Summary
Small RNAs are principal elements of bacterial gene regulation and physiology. Two small RNAs in Brucella abortus, AbcR1 and AbcR2, are required for wild-type virulence. Examination of the abcR loci revealed the presence of a gene encoding a LysR-type transcriptional regulator flanking abcR2 on chromosome 1. Deletion of this lysR gene (bab1_1517) resulted in the complete loss of abcR2 expression while no difference in abcR1 expression was observed. The B. abortus bab1_1517 mutant strain was significantly attenuated in macrophages and mice, and bab1_1517 was subsequently named vtlR for virulence-associated transcriptional LysR-family regulator. Microarray analysis revealed three additional genes encoding small hypothetical proteins also under the control of VtlR. Electrophoretic mobility shift assays demonstrated that VtlR binds directly to the promoter regions of abcR2 and the three hypothetical protein-encoding genes, and DNase I footprint analysis identified the specific nucleotide sequence in these promoters that VtlR binds to and drives gene expression. Strikingly, orthologs of VtlR are encoded in a wide range of host-associated α-proteobacteria, and it is likely that the VtlR genetic system represents a common regulatory circuit critical for host–bacterium interactions.
Introduction
Brucella spp. are Gram-negative α2-proteobacteria that are facultative intracellular pathogens capable of residing within both professional and non-professional phagocytic cells (Köhler et al., 2003; Roop et al., 2004). The brucellae can infect a variety of wild and domesticated animals, leading to abortions and sterility, and furthermore, Brucella infections in humans result in a debilitating condition characterized most commonly by a chronic relapsing fever (Corbel, 1997; Pappas et al., 2006). Owing to the zoonotic nature of Brucella, humans often acquire this infection by consumption of unpasteurized dairy products, direct contact with mucosal secretions from an infected animal, or by occupational exposure (i.e., veterinarians and laboratory workers) (Pappas et al., 2005). Currently, brucellosis remains one of the most widespread and economically devastating zoonotic diseases worldwide with an estimated 500 000 human cases per year (Pappas et al., 2006; Seleem et al., 2010).
Upon entering the host, the brucellae are engulfed by cells of the reticuloendothelial system, particularly macrophages, and following trafficking of the bacteria through the host cell, the brucellae ultimately reside in an intracellular vacuole called the replicative Brucella-containing vacuole, which is associated with the endoplasmic reticulum (Arenas et al., 2000; Celli, 2006). While trafficking through the macrophage, the bacteria encounter many potentially harmful conditions, including exposure to reactive oxygen and nitrogen species, destructive enzymes, low pH, nutrient deprivation and diminished oxygen levels (Aderem, 2003; Roop et al., 2009). To combat the harsh intracellular environment of the macrophage, the brucellae encode an arsenal of strategies, including the VirB type IV secretion system (O’Callaghan et al., 1999; Lacerda et al., 2013), as well as other stress response mechanisms (Roop et al., 2009). Nonetheless, much remains to be elucidated about the regulatory components controlling gene expression during the intracellular life of Brucella.
LysR-type transcriptional regulators (LTTRs) are the most common type of prokaryotic DNA-binding protein, and these regulatory proteins can function as either activators or repressors of gene expression (Schell, 1993). As a family, LTTRs are highly conserved in protein structure, and these regulators are composed of an N-terminal helix–turn–helix DNA-binding domain and a C-terminal co-inducer binding domain (Maddocks and Oyston, 2008). Given the broad conservation of LTTRs among diverse groups of bacteria, it is not surprising that these regulators control expression of a wide variety of genes, including those involved in functions related to virulence (Russell et al., 2004; Doty et al., 1993), quorum sensing (O’Grady et al., 2011), motility (Heroven and Dersch, 2006) and metabolism (Hartmann et al., 2013). Brucella spp. encode several LysR proteins, few of which have been linked to virulence and, in general, very little is known about the role of LysR regulators in the brucellae (Haine et al., 2005).
Another prominent and widely distributed class of bacterial genetic regulators is small regulatory RNAs (sRNAs). sRNAs are comparatively short (i.e., ~50–300 nucleotides) regulatory RNA molecules that often control gene expression at the post-transcriptional level by binding directly to target mRNAs to alter their structure, stability and/or access to the ribosome-binding site. Depending on the nature of the sRNA–mRNA interaction, sRNAs may exert positive or negative effects on the expression of a given gene (Waters and Storz, 2009). Not surprisingly, bacterial sRNAs regulate myriad cellular processes, including stress response, metabolism, quorum sensing and virulence (Bejerano-Sagie and Xavier, 2007; Hoe et al., 2013; Papenfort and Vogel, 2014; Murphy and Payne, 2007). The brucellae encode two highly related sRNAs called AbcR1 and AbcR2, and these sRNAs are required for wild-type virulence of Brucella in macrophages and mice (Caswell et al., 2012b). However, to date, nothing is known about the mechanisms controlling abcR expression in Brucella.
The present study describes a LTTR in B. abortus that activates the expression abcR2, but not abcR1, and more importantly, this regulator is essential for the capacity of the bacteria to successfully infect macrophages and mice. In addition, this LysR regulator, named VtlR for virulence-associated transcriptional LysR-family regulator, also directly activates the expression of three genes putatively encoding small hypothetical proteins of ~50 amino acids in length. Most striking is the fact that numerous host-associated α-proteobacteria appear to encode a protein orthologous to VtlR, as well as cognate orthologous small hypothetical proteins. The VtlR system may represent a common genetic regulatory mechanism required for efficient host–bacterium interactions among the α-proteobacteria.
Results
A LysR-type transcriptional regulator, VtlR, controls expression of the small RNA-encoding gene, abcR2 in Brucella abortus
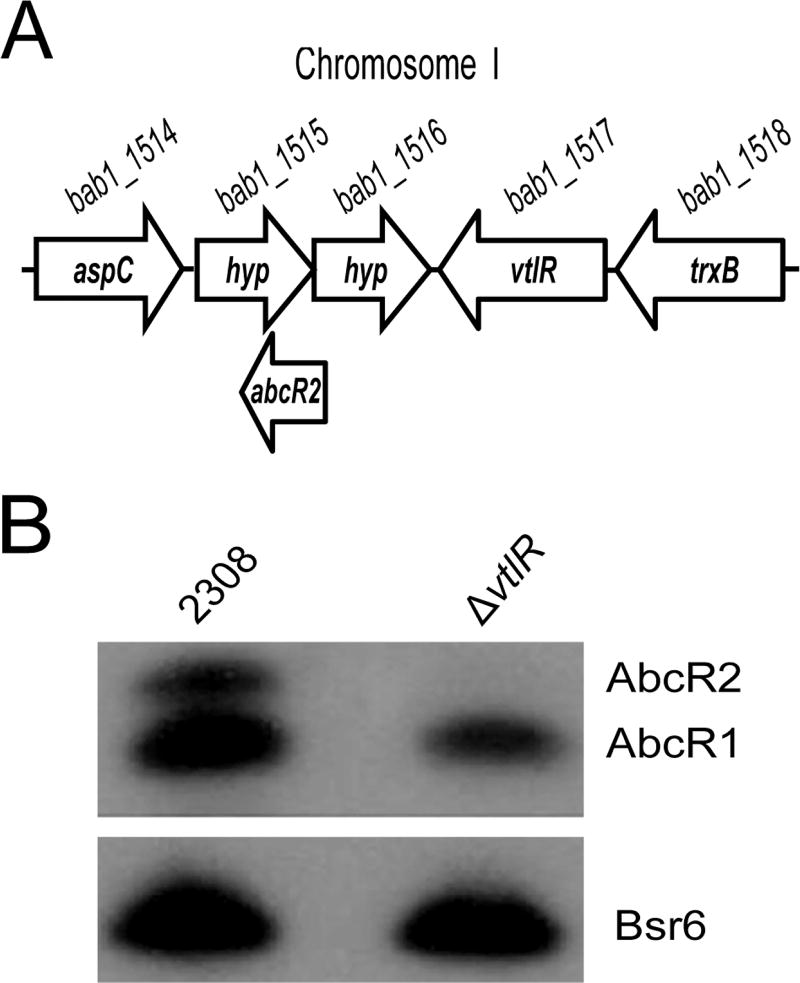
Two highly similar small RNAs, AbcR1 and AbcR2, are essential for the wild-type virulence of B. abortus (Caswell et al., 2012b), but to date, there is no information regarding the regulation of abcR1 and abcR2 expression. Flanking the abcR2 gene in B. abortus 2308 is a gene designated vtlR (bab1_1517) predicted to encode a LysR-type transcriptional regulatory protein of approximately 33 kDa in size (Fig. 1A). Because LysR regulators are often encoded in close proximity to the genes they regulate (Schell, 1993), it was hypothesized that VtlR controls expression of one or both of the abcR genes. To test this hypothesis, an in-frame, unmarked gene deletion of vtlR was constructed, and the expression of the AbcR sRNAs was assessed (Fig. 1B). Northern blot analyses demonstrated that AbcR2 production was abolished by deletion of vtlR; however, there was no effect on the levels of AbcR1 expression in the ΔvtlR mutant strain compared with the parental strain 2308. As a control for RNA production in the strains tested, there were no differences observed for the production of the Brucella sRNA Bsr6. Taken together, these data demonstrate that VtlR is an activator of abcR2 expression in B. abortus 2308.
