Summary
Certain genes show rapid reactivation after several generations of repression, a conserved phenomenon called epigenetic transcriptional memory. Following previous growth in galactose, GAL gene transcriptional memory confers a strong fitness benefit in Saccharomyces cerevisiae adapting to growth in galactose for up to 8 generations. A genetic screen for mutants defective for GAL gene memory revealed new insights into the molecular mechanism, adaptive consequences and evolutionary history of memory. A point mutation in the Gal1 coactivator that disrupts the interaction with the Gal80 inhibitor specifically and completely disrupted memory. This mutation confirms that cytoplasmically-inherited Gal1 produced during previous growth in galactose directly interferes with Gal80 repression to promote faster induction of GAL genes. This mitotically heritable mode of regulation is recently evolved; in a diverged Saccharomyces species, GAL genes show constitutively faster activation due to genetically-encoded basal expression of Gal1. Thus, recently diverged species utilize either epigenetic or genetic strategies to regulate the same molecular mechanism. The screen also revealed that the central domain of the Gal4 transcription factor both regulates the stochasticity of GAL gene expression and potentiates stronger GAL gene activation in the presence of Gal1. The central domain is critical for GAL gene transcriptional memory; Gal4 lacking the central domain fails to potentiate GAL gene expression and is unresponsive to previous Gal1 expression.
Keywords: Epigenetics, transcriptional memory, GAL genes, stochastic expression, adaptive fitness, evolution, trade-offs
Introduction
Transcriptional adaptation to fluctuations in nutrient availability is critical for fitness [1]. In response to previous experiences, certain inducible genes show a mitotically heritable increase in the rate of transcription [2–12]. This epigenetic phenomenon, referred as transcriptional memory, is observed in yeast, Drosophila and humans [5, 7, 13]. However, its evolutionary history and adaptive impact have not been explored. Also, while some aspects of transcriptional memory are deeply conserved, gene-specific features also occur [4, 6, 8, 14], suggesting that gene-specific regulatory systems can be regulated by transcriptional memory.
In S. cerevisiae, GAL genes exhibit transcriptional memory. These genes are specifically induced in galactose to mediate galactose utilization [15, 16]. When cells are shifted from glucose to galactose, the initial rate of induction of GAL genes is very slow; the Gal1 protein reaches steady state levels ~10h after shifting from glucose to galactose (Figure 1B; compare with the doubling time of budding yeast, 1.5–2h). However, in cells that have been previously grown in galactose, GAL genes are rapidly reactivated. This phenomenon persists in the population for 7–8 cell divisions after repression in glucose [2, 11, 14, 17]. Like other genes that show memory, GAL transcriptional memory is also associated with changes in chromatin structure, leading to a state that is poised for faster reactivation [14].
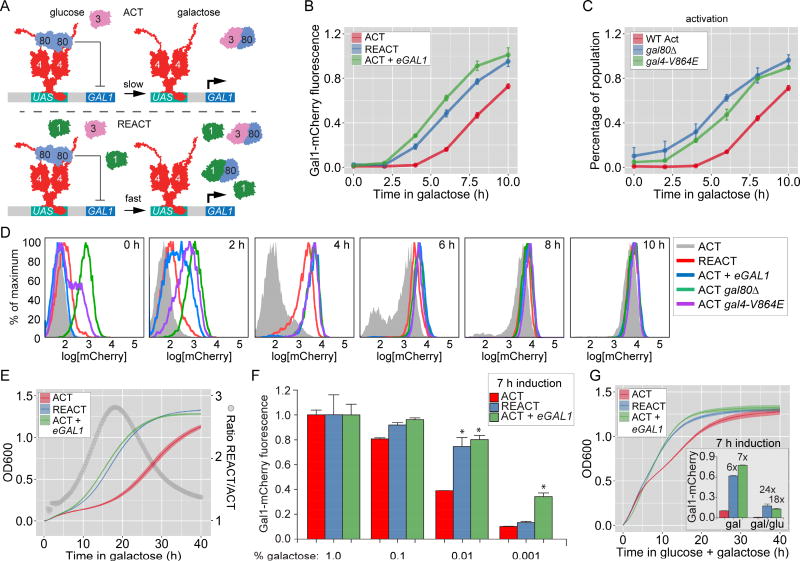
Figure 1. Expression changes during GAL memory and their fitness effect.
A. Model for GAL1 regulation and memory. Top panel: during activation, Gal3 sequesters the Gal80 repressor from the Gal4 activator, leading to GAL gene expression. Bottom panel: during reactivation, residual Gal1 augments Gal3 co-activation, leading to faster expression kinetics. B – G. Naïve cells (ACT), naïve cells expressing ectopic Gal1 (ACT + eGAL1), or cells that were grown in galactose overnight and shifted to glucose for 12 hours (REACT) were shifted to media containing galactose and either GAL1-mCherry fluorescence (B–D & F) or OD600 (E and G) were measured. GAL1-mCherry fluorescence was measured relative to constitutively expressed CFP using flow cytometry. C & D. Effect of Gal80 inhibition on Gal1-mCherry expression. D. Overlay of histograms for Gal1-mCherry from corresponding strains at the indicated times in B and C. E. At time = 0, all cultures were diluted to an OD600 of 0.1 in galactose and OD600 was measured every 20 minutes using 96-well plate reader. Open circles represent the ratio of OD600 between REACT and ACT. F. Gal1-mCherry levels relative to CFP control after 7 hours in different concentration of galactose, plotted as fraction of expression in 1% galactose. G. Growth and Gal1-mCherry expression (inset) in 0.2% glucose + 1.8% galactose. * = p ≤ 0.05, Student’s t test. Error bars for Gal1mCherry fluorescence represent SEM from ≥ 3 biological replicates. The line and the bounding envelope for the OD600 measurements is the mean and SEM, respectively, from ≥ 5 biological replicates. The yeast strains and the number of biological replicates for all experiments are listed in Table S1 and S2, respectively.
The initial induction of GAL genes is slow in part because, in most cells, there are fewer molecules of the Gal3 co-activator than the Gal80 inhibitor [11, 18]. Consequently, the expression is initially heterogeneous, with some cells responding and others not [18]. On the other hand, during memory. reactivation is more uniform [18]. GAL memory requires the Gal1 galactokinase, a paralog of Gal3 [2, 11, 14, 17]. Gal1 is abundantly produced in galactose, extremely stable and diluted very slowly after repression in glucose [14]. A few hundred molecules of Gal1 are necessary and sufficient to produce the effects of memory, including the changes in chromatin structure over the GAL1 promoter and uniformity of expression [11, 14]. Because both Gal1 and Gal3 can interact with Gal80 to relieve inhibition of the Gal4 activator [19–23], this leads to a model whereby residual Gal1 allows cells to more rapidly overcome Gal80 repression and rapidly induce GAL genes during memory (Figure 1A).
Here, we explored the adaptive value, evolutionary history and molecular mechanism of GAL gene transcriptional memory. In S. cerevisiae, GAL transcriptional memory confers a strong adaptive advantage, allowing much faster adaptation to galactose and better utilization of mixed sugars. In contrast, S. uvarum, a divergent Saccharomyces species, does not benefit from previous growth in galactose but instead shows constitutively fast GAL gene activation due to basal GAL1 expression. Replacing the GAL1 promoter in S. cerevisiae with that from S. uvarum recapitulated this difference in behavior, confirming that the basal expression of Gal1 and the resulting fast activation of other GAL genes are genetically encoded by the promoter. Using a point mutation in Gal1 that disrupts the interaction with Gal80, we demonstrate that fast induction of GAL genes in both species results from a physical interaction between Gal1 and Gal80. A screen for mutants that block GAL memory downstream of Gal1 identified a mutation in Gal4 central domain (CD), revealing a critical role for this domain in transcriptional memory downstream of both Gal1 and Gal80. The CD serves to potentiate Gal4 activation and this function is regulated by Gal80. Ectopically expressed CD complemented deletion of the central domain in trans and was recruited to chromatin-bound Gal4Δcd, suggesting that potentiation is the result of inter-domain interactions within Gal4. Thus, recently diverged species employ either epigenetic or genetic strategies to alter the transcriptional potency of a transcription factor, promoting faster adaptation to changes in carbon source.
