.033319 |
| Cell morphogenesis | 24 | 1.96E-05 | 0.034027 |
| Positive regulation of cell motion | 12 | 2.46E-05 | 0.042821 |
| Negative regulation of biosynthetic process | 32 | 2.51E-05 | 0.04366 |
| Cellular component | |||
| Nuclear lumen | 54 | 1.14E-05 | 0.015418 |
| Nuclear envelope | 16 | 1.76E-05 | 0.023821 |
| Molecular function | |||
| Transcription regulator activity | 70 | 6.80E-07 | 9.94E-04 |
| DNA binding | 94 | 2.12E-06 | 0.0031 |
| Transcription factor activity | 48 | 1.39E-05 | 0.020262 |
FDR, false discovery rate.
Figure 14.
GO analysis for target genes of miR-23b-3p. The length of horizontal column represents the number of genes and their color stand for P-value.
Figure 15.
Network analysis with the prospective target genes of miR-23b-3p. Network nodes represent proteins. Colored nodes, query proteins and first shell of interactors; white nodes, second shell of interactors. Small nodes, protein of unknown 3D structure; large nodes, some 3D structure is known or predicted.
Discussion
HCC is one of the most prevalent forms of liver cancer, and nearly one million cases of HCC occur annually. Thus, it is urgent to explore the mechanism of HCC tumorigenesis and progression and to determine treatment and prevention strategies. Exploiting a new self-assembled cell microarray, Zhang et al carried out high-throughput screening for miRNAs that are involved in cell transplantation. They discovered miR-23b-3p, which functioned as a tumor suppressor in human colon cancer (27). Since then, substantial research has studied the functions of miR-23b-3p in many physiological and pathological processes, especially tumor progression. As noted above, several studies showed that miR-23b-3p played a dual role in various cancers; these studies also investigated the mechanisms of miR-23b-3p. This study focused on determining the role of miR-23b-3p in HCC and its effect on patient prognosis.
In the meta-analysis of data downloaded from GEO data-sets, the pooled SMD indicated that miR-23b-3p expression showed no remarkable difference between HCC patients and normal subjects. No heterogeneity was noted. Additionally, based on GEO datasets, we investigated the association between miR-23b-3p and the clinicopathological aspects of HCC and observed that miR-23b-3p was significantly correlated with invasion. We failed to prove the relevance of miR-23b-3p and other clinical parameters. Therefore, diagnosis and targeted therapy based on the miR-23b-3p expression level required further research.
We identified miR-23b-3p as a HCC biomarker by analyzing TCGA data. miR-23b-3p expression was obviously down-regulated in HCC tissues compared to normal liver tissues. Additionally, miR-23b-3p expression was lower (P<0.05) in patients with hepatitis B, smoking history, vascular invasion and lymphatic metastasis. However, only hepatitis B history and lymphatic metastasis of HCC revealed diagnostic values. HCC patients with high miR-23b-3p expression presented longer survival; however, this finding was non-significant.
According to the results above, we propose that miR-23b-3p may play a crucial part in HCC tumorigenesis and development. Therefore, we recruited 101 fresh HCC tissues and counterpart adjacent non-cancerous HCC to inspect the expression of miR-23b-3p in HCC tissues and its association with the clinicopathological aspects and prognosis of HCC patients.
However, in contrast with the counterpart adjacent non-cancerous HCC, we observed that miR-23b-3p was aberrantly downregulated in HCC tissue (P<0.001). This consequence was supported by a statistical analysis of TCGA data but in contrast to the microarray meta-analysis. Additionally, HCC patients with multiple tumor nodes showed a lower level of miR-23b-3p expression than those with single tumor nodes (P<0.05). In addition to other parameters that indicated HCC progression, the deterioration of metastasis and the portal vein tumor embolus were strongly associated with a decrease in miR-23b-3p (P<0.001). In line with the aforementioned findings, miR-23b-3p was closely linked to HCC aggressiveness and metastasis. Additionally, Salvi et al transfected miR-23b-3p into HCC cells (SKHeplC3), which resulted in a reduction in motility and proliferative potential; they also found that miR-23b-3p was deregulated in HCC (28). To some extent, our study also revealed that increased miR-23b-3p was associated with an increased time-to-recurrence (months) in HCC patients. However, there was no obvious alteration between the two groups concerning the levels of miR-23b-3p. The lack of a study population may explain this result. Thus, a larger cohort of patients is required to study this in the near future. Additionally, only tumor nodes, metastasis portal vein tumor embolus and EGFR expression of HCC were found to correlate with miR-23b-3p expression and presented diagnostic values.