Fig. 1.
VtlR, a LysR-type transcriptional regulator, is an activator of the small RNA-encoding gene abcR2
A. Diagram of the abcR2 locus on chromosome 1 of Brucella abortus 2308. In this study, the gene vtlR (bab1_1517) encoding a LysR-type regulator was hypothesized to control the transcription of the small RNA, abcR2.
B. Northern blot analysis assessing the levels of the AbcR sRNAs in the parental strain B. abortus 2308 and in the B. abortus ΔvtlR mutant strain. The Brucella sRNA, Bsr6, served as a loading control for this experiment.
VtlR is essential for the wild-type virulence of B. abortus 2308
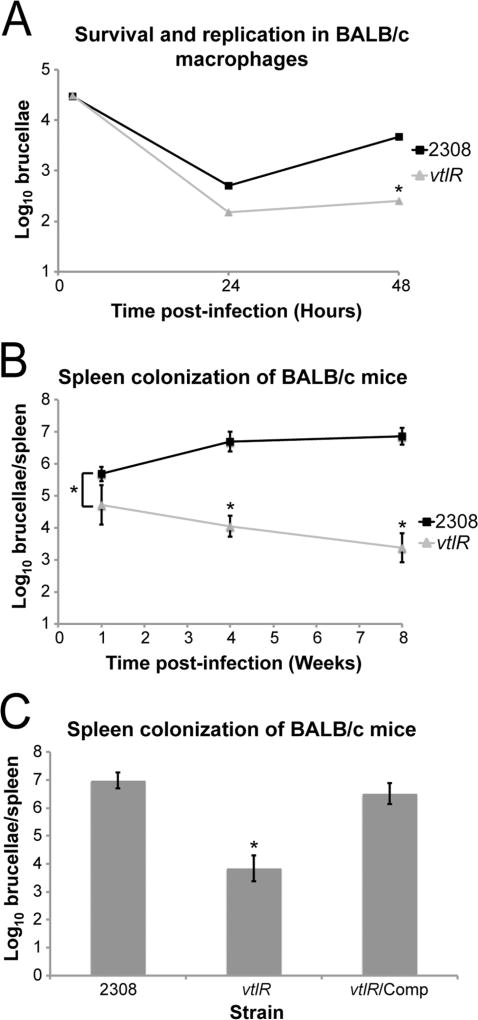
To study the role of VtlR in the pathogenesis of Brucella, a series of in vitro and in vivo experiments were performed with the ΔvtlR mutant strain compared with the parental strain 2308 (Fig. 2). Primary murine macrophages were infected with the parental strain 2308 and the B. abortus ΔvtlR strain (Fig. 2A). The ΔvtlR mutant exhibited a reduced ability to survive and replicate within macrophages compared with the parental strain 2308. The B. abortus ΔvtlR mutant strain was also assessed by its ability to establish a chronic infection in a mouse model (Fig. 2B). Female BALB/c mice were infected via the intraperitoneal route with either the ΔvtlR mutant strain or the parental strain 2308. The ΔvtlR mutant strain was significantly attenuated compared with the parental strain 2308. Furthermore, reconstruction of the ΔvtlR locus (vtlR/Comp) restored spleen colonization to levels similar to the parental strain 2308 (Fig. 2C). To assess the potential pleiotropic effect of deleting vtlR in B. abortus, in vitro growth of the parental strain 2308 and the ΔvtlR strain in both nutrient-rich (brucella broth) or nutrient-deficient Gerhardt’s minimal medium (GMM) medium was assessed (Fig. S1). No differences were observed between the strains tested in either growth conditions. Overall, these results demonstrate that VtlR is required for the full virulence of B. abortus 2308 in vitro as well as in vivo.
Fig. 2.
VtlR is essential for virulence of Brucella abortus 2308. A. Survival and replication of the B. abortus ΔvtlR mutant in primary murine macrophages. Primary macrophages were collected from the peritoneal cavity of female BALB/c mice and infected with the parental B. abortus strain 2308 or the ΔvtlR mutant strain. At 2, 24 and 48 h post-infection, macrophages were lysed, and serial dilutions of the lysates were plated on blood agar to determine the number of intracellular brucellae. Statistically significant differences (one-way ANOVA; P < 0.005) are indicated by an asterisk (*).
B. Spleen colonization of BALB/c mice infected with the B. abortus ΔvtlR mutant strain. Six week old female BALB/c mice were infected intraperitoneally with B. abortus 2308 or the ΔvtlR mutant strain. Five mice were used for each Brucella strain at each time point. At 1, 4 and 8 weeks post-infection, mice were sacrificed, and the spleens were removed and homogenized. Serial dilutions of the spleen homogenates were plated onto blood agar to enumerate the number of brucellae per spleen. Data are presented as average brucellae per spleen +/− the standard deviation from the five mice infected with each Brucella strain. Statistically significant differences (t-test; P < 0.05) are indicated by an asterisk (*).
C. Spleen colonization of BALB/c mice infection with the reconstructed ΔvtlR mutant strain (ΔvtlR/Comp). This experiment was performed as described above in Fig. 2B except spleen colonization was evaluated after a single time-point of 4 weeks post-infection. Data is presented as average brucellae per spleen +/− the standard deviation from the five mice infected with each Brucella strain. Statistically significant differences (t-test; P < 0.05) are indicated by an asterisk (*).
Microarray analysis reveals the VtlR regulon
Although VtlR plays a critical role in the virulence of Brucella, the absence of abcR2 expression does not fully explain the attenuation of the ΔvtlR mutant strain. Caswell et al. demonstrated that AbcR1 and AbcR2 are functionally redundant sRNAs in Brucella, as attenuation of the brucellae was only observed when both abcR1 and abcR2 were deleted, but not when either abcR1 or abcR2 were individually deleted (Caswell et al., 2012b). Therefore, we hypothesized that VtlR must regulate the expression of other genes required for survival and replication of the bacteria in the host. To determine the full genetic regulon of VtlR in B. abortus, microarray analysis was employed using RNA isolated from the parental strain 2308 and the vtlR mutant strain grown in rich medium (Table 1; Geo Accession GSE69564). Eight genes, including bab1_0310, bab1_0313, bab1_0314 and bab1_1794, which are known targets of the AbcR sRNAs, were shown to be significantly elevated in the ΔvtlR mutant strain. Moreover, three genes encoding putative small hypothetical proteins, bab1_0914, bab2_0512 and bab2_0574, exhibited diminished levels in the ΔvtlR mutant strain compared with the parental strain 2308.
