Abstract
The TGF-β superfamily signaling is involved in a variety of biological processes during embryogenesis and in adult tissue homeostasis. Faulty regulation of the signaling pathway that transduces the TGF-β superfamily signals accordingly leads to a number of ailments, such as cancer and cardiovascular, metabolic, urinary, intestinal, skeletal, and immune diseases. In recent years, a number of studies have elucidated the essential roles of TGF-βs and BMPs during neuronal development in the maintenance of appropriate innervation and neuronal activity. The new advancement implicates significant roles of the aberrant TGF-β superfamily signaling in the pathogenesis of neurological disorders. In this review, we compile a number of reports implicating the deregulation of TGF-β/BMP signaling pathways in the pathogenesis of cognitive and neurodegenerative disorders in animal models and patients. We apologize in advance that the review falls short of providing details of the role of TGF-β/BMP signaling or mechanisms underlying the pathogenesis of neurological disorders. The goal of this article is to reveal a gap in our knowledge regarding the association between TGF-β/BMP signaling pathways and neuronal tissue homeostasis and development and facilitate the research with a potential to develop new therapies for neurological ailments by modulating the pathways.
Keywords: cognitive disease, neurodegenerative disease, TGF-β, BMP, GDFs, neurons
Introduction
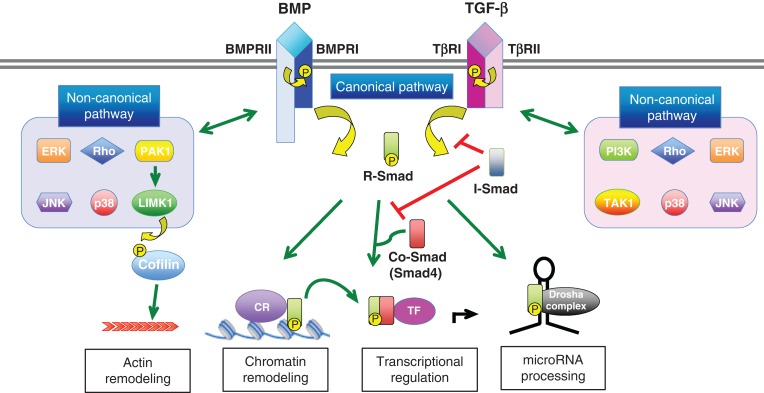
The transforming growth factor-β (TGF-β) superfamily of growth factors comprises approximately 20 evolutionarily conserved cytokines subdivided into several families, including TGF-βs, bone morphogenetic proteins (BMPs), activins, inhibins, nodals, and growth and differentiation factors (GDFs) (Table 1) [1]. They bind to two sets of ligand-specific receptors (Types I and II), which contain serine/threonine kinases. Receptor activation is transduced to the nucleus by Smad proteins, and this short cascade is known as the canonical pathway (Fig. 1) [2,3]. Smads 1, 5, and 8 are substrates of the Type I BMP receptor kinases, while Smads 2 and 3 respond to the Type I TGF-β receptor; together they are known as ‘receptor-specific’ R-Smads (Table 1). Type II receptors phosphorylate the Type I receptors, which then phosphorylate the R-Smads (Fig. 1). The activated R-Smads bind Smad4 (Co-Smad), and then the complex translocates to the nucleus and activates (or inhibits) transcription by binding to DNA-binding transcription factors (Fig. 1). The Smad-dependent signal can be negatively regulated by inhibitory Smads (I-Smads), Smad6 and Smad7 (Table 1 and Fig. 1). It is also known that the TGF-β/BMP signal can be transduced through a variety of intracellular Smad-independent pathways, including LIM domain kinase 1 (LIMK1)-actin depolymerizing factor (ADF)-cofilin and mitogen activated protein kinase pathways (known as ‘non-canonical’) [4] (Fig. 1). In this review article, we try to discuss the roles of TGF-β signaling pathways in neuronal diseases and to reveal a gap in our knowledge regarding the association between TGF-β/BMP signaling pathways and neuronal tissue homeostasis and development.
Table 1.