Results
Transcriptional memory enhances fitness by promoting uniform, rapid activation of GAL genes
Because Gal1 is both necessary and sufficient to promote faster induction of GAL genes during memory, Gal1 likely interacts with Gal80 to allow rapid de-repression of GAL genes (Figure 1A). The relative rates of GAL1 transcription can be compared by measuring Gal1-mCherry fluorescence expressed using flow-cytometry [11, 14]. In cells that had not been previously exposed to galactose (i.e. naïve cells), Gal1-mCherry was undetectable for the first 4h after shifting from glucose to galactose (Figure 1B & 1D; ACT). Between 4h and 8h after switching cells to galactose, expression of Gal1-mCherry was apparent in a subset of cells in the population (i.e. bimodal expression; Figure 1B & 1D, ACT). After 10h in galactose, the entire population expressed Gal1-mCherry (Figure 1D; ACT). In contrast, in cells that were previously grown in galactose and then repressed for 12 hours (~7–8 cell divisions; i.e. memory), the population responded uniformly and rapidly; Gal1-mCherry fluorescence was measurable within 4h after shifting back to galactose (unimodal expression; Figure 1B and D, REACT). Likewise, ectopic expression of GAL1 promoted both rapid and unimodal accumulation of Gal1-mCherry (Figure 1B and D; ACT + eGAL1; refs 11, 14, 17, 24). Furthermore, consistent with the model in Figure 1A, either loss of Gal80 or a point mutation in Gal4 (V864E) that disrupts the interaction with Gal80 [25] also resulted in rapid, unimodal expression of Gal1-mCherry (Figure 1C and 1D). Thus, memory leads to faster and more uniform GAL1 transcriptional activation, likely by promoting rapid and uniform relief of Gal80 repression.
To quantify the adaptive effect of faster reactivation of GAL genes during memory, we followed the growth kinetics upon shifting cells from glucose to galactose (Figure 1E). Naïve cells exhibited a long growth lag before entering exponential phase (Figure 1E; ACT). In contrast, during memory or in cells ectopically expressing Gal1, adaptation was much faster (Figure 1E; REACT, ACT + eGAL1). Although the growth rates were ultimately similar once cells reached exponential phase, memory confers a large fitness benefit by decreasing the growth lag after shifting cells from glucose to galactose (Figure 1E, grey circles).
Rapid GAL gene activation in fungal species is also associated with increased responsiveness to low concentrations of galactose [26–28]. During memory or in cells expressing ectopic Gal1, Gal1-mCherry was expressed at higher levels in media with low concentrations of galactose (Figure 1F). Because yeast cells are likely exposed to mixtures of sugars in nature, we asked if this higher sensitivity for galactose also impacts the expression of Gal1-mCherry in the presence of glucose. S. cerevisiae normally does not induce GAL genes in the presence of low levels of glucose (0.2% glucose and 1.8% galactose; Figure 1G, inset). However, during memory or in the presence of ectopic Gal1, Gal1-mCherry expression was observed in the presence of glucose (Figure 1G; inset). This Gal1-mCherry expression correlated with a fitness benefit in 0.2% glucose + 1.8% galactose medium. In this medium, once glucose is exhausted after ~7h of growth, naïve cells exhibited a significant lag before adapting to galactose (Figure 1G; refs 27, 29, 30). However, during memory or in the presence of ectopic Gal1, this lag was absent and cells adapted immediately to galactose (Figure 1G). Thus, transcriptional memory provides a strong adaptive advantage in both galactose and glucose-galactose mixtures.
Gal1-D117V disrupts the interaction with Gal80, specifically blocking GAL transcriptional memory
To explore the molecular basis of faster reactivation of GAL genes during memory, we performed a genetic screen based on fluorescence activated cell sorting (FACS). After 4 hours in galactose, strong expression of Gal1-mCherry occurs during reactivation but not during activation (Figure 1D & 2A). We exploited this difference to sort for GAL memory mutants: UV-mutagenized cells that failed to express Gal1-mCherry after 4h of reactivation (Figure 2A; sort I) but expressed Gal1-mCherry after 12h in galactose (Figure 2A; sort II). This second sort removed Gal− mutants or those that had lost Gal1-mCherry expression. The recovered cells were colony-purified and screened by flow cytometry to identify those that specifically lost rapid GAL1 reactivation during memory.
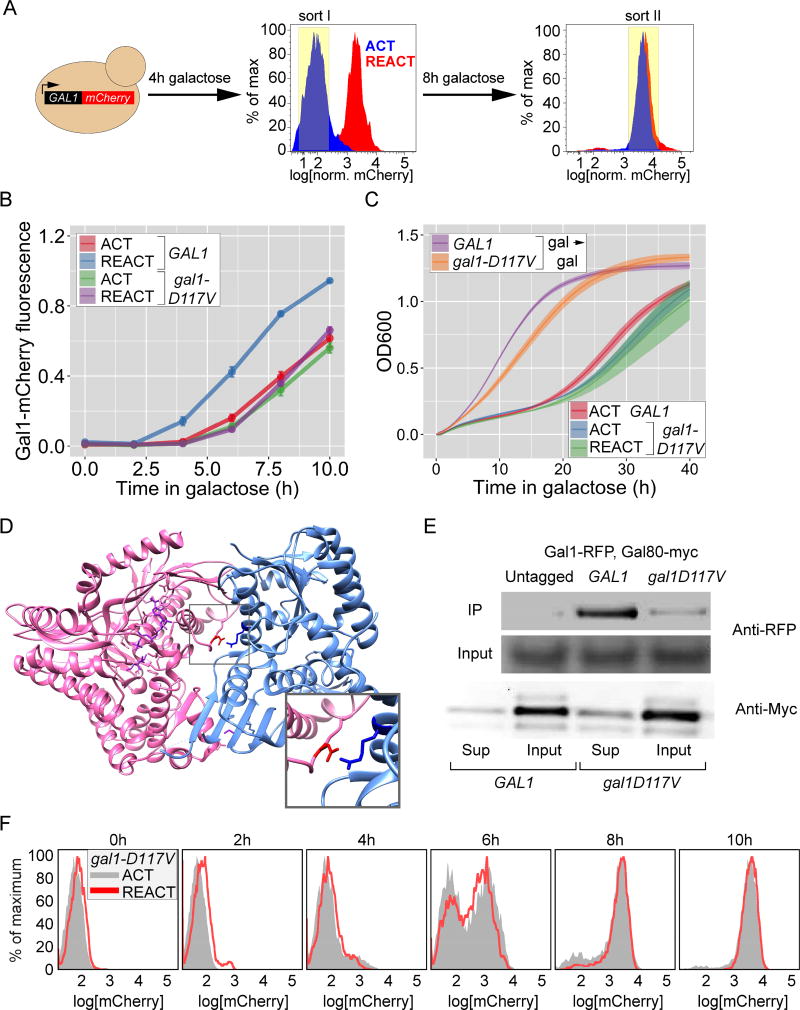
Figure 2. Genetic screen for mutants defective for GAL memory identifies gal1-D117V.
A. Schematic of the 2-step FACS based screen (see Methods for details). B. Gal1-mCherry intensity relative to CFP internal control in wild-type and gal1-D117V mutant, measured by flow-cytometry. Cells were shifted from glucose to galactose for activation (ACT) or grown in galactose overnight, shifted to glucose for 12 hours and then shifted to galactose for reactivation (REACT). Error bar represents SEM from ≥ 4 biological replicates. C. Growth of wild-type and gal1-D117V mutant cells assayed by measuring OD600 every 20 minutes during continuous growth in galactose (gal → gal), during activation (ACT) or reactivation (REACT) after 12hours of repression. The line represents the mean and the envelope represent the SEM from ≥ 4 biological replicates. D. Co-crystal structure between Gal3 (pink) and Gal80 (blue), highlighting the salt bridge between the Gal3-Asp111 and Gal80-Arg367 (inset). E. Lysates from strains expressing Gal80-13xmyc and Gal1-mCherry were subjected to co-immunoprecipitation using anti-myc antibody. The immunoprecipitated fractions (IP; top), the input fractions (middle) and the supernatant fraction after immunodepletion (bottom) were resolved by SDS PAGE and immunoblotted against either mCherry (top two panels) or the myc epitope tag (bottom panels). F. Overlay of histograms for ACT and REACT of gal1D117V in B. The yeast strains and the number of biological replicates for all experiments are listed in Table S1 and S2, respectively. Additional characterization of gal1-D117V mutant, Figure S1.
Based on the model in Figure 1A, we expected to identify null alleles of Gal1. We removed many such mutants by focusing on mutants that were able to grow on galactose (Gal1 is required to grow on galactose; not shown). The screen also identified an allele of GAL1 that was Gal+ but specifically blocked memory; gal1-D117V reduced the rate of Gal1-mCherry reactivation during memory, without altering the rate of activation of Gal1-mCherry (Figure 2B). Reconstruction of the gal1-D117V mutation into the GAL1 locus recapitulated these phenotypes (not shown), confirming that this mutation is causative. gal1-D117V cells also lost the fitness benefit of memory; the growth of gal1-D117V during reactivation closely resembled the growth of naïve wild-type cells during activation (Figure 2C). This mutation had no effect on Gal1-mCherry stability (Figure S1B) or the rate of activation (Figure 2B) and only slightly affected the rate of exponential growth in galactose (Figure 2C). Finally, the effects of this mutation were recessive because ectopic expression of Gal1 in gal1-D117V led to faster Gal1-mCherry expression (Figure S1C) and rapid adaptation to growth in galactose (Figure S1D). Thus, the gal1-D117V mutation specifically disrupts memory without significantly affecting other functions of Gal1.