Table 1.
Genes greater than twofold up- or downregulated in Brucella abortus vtlR mutant.
| BAB # | Fold decrease in ΔvtlR strain | Description of gene product |
|---|---|---|
| bab1_0914 | 10.1 | Hypothetical protein |
| bab1_1517 | 14.5 | Transcriptional regulator (VtlR) |
| bab2_0512 | 8.4 | Hypothetical protein |
| bab2_0574 | 15.7 | Hypothetical protein |
| bab2_0801 | 2.0 | Hypothetical protein |
|
| ||
| Fold increase in ΔvtlR strain | ||
|
| ||
| bab1_0310 | 2.3 | Allantoate amidohydrolase |
| bab1_0313 | 3.0 | Dihydropyrimidine dehydrogenase |
| bab1_0314 | 3.5 | Putative oxidoreductase |
| bab1_0355 | 2.2 | LrgA family protein |
| bab1_0356 | 2.2 | LrgB family protein |
| bab1_1794 | 2.2 | Leu/Ile/Val-binding family protein |
| bab2_0348 | 2.7 | Protein kinase |
| bab2_0961 | 2.0 | Dihydrodipicolinate synthetase |
List of genes differentially expressed in the B. abortus ΔvtlR mutant strain compared with the parental strain 2308. Microarray analysis was performed with total cellular RNA from Brucella strains grown to late exponential phase in nutrient-replete brucella broth. The genes listed are more than twofold decreased in the ΔvltR mutant strain, or more than twofold increased in the ΔvltR mutant strain. The complete microarray analysis can be found at GEO Accession GSE69564.
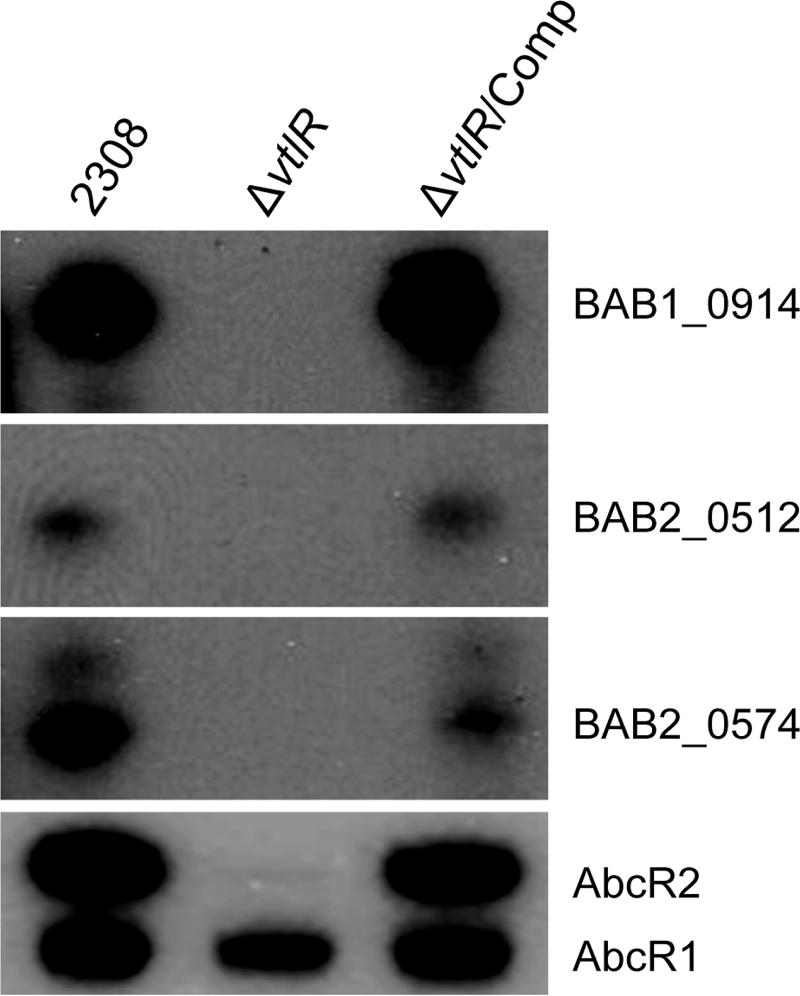
To confirm the regulation of these hypothetical protein-encoding genes by VtlR, northern blot analyses were performed with RNA isolated from the parental strain 2308, the ΔvtlR mutant strain and the reconstructed vtlR mutant strain (vtlR/Comp) (Fig. 3). The vtlR mutant exhibited no production of the BAB1_0914, BAB2_0512, BAB2_0574 or AbcR2 RNAs. Importantly, AbcR1 production was maintained in the ΔvtlR mutant strain compared with strain 2308. Complementation of vtlR expression in the ΔvtlR mutant strain restored production of AbcR2, BAB1_0914, BAB2_0512 and BAB2_0574 RNAs. These data confirm that VtlR activates the expression of these genes in B. abortus 2308.
Fig. 3.
VtlR activates the expression of abcR2 and three hypothetical protein-encoding genes. Northern blot analyses evaluating expression of the genes encoding the AbcR sRNA, and the genes encoding BAB1_0914, BAB2_0512 and BAB2_0574. RNA isolated from the parental strain B. abortus 2308, the vtlR mutant strain, and the genetically complemented vtlR mutant strain (vtlR/Comp) were separated by denaturing polyacrylamide gel electrophoresis, and following transfer to a positively-charged membrane, the membranes were probed for RNAs corresponding to the AbcR sRNAs (a common probe that recognizes AbcR1 and AbcR2), BAB1_0914, BAB2_0512 or BAB2_0574.
VtlR binds directly to the promoter regions of abcR2, bab1_0914, bab2_0512 and bab2_0574
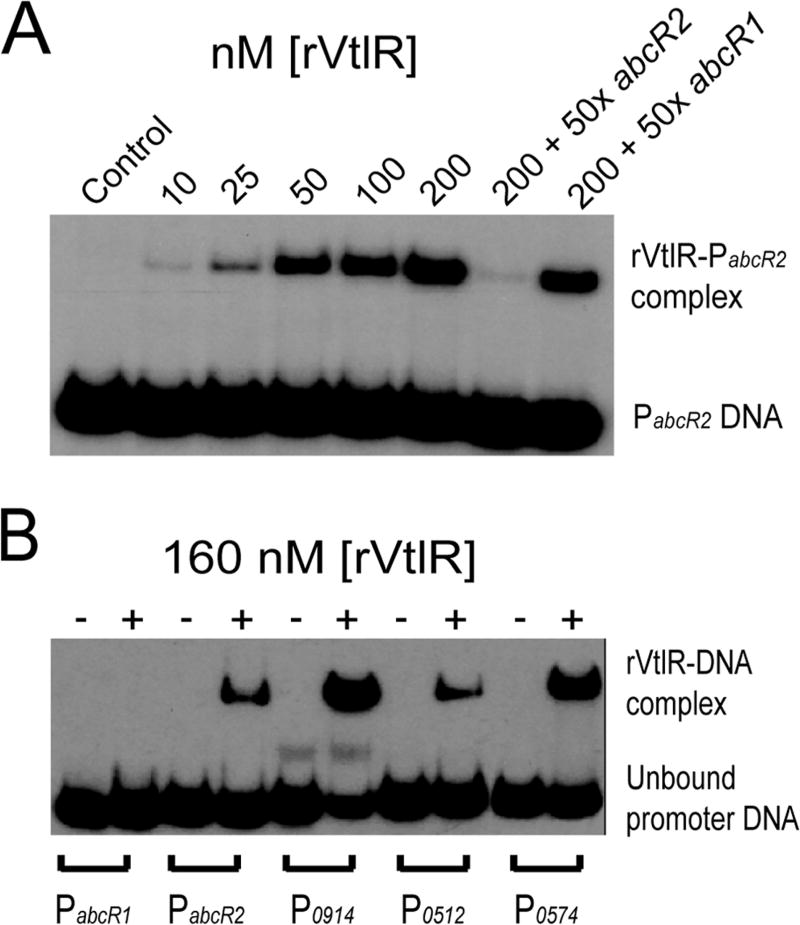
Electrophoretic mobility shift assays (EMSAs) were employed to assess the direct regulation of abcR2, bab1_0914, bab2_0512 and bab2_0574 by VtlR in B. abortus 2308 (Fig. 4). First, recombinantly purified B. abortus VtlR (rVtlR) was incubated with and subsequently bound to radiolabeled DNA corresponding to the promoter region of abcR2 in a concentration-dependent manner (Fig. 4A). Importantly, specific, unlabeled DNA (i.e., abcR2 promoter region DNA) abolished binding between rVtlR and the labeled abcR2 promoter DNA, but non-specific, unlabeled DNA (i.e., abcR1 promoter region DNA) did not affect interactions between rVtlR for the labeled abcR2 promoter. Subsequent EMSA experiments demonstrated that, in addition to the abcR2 promoter, VtlR also binds directly to the promoter regions of bab1_0914, bab2_0512 and bab2_0574, but not to the abcR1 promoter (Fig. 4B). Overall, these experiments confirm the B. abortus VtlR protein interacts directly with the promoters of abcR2, bab1_0914, bab2_0512 and bab2_0574 to activate gene transcription.