Molecules in the BMP, TGF-β, and activin signaling pathway in mammals and Drosophila
| Mammals | Drosophila | |
|---|---|---|
| Ligands | ||
| BMP | BMP2, BMP4 | Dpp |
| GDF5, GDF6, GDF7 | Gbb | |
| BMP5, BMP6, BMP7, BMP8 | ||
| GDF1, GDF2, GDF3 | Scw | |
| BMP9, BMP10 | ||
| BMP3, GDF10 | ||
| Nodal | ||
| TGF-β | TGF-β1, TGF-β2, TGF-β3 | Mav |
| Lefty1, Lefty2 | ||
| Activin | ActivinA, ActivinB, ActivinC, ActivinE | Act-β, Daw |
| InhivinA, InhibinC, InhibinE | Myo | |
| Type I receptors | ||
| BMP | ACVRL1 (ALK1), ACVRI (ALK2) | Sax |
| BMPR1A (ALK3), BMPR1B (ALK6) | Tkv | |
| TGF-β | TβRI (ALK5), ACVRL1 (ALK1) | BaboA, BaboB, BaboC |
| Activin | ACVR1B (ALK4), ACVR1C (ALK7) | BaboA, BaboB, BaboC |
| Type II receptors | ||
| BMP | BMPR2 | Wit |
| ACVRIIA (ACTRIIA), ACVRIIB (ACTRIIB) | Punt | |
| TGF-β | TβRII | Punt |
| Activin | ACVRIIA (ACTRIIA), ACVRIIB (ACTRIIB) | Punt |
| R-Smads | ||
| BMP | Smad1, Smad5, Smad8 | Mad |
| TGF-β | Smad2, Smad3 | Smox |
| Activin | Smad2, Smad3 | Smox |
| Co-Smads | ||
| BMP | Smad4 | Medea |
| TGF-β | Smad4 | Medea |
| Activin | Smad4 | Medea |
| I-Smads | ||
| BMP | Smad6, Smad7 | Dad |
| TGF-β | Smad7 | Dad |
| Activin | Smad7 | Dad |
Figure 1.
Signal transduction by TGF-βs and BMPs TGF-β and BMP ligands induce formation of heteromeric complex between specific Type II and Type I receptors. The Type II receptors transphosphorylate the Type I receptors and activate the Type I receptor kinases. The Type I receptor transmits the signal to the cell by phosphorylating receptor-regulated (R)-Smads, which form heteromeric complexes with Smad4 (common (Co)-Smad) and translocate in the nucleus where by interacting with other transcription factors regulate gene transcriptional responses, chromatin remodeling, and/or control of microRNA processing [‘canonical (or Smad-dependent) pathway’]. Inhibitory (I)-Smads; Smad6 and Smad7 inhibit activation of R-Smads. In addition, the activated Type I receptors can activate ‘non-canonical (or non-Smad) pathway’ via different effectors, such as extracellular signal-regulated kinases (ERK), c-Jun N-terminal kinase (JNK), p38, Rho, phosphoinositide 3-kinase (PI3K), transforming growth factor beta-activated kinase 1 (TAK1), p21 (RAC1) activated kinase 1 (PAK1), and LIM domain kinase 1 (LIMK1). CR and TF stand for chromatin remodeling protein and transcription factor, respectively.