The structures of Gal1, Gal3 and Gal3–Gal80 are known [20, 31]. Gal1 and Gal3 show 74% sequence identity and are structurally superimposable with a root mean square deviation of ~ 1.1 Angstroms (Figure S1A; refs 20, 31). Aspartate 117 maps to the predicted interaction surface between Gal1 and Gal80. In the Gal3–Gal80 structure, Gal3-Asp111 is at the structurally equivalent position to Gal1-Asp117 and forms an ionic bond with Gal80-Arg367 (Figure 2D and S1A; ref 20). To test if disrupting this salt bridge reduces the affinity between Gal1 and Gal80, we performed co-immunoprecipitation of wild-type and D117V Gal1-mCherry with Gal80-myc. Although these proteins were expressed at similar levels, immunoprecipitation of Gal80 recovered only ~20% of Gal1-D117V compared with wild-type Gal1 (Figure 2E). This reduced affinity for Gal80 lead to slow, bimodal expression of Gal1-mCherry during both activation and reactivation (Figure 2F). Furthermore, a complementary mutant in Gal80 (R367L) predicted to disrupt the salt bridge between Gal80 and both Gal3 and Gal1 led to a Gal− phenotype (not shown). Thus, interaction between Gal1 and Gal80 plays a critical role in GAL gene transcriptional memory.
Constitutively fast GAL expression in S. uvarum is due to higher basal expression of Gal1
S. uvarum diverged from S. cerevisiae ~20 million years ago and has evolved a distinct strategy for adapting to growth in galactose (Figure S2A; refs 27, 32, 33). We asked if this species benefits from previous growth in galactose. Although the rate of Gal1-mCherry reactivation during memory was slightly faster than the rate of activation in S. uvarum (Figure 3A & S2B), this difference was much smaller than that observed in S. cerevisiae (Figure 3B). Moreover, in S. uvarum, previous growth in galactose did not lead to a fitness benefit (Figure 3C). This, suggests that the rapid initial GAL gene induction in S. uvarum is sufficient to provide maximal fitness benefit and that increasing this rate further provides no additional effect.
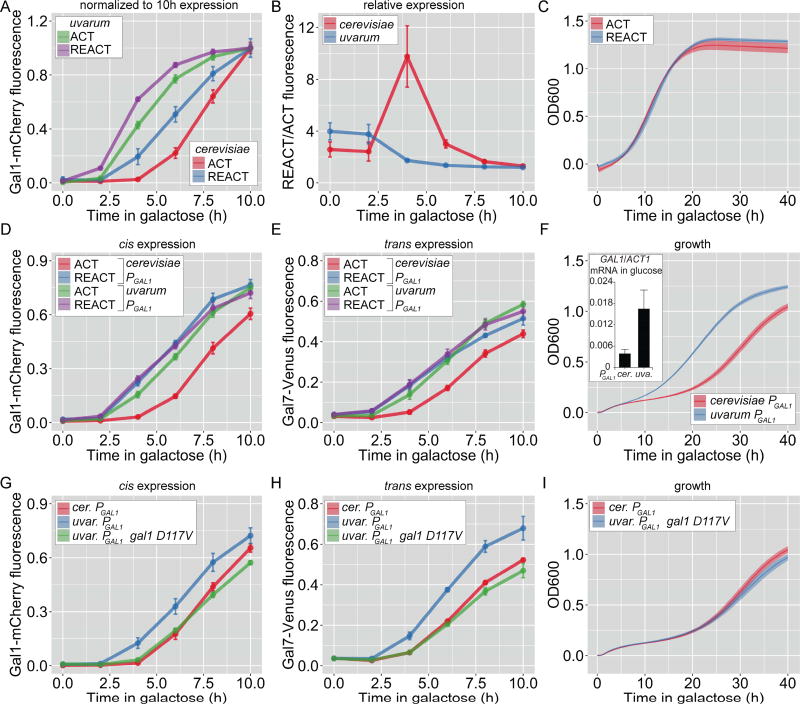
Figure 3. Recently diverged Saccharomyces species utilize genetic and epigenetic switches to adapt to growth in galactose.
A–I. Cells were shifted from glucose to galactose for activation (ACT) or grown in galactose overnight, repressed for 12h (S. cerevisiae) or 18h (S. uvarum) in glucose and then shifted to galactose for reactivation (REACT). A. Gal1-mCherry fluorescence during activation and reactivation in S. cerevisiae and S. uvarum, normalized to expression at 10 h. B. Ratio of reactivation to activation from the data in B. C. OD600 of S. uvarum during activation and reactivation. D–F. The GAL1 promoter from S. uvarum was introduced in place of the endogenous GAL1 promoter in S. cerevisiae. Gal1-mCherry (D) and Gal7-Venus (E) fluorescence relative to CFP and OD600 (F) were measured during activation (ACT) and reactivation (REACT). Inset: Basal GAL1 mRNA, relative to ACT1, transcribed from the PGAL1 from S. cerevisiae and S. uvarum in glucose media. G–I. The GAL1 promoter from S. uvarum driving expression of GAL1 or gal1-D117V was introduced in place of the endogenous GAL1 gene in S. cerevisiae. Gal1-mCherry (G) and Gal7-Venus (H) fluorescence relative to CFP and OD600 (I) was measured during activation (ACT) and reactivation (REACT). Error bars represent SEM from ≥ 3 biological replicates for Gal1-mCherry fluorescence, ≥5 biological replicates for OD600 and 12 replicates for RNA estimation. The yeast strains and the number of biological replicates for all experiments are listed in Table S1 and S2, respectively. GAL1 expression from PGAL1 from other Saccharomyces species, Figure S2; Growth trade-off for basal GAL1 expression, Figure S3.
Several differences between S. uvarum and S. cerevisiae might explain the difference in their response to previous growth in galactose; S. uvarum has higher basal expression of the key activators GAL1, GAL3 and GAL4 and lower expression of the GAL80 inhibitor [27, 33]. Thus, differences in cis-acting elements in the promoter, trans-acting factors or both could lead to constitutive fast GAL gene expression. To investigate these possibilities, we substituted the GAL1 promoter (PGAL1) in S. cerevisiae with PGAL1 from S. uvarum. In this strain, induction of Gal1-mCherry during both activation and reactivation was as fast as reactivation in wild-type cells (Figure 3D). Thus, PGAL1 from S. uvarum is sufficient to induce constitutively fast GAL1 expression in S. cerevisiae without any other uvarum factors.
The effects of PGAL1 from S. uvarum are consistent with this promoter being more easily induced. Hybrid cerevisiae-uvarum promoters suggest that this effect is largely explained by differences in the UASGAL elements and GAL1-proximal sequences (Figure S2C–E; ref 34). However, because epigenetic GAL gene transcriptional memory in S. cerevisiae requires only a few hundred molecules of Gal1 per cell [14], very low basal expression of Gal1 by PGAL1 from S. uvarum might also cause this faster induction through a positive feedback mechanism. To distinguish between these possibilities, we asked if S. cerevisiae with the S. uvarum PGAL1 also promoted faster activation of other GAL genes in trans. In cells bearing the S. uvarum PGAL1, the rate of Gal7-Venus activation and reactivation was as fast as that observed during memory in wild-type S. cerevisiae (Figure 3E). Likewise, S. uvarum PGAL1 promoted faster adaptation to galactose (Figure 3F). Thus, the S. uvarum PGAL1 is sufficient to induce constitutive fast activation of GAL genes in trans and faster adaptation to galactose, likely through basal Gal1 production.
While the levels of basal Gal1 protein in cells having the PGAL1 from S. uvarum were below the level of detection using either flow cytometry or immunoblot, RT qPCR revealed that this promoter led to a significant increase in GAL1 mRNA in cells grown in glucose (p = 0.03; Student’s t-test; Figure 3F, inset; [27]). If this basal expression were causative, then disrupting the interaction between Gal1 and Gal80 should block this effect. Indeed, introduction of the gal1-D117A mutation into the S. cerevisae strain harboring the S. uvarum PGAL1 blocked the cis and trans effects of this promoter on expression (Figure 3G & H) and growth (Figure 3I). Thus, constitutive fast GAL genes induction conferred by the S. uvarum PGAL1 is due to genetically encoded basal expression of Gal1 that impinges upon the same molecular mechanism employed during epigenetic transcriptional memory in S. cerevisiae.