Fig. 4.
VtlR binds to the promoter regions of the small RNA abcR2, as well as three genes bab1_0914, bab2_0512 and bab2_0574 encoding hypothetical proteins.
A. Recombinant VtlR (rVtlR) protein was evaluated for binding to the abcR2 promoter region using electrophoretic mobility shift assays (EMSAs). Increasing concentrations of rVtlR were incubated with radiolabeled abcR2 promoter region DNA. In some binding reactions, either unlabeled specific (abcR2 promoter region DNA) or unlabeled non-specific (abcR1 promoter region DNA) competitor DNA fragments were added to the binding reactions.
B. EMSAs of rVtlR protein binding to the promoter regions of abcR1, abcR2, bab1_0914, bab2_0512 and bab2_0574. Radiolabeled promoter DNA fragments (approximately 200 bp) were either incubated with (+) or without (−) 160 nM rVtlR.
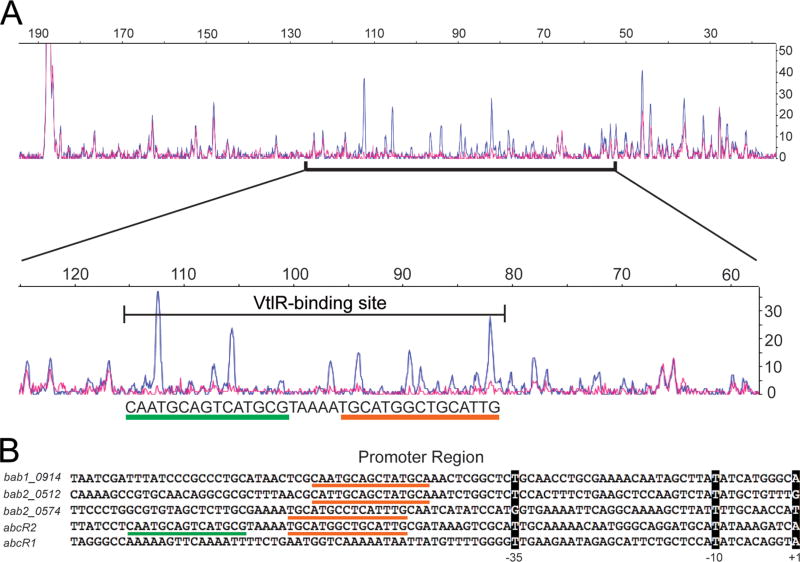
DNase I footprinting was employed to determine the specific DNA motif within the abcR2 promoter region that is bound by VtlR in B. abortus (Fig. 5). An approximately 200 bp 6-Fluorescein amidite (6FAM)-labeled DNA fragment corresponding to the promoter region of abcR2 was incubated with or without rVtlR, and the complexes were treated with DNase. Following DNA fragment analysis by capillary electrophoresis, it was determined that a 35 bp region in the abcR2 promoter was protected from DNase digestion by VtlR (Fig. 5A). DNA sequencing analyses determined the sequence of the protected region to be CAATGCAGCCATGCATTTTACGCATGACTGCATTG. This sequence is composed of an inverted repeat of CAAT GCAGCCATGCA separated by anAT rich region, suggesting that VtlR may bind as a dimer to the abcR2 promoter. Moreover, the CAATGCAGCCATGCA motif is only found in the promoter regions of abcR2, bab1_0914, bab2_0512 and bab2_0574 and not in any other location of the B. abortus 2308 chromosomes (Fig. 5B). Taken together, these data demonstrate the specific nucleotide sequence utilized by VtlR to activate gene expression in B. abortus 2308.
Fig. 5.
DNase I footprinting analysis of VtlR binding site in the abcR2 promoter.
A. Overlaid DNase digestion pattern of the abcR2 200 bp promoter region with (pink line) or without (blue line) 160 nM rVtlR. The protected region of abcR2 is indicated by the black horizontal line labeled ‘VtlR-binding site’, and the nucleotide sequence is indicated underneath of the digestion pattern. Orange underlining beneath nucleotides indicates the conserved sequence, whereas the green underlining beneath nucleotides indicates the inverted repeat within the 35 bp VtlR-binding site of PabcR2.
B. The promoter regions (Pgene) of the VtlR-regulated genes, bab1_0914, bab2_0512, bab2_0574, and abcR2, and abcR1 as a negative control. The transcriptional start site (+1), as well as the −10 and −35 regions are indicated by nucleotides highlighted in black. The conserved nucleotide sequence is indicated by the orange underline. As in panel B, the inverted repeat found in PabcR2 is indicated by the green underline.
Deletion of four VtlR-regulated genes does not lead to attenuation of B. abortus in macrophages
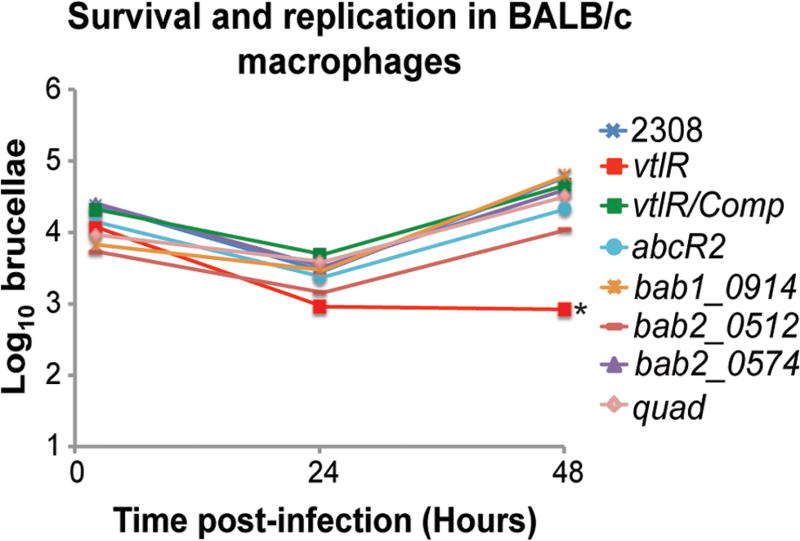
As demonstrated in Fig. 2, a B. abortus vtlR mutant exhibits a decreased ability to survive and replicate within macrophages compared with the parental strain 2308 (Fig. 2A). To begin to elucidate the role of the VtlR-regulated genes (i.e., abcR2, bab1_0914, bab2_0512, bab2_0574) in B. abortus virulence, isogenic mutant strains, as well as a strain with all four VtlR-regulated genes deleted, were constructed. Primary naïve macrophages were isolated from BALB/c mice and infected with either B. abortus 2308, ΔvtlR, ΔvltR with a reconstructed locus (vtlR/Comp), ΔabcR2, Δbab1_0914, Δbab2_0512, Δbab2_0574 or ΔabcR2/bab1_0914/bab2_0512/bab2_0574 (quad). As expected, the ΔvtlR mutant strain was attenuated compared with strain 2308, and reconstruction of the vtlR locus in the mutant restored intracellular levels of brucellae to those of the parental strain (Fig. 6). However, B. abortus strains with individual deletions of abcR2, bab1_0914, bab2_0512 or bab2_0574 did not exhibit decreased capacities to survive and replicate in macrophages, and likewise, a strain with all four of the genes deleted was not attenuated in this model.