TGF-β Superfamily in Neuronal Development and Function
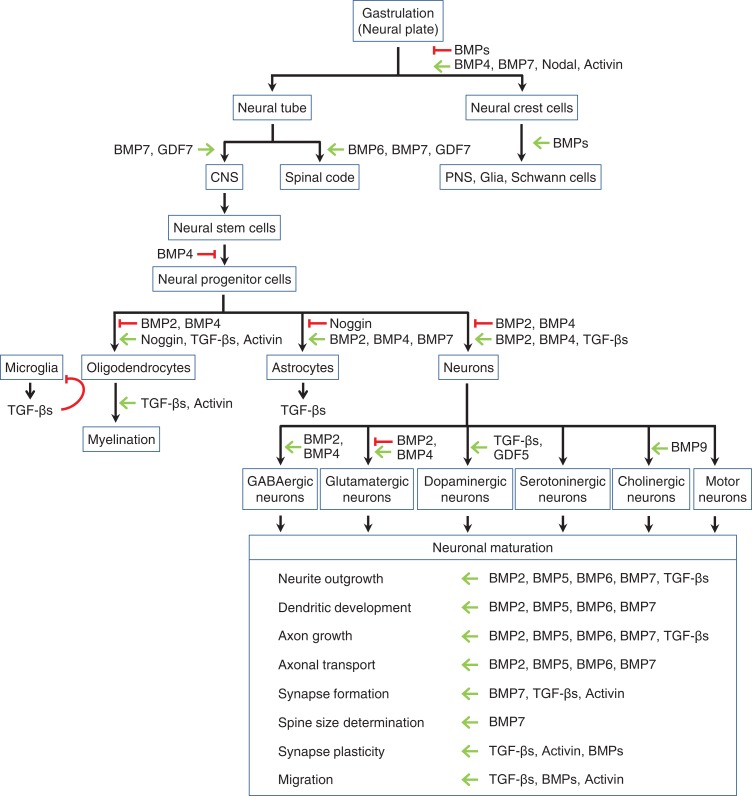
The BMP signaling pathway instructs key developmental events during early development of the nervous system and cell fate specification [5,6] (Fig. 2). The activities of BMPs and downstream effectors are dynamically and carefully regulated to play key roles during gastrulation and dorso-ventral patterning within the neural tube, the embryonic precursor of the brain and the spinal cord as well as the adult brain homeostasis and functions [7]. For example, an important cell type in embryos is the neural crest, which originates from the dorsal most region of the neural tube [8]. Neural crest cells give rise to several cell types in the peripheral nervous system (PNS), including glial and Schwann cells [8]. An intermediate level of BMP signaling is required for neural crest cell generation and cell fate choice [7] (Fig. 2). BMPs also control patterning of the dorsal spinal cord [6,9,10], while it has been reported that molecules in the canonical BMP signaling pathway, such as ligands (GDF7 and BMP7), receptors (BMPR1A/B), and signal transducers (Smad1/5/8) are essential for patterning of the ventral spinal code, dorsal spinal code neural axonal guidance, forebrain development, and cerebellar granule neuron development [6,9,10] (Fig. 2). In the central nervous system (CNS), BMPs, activins, TGF-βs, and GDFs contribute to the differentiation of neural stem cells and neural progenitor cells (NPCs) [5] (Fig. 2). For example, the BMP signaling promotes astrogliogenesis, and simultaneously inhibits oligodendroglial and neuronal lineages by activating downstream signals [7] (Fig. 2). Oligodendrogenesis is inhibited by BMP signaling and promoted by noggin [5] (Fig. 2). It has been shown that BMP signaling inhibits myelination through the inhibition of oligodendrogenesis [11,12] (Fig. 2). On the contrary, TGF-β and activin signaling promote both oligodendrogenesis and myelination [5,13] (Fig. 2). Thus, the TGF-β superfamily orchestrates patterning and determination of cell fate in the CNS, as well as the PNS.
Figure 2.
Roles of TGF-βs and BMPs in the neural development and function Signaling by the TGF-β family is required for proper neural development and function. Both inductive (green arrows) and inhibitory signals (red lines) of different ligands at various steps of neural differentiation are shown. CNS and PNS stand for central nervous system and peripheral nervous system, respectively.
The BMP signaling pathway regulates neurite outgrowth, dendritic development, and axon growth in neurons via various Smad-dependent and -independent signaling pathways, such as those involving LIMK1−cofilin, repulsive guidance molecule (RGM), and neurogenin [5,14,15] (Fig. 1). BMP signaling also facilitates axonal transport and organization of the microtubule network in neurons [16] (Fig. 2). The BMP receptors undergo endocytosis and dynein-dependent retrograde transport along the axon [17], implying that the trafficking of BMP receptor complexes might affect the cytoskeletal dynamics and axonal development (Fig. 2). TGF-β1−3 are found to increase the number and length of neurites [18] (Fig. 2).