Fitness costs of constitutive GAL1 expression
If faster GAL genes expression promotes adaptation to galactose, why is it restricted to memory in S. cerevisiae? Basal Gal1 expression is detrimental to growth in glucose-galactose mixtures because the galactose-1-phosphate generated by the galactokinase activity of Gal1 inhibits phosphoglucomutase and slows glycolysis [27, 35]. Consistent with this model, S. cerevisiae expressing ectopic GAL1 or S. uvarum showed a measurable growth disadvantage when adapting from glucose to a glucose – galactose mixture (1:1 ratio, 1% each sugar; Figure S3A & B). Competitive growth experiments between wild-type S. cerevisiae and S. cerevisiae harboring the S. uvarum PGAL1 also showed a fitness cost to basal expression of Gal1 (Figure S3C). Thus, consistent with previous studies, basal GAL1 expression can have both positive and negative effects on fitness; it promotes adaptation from glucose to galactose but is detrimental in glucose-galactose mixtures [29, 36].
The Gal4 central domain promotes stronger transcription during GAL memory
In addition to the gal1-D117V mutant, which showed specific loss of memory without strong effects on activation, the flow cytometry screen also identified a mutation in Gal4 (L282P) that both blocked memory and led to defective activation of Gal1-mCherry (Figure S4A). This mutation likely destabilizes the Gal4 protein, leading to lower protein levels (Figure S4A, inset). However, this mutation was interesting because it was resistant to ectopic expression of Gal1 (Figure S4A), confirming that the loss of memory in gal4-L282P cells was not simply an effect of lower levels of Gal1 during reactivation. Thus, Gal4-L282P both causes a defect in activation and blocked memory downstream of Gal1.
The gal4-L282P mutation lies within the central domain of Gal4 (CD; Gal4-239–767, Figure 4A) [37]. In other members of the zinc binuclear cluster transcription factor family, the central domain has been proposed to have a regulatory function [38–41]. However, the role of CD is unclear; deletion of this domain produces a largely functional Gal4 activator but mutations in this domain disrupt Gal4 function [37, 42].
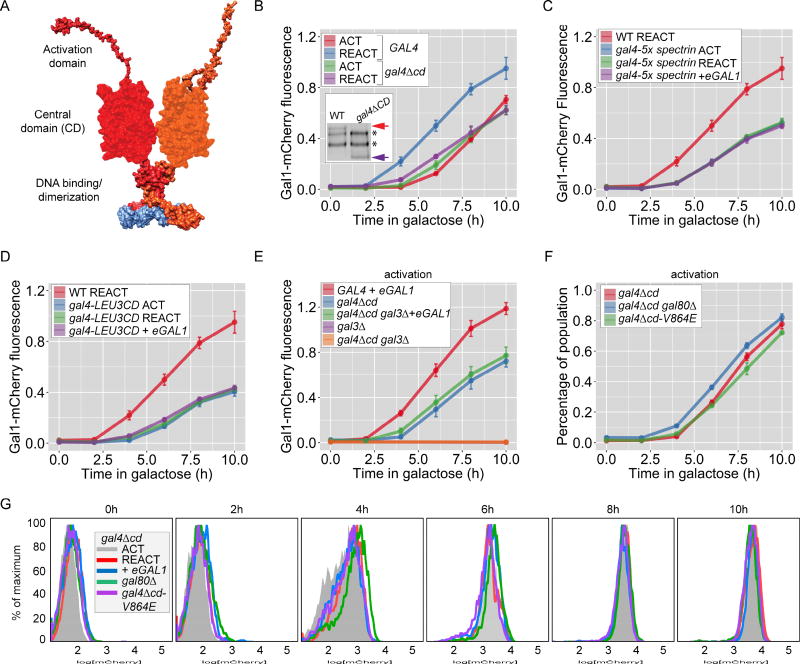
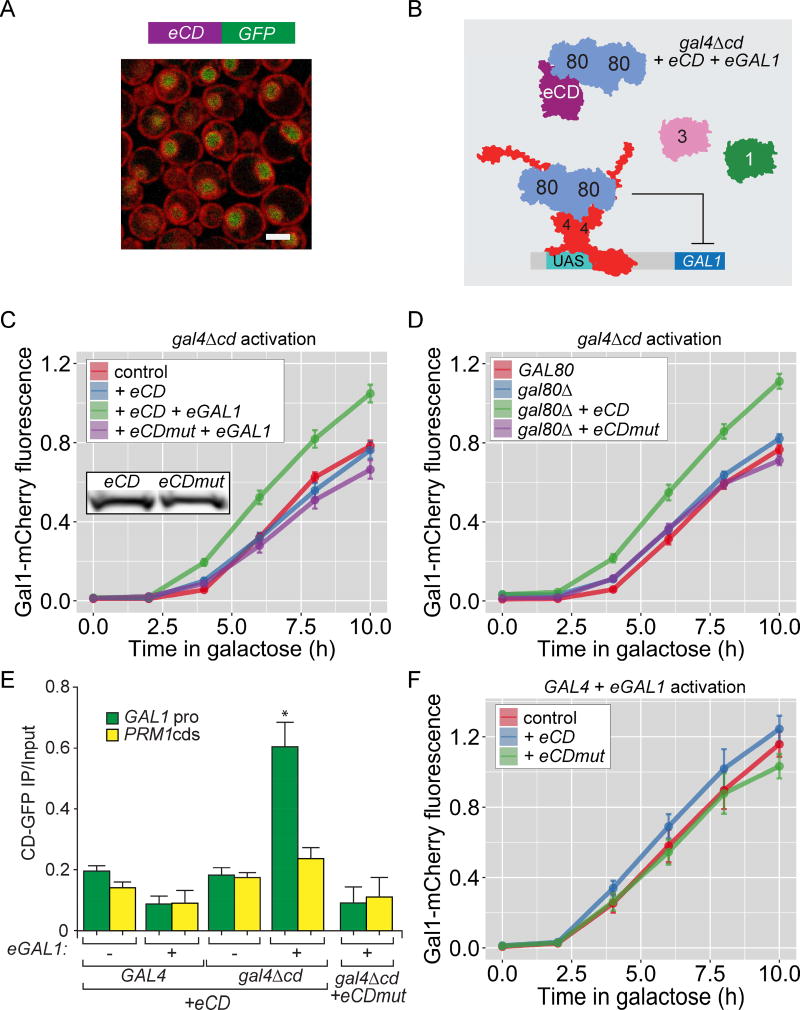
Figure 4. The Gal4 central domain is required for GAL memory.
A. Schematic of the putative domain organization with a large central domain of Gal4 (based on a structural prediction), between the N-terminal DNA binding domain and unstructured C-terminal activation domain. B–F. Naïve cells (ACT), naïve cells expressing ectopic GAL1 (ACT+eGAL1), or cells that were grown in galactose overnight and shifted to glucose for 12 hours, were shifted to galactose (REACT) to assay the Gal1-mCherry fluorescence relative to constitutively expressed CFP. B. Wild-type and gal4Δcd mutant. Inset: immunoblot of Gal4-myc immunoprecipitated from wild-type and gal4Δcd mutant cells; arrows: Gal4, * = non-specific bands. C. and D. Central domain of Gal4 was replaced with either 5-tandem repeats of β-spectrin domain (C) or the central domain from Leu3 (D). E. Wild-type, gal3Δ, gal4Δcd and gal4Δcd gal3Δ strains with or without eGAL1. Only the 0h and 10h time points are plotted for gal3Δ and gal4Δcd gal3Δ mutants. F. gal4Δcd strains with and without gal80Δ and gal4V864E mutation. G. Overlay of histograms of biological replicates from the indicated strains and time points in B and F. Error bars represent SEM from ≥ 3 biological replicates. The yeast strains and the number of biological replicates for all experiments are listed in Table S1 and S2, respectively. Additional characterization of gal4Δcd mutant, Figure S4.
To explore the role of the Gal4 central domain, we tested how loss of this domain affected memory and the response to Gal1. Unlike Gal4-L282P, Gal4Δcd protein levels were similar to full length Gal4 protein levels (Figure 4B; Inset) and the rate of Gal1-mCherry activation was similar in gal4Δcd and wild-type cells (Figure 4B; refs 37). However, cells lacking the central domain showed no memory (Figure 4B) and were unaffected by ectopic expression of either Gal1 (Figure S4B) or Gal3 (not shown). Further, dimethylation of lysine 4 on histone H3 (H3K4me2), a chromatin mark that is associated with GAL transcriptional memory and is induced by ectopic expression of Gal1 [14], was still observed in cells lacking the Gal4 central domain (Figure S4D). Therefore, the gal4Δcd mutant blocked memory downstream of both Gal1 and chromatin changes associated with memory (Figure S4C).