Fig. 6.
Assessment of the role of VtlR-regulated genes in Brucella abortus virulence in macrophages. Survival and replication of B. abortus 2308, ΔvtlR, reconstructed ΔvtlR locus (vtlR/Comp), ΔabcR2, Δbab1_0914, Δbab2_0512, Δbab2_0574, and a quadruple mutant of abcR2, bab1_0914, bab2_0512 and bab2_0574 (quad) in primary murine macrophages. Macrophages were collected from the peritoneal cavity of female BALB/c mice and infected with the Brucella abortus strains. At 2, 24 and 48 h post-infection, macrophages were lysed, and serial dilutions of the lysates were plated on blood agar to determine the number of intracellular brucellae. Statistically significant differences (one-way ANOVA; P < 0.005) compared with the wild-type are indicated by an asterisk (*).
Extensive conservation of VtlR in the α-proteobacteria
Putative orthologs of VtlR are present in multiple α-proteobacteria, including the soil dwelling bacterium Ochrobactrium anthropi, the human pathogen Bartonella henselae, the plant symbiont Sinorhizobium meliloti and the plant pathogen Agrobacterium tumefaciens (Fig. S2). Although LTTRs are the most common prokaryotic transcriptional regulators (Schell, 1993; Maddocks and Oyston, 2008), VtlR orthologs found in α-proteobacteria exhibit a strikingly high degree of amino acid identity and similarity, suggesting a common function across this group of bacteria. Furthermore, orthologs of VtlR have been demonstrated to play an important role in host–bacterium interactions in other α-proteobacteria. For instance, in S. meliloti 1021, the ortholog of VtlR, designated LsrB, is critical for the successful symbiosis between S. meliloti and alfalfa plants (Luo et al., 2005). An S. meliloti ΔlsrB mutant is deficient in both establishing an effective symbiosis and fixing nitrogen compared to the parental strain 1021. Altogether, these data suggest that the VtlR regulatory system is paramount for the host–bacterium relationship of α-proteobacteria and their respective hosts.
Discussion
The VtlR protein (BAB1_1517) is required for B. abortus virulence (Fig. 2), and the link between Brucella virulence and VtlR has been observed previously in B. suis (BR1498; Foulongne et al., 2000) and in B. melitensis (BMEI0513; Wu et al., 2006). However, although these previous studies reported observations of attenuation of strains generated by transposon mutagenesis, the present work defines the role of VtlR in Brucella pathogenesis. Moreover, this study sheds light on the molecular basis for the role of VtlR in Brucella virulence. Specifically, VtlR directly activates the expression of abcR2, encoding an important small regulatory RNA, as well as three genes encoding novel small hypothetical proteins (Figs 1, 3–5, and Table 1; Geo Accession GSE69564). Although VtlR was also linked to the expression of other genes (Table 1; Geo Accession GSE69564), the promoter regions of these genes do not contain the putative VtlR-binding motif CAATGCAGCCATGCA and thus, we conclude that these genes are indirectly regulated by VtlR. For example, several of the genes exhibiting increased expression in the B. abortus ΔvtlR mutant strain (i.e., bab1_0310, bab1_0313, bab1_0314 and bab1_1794) are AbcR sRNA target genes, and expression of these genes is known to be elevated when the AbcR sRNAs are deleted (Caswell et al., 2012b). In the same vein, the genes bab1_0355, bab1_0356, bab2_0348 and bab2_0961 are likely indirectly regulated by VtlR, but presently, the link between VtlR and the expression of these genes remains to be elucidated.
The attenuation of the B. abortus ΔvtlR mutant is striking (Fig. 2), but given the fact that both abcR1 and abcR2 are required for wild-type virulence of B. abortus 2308 (Caswell et al., 2012b), the absence of abcR2 in this mutant strain cannot fully account for the virulence defect exhibited by the B. abortus vtlR mutant. Interestingly, the VtlR-regulated genes bab1_0914, bab2_0512 and bab2_0574 also do not appear to play a role in the wild-type survival and replication of B. abortus in murine macrophages (Fig. 6). Additional work aimed at dissecting the full VtlR regulon (i.e., both directly and indirectly regulated genes), as well as animal model experiments, are needed to fully elucidate the biological role(s) of bab1_0914, bab2_0512 and bab2_0574 in B. abortus. Interestingly, genes encoding other putative small proteins in Brucella have been linked to the virulence of Brucella strains (Köhler et al., 2002; Lavigne et al., 2005; Sun et al., 2012).
To date, bioinformatics approaches have failed to provide insight into the biological function(s) of BAB1_0914, BAB2_0512 or BAB2_0574 in Brucella, but ongoing work in our laboratory is aimed at defining the role of these hypothetical proteins in the biology of Brucella (Budnick, unpublished data). Nonetheless, the putative proteins encoded by bab1_0914, bab2_0512 and bab2_0574, as well as the regulator VtlR, are all highly conserved among the α-proteobacteria. Although it is currently unknown if these genes encode authentic small proteins in Brucella, we have shown that the RNAs encoded by bab1_0914, bab2_0512 and bab2_0574 are stably produced in a B. abortus hfq mutant (Fig. S3), indicating that these RNAs are not prototypical small regulatory RNAs and likely encode proteins. Our laboratory is currently interrogating B. abortus 2308 to determine if BAB1_0914, BAB2_0512 and/or BAB2_0574 are bona fide proteins produced in this strain.
VtlR orthologs are encoded flanking the abcR genes of A. tumefaciens (Wilms et al., 2011) and S. meliloti (Torres-Quesada et al., 2013). Unlike the situation in the brucellae, the abcR genes in A. tumefaciens and S. meliloti are encoded in tandem, and thus, it is likely that the VtlR orthologs in these bacteria control the expression of one or both of the abcR genes in these bacteria. In fact, a sequence highly similar to the CAATGCAGCCATGCA VtlR-binding motif of the Brucella abcR2 promoter is present in the promoter region of the abcR1 in S. meliloti (Fig. S4). Orthologs of the other three genes directly regulated by VtlR in B. abortus (i.e., bab1_0914, bab2_0512 and bab2_0574) are also highly conserved in the α-proteobacteria, and the promoter regions of these orthologs also contain a putative VtlR-like binding site (Fig. S4). Therefore, it is likely that the VtlR system activates the expression of the abcR genes, as well as three hypothetical protein-encoding genes in other members of the α-proteobacteria.