Altogether, the data derived from GEO datasets, TCGA datasets and qPCR were included to perform a comprehensive meta-analysis. The pooled SMD indicated that pronounced downregulation of miR-23b-3p expression was noted in HCC patients as compared to healthy subjects, which indicated that low level of miR-23b-3p may play an important role in HCC tumorigenesis and diagnosis. Nevertheless, high heterogeneity indicated that this conclusion was less reliable. According to sensitivity analysis, the data from TCGA and qPCR contributed to the high heterogeneity. The difference of source of specimen and the detection methods of miR-23b-3p expression may be the major factors.
Ten prediction algorithms (utilizing different matching criteria and computational algorithms) were employed in miR-23b-3p target gene prediction. In contrast to the algorithms that provided validated targets and literature extraction, the prediction algorithms could not predict with accuracy. Some factors, such as varying approaches and rules for miRNA targeting, can affect the target prediction results. To reduce the defects associated with target prediction, we combined the results of these algorithms and stipulated that only the genes that appeared no fewer than five times would be involved in the next analysis.
In addition to targets based on experimental validation, prediction targets combined with algorithms were enrolled for GO analysis, pathways analysis and network interaction. A bioinformatics analysis predicted that miR-23b-3p was involved in extensive target regulation. Because these targets took part in multiple phases of tumorigenesis and tumor progression, we speculated that miR-23b-3p was dysregulated in various tumors and led to changes in biological characteristics by regulating a series of target genes in different phases. In a GO analysis, the targets of miR-23b-3p were involved in biological processes (like the regulation of transcription and transcription), cellular components (such as the nuclear lumen and nuclear envelope) and molecular functions (such as transcription regulator activity and DNA binding). These targets play a vital role in major cell processes, and a number of them participate in the carcinogenesis of various cancers, including HCC. According to KEGG analysis, miR-23b-3p participated in the hepatitis B pathway, which was recognized as a key factor of hepatic cirrhosis. Moreover, hepatic cirrhosis could result in hepatic carcinoma. We also found that miR-23b-3p was correlated with hepatitis B in the HCC patients who were included in TCGA. Thus, we speculated that miR-23b-3p may play an important role in HCC tumorigenesis.
A network analysis was ultimately conducted to predict the interaction networks of potential targets. Highly connected genes were regarded to play critical roles in stabilization, interaction, and gene network regulation. A connectivity analysis showed the highest connectivity between SRC and AKT1. SRC is a proto-oncogene encoding a tyrosine-protein kinase that belongs to Src family kinases, which are considered vital to cell proliferation, differentiation, apoptosis and invasion (29,30). Src family kinases have been shown to play a vital role in several malignancies, including breast cancer (30), colon cancer (31), ovarian cancer (32) and HCC (33), as well as melanoma, glioma, and various types of sarcoma (34). Akt1 or protein kinase B (PKB)α, which is regularly activated in human cancers, can phosphorylate downstream molecules and cope with a wide range of cell processes (35). Moreover, Akt1 was directly or indirectly controlled by Src kinase activity (35,36). We conjectured that Src kinase and Akt1 may function in the same signal transduction pathways that result in carcinogenesis and development. This suggests that researchers should consider factors related to the hub genes for improving diagnosis and treatment.
Inquiries in various cancers indicated that miR-23b-3p, functioning as an oncogenic miRNA, can accelerate tumor processes such as proliferation and invasion. Jin et al reported that miR-23b/27b expression was elevated in breast cancer and that it indicated a poor prognosis (12). Nischarin partially participated in reducing the level of miR-23b/27b expression by inhibiting NF-κB phosphorylation and disturbing cancer aggressiveness in vivo (2). Additionally, the AKT/NF-κB pathway, which responds to TNFα and EGF, is the pivotal signaling cascade for Her2/neu-dependent miR-23b/27b in vitro (2). Li et al demonstrated that Fas mRNA, which enhances the cell multiplication and apoptosis rates, was directly silenced by miR-23b-3p upregulation in thymic lymphoma cells (37).
Many studies also noted that the role of miR-23b-3p as a tumor suppressor was downregulated in several cancers. Majid et al discovered that miR-23b-3p was downregulated in prostate cancer, and its decreasing function of directly inhibiting proto-oncogene Src kinase may lead to the acceleration of neoplasm growth (38). The same author in another study stated that miR-23b-3p played a key role in migration of bladder cancer cells via posttranscriptionally directly regulating at Zebl, which could modulate the epithelial-to-mesenchymal transition (EMT) (39). All of the mechanisms by which miR-23b-3p acts as a tumor suppressor may contribute to HCC aggressiveness; further studies are required.