Given the weak sequence conservation of the central domain, we asked if CD promotes memory by acting as a spacer to increase the access of the activation domain to co-activators. The CD was replaced either with domains 12–16 of human β-spectrin, which should function as a spacer of similar size to the CD [43] or with the central domain from Leu3, a related transcription factor [40, 44]. Although these hybrid proteins supported Gal1-mCherry expression, they blocked memory and were unresponsive to Gal1 (Figures 4C and D). Thus, the Gal4 central domain has a sequence-specific function in potentiating expression and is neither a simple spacer, nor a generic, swappable domain.
Because Gal3 has a higher affinity for Gal80 than Gal1 [23], loss of memory could result if Gal4Δcd is de-repressed normally by Gal3, but is unresponsive to Gal1 (Figure 1A). To test this hypothesis, we asked if Gal1 could replace Gal3 to promote activation of Gal1-mCherry. In cells lacking Gal3, Gal1-mCherry is not expressed (Figure 4E; gal3Δ and gal4Δcd gal3Δ). However, ectopic expression of Gal1 complemented this defect in gal4Δcd cells, allowing Gal1-mCherry expression (Figure 4E), but at levels observed during initial activation. This argues that Gal4Δcd responds to both Gal1 and Gal3, but is limited in its activity, leading to slower/lower expression of Gal1-mCherry.
The Gal4 central domain is a target of Gal80 repression
Loss of the Gal4 central domain also altered Gal80 repression. During both activation and reactivation, gal4Δcd cells showed unimodal Gal1-mCherry expression (Figures 4G & S4E). Hence, loss of the central domain had two effects: it both reduced the strength of Gal1-mCherry expression during reactivation (as measured by average expression in the population; Figure 4B) and led to a more uniform activation of the population (Figures 4G & S4E). Because both loss of Gal80 and transcriptional memory also led to unimodal activation (Figure 1D), this implied that the central domain is required for proper Gal80 repression. If so, then loss of Gal80 might not further increase the rate of activation. Indeed, neither loss of Gal80 nor disruption of the Gal4–Gal80 interaction (gal4-V864E) significantly increased the overall rate of activation in the gal4Δcd cells (Figure 4F & G). Thus, the Gal4 central domain operates downstream of Gal80 to regulate bi-modal expression and potentiates maximal expression during memory.
If loss of the Gal4 central domain completely blocked Gal80 repression, it should lead to expression of Gal1-mCherry in raffinose medium since Gal80 is the sole repressor of GAL genes expression in raffinose. In raffinose, while either loss of Gal80 or loss of the interaction between Gal4 and Gal80 (gal4-V864E; ref 25) led to de-repression of Gal1-mCherry, deletion of the central domain alone did not (Figure S4C). Thus, loss of the Gal4 central domain does not disrupt Gal80 repression but leads to faster relief of Gal80 repression.
An inter-domain interaction potentiates Gal4 activation
In the other members of the Gal4 transcription factor family, the central domain directly interacts with the activation domain to allosterically regulate activation [38, 40, 41]. To test if the central domain (CD) interacts with the rest of Gal4, we asked if this domain could potentiate Gal4Δcd activation in trans (Figure 5B; schematic). Ectopically expressed CD localized in the nucleus (Figure 5A), independent of Gal4 (Figure S5A). Ectopic CD increased the rate of Gal1-mCherry activation in gal4Δcd strains (Figure 5C and D). However, this effect required either expression of ectopic Gal1 (Figure 5C) or loss of Gal80 (Figure 5D). ChIP experiments revealed that ectopic CD was recruited to the GAL1 promoter by Gal4Δcd (Figure 5E). This suggests that CD physically interacts with Gal4Δcd to potentiate activation and this interaction is regulated by Gal1–Gal80.
Figure 5. An inter-domain interaction that is regulated by Gal80 potentiates Gal4 activation.
A. Confocal micrograph showing nuclear localization of the Gal4 central domain-GFP expressed from PADH1 (eCD-GFP) in cells having the nuclear envelope/endoplasmic reticulum marked with a RFP tagged protein. B. Schematic for the experimental setup in C–F. Gal80 dimer binds to the activation domain of Gal4Δcd and to eCD. C, D & F. Gal1-mCherry fluorescence relative to CFP in presence of eCD and eCDmut (L282P). C. gal4Δcd mutant with or without eGAL1. Inset: immunoblot of eCD-GFP and eCDmut-GFP. D. gal4Δcd or gal4Δcd gal80Δ mutants. E. ChIP against eCD-GFP or eCDmut-GFP in the indicated strains. Recovery of the GAL1 promoter and a control locus, PRM1, were quantified relative to input by real time quantitative PCR. * p ≤ 0.05 (Student’s t-test) relative to the ChIP enrichment of PRM1. F. Wild-type cells with and without eGAL1. Error bars represent SEM from ≥ 3 biological replicates for Gal1-mCherry fluorescence and ≥ 3 replicated for ChIP experiments. The yeast strains and the number of biological replicates for all experiments are listed in Table S1 and S2, respectively. Further characterization of eCD, Figure S5. See also Figure S4.
This effect was highly specific; in the absence of ectopic Gal1, ectopic CD neither upregulated Gal1-mCherry expression nor bound to the GAL1 promoter (Figure 5C, 5E, & S5B). Furthermore, ectopic CD neither bound nor potentiated activation from full-length Gal4 (Figure 5E, 5F, S5C). Finally, ectopic L282P mutant CD (CDmut, expressed at similar levels to wild-type CD; Figure 5C, inset) was not recruited to Gal4Δcd (Figure 5E). Thus, the L282P mutation in the Gal4 CD disrupts the interaction with other domains of Gal4, blocking potentiation.
Discussion
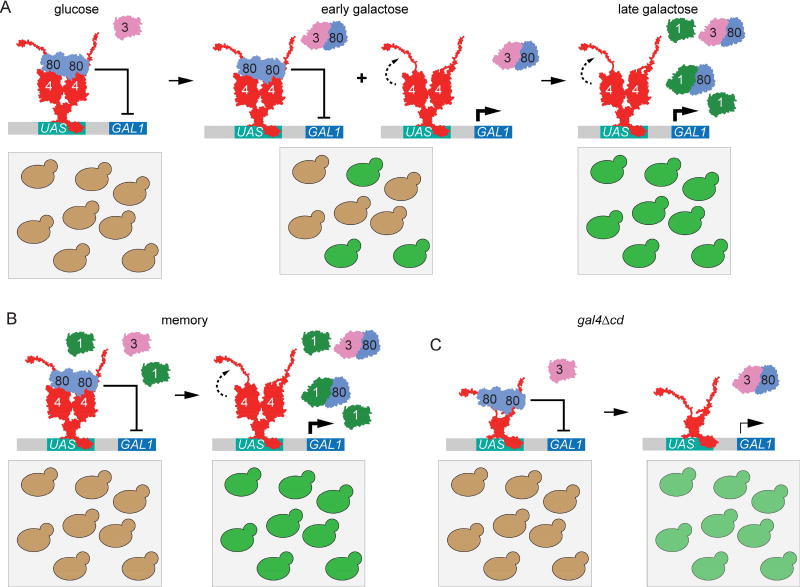
This study provides important new insights into both the molecular mechanism of epigenetic GAL gene transcriptional memory in S. cerevisiae and an illustration of the evolutionary logic whereby the same molecules can produce either conditional, epigenetic mechanisms of faster reactivation or constitutive, genetic mechanisms of fast activation. Our current model for both is shown in Figure 6. Gal80 physically interacts with both the activation and central domains of Gal4, which may explain why ectopic CD is only able to complement the gla4Δcd phenotype in either the absence of Gal80 or in the presence of ectopic Gal1 [21, 45–48]. Although it is not yet clear if the Gal80 that binds to the Gal4 activation domain is the same molecule as the Gal80 that interacts with the central domain, our results suggest that both interactions are required for proper repression. Early during activation, Gal3 interacts with Gal80, permitting Gal4-mediated transcriptional activation in a subset of the cells in the population (Figure 6A). In these cells, the central domain potentiates activation, leading to high-level expression. During memory, or in S. uvarum, the population shows uniform, rapid transition to high-level expression of GAL genes because of elevated concentrations of the Gal1 coactivator (Figure 6B). However, in cells lacking the Gal4 central domain, the population responds uniformly, but the level of expression is low. These cells do not show memory both because they are less well repressed by Gal80 (and therefore do not benefit from previous expression of Gal1) and because they are unable to achieve full activation.
Figure 6. Model for epigenetic potentiation of Gal4 activation through interdomain potentiation.