The TGF-β superfamily also enhances synapse formation [19]. BMP7 and activin promote synapse development in hippocampal neurons [20] (Fig. 2). In addition, BMP signaling determines synaptic size [21], while astrocyte-derived TGF-β promotes synapse formation (Fig. 2) [22]. The roles of the TGF-β superfamily pathways in synapse formation have been characterized in the development of neuromuscular junctions (NMJ) in Drosophila [23]. Glass bottom boat (Gbb), a Drosophila ortholog of BMP (Table 1) secreted from muscle cells, promotes pre-synaptic formation at the NMJ [23]. Gbb binds the Type II receptor wishful thinking (Wit) and the Type I receptor thickvein (Tkv) or Saxophone (Sax) (Table 1) and regulates synaptogenesis via Smad1/5/8 (Mothers against Dpp or Mad) (Table 1) and Drosophila ortholog of LIMK1 (Limk). Maverick (Mav), a Drosophila ortholog of TGF-β, is secreted by glia, binds to Punt, a Type II receptor (Table 1) expressed on the surface of muscle cells, and potentiates Gbb-depending signaling, hence promoting synaptic growth at the NMJ [24]. The TGF-β signaling pathway also regulates synaptic plasticity and memory [22]. TGF-β2 enhances synaptic transmission in primary cultured hippocampus neurons [25]. Central nervous-specific knockout (KO) of TGF-β1 in mouse exhibits a reduction of dendritic spine density, impaired long-term potentiation (LTP), and facilitated long-term depression (LTD) in CA1, CA2, and CA3 synapses in the hippocampus [26]. Bmpr1a and Bmpr1b double KO mice show reduced synaptic transmission [21,27,28]. Activin increases the number of synaptic contacts and the length of dendritic spine necks [20], suggesting that activin improves synaptic plasticity and LTP by modifying synaptic morphology [29]. Based on a transgenic mouse expressing a dominant negative mutant of the activin receptor Type 1B (Acvr1b) (also known as ALK4), it has been suggested that activin signals suppress inhibitory synaptic transmission [30,31]. In summary, multiple conserved members of the TGF-β superfamily of cytokines play key roles both in synaptogenesis and in plasticity of the nervous system. In the next paragraph, we list the instances in which TGF-β superfamily pathways have been linked to the pathogenesis of different neurological disorders.
TGF-β/BMP Signaling in Developmental Disorders
Fragile X syndrome
Fragile X syndrome (FXS) (OMIM No. 300624) is the most common cause of inherited intellectual disability and the genetic cause of autism spectrum disorders (ASDs). Inactivating mutation of the fragile X mental retardation 1 (FMR1) gene, which locates at Xq27.3, a ‘fragile site’, or the expansion of CGG repeats (more than 200 repeats) in the 5′ UTR of FMR1 causes FXS [32]. It is estimated that 1 in 4000 men and 1 in 8000 women are affected worldwide [32,33]. It is thought that the X chromosome with the fragile X site is more often inactivated compared with the nonaffected X chromosome [33]. FXS is diagnosed within the first 3 years of life by genetic testing. The FXS patients exhibit cognitive impairment, anxiety, hyperactivity, and autistic behavior as well as characteristic physical features, such as large and prominent ears, macroorchidism, and ovarian insufficiency [32,33]. Females with FXS tend to present less affected but wider range of phenotypic characteristics than males, depending on the inactivation ratio of the fragile X site [32]. The FMR1 gene encodes fragile X mental retardation protein (FMRP), an RNA binding protein that, in a majority of cases, represses translation [34]. Several genome-wide analyses to identify FMRP target mRNAs have been performed [35,36]. Based on the identification of FMRP targets, various pharmacological or genetic strategies to reduce the expression or activity of FMRP targets and rescue the pathogenesis have been employed in FXS model animals [34]. However, currently there is no effective therapy for FXS. Recently, the transcript of the Type 2 BMP receptor (BMPR2) gene has been identified as a novel target of FMRP [37] (Table 2). In FXS patients- and Fmr1-KO mice-derived brains and neurons, BMPR2 protein amount is increased [37], resulting in LIMK1 activation and increased phosphorylation of cofilin [37]. The BMP-LIMK1-cofilin pathway regulates