The conservation of VtlR orthologs is high in the α-proteobacteria (Fig. S2). While we propose that the VtlR systems are important in each of their respective bacteria for interactions in their specific environment, there is evidence of evolutionary adaptation of each unique protein to each niche. The VtlR ortholog in S. meliloti has been identified and characterized, and this protein was designated LsrB for LysR-type symbiosis regulator (Luo et al., 2005). Interestingly, lsrB is required for efficient symbiosis of S. meliloti with alfafa plants, and it was one of two LTTRs (from a total of 90 screened) necessary for symbiosis and nitrogen fixation. Heterologous complementation of the B. abortus ΔvtlR mutant strain with the lsrB gene from S. meliloti was unable to restore the capacity of the ΔvtlR mutant to colonize mice at wild-type levels (data not shown), which alludes to the fact that these LTTRs having evolved over time to suit their specific environment and host, more likely by altering the C-terminal co-inducer binding domain. More recently, LsrB was shown to play a role in the LPS biosynthesis in S. meliloti (Tang et al., 2014). To date, there is no evidence linking VtlR to LPS biosynthesis in Brucella.
To summarize, the findings in this paper reveal VtlR as the LTTR activator of the sRNA-encoding gene abcR2, as well as three additional genes, bab1_0914, bab2_0512 and bab2_0574, encoding small hypothetical proteins. VtlR was also found to play a critical role in the survival of B. abortus in both macrophage and mouse models of infection. The conservation of VtlR, along with the three hypothetical proteins, in α-proteobacteria seems to be a common genetic system necessary for the interaction of the bacteria with its host, regardless of whether this interaction is pathogenic or symbiotic, and thus, the VtlR system is likely a common genetic determinant of host– bacterium interactions in the α-proteobacteria.
Experimental procedures
Bacterial strains and growth conditions
Brucella abortus 2308 and derivative strains were routinely grown on Schaedler agar (BD, Franklin Lakes, NJ, USA) containing5%defibrinated bovine blood (Quad Five, Ryegate, MT, USA) (SBA), or in brucella broth (BD). For cloning and recombinant protein expression, Escherichia coli strains (DH5α and BL21) were grown routinely on tryptic soy agar (BD) or in Luria–Bertani (LB) broth. When appropriate, growth media were supplemented with ampicillin (100 µg ml−1) or kanamycin (45 µg ml−1). All cultures of and experiments using Brucella strains were carried out in a Biosafety Level-3 (BSL-3) laboratory.
Construction of B. abortus mutants and genetic complementation
The vtlR gene (bab1_1517) was mutated using an unmarked gene excision strategy described previously (Caswell et al., 2012a). An approximately 1 kb fragment of the upstream region of the gene was amplified by PCR using the primers BAB1_1517-Up-For and BAB1_1517-Up-Rev, genomic DNA from B. abortus 2308 as a template, and Platinum® Pfx DNA Polymerase (Invitrogen, Carlsbad, CA, USA). Likewise, an approximately 1 kb fragment of the downstream region of the gene was amplified by PCR using the primers BAB1_1517-Down-For and BAB1_1517-Down-Rev. The sequences of all oligonucleotides used in this study are in Table S1. The upstream fragment was digested with the restriction enzyme BamHI, whereas the downstream fragment was digested with PstI, and both fragments were treated with polynucleotide kinase (Monserate Biotechnology Group, San Diego, CA, USA). The fragments were combined in a single ligation mix with BamHI/PstI-digested pNPTS138 (Spratt et al., 1986), which has a kanamycin resistance marker and sacB gene for counter-selection with sucrose, and T4 DNA ligase (Monserate Biotechnology Group). The resulting plasmid, pC3038, was introduced into B. abortus 2308, and a merodiploid clone was obtained by selection on SBA + kanamycin. Kanamycin-resistant transformants were grown for 6–8 h in brucella broth and then plated onto SBA plates containing 10% sucrose. Genomic DNA from sucrose-resistant, kanamycin-sensitive colonies was isolated and screened by PCR for loss of the vtlR gene. The isogenic vtlR mutant derived from B. abortus 2308 was named CC126. All plasmids generated in the study can be found on Table S2.
Plasmids for isogenic mutagenesis of Δbab1_0914, Δbab2_0512 and Δbab2_0574 were constructed as described above, and all primers and plasmids used can be found in Table S1 and Table S2 respectively. The quadruple mutant of abcR2, bab1_0914, bab2_0512 and bab2_0574 was constructed by sequential mutagenesis of B. abortus 2308 using the constructed plasmids used for generation of the isogenic mutant strains.
Reconstruction of the vtlR loci was done using the sacB counter-selection strategy described above but with modification. The wild-type vtlR locus was amplified using primers BAB1_1517-Up-For and BAB1_1517-Down-Rev, genomic DNA from B. abortus 2308 as a template, and Taq polymerase (Monserate Biotechnology Group). The approximately 3 kb fragment was digested with BamHI and PstI and ligated into BamHI/PstI-digested pNPTS138. The resulting plasmid, pLS003, was introduced into B. abortus CC126 (vtlR mutant), and selection and screening were carried out as described above. The strain carrying the reconstructed vtlR locus was named LS002.
Northern blot analysis
RNA was isolated from Brucella cultures as described previously (Caswell et al., 2012a). Ten micrograms of RNA was separated on a denaturing 10% polyacrylamide gel containing 7 M urea and 1 × TBE (89 Tris-base, 89 mM boric acid and 2 mM EDTA). A low-molecular-weight DNA ladder (New England BioLabs, Ipswich, MA, USA) was labeled with [γ-32P]-ATP (Perkin-Elmer, San Jose, CA, USA) and polynucleotide kinase. This radiolabeled ladder was also separated on the polyacrylamide gel. Following electrophoresis in 1 × TBE buffer, the ladder and RNA samples were transferred to an Amersham Hybond™-N+ membrane (GE Healthcare, Piscataway, NJ, USA) by electroblotting in 1 × TBE buffer. The samples were UV-cross-linked to the membrane and the membrane was pre-hybridized in ULTRAhyb®-Oligo Buffer (Ambion, Austin, TX, USA) for 2 h at ~45°C in a rotating hybridization oven. The oligonucleotide probes [AbcR-Northern (for detection of both AbcR1 and AbcR2), 0914-Northern, 0512-Northern, 0574-Northern, and Bsr6-Northern] were end-labeled with [γ-32P]-ATP and polynucleotide kinase. The radiolabeled probes were incubated with the prehybridized membranes at ~45°C in a rotating hybridization oven overnight (~12 h). The next day, membranes were washed four times for 30 min each with 2 × SSC (300 mM sodium chloride and 30 mM sodium citrate), 1 × SSC, 0.5 × SSC and 0.025 × SSC at ~45°C in a rotating hybridization oven. Each SSC wash buffers contained 0.1% sodium dodecyl sulphate (SDS). The membranes were then exposed to X-ray film and visualized by autoradiography.
Microarray analysis
RNA was isolated from Brucella cultures grown to late exponential phase in brucella broth (Caswell et al., 2012a), and contaminating genomic DNA was removed by treatment with RNase-free DNase I. Ten micrograms of each RNA sample, B. abortus 2308 and B. abortus vtlR mutant, were reverse transcribed, fragmented and 3′ biotinylated as previously described (Beenken et al., 2004). The labeled cDNA (1.5 µg) was hybridized to custom-made B. abortus GeneChips (PMD2308a520698F) according to the manufacturer’s recommendations for antisense prokaryotic arrays (Affymetrix, Santa Clara, CA, USA). Signal intensities were normalized to the median signal intensity value for each GeneChip, averaging and analyzed with GeneSpring Software X. RNA species exhibiting ≥ 2-fold change in expression, as determined by Affymetrix algorithms to be statistically differentially expressed (t-test; P < 0.05), between B. abortus 2308 and the vtlR mutant strain were stated. The microarrays used in this study were developed based on B. melitensis biovar abortus 2308 and all B. abortus GenBank entries that were available at the time of design. In total, predicted open reading frames and intergenic regions were represented on PMD2308a520698F. The GenBank accession number for the microarray data is GSE69564.