A. In wild-type cells during early activation, Gal80 repression is relieved in subset of population, leading to lower-level expression. Inter-domain interaction between central domain and activation domain potentiates higher activation levels in cells relieved of Gal80. B. During memory (or in the presence of basal Gal1 expression), Gal80 repression is relieved early in whole population leading to unimodal, fully potentiated GAL gene expression. C. gal4Δcd cells show uniform, low-level activation.
Slight differences in the degree of repression of GAL genes among Saccharomyces species leads to two different strategies that favor growth under different conditions. Low-level basal GAL1 expression in S. uvarum leads to rapid adaptation to galactose but also encumbers a fitness cost in glucose-galactose mixtures [27, 29, 36]. On the other hand, tight GAL1 repression restricts fitness during initial induction in galactose, but leads to optimal utilization of glucose in the presence of other sugars. Glucose is the most efficiently utilized sugar through glycolysis and S. cerevisiae has a clear preference for it; expression of several genes is optimized for growth in glucose over other carbon sources [49]. Epigenetic memory in S. cerevisiae allows cells to benefit from recent growth in galactose without compromising the preference for glucose over longer time scales.
A whole-genome duplication during Saccharomyces evolution has led to specialization of function between duplicated paralogs Gal1 and Gal3 [26, 28, 32–34]. The sub-functionalization of these proteins in different species have led to different evolutionary paths. K. lactis and C. albicans, which diverged from Saccharomyces before the whole genome duplication, also exhibit constitutive fast induction of GAL genes [26, 28, 32]. This is because these species lack Gal3, and therefore must express higher basal levels of Gal1 to allow expression of the GAL genes. This implies that basal GAL1 expression is the ancestral regulatory scheme that has been maintained in S. uvarum, in part through promoter-driven basal expression of Gal1 [26, 34, 50]. Replacing PGAL1 in S. cerevisiae with the PGAL1 from the more closely related Saccharomyces species S. mikatae and S. paradoxus did not lead to faster induction of Gal1 (Figure S2A & B). This suggests that basal GAL1 expression due to promoter differences persisted in S. uvarum, but was lost in S. cerevisiae, S. paradoxus and S. mikitae. Tighter GAL1 repression has been accompanied by specialization of GAL3 as a co-activator: Gal3 from S. cerevisiae has lost galactokinase activity and has 10-fold higher affinity for Gal80 repressor than Gal1 [21, 23, 27, 33, 34]. Taken together, our results suggest that GAL transcriptional memory in S. cerevisiae is a product of tighter GAL1 repression and specialization of GAL3 as a co-activator. Thus, GAL memory may be an example where an epigenetic mechanism for faster reactivation evolved from an ancestral state of genetically encoded fast activation.
Using a FACS-based genetic screen, we identified two mutations that provide important insight into the molecular mechanism of GAL transcriptional memory. The gal1-D117V mutation maintains galactokinase function but reduces affinity for Gal80, specifically disrupting memory. Furthermore, gal1-D117V blocked fast GAL gene activation caused by the S. uvarum PGAL1, confirming that these effects are mediated by low level expression of Gal1.
The screen also identified gal4-L282P, a mutation in the central domain of Gal4 that blocks the ability of Gal4 to respond to Gal1. Deletion of the central domain disrupted memory without strongly altering Gal4 protein levels or the rate of activation. The central domain has two functions: it promotes tighter Gal80 repression and it promotes stronger Gal4 activity (Figure 6C). Disrupting these functions resulted in a qualitative change in the GAL1 transcriptional output, leading to a more uniform population of cells that transitioned to a weaker level of expression (Figure 6C). In other words, unimodal induction is necessary, but not sufficient, for the rapid expression observed during memory. Because Gal80 interacts with both the central domain and the activation domain [21, 45–48], we propose that the central domain either enhances Gal80 recruitment to Gal4 or inhibits dissociation of Gal80 from Gal4. If so, then Gal80 would likely also regulate the potentiation of Gal4 activation by the central domain. We envision that the central domain physically interacts with the activation domain, allosterically altering its ability to promote transcription. Consistent with this notion, the CD can interact with Gal4Δcd in trans to increase the rate of activation, and this interaction is both regulated by Gal80 and disrupted by substitution of Proline for Leucine 282. Thus, the central domain plays a critical role in promoting both stronger repression by Gal80 and stronger transcription.
GAL transcriptional memory is a manifestation of ongoing resolution of expression levels of the partially redundant paralogs, Gal1 and Gal3. Among different Saccharomyces species, the degree of repression of GAL1 in glucose dictates whether faster adaptation to galactose is a hardwired, genetic mechanism or a conditional, epigenetic mechanism. Because basal GAL1 expression compromises fitness in mixtures of sugars, S. cerevisiae has traded faster kinetics of GAL gene activation for optimal growth in glucose-galactose mixtures. But during memory, S. cerevisiae can more rapidly adapt to a challenge that they have experienced recently by switching from a heterogeneous to uniform behavior and employing an inter-domain potentiation of Gal4 activation.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jason Brickner (j-brickner@northwestern.edu)
Experimental model and subject details
All S. cerevisiae strains (W303 background) were generated from either CRY1 (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) or CRY2 (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1; [51]) parent strain. S. uvarum strains were generated from JRY8153 strain from the Hittinger lab [52]. Yeast strains used in this study appear in Table S1. For flow cytometric estimation of GAL1 expression, Gal1 was C-terminally tagged with mCherry. A constitutively expressed CFP or Venus was used as an internal reference.
Method details
Plasmids, yeast strains, and molecular biology
Plasmids for constitutive expression of GAL1, GAL3 and central domain of Gal4 (CD) were generated by amplifying these inserts with appropriate restriction site overhangs and cloning downstream of ADH1 promoter (PADH) into pRS304 and pRS306 [53]. The resulting plasmids were linearized for insertion at TRP1 and URA3 loci, respectively. CFP and Venus fluorophores for normalization of GAL1-mCherry expression were cloned downstream of PTDH in pRS306 vector and integrated at the URA3 locus. Gene deletion and C-terminal tagging was performed as described previously [54]. Briefly, primers amplifying the deletion cassettes or fluorescent tags were guided by overhangs homologous to the target site and transformants were selected on the appropriate selection media. GAL1, GAL4 and promoter mutants were generated by first inserting a URA3-SUP4-o cassette into the coding sequence, followed by transformation with a mutant PCR product and selection of FOA, as described previously [55]. The hybrid promoters were generated by stitching together promoter fragments through overlapping PCR. Cells were grown in Synthetic Dextrose Complete (SDC), Synthetic Galactose Complete (SGC) or Synthetic Raffinose Complete (SRC) at 25°C for growth rate studies. For expression and ChIP experiments cells were grown in Yeast Peptone Glucose (YPD) or Yeast Peptone Galactose (YPG), with the exception of gal80Δ, which was growth SDC prior to induction with rich galactose media. In all media, except in figure 1E, the final concentration of total sugar was 2%.
Flow cytometry
Cells having GAL1 tagged with mCherry were shifted from YPD to YPG and maintained between 0.05 to 0.3 OD600 throughout the induction. 1 ml of culture was harvested at different times of induction and the cells were frozen and stored in 10% glycerol at −80°C. Cells were thawed on ice and passed through a BD LSRII flow-cytometer in the Robert Lurie Comprehensive Cancer Center Flow Cytometry Core Facility. mCherry, CFP and Venus were excited with 561nm, 405nm and 488nm lasers, respectively. For detecting mCherry emission a 600nm long pass dichroic mirror and 610/20nm band pass filter set, for CFP emission 505nm long pass dichroic mirror and 525/50 band pass filter set and for Venus emission 530/30 band pass filter set was used. Roughly 5000 cells were analyzed to obtain the average intensities. The constitutively expressed CFP (PTDH-CFP) and Venus (PTDH-VENUS) served as a normalization control for Gal1-mCherry fluorescence in S. cerevisiae and S. uvarum respectively; Gal1 fluorescence intensity was expressed as ratio of Gal1-mCherry to CFP or Venus. Biological replicates for all GAL1-mCherry induction were performed at least three times using independent cultures on different days. The data analysis did not necessitate either randomization or blinding at any stage.
Genetic Screen
Exponentially growing wild-type cells in SGC were mutagenized by exposure to 254nm ultra violet (UV) light, using a hand-held lamp (UVGA-25, UVP Inc). 10ml of cells at OD600nm of 0.1 were placed in a 10cm Petri Dish. These cells were exposed to UV for 60sec from a distance of 15cm to the UV source, which killed 30% of the cells [56, 57]. The mutagenized cells were transferred to YPD, grown for 12h and then shifted to YPG for 4h. Fluorescence activated cell sorting for non-fluorescent cells was done using the BD FACSAria SORP 5 at the Northwestern Flow Cytometry Core Facility. Approximately two million cells were harvested in YPG. Cells were pelleted and then resuspended in fresh YPG for additional 8h. Sorting was then performed for cells that have wild-type Gal1-mCherry expression levels. Cells collected from the second sort were plated for single colonies on galactose plates. GAL1-mCherry activation and reactivation kinetics was individually assayed for each colony. Complementation with wild-type GAL genes was used for mapping mutations that lead to specific reactivation defects followed by subsequent Sanger sequencing of the mutant loci to identify the mutation.