Furthermore, extensive study suggested that miR-23b-3p was associated with other clinicopathological features in HCC. Bisio et al confirmed the cis-mediated regulation of miR-23 by p53 in the MCF7 cell line (40). However, we detected the reverse (upregulated miR-23b-3p) in p53-positive tissue (P>0.05). There were two possible reasons for this. First, miR-23b-3p served as oncogenic miRNA in the MCF7 cell line but acted as a tumor suppressor in HCC. Notably, the specific mechanism remained unclear. Secondly, our data was less trustworthy due to its lack of significance. Chen et al reported that the downregulated expression of miR-23b-3p inhibited HIF-1α/VEGF and β-catenin/Tcf-4 signaling by enhancing the level of VHL (41). The quantity of VEGF in our study was also identical with miR-23b-3p expression; however, this was without significance (P>0.05). Patients with occult hepatitis virus B infection (OBI), who are generally negative for HBsAg, were discovered to have a differential expression of miR-23b-3p (42). Notably, as Salvi et al reported, miR-23b-3p overexpression contributed to a decrease in urokinase-type plasminogen (uPA) and c-met. This was determined by identifying the 3′-UTR of uPA and c-met in two and four sites, respectively, as well as by observing a reduction in cell proliferation and metastasis in HCC (28). The present study partially supports the above study. To our knowledge, the present study is also the first one to explore the clinicopathological significance of miR-23b-3p and contributed to a more comprehensive understanding of the correlation between miR-23b-3p and HCC progression.
Expression of miR-23b-3p was studied in HCC and their relationship explored. A preliminary analysis of the biological characteristics and function of miR-23b-3p was performed via bioinformatical methods. Despite existing limitations, miR-23b-3p was thought to be a tumor suppressor affecting the carcinogenesis and aggressiveness of HCC and may represent a predictive biomarker and therapeutic target for HCC. These results may contribute to a theoretical basis for HCC diagnosis and therapy.
Acknowledgments
The authors thank GEO and TCGA for the public available data.
Footnotes
Funding
This study was supported in part by the Natural Science Foundation of Guangxi, China (2015GXNSFBA139157 and 2017GXNSFAA198026) and Youth Science Foundation of Guangxi Medical University (WLXSZX18001).
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Authors' contributions
RQH and PRW designed the study, did the experiments, analyzed and interpreted the data, and wrote the manuscript. They contributed equally to this work. XLX, XY and HWL performed the statistical analysis, designed and filled-out the figures and tables. XHQ, LHY, ZGP recruited the specimens, carried out the miRNA isolation and real-time RT-qPCR. LHY, ZGP and GC participated in designing the study, supervised all experiments and corrected the manuscript. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
All experiments referred to patient tissues were authorized by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (no. 2016-KY-NSFC-094). This study was a sub-study of the project mentioned in ethics approval. Method and participants are utilized in the same way.
Consent for publication
The consents for publication were obtained from all the patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.de Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology. 2015;62:1190–1200. doi: 10.1002/hep.27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Yun EH, Lim MK, Oh JK, Park JH, Shin A, Sung J, Park EC. Combined effect of socioeconomic status, viral hepatitis, and lifestyles on hepatocelluar carcinoma risk in Korea. Br J Cancer. 2010;103:741–746. doi: 10.1038/sj.bjc.6605803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M'Bengue AK, Doumbia M, Denoman SR, Ouattara DN, Adoubi I, Pineau P. A major shift of viral and nutritional risk factors affects the hepatocellular carcinoma risk among Ivorian patients: A preliminary report. Infect Agent Cancer. 2015;10:18. doi: 10.1186/s13027-015-0013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Hara SP, Mott JL, Splinter PL, Gores GJ, LaRusso NF. MicroRNAs: Key modulators of posttranscriptional gene expression. Gastroenterology. 2009;136:17–25. doi: 10.1053/j.gastro.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Francis P, Moon SY, Bilke S, Zhu YJ, Meltzer PS. Role of the microRNA-23 27 24 clusters in osteosarcoma. Cancer Res. 2012;72(Suppl 8):1113. doi: 10.1158/1538-7445.AM2012-1113. [DOI] [Google Scholar]