Production and purification of recombinant proteins
For EMSAs in Fig. 4A, the Strep-tag II system (IBA, Göttingen, Germany) was used to produce a recombinant version of the Brucella VtlR protein in E. coli strain BL21. The coding region of the vtlR gene (bab1_1517) was amplified using the primers rVtlR-For and rVtlR-Rev, B. abortus 2308 chromosomal DNA as a template, and Taq polymerase. The DNA fragment was digested with BbsI and ligated into BsaI-digested pASK-IBA6, which encodes an amino-terminal Strep-tag on the protein of interest that is localized to the periplasmic space of the host cell. The resulting plasmid, pLS014 (rVtlR-pIBA6), was transformed into E. coli strain BL21, and the strains harboring the recombinant protein expression plasmid were grown to an optical density at 600 nm of approximately 0.6 in LB broth before the production of the recombinant protein was induced by adding anhydrotetracycline (200 µg ml−1 final concentration). Following incubation for 2 h at 37°C with shaking, the cells were collected by centrifugation (4,200 × g for 10 min at 4°C) and lysed by treatment with CelLyticB (Sigma, St Louis, MO, USA) in the presence of the protease inhibitor phenylmethanesulfonyl fluoride. The supernatant from the suspension of lysed cells was cleared by centrifugation (14 000 × g for 10 min at 4°C) and passed through an affinity column packed with Strep-Tactin Sepharose. The column was washed extensively with Buffer W (100 mM Tris-HCl, 300 mM NaCl, pH 8.0) and the recombinant protein was eluted with 2.5 mM desthiobiotin in Buffer W. The degree of purity of the recombinant VtlR was high as judged by visualization of a single, major band on SDS-PAGE.
For EMSAs in Fig. 4B and DNase I footprinting in Fig. 5, the Strep-tag II system (IBA, Göttingen, Germany) was used as above to generate a recombinant Brucella VtlR, but with modification. The vtlR DNA fragment was digested with BbsI and ligated into BsaI-digested pASK-IBA7, which incorporates an amino-terminal Strep-tag on the protein of interest. The protein is then localized in the cytosol of the cell. The resulting plasmid, rVtlR-pIBA7, was transformed into E. coli strain BL21, and the strains harboring the recombinant protein expression plasmid were grown and induced as described above. Following incubation for 2 h at 37°C with shaking, the cells were collected by centrifugation (4,200 × g for 10 min at 4°C) and lysed by treatment with CelLyticB (Sigma) and DNase I in the presence of the protease inhibitor phenylmethanesulfonyl fluoride. Subsequent purification steps of rVtlR was followed as described above.
EMSAs
All rVtlR EMSA experiments were carried out in a final volume of 20 µl containing binding buffer composed of 10 mM Tris-HCl (pH 7.4), 50 mM KCl, 1 mM dithiothreitol, 6% glycerol, 0.5 mM EDTA, 50 µg ml−1 bovine serum albumin and 50 µg ml−1 salmon sperm DNA. Approximately 200 bp DNA fragments of the abcR1, abcR2, bab1_0914, bab2_0512 and bab2_0574 promoter regions were amplified by PCR using primers: abcR1-EMSA-For, abcR1-EMSA-Rev; abcR2-EMSA-For, abcR2-EMSA-Rev; 0914-EMSA-For3, 0914-EMSA-Rev3; 0512-EMSA-For3, 0512-EMSA-Rev3; 0574-EMSA-For3, 0574-EMSA-Rev3. Chromosomal DNA from B. abortus 2308 was used as a template in all PCR reactions. The amplified fragments were purified by agarose gel electrophoresis, and the fragments were end-labeled with [γ-32P]ATP and polynucleotide kinase. In Fig. 4A, increasing amounts of rVtlR were mixed with the radiolabeled abcR2 fragment in binding buffer. As controls, 50 × molar concentrations of non-radiolabeled abcR2 promoter region DNA (specific competitor) or nonradiolabeled abcR1 promoter region DNA (non-specific competitor) were added to some reactions. In Fig. 4B, 160 nM of rVtlR was either added (+) or not added (−) to binding reactions with the promoter fragments of abcR1, abcR2, bab1_0914, bab2_0512 and bab2_0574. All binding reactions were incubated at room temperature for 20 min and then subjected to electrophoresis on 6% native polyacrylamide gels in 0.5 × TB (Tris-borate) running buffer for approximately 1 h at 4°C. Following electrophoresis, gels were dried onto 3MM Whatman® paper using a vacuum gel dryer system and visualized by autoradiography.
DNase I footprinting
DNase I footprinting was performed as previously described by Zianni et al. (2006). The approximately 200 bp abcR2 promoter fragment was labeled with a 5′-6FAM modification and amplified by PCR using primers abcR2-6FAM-For and abcR2-EMSA-Rev, Taq polymerase, and B. abortus 2308 chromosomal DNA. Following gel purification, the FAM-labeled abcR2 probe was mixed in EMSA binding buffer with or without 160 nM of rVtlR, to a total volume of 20 µl, and incubated at room temperature for 20 min. All reactions were then treated with 0.50 U DNase I (Ambion) for 4 min at room temperature. The DNase I reactions were terminated by addition of 0.5 M EDTA and incubation at 90°C for 5 min. The DNA fragments were purified using phenol-chloroform extraction and ethanol precipitation. Fragment analysis was performed with a Genescan™-500-Liz® size standard on an Applied Biosystems 3130 (Applied Biosystems, Grand Island, NY, USA) at the Virginia Bioinformatics Institute (Blacksburg, VA, USA). To determine the sequence of the VtlR-binding site, sequencing reactions with the 5′-FAM-labeled abcR2 oligonucleotide and Thermo Sequenase Dye Primer Manual Cycle Sequencing Kit (Affymetrix) were used according to the manufacturer’s directions.
Virulence of Brucella mutant strains in murine macrophages and mice
Experiments to test the virulence of Brucella mutant strains in primary, peritoneal murine macrophages were carried out as described previously (Gee et al., 2005). Macrophages were harvested from mice and seeded into 96-well plates in Dulbecco’s modified Eagle’s medium with 5% fetal bovine serum. The following day, macrophages were infected with opsonized brucellae at a multiplicity of infection of 50:1. After 2 h, the infected macrophages were treated with gentamicin (50 µg ml−1) for 1 h. The macrophages were then lysed with 0.1% deoxycholate in PBS, and serial dilutions were plated on Schaedler blood agar (SBA). For the 24 and 48 h time points, macrophages were washed with PBS following gentamicin treatment and fresh cell culture medium containing gentamicin (20 µg ml−1) was added to the monolayer. At the indicated time points, macrophages were lysed and serial dilutions were plated on SBA in triplicates.
The infection and colonization of mice by Brucella vtlR mutant was performed as described previously (Gee et al., 2005). Briefly, 6-week-old female BABL/c mice (five mice per Brucella strain) were infected intraperitoneally with ~5 × 104 CFU of each Brucella strain in sterile PBS. The mice were sacrificed at 1, 4 and 8 weeks post-infection and serial dilutions of spleen homogenates were plated on SBA. All data was analyzed using the statistical software JMP 11.0.0 (SAS Institute, Cary, NC, USA).
Supplementary Material
Acknowledgments
We would like to thank Hardik M. Zatakia for his help with DNase I footprinting. We thank the Virginia-Maryland College of Veterinary Medicine and the Fralin Life Sciences Institute at Virginia Tech for providing funding for this project.