Growth Assay
Exponentially growing cells were diluted to an OD600 = 0.1, washed with media containing no sugar and then re-suspended in the media with the desired sugar. About 350µl of each culture was added to a well in a 96 well flat bottom plate. Growth was monitored by measuring OD600 every 20 minutes for 24h – 40h using a 96-well plate reader (BioTek Synergy™), normalized to media without cells. The cultures were not shaken at any time during the growth assay. The The cell density at t = 0 was subtracted from all measurements. At least three biological replicates were done for each condition each time and repeated on different days.
Chromatin Immunoprecipitation
ChIP was performed as described previously with slight modifications [3, 8, 58, 59]. Approximately 100ml culture of exponentially growing cells at OD600nm of 0.8 was fixed with 1% formaldehyde for 15min at room temperature. Formaldehyde was quenched with a final concentration of 0.15M Glycine. Cells were then washed with 0.1M Tris pH 7.5 and collected through filtration. From this point up to elution, the cells were kept at ~ 4°C. The cells were suspended in 600µl cold lysis buffer (50mM HEPES pH 7.5, 140mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% Sodium deoxycholate) and vortexed with 600µl of glass beads using Vortex Genie 2. The cells were vortexed five times at max speed for 1min, with 1min intervals. The lysed cells were harvested and spun at 7000g for 10min. The supernatant was aspirated and the pellet containing the chromatin was re-suspended in 1ml of lysis buffer. The re-suspended pellet was sonicated using a Branson Sonifier 450, for 15 pulses of 10sec, with 10sec intervals. The sonicated lysate was spun at 15000g for 10min. The supernatant was harvested and diluted to ~4mg/ml using lysis buffer. 50µl of supernatant was taken as input control from each sample and mixed with 200µl of 1%SDS. For immuno-precipitation, 8µl of anti-Rabbit IgG Dynabeads were mixed with 2µl of either anti-GFP or anti-H3K4me2, for each sample. The supernatant was incubated overnight at 4°C with the antibody bound beads. The beads were isolated using a magnetic separator and washed five times with 1ml of lysis buffer. For elution of immuno-precipitated DNA, beads were incubated with 125µl of elution buffer (50mM Tris pH 7, 10mM EDTA, 1% SDS) at 65°C for 15min. This was repeated twice and the two eluates were pooled. Both the input and the eluate were treated with 2µl of 10mg/ml RNAseA at 37°C for 30min, followed by 2µl of 20mg/ml of Proteinase K at 42°C for 2 hours. Un-crosslinking was done at 65°C for 8h and DNA fragments were eluted in 1× TE using Qiagen PCR clean-up kit. The input and immuno-precipitated samples were diluted 400-fold and 10-fold, respectively. The recovery of the fragments from GAL1, ACT1 and PRM1 loci, by ChIP, were quantified by qPCR using primers specified in the key resource table. The enrichment was quantified as ratio of Immuno-precipitated DNA over input DNA (IP/Input). We excluded Ct values (qPCR) that were more than two cycles apart from the average for the same DNA samples.
Microscopy
The cells containing CD-GFP and ER/nuclear envelope targeted mCherry were grown in SDC overnight. The OD600 was kept below 0.5 and a sample of this culture was directly spread on a slide for imaging. The SP5 Line Scanning Confocal Microscope (Leica Biosystems) at the Northwestern University Biological Imaging Facility was used for imaging as described earlier [60]. The Argon 488nm and Diode pumped solid-state 561nm lasers at ~10% power were used for exciting the CD-GFP and nuclear directed Cherry, respectively. The images were acquired for 150µm × 150µm field at 2048 × 2048 pixel resolution for ten z-stacks of 0.73 µm with 0.34µm step size through 100 × 1.44 NA objectives. The images from the GFP and Cherry channels were merged using the LAS AF Lite software from Leica.
Competitive growth assay
Two exponentially growing S. cerevisiae strains, in YPD, containing either PGAL1uvarum or PGAL1cerevisiae were mixed together in equal ratio. The mixture was co-cultured in YP media containing 1% glucose and 1% galactose at 25°C for 36h with periodic dilutions. The OD600 for the cultures were kept below 0.5 at all times. Constitutively expressed Venus in one strain was used for flow-cytometric estimation of changes in relative fraction of the two populations over time. Parallel competitive assays were performed with Venus expressed in either one of the strain to control for the effect of Venus on growth. Malthusian fitness coefficients (M) were calculated as M = ln(10log[(GFPend/BFPend)/(GFPstart/BFPstart)]/t) as described previously [34].
Quantitation and Statistical analysis
At least 3 biological replicates were performed for each experiment to plot the average and standard error of the means. The replicates were performed from cultures grown independently on different days. The data was plotted using the GG-plot. To evaluate the significance of difference between strains or treatments with respect to the reference, an unpaired, two-tailed Student’s t-test was performed. We did not perform additional tests for applicability of Student’s t-test to our data.
Supplementary Material
Acknowledgments
The authors thank members of the Brickner laboratory and Professors Richard Gaber and Erik Andersen for helpful comments on the manuscript, Professor Todd Hittenger (University of Wisconsin, Madison) for sharing S. uvarum strains, Meihua Christina Kuang for help with the growth assay and Professor Mark Ptashne (Memorial Sloan Kettering Cancer Institute) for a fruitful discussion. This work was supported by NIH grant R01 GM118712 and an American Heart Association predoctoral fellowship (VS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
JHB and VS conceived the project and VS carried out the experiments.
The authors declare no conflict of interests.
References
- 1.Lopez-Maury L, Marguerat S, Bahler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nature reviews. Genetics. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 2.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS biology. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Urso A, Takahashi YH, Xiong B, Marone J, Coukos R, Randise-Hinchliff C, Wang JP, Shilatifard A, Brickner JH. Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. eLife. 2016;5 doi: 10.7554/eLife.16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding Y, Fromm M, Avramova Z. Multiple exposures to drought 'train' transcriptional responses in Arabidopsis. Nature communications. 2012;3:740. doi: 10.1038/ncomms1732. [DOI] [PubMed] [Google Scholar]
- 5.Gialitakis M, Arampatzi P, Makatounakis T, Papamatheakis J. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol Cell Biol. 2010;30:2046–2056. doi: 10.1128/MCB.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan Q, Haroon S, Bravo DG, Will JL, Gasch AP. Cellular memory of acquired stress resistance in Saccharomyces cerevisiae. Genetics. 2012;192:495–505. doi: 10.1534/genetics.112.143016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Molecular cell. 2010;40:112–125. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Light WH, Freaney J, Sood V, Thompson A, D'Urso A, Horvath CM, Brickner JH. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS biology. 2013;11:e1001524. doi: 10.1371/journal.pbio.1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seong KH, Li D, Shimizu H, Nakamura R, Ishii S. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011;145:1049–1061. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 10.Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nature genetics. 2006;38:706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- 11.Zacharioudakis I, Gligoris T, Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Current biology : CB. 2007;17:2041–2046. doi: 10.1016/j.cub.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 12.Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 13.Pascual-Garcia P, Debo B, Aleman JR, Talamas JA, Lan Y, Nguyen NH, Won KJ, Capelson M. Metazoan Nuclear Pores Provide a Scaffold for Poised Genes and Mediate Induced Enhancer-Promoter Contacts. Molecular cell. 2017;66:63–76. e66. doi: 10.1016/j.molcel.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sood V, Cajigas I, D’Urso A, Light WH, Brickner JH. GAL gene epigenetic transcriptional memory in Saccharomyces cerevisiae depends on growth in glucose and the Tup1 transcription factor. Submitted. 2017 doi: 10.1534/genetics.117.201632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 16.Traven A, Jelicic B, Sopta M. Yeast Gal4: a transcriptional paradigm revisited. EMBO reports. 2006;7:496–499. doi: 10.1038/sj.embor.7400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kundu S, Peterson CL. Dominant role for signal transduction in the transcriptional memory of yeast GAL genes. Mol Cell Biol. 2010;30:2330–2340. doi: 10.1128/MCB.01675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biggar SR, Crabtree GR. Cell signaling can direct either binary or graded transcriptional responses. The EMBO journal. 2001;20:3167–3176. doi: 10.1093/emboj/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egriboz O, Goswami S, Tao X, Dotts K, Schaeffer C, Pilauri V, Hopper JE. Self-association of the Gal4 inhibitor protein Gal80 is impaired by Gal3: evidence for a new mechanism in the GAL gene switch. Mol Cell Biol. 2013;33:3667–3674. doi: 10.1128/MCB.00646-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavy T, Kumar PR, He H, Joshua-Tor L. The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation. Genes & development. 2012;26:294–303. doi: 10.1101/gad.182691.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Platt A, Reece RJ. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. The EMBO journal. 1998;17:4086–4091. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timson DJ, Ross HC, Reece RJ. Gal3p and Gal1p interact with the transcriptional repressor Gal80p to form a complex of 1:1 stoichiometry. The Biochemical journal. 2002;363:515–520. doi: 10.1042/0264-6021:3630515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavy T, Yanagida H, Tawfik DS. Gal3 Binds Gal80 Tighter than Gal1 Indicating Adaptive Protein Changes Following Duplication. Molecular biology and evolution. 2016;33:472–477. doi: 10.1093/molbev/msv240. [DOI] [PubMed] [Google Scholar]