dendritogenesis and axon growth in CNS neurons [14,38]. It has been also reported that overexpression of wild-type or a constitutively active mutation in LIMK1 causes increased growth cone formation, axonal outgrowth, abnormal dendritogenesis, and impaired neural migration in primary neurons and in transgenic mice expressing a constitutively active Limk1 [39,40]. Interestingly, both Fmr1-KO mice and human FXS patients exhibit an increase of the spine density and the number of immature spines [41–44]. Genetic or pharmacological inhibition of the Smad-independent BMP-BMPR2-LIMK1-cofilin pathway rescues the neuron morphological and behavioral abnormalities in Fmr1-KO mice [37,45]. Retrograde BMP signaling also plays an important role in synapse growth and stability via both Smad-dependent and -independent (LIMK1-cofilin-dependent) pathways at the Drosophila NMJ [46,47]. Like FXS patients and Fmr1-KO mice, Drosophila Fmr1 (dFmr1) mutants have an increased number of synaptic boutons and branches at the NMJ [48–50]. These morphological changes of the synapses at the NMJ are accompanied by an increase in the crawling velocity of larvae [37]. Genetic or pharmacological inhibition of the Wit-Limk pathway in larvae ameliorates the neuromorphological and locomotive abnormalities in dFmr1 mutants [37,45], suggesting that FMRP-dependent regulation of the BMPR2-LIMK1-cofilin axis is an evolutionarily conserved pathway important for synapse development, and that aberrant activation of this pathway is one of the underlying causes of the FXS pathogenesis.
Table 2.
Deregulation of TGF-β signaling pathway associated with various neurological disorders
| Disease | Affected pathway | Mechanism | Impact on pathogenesis | Reference |
|---|---|---|---|---|
| Fragile X syndrome | BMP signaling (U) | Decreased translational inhibition of BMPR2 by FMRP | Increased dendritogenesis and spine number | [37] |
| Williams syndrome | BMP signaling (D?) | Deletion of LIMK1 | Abnormal synapse morphology and function | [51] |
| Angelman syndrome | BMP signaling (U) | Decreased BMPR1A degradation by UBE3A | Abnormal spine formation | [52] |
| Mowat-Wilson syndrome | BMP signaling (U or D) | Deregulation of SIP1 | Defective neural crest formation | [53,54] |
| Troyer syndrome | BMP signaling (U) | Decreased endocytotic degradation of BMPR2 by SPARTIN | Increased synapse formation and neurodegeneration | [55,56] |
| SPG3A | BMP signaling (U) | Decreased endocytotic degradation of BMPR2 by ATL1 | Abnormal microtubule dynamics and axon guidance? | [57–59] |
| SPG4 | BMP signaling (U) | Decreased endocytotic degradation of BMPR2 by SPAST | Abnormal microtubule dynamics and axon guidance? | [57,60] |
| SPG6 | BMP signaling (U) | Decreased endocytotic degradation of BMPR2 by NIPA1 | Abnormal synapse formation, microtubule dynamics, and axon guidance | [16,57,60,61] |
| Alzheimer’s disease | TGF-β signaling (U or D) | Deregulation of TGF-β1, Smad7, nuclear R-Smad/Co-Smad. | Aβ accumulation, aberrant microglia activation, and increased neurodegeneration | [62–71] |
| BMP signaling (U) | Elevation of BMP4 and BMP6 | Inhibition of neurogenesis | [72–75] | |
| Parkinson’s disease | TGF-β signaling (U or D) | Deregulation of ligands and receptors | Degeneration of DA neuron | [76–79] |
| BMP signaling (D) | Deregulation of ligands and receptors | Inhibition of DA neuron differentiation and protection from neurodegeneration | [80–87] | |
| Huntington’s disease | TGF-β signaling (U or D) | Deregulation of neuronal and circulating TGF-β1 | Increased neurodegeneration? | [88–92] |
| BMP signaling (U) | N.D. | Increased dendritic branching and synapse size | [93] | |
| Amyotrophic lateral sclerosis | TGF-β signaling (U or D) | Deregulation of ligands and Smad2/3 protein stability | Inhibition of neuroprotection and increased axon degeneration | [94–98] |
| BMP signaling (U or D) | Deregulation of receptor trafficking | Impaired synapse growth | [99–102] | |
| Multiple sclerosis | TGF-β signaling (U) | N.D. | Inflammation? | [103–105] |
| BMP signaling (U) | Deregulation of BMP2, BMP4, BMP5 and noggin | Inhibition of neuronal differentiation and myelination | [106–112] |
U, D, and N.D. stand for ‘Up’, ‘Down’ and ‘not determined’, respectively.