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Aderem A. Phagocytosis and the inflammatory response. J Infect Dis. 2003;187:S340–S345. doi: 10.1086/374747. [DOI] [PubMed] [Google Scholar]
- Arenas GN, Staskevich AS, Aballay J, Mayorga LS. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect Immun. 2000;68:4255–4263. doi: 10.1128/iai.68.7.4255-4263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ. Global gene expression in Staphlococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M, Xavier KB. The role of small RNAs in quorum sensing. Curr Opin Microbiol. 2007;10:189–198. doi: 10.1016/j.mib.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Caswell CC, Gaines JM, Roop RMII. The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308. J Bacteriol. 2012a;194:3–14. doi: 10.1128/JB.05623-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, et al. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol. 2012b;85:345–360. doi: 10.1111/j.1365-2958.2012.08117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J. Surviving inside a macrophage: the many ways of Brucella. Res Microbiol. 2006;157:93–98. doi: 10.1016/j.resmic.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Corbel MJ. Brucellosis: an overview. Emerg Infect Dis. 1997;3:213–221. doi: 10.3201/eid0302.970219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty SL, Chang M, Nester EW. The chromosomal virulence gene, cheV, of Agrobacterium tumefaciens is regulated by a LysR family member. J Bacteriol. 1993;175:7880–7886. doi: 10.1128/jb.175.24.7880-7886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulongne V, Bourg G, Cazevieille C, Michaux-Charachon S, O’Callaghan D. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect Immun. 2000;68:1297–1303. doi: 10.1128/iai.68.3.1297-1303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee JM, Valderas MW, Kovach ME, Grippe VK, Robertson GT, Ng WL, et al. The Brucella abortus Cu,Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect Immun. 2005;73:2873–2880. doi: 10.1128/IAI.73.5.2873-2880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haine V, Sinon A, Van Steen F, Rousseau S, Dozot M, Lestrate P, et al. Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect Immun. 2005;73:5578–5586. doi: 10.1128/IAI.73.9.5578-5586.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T, Zhang B, Baronian G, Schulthess B, Homerova D, Grubmüller S, et al. Catabolite control protein E (CcpE) is a LysR-type transcriptional regulator of tricarboxylic acid cycle activity in Staphylococcus aureus. J Biol Chem. 2013;288:36116–36128. doi: 10.1074/jbc.M113.516302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heroven AK, Dersch P. RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis. Mol Microbiol. 2006;62:1469–1483. doi: 10.1111/j.1365-2958.2006.05458.x. [DOI] [PubMed] [Google Scholar]
- Hoe CH, Raabe CA, Rozhdestvensky TS, Tang TH. Bacterial sRNAs: regulation in stress. Int J Med Microbiol. 2013;303:217–229. doi: 10.1016/j.ijmm.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Köhler S, Foulongne V, Ouahrani-Bettache S, Bourg G, Teyssier J, Ramuz M, Liautard JP. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc Natl Acad Sci USA. 2002;99:15711–15716. doi: 10.1073/pnas.232454299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Michaux-Charachon S, Porte F, Ramuz M, Liautard JP. What is the nature of the replicative niche of a stealthy bug named Brucella? Trends Microbiol. 2003;11:215–219. doi: 10.1016/s0966-842x(03)00078-7. [DOI] [PubMed] [Google Scholar]
- Lacerda TL, Salcedo SP, Gorvel JP. Brucella T4SS: the VIP pass inside host cells. Curr Opin Microbiol. 2013;16:45–51. doi: 10.1016/j.mib.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Lavigne JP, Patey G, Sangari FJ, Bourg G, Ramuz M, O’Callaghan D, Michaux-Charachon S. Identification of a new virulence factor, BvfA, in Brucella suis. Infect Immun. 2005;73:5524–5529. doi: 10.1128/IAI.73.9.5524-5529.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Yao SY, Becker A, Rüberg S, Yu GQ, Zhu JB, Cheng HP. Two new Sinorhizobium meliloti LysR-type transcriptional regulators required for nodulation. J Bacteriol. 2005;187:4562–4572. doi: 10.1128/JB.187.13.4562-4572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- Murphy ER, Payne SM. RhyB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infect Immun. 2007;75:3470–3477. doi: 10.1128/IAI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan D, Cazevielle C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, et al. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- O’Grady EP, Nguyen DT, Weisskopf L, Eberl L, Sokol PA. The Burkholderia cenocepacia LysR-type transcriptional regulator ShvR influences expression of quorum-sensing, protease, type II secretion, and afc genes. J Bacteriol. 2011;193:163–176. doi: 10.1128/JB.00852-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Vogel J. Small RNA functions in carbon metabolism and virulence of enteric pathogens. Front Cell Infect Microbiol. 2014;4:91. doi: 10.3389/fcimb.2014.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas G, Akritidis N, Bosilkovski M, Tsianos E. Brucellosis. N Engl J Med. 2005;352:2325–2336. doi: 10.1056/NEJMra050570. [DOI] [PubMed] [Google Scholar]
- Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos E. The new global map of human brucellosis. Lancet Infect Dis. 2006;6:91–99. doi: 10.1016/S1473-3099(06)70382-6. [DOI] [PubMed] [Google Scholar]
- Roop RM, II, Bellaire BH, Valderas MW, Cardelli JA. Adaptation of the brucellae to their intracellular niche. Mol Microbiol. 2004;52:621–630. doi: 10.1111/j.1365-2958.2004.04017.x. [DOI] [PubMed] [Google Scholar]
- Roop RM, II, Gaines JM, Anderson ES, Caswell CC, Martin DW. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol. 2009;198:221–238. doi: 10.1007/s00430-009-0123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DA, Byrne GA, O’Connell EP, Boland CA, Meijer WG. The LysR-type transcriptional regulator VirR is required for expression of the virulence gene vapA of Rhodococcus equi ATCC 33701. J Bacteriol. 2004;186:5576–5584. doi: 10.1128/JB.186.17.5576-5584.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- Seleem MN, Boyle SM, Sriranganathan N. Brucellosis: a re-emerging zoonosis. Vet Microbiol. 2010;140:392–398. doi: 10.1016/j.vetmic.2009.06.021. [DOI] [PubMed] [Google Scholar]
- Spratt BG, Hedge PJ, te Heesen S, Edelman A, Broome-Smith JK. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- Sun YH, de Jong MF, den Hartigh AB, Roux CM, Rolán HG, Tsolis RM. The small protein CydX is required for function of cytochrome bd oxidase in Brucella abortus. Front Cell Infect Microbiol. 2012;2:47. doi: 10.3389/fcimb.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Wang Y, Luo L. Transcriptional regulator LsrB of Sinorhizobium meliloti positively regulates the expression of genes involved in lipopolysaccharide biosynthesis. Appl Environ Microbiol. 2014;80:5265–5273. doi: 10.1128/AEM.01393-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Quesada O, Millán V, Nisa-Martínez R, Bardou F, Crespi M, Toro N, Jiménez-Zurdo JI. Independent activity of the homologous small regulatory RNAs AbcR1 and AbcR2 in the legume symbiont Sinorhizobium meliloti. PLoS ONE. 2013;8:e68147. doi: 10.1371/journal.pone.0068147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilms I, Voss B, Hess WR, Leichert LI, Narberhaus F. Small RNA-mediated control of the Agrobacterium tumefaciens GABA binding protein. Mol Microbiol. 2011;80:492–506. doi: 10.1111/j.1365-2958.2011.07589.x. [DOI] [PubMed] [Google Scholar]
- Wu Q, Pei J, Turse C, Ficht TA. Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized roles in virulence and survival. BMC Microbiol. 2006;6:102. doi: 10.1186/1471-2180-6-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zianni M, Tessanne K, Merighi M, Laguna R, Tabita FR. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech. 2006;17:103–113. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.