- 24.Bhat PJ, Hopper JE. Overproduction of the GAL1 or GAL3 protein causes galactose-independent activation of the GAL4 protein: evidence for a new model of induction for the yeast GAL/MEL regulon. Mol Cell Biol. 1992;12:2701–2707. doi: 10.1128/mcb.12.6.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salmeron JM, Jr, Leuther KK, Johnston SA. GAL4 mutations that separate the transcriptional activation and GAL80-interactive functions of the yeast GAL4 protein. Genetics. 1990;125:21–27. doi: 10.1093/genetics/125.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalal CK, Zuleta IA, Mitchell KF, Andes DR, El-Samad H, Johnson AD. Transcriptional rewiring over evolutionary timescales changes quantitative and qualitative properties of gene expression. eLife. 2016;5 doi: 10.7554/eLife.18981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roop JI, Chang KC, Brem RB. Polygenic evolution of a sugar specialization trade-off in yeast. Nature. 2016;530:336–339. doi: 10.1038/nature16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubio-Texeira M. A comparative analysis of the GAL genetic switch between not-so-distant cousins: Saccharomyces cerevisiae versus Kluyveromyces lactis. FEMS yeast research. 2005;5:1115–1128. doi: 10.1016/j.femsyr.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Atolia E, Hua B, Savir Y, Escalante-Chong R, Springer M. Natural variation in preparation for nutrient depletion reveals a cost-benefit tradeoff. PLoS biology. 2015;13:e1002041. doi: 10.1371/journal.pbio.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee KB, Wang J, Palme J, Escalante-Chong R, Hua B, Springer M. Polymorphisms in the yeast galactose sensor underlie a natural continuum of nutrient-decision phenotypes. PLoS genetics. 2017;13:e1006766. doi: 10.1371/journal.pgen.1006766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thoden JB, Sellick CA, Timson DJ, Reece RJ, Holden HM. Molecular structure of Saccharomyces cerevisiae Gal1p, a bifunctional galactokinase and transcriptional inducer. The Journal of biological chemistry. 2005;280:36905–36911. doi: 10.1074/jbc.M508446200. [DOI] [PubMed] [Google Scholar]
- 32.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 33.Kuang MC, Hutchins PD, Russell JD, Coon JJ, Hittinger CT. Ongoing resolution of duplicate gene functions shapes the diversification of a metabolic network. eLife. 2016;5 doi: 10.7554/eLife.19027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449:677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- 35.de Jongh WA, Bro C, Ostergaard S, Regenberg B, Olsson L, Nielsen J. The roles of galactitol, galactose-1-phosphate, and phosphoglucomutase in galactose-induced toxicity in Saccharomyces cerevisiae. Biotechnology and bioengineering. 2008;101:317–326. doi: 10.1002/bit.21890. [DOI] [PubMed] [Google Scholar]
- 36.Venturelli OS, Zuleta I, Murray RM, El-Samad H. Population diversification in a yeast metabolic program promotes anticipation of environmental shifts. PLoS biology. 2015;13:e1002042. doi: 10.1371/journal.pbio.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 38.Des Etages SA, Saxena D, Huang HL, Falvey DA, Barber D, Brandriss MC. Conformational changes play a role in regulating the activity of the proline utilization pathway-specific regulator in Saccharomyces cerevisiae. Molecular microbiology. 2001;40:890–899. doi: 10.1046/j.1365-2958.2001.02432.x. [DOI] [PubMed] [Google Scholar]
- 39.Rohde JR, Trinh J, Sadowski I. Multiple signals regulate GAL transcription in yeast. Mol Cell Biol. 2000;20:3880–3886. doi: 10.1128/mcb.20.11.3880-3886.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D, Hu Y, Zheng F, Zhou K, Kohlhaw GB. Evidence that intramolecular interactions are involved in masking the activation domain of transcriptional activator Leu3p. The Journal of biological chemistry. 1997;272:19383–19392. doi: 10.1074/jbc.272.31.19383. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. The EMBO journal. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Molecular cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- 43.Bhattacharyya S, Renn JP, Yu H, Marko JF, Matouschek A. An assay for 26S proteasome activity based on fluorescence anisotropy measurements of dye-labeled protein substrates. Analytical biochemistry. 2016;509:50–59. doi: 10.1016/j.ab.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou KM, Kohlhaw GB. Transcriptional activator LEU3 of yeast. Mapping of the transcriptional activation function and significance of activation domain tryptophans. The Journal of biological chemistry. 1990;265:17409–17412. [PubMed] [Google Scholar]
- 45.Abramczyk D, Holden S, Page CJ, Reece RJ. Interplay of a ligand sensor and an enzyme in controlling expression of the Saccharomyces cerevisiae GAL genes. Eukaryotic cell. 2012;11:334–342. doi: 10.1128/EC.05294-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes & development. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leuther KK, Johnston SA. Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science. 1992;256:1333–1335. doi: 10.1126/science.1598579. [DOI] [PubMed] [Google Scholar]
- 48.Sil AK, Alam S, Xin P, Ma L, Morgan M, Lebo CM, Woods MP, Hopper JE. The Gal3p-Gal80p-Gal4p transcription switch of yeast: Gal3p destabilizes the Gal80p-Gal4p complex in response to galactose and ATP. Mol Cell Biol. 1999;19:7828–7840. doi: 10.1128/mcb.19.11.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keren L, Hausser J, Lotan-Pompan M, Vainberg Slutskin I, Alisar H, Kaminski S, Weinberger A, Alon U, Milo R, Segal E. Massively Parallel Interrogation of the Effects of Gene Expression Levels on Fitness. Cell. 2016;166:1282–1294. e1218. doi: 10.1016/j.cell.2016.07.024. [DOI] [PubMed] [Google Scholar]
- 50.Meyer J, Walker-Jonah A, Hollenberg CP. Galactokinase encoded by GAL1 is a bifunctional protein required for induction of the GAL genes in Kluyveromyces lactis and is able to suppress the gal3 phenotype in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5454–5461. doi: 10.1128/mcb.11.11.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brickner JH, Fuller RS. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. The Journal of cell biology. 1997;139:23–36. doi: 10.1083/jcb.139.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scannell DR, Zill OA, Rokas A, Payen C, Dunham MJ, Eisen MB, Rine J, Johnston M, Hittinger CT. The Awesome Power of Yeast Evolutionary Genetics: New Genome Sequences and Strain Resources for the Saccharomyces sensu stricto Genus. G3. 2011;1:11–25. doi: 10.1534/g3.111.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Longtine MS, McKenzie A, Demarini DJ, 3rd, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 55.Brickner DG, Sood V, Tutucci E, Coukos R, Viets K, Singer RH, Brickner JH. Subnuclear positioning and interchromosomal clustering of the GAL1-10 locus are controlled by separable, interdependent mechanisms. Molecular biology of the cell. 2016;27:2980–2993. doi: 10.1091/mbc.E16-03-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oftedal P. A theoretical study of mutant yield and cell killing after treatment of heterogeneous cell populations. Hereditas. 1968;60:177–210. doi: 10.1111/j.1601-5223.1968.tb02201.x. [DOI] [PubMed] [Google Scholar]
- 57.Eckardt F, Haynes RH. Kinetics of mutation induction by ultraviolet light in excision-deficient yeast. Genetics. 1977;85:225–247. doi: 10.1093/genetics/85.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nature cell biology. 2010;12:111–118. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS biology. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Egecioglu DE, D'Urso A, Brickner DG, Light WH, Brickner JH. Approaches to studying subnuclear organization and gene-nuclear pore interactions. Methods in cell biology. 2014;122:463–485. doi: 10.1016/B978-0-12-417160-2.00021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.