- 8.Goto Y, Nishikawa R, Kojima S, Sakamoto S, Kawamura K, Imamoto T, Chiyomaru T, Enokida H, Kinoshita T, Naya Y, et al. The functional significance and its regulated molecular targets of microrna-23b/27b/24-1 cluster in prostate cancer. J Urol. 2014;191:e456–e457. doi: 10.1016/j.juro.2014.02.1233. [DOI] [Google Scholar]
- 9.Ishihara T, Chiyomaru T, Inoguchi S, Enokida H, Seki N, Nakagawa M. The clustered microRNA-23b/27b function as tumor suppressors and useful prognostic markers in renal cell carcinoma. J Urol. 2014;191:e243. doi: 10.1016/j.juro.2014.02.870. [DOI] [Google Scholar]
- 10.Jiang W, Min J, Sui X, Qian Y, Liu Y, Liu Z, Zhou H, Li X, Gong Y. MicroRNA-26a-5p and microRNA-23b-3p up-regulate peroxiredoxin III in acute myeloid leukemia. Leuk Lymphoma. 2015;56:460–471. doi: 10.3109/10428194.2014.924115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han L, Chen L, Zhang K, Shi Z, Zhang J, Zhang A, Wang Y, Song Y, Zheng Y, Jiang T, et al. MicroRNA-23b expression is regulated by VHL and effects on glioma cell survival and invasion. Cancer Res. 2012;72:8. doi: 10.1158/1538-7445.AM2012-145. [DOI] [Google Scholar]
- 12.Jin L, Wessely O, Marcusson EG, Ivan C, Calin GA, Alahari SK. Prooncogenic factors miR-23b and miR-27b are regulated by Her2/Neu, EGF, and TNF-α in breast cancer. Cancer Res. 2013;73:2884–2896. doi: 10.1158/0008-5472.CAN-12-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma G, Dai W, Sang A, Yang X, Gao C. Upregulation of microRNA-23a/b promotes tumor progression and confers poor prognosis in patients with gastric cancer. Int J Clin Exp Pathol. 2014;7:8833–8840. [PMC free article] [PubMed] [Google Scholar]
- 14.Rogler CE, Levoci L, Ader T, Massimi A, Tchaikovskaya T, Norel R. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50:575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- 15.Yuan B, Dong R, Shi D, Zhou Y, Zhao Y, Miao M, Jiao B. Down-regulation of miR-23b may contribute to activation of the TGF-β1/Smad3 signalling pathway during the termination stage of liver regeneration. FEBS Lett. 2011;585:927–934. doi: 10.1016/j.febslet.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 16.Salvi A, Sabelli C, Moncini S, Venturin M, Arici B, Riva P, Portolani N, Giulini SM, De Petro G, Barlati S. MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 2009;276:2966–2982. doi: 10.1111/j.1742-4658.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- 17.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 18.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: A software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan N. Meta-analysis: A quantitative approach of data pooling. Pak Oral Dental J. 2000;20:214–221. [Google Scholar]
- 21.Budhu A, Roessler S, Zhao X, Yu Z, Forgues M, Ji J, Karoly E, Qin LX, Ye QH, Jia HL, et al. Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes. Gastroenterology. 2013;144:1066–1075.e1. doi: 10.1053/j.gastro.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Xie L, He X, Li J, Tu K, Wei L, Wu J, Guo Y, Ma X, Zhang P, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616–1622. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- 23.Sato F, Hatano E, Kitamura K, Myomoto A, Fujiwara T, Takizawa S, Tsuchiya S, Tsujimoto G, Uemoto S, Shimizu K. MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria. PLoS One. 2011;6:e16435. doi: 10.1371/journal.pone.0016435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 25.Shen J, LeFave C, Sirosh I, Siegel AB, Tycko B, Santella RM. Integrative epigenomic and genomic filtering for methylation markers in hepatocellular carcinomas. BMC Med Genomics. 2015;8:28. doi: 10.1186/s12920-015-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murakami Y, Kubo S, Tamori A, Itami S, Kawamura E, Iwaisako K, Ikeda K, Kawada N, Ochiya T, Taguchi YH. Comprehensive analysis of transcriptome and metabolome analysis in intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Sci Rep. 2015;5:16294. doi: 10.1038/srep16294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Hao Y, Yang J, Zhou Y, Li J, Yin S, Sun C, Ma M, Huang Y, Xi JJ. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat Commun. 2011;2:554. doi: 10.1038/ncomms1555. [DOI] [PubMed] [Google Scholar]
- 28.Salvi A, Sabelli C, Moncini S, Venturin M, Arici B, Riva P, Portolani N, Giulini SM, De Petro G, Barlati S. MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 2009;276:2966–2982. doi: 10.1111/j.1742-4658.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- 29.Dehm SM, Bonham K. SRC gene expression in human cancer: The role of transcriptional activation. Biochem Cell Biol. 2004;82:263–274. doi: 10.1139/o03-077. [DOI] [PubMed] [Google Scholar]
- 30.Finn RS. Targeting Src in breast cancer. Ann Oncol. 2008;19:1379–1386. doi: 10.1093/annonc/mdn291. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y, Scott A, Zhang P, Hao Y, Feng X, Somasundaram S, Khalil AM, Willis J, Wang Z. Regulation of paxillin-p130-PI3K-AKT signaling axis by Src and PTPRT impacts colon tumorigenesis. Oncotarget. 2017;8:48782–48793. doi: 10.18632/oncotarget.10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun L, Wang D, Li X, Zhang L, Zhang H, Zhang Y. Extracellular matrix protein ITGBL1 promotes ovarian cancer cell migration and adhesion through Wnt/PCP signaling and FAK/SRC pathway. Biomed Pharmacother. 2016;81:145–151. doi: 10.1016/j.biopha.2016.03.053. [DOI] [PubMed] [Google Scholar]
- 33.Xiong J, Wu JS, Mao SS, Yu XN, Huang XX. Effect of saracatinib on pulmonary metastases from hepatocellular carcinoma. Oncol Rep. 2016;36:1483–1490. doi: 10.3892/or.2016.4968. [DOI] [PubMed] [Google Scholar]
- 34.Shor AC, Keschman EA, Lee FY, Muro-Cacho C, Letson GD, Trent JC, Pledger WJ, Jove R. Dasatinib inhibits migration and invasion in diverse human sarcoma cell lines and induces apoptosis in bone sarcoma cells dependent on SRC kinase for survival. Cancer Res. 2007;67:2800–2808. doi: 10.1158/0008-5472.CAN-06-3469. [DOI] [PubMed] [Google Scholar]
- 35.Vojtechová M, Turecková J, Kucerová D, Sloncová E, Vachtenheim J, Tuhácková Z. Regulation of mTORC1 signaling by Src kinase activity is Akt1-independent in RSV-transformed cells. Neoplasia. 2008;10:99–107. doi: 10.1593/neo.07905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park S, Kim D, Kaneko S, Szewczyk KM, Nicosia SV, Yu H, Jove R, Cheng JQ. Molecular cloning and characterization of the human AKT1 promoter uncovers its up-regulation by the Src/Stat3 pathway. J Biol Chem. 2005;280:38932–38941. doi: 10.1074/jbc.M504011200. [DOI] [PubMed] [Google Scholar]
- 37.Li B, Sun M, Gao F, Liu W, Yang Y, Liu H, Cheng Y, Liu C, Cai J. Up-regulated expression of miR-23a/b targeted the pro-apoptotic Fas in radiation-induced thymic lymphoma. Cell Physiol Biochem. 2013;32:1729–1740. doi: 10.1159/000356607. [DOI] [PubMed] [Google Scholar]
- 38.Majid S, Dar AA, Saini S, Arora S, Shahryari V, Zaman MS, Chang I, Yamamura S, Tanaka Y, Deng G, et al. miR-23b represses proto-oncogene Src kinase and functions as methylation-silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer Res. 2012;72:6435–6446. doi: 10.1158/0008-5472.CAN-12-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majid S, Dar AA, Saini S, Deng G, Chang I, Greene K, Tanaka Y, Dahiya R, Yamamura S. MicroRNA-23b functions as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS One. 2013;8:e67686. doi: 10.1371/journal.pone.0067686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bisio A, De Sanctis V, Del Vescovo V, Denti MA, Jegga AG, Inga A, Ciribilli Y. Identification of new p53 target microRNAs by bioinformatics and functional analysis. BMC Cancer. 2013;13:552. doi: 10.1186/1471-2407-13-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Han L, Zhang K, Shi Z, Zhang J, Zhang A, Wang Y, Song Y, Li Y, Jiang T, et al. VHL regulates the effects of miR-23b on glioma survival and invasion via suppression of HIF-1α/VEGF and β-catenin/Tcf-4 signaling. Neuro Oncol. 2012;14:1026–1036. doi: 10.1093/neuonc/nos122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Li L, Zhou Z, Wang N, Zhang CY, Zen K. A pilot study of serum microRNA signatures as a novel biomarker for occult hepatitis B virus infection. Med Microbiol Immunol. 2012;201:389–395. doi: 10.1007/s00430-011-0223-0. [DOI] [PubMed] [Google Scholar]