Williams syndrome
Williams syndrome (WS) (OMIM No. 194050) is a developmental disorder characterized by mental retardation or learning difficulties, cardiovascular problems, particular facial features, and a typical personality that includes overfriendliness and anxiety [113]. The prevalence is 1 in 20,000, occurring sporadically within the general population [114]. Post-mortem layer V/VI cortical neurons in WS patients’ brain and iPSC-derived neurons show longer dendrites and an increased number of spines and synapses [115]. WS is caused by a deletion of the chromosome 7p11.23 locus, which comprises ~25 protein coding genes, including CYLN2, ELN, GTF2I, GTF2IRD1, FZD9, and LIMK1 [116]. LIMK1 is considered as one of the critical genes in the pathogenesis of WS [51] because Limk1-KO mice exhibit abnormalities in synapse morphology and function, enhanced hippocampus LTP, increased locomotor activities, and impaired leaning [117]. Furthermore, both Limk1-KO mice and humans with the WS genetic mutation present spine abnormalities and cognitive impairment [117–119]. Additionally, overexpression of a dominant negative form of Limk1 causes aberrant axonal guidance and neuronal migration in the embryonic mouse brain [40]. As described in the FXS section above, aberrant activation of the BMPR2-LIMK1 pathway is implicated in FXS, while inhibition or inactivation of LIMK1 leads to WS [37] (Table 2). It is intriguing to speculate that restoring the BMP-BMPR2-LIMK1-cofilin activity might be able to ameliorate the pathogenesis of WS.
Angelman syndrome
Angelman syndrome (AS) (OMIM No. 105830) is a neurodevelopment disorder resulted mainly from deficient expression or function of the maternally inherited allele of UBE3A gene on the chromosome 15q11.2-q13 locus. The incidence of AS is between 1 in 15,000 and 1 in 20,000 [120]. The clinical features of AS are microcephaly, severe intellectual deficit, speech impairment, epilepsy, electroencephalogram abnormalities, ataxic movements, tongue protrusion, paroxysms of laughter, abnormal sleep patterns, and hyperactivity [121]. UBE3A encodes an E3 ubiquitin ligase which plays an important role in the neuronal ubiquitin-proteasome pathway and synaptic development [122–125]. Several proteins have been identified as targets of UBE3A, such as ECT2, p53, p27, HR23A, Arc, and ephexin-5 [121]. AS model mice show reduced synapse formation and experience-dependent synapse remodeling [124,126]. Mice with maternal mutation in Ube3a develop defects in context-dependent learning and LTP [127]. The neurons from the Ube3a maternal-deficient mice exhibit shorter spines and reduced dendritic spine density [125]. dUbe3a-null mutants show a reduced dendritic branching and growth of the terminal dendritic processes of sensory neurons, as well as a decreased number of satellite boutons at the NMJ in Drosophila [52,128]. Along with morphological abnormalities, deficits in locomotor behavior, circadian rhythms, and long-term memory are also observed in the dUbe3a mutants [128]. Recently, Tkv, Drosophila ortholog of BMPR1A (Table 1), has been identified as a dUbe3a target [52]. The increased activation of the Smad pathway and bouton formation in Ube3a mutants are rescued by Tkv mutation, suggesting that aberrant activation of the BMP signaling pathway is responsible for the synaptic abnormalities in AS [52]. BMPR1A (ALK3) was also identified as a target of UBE3A in human [52] (Table 2). UBE3A knockdown (KD) results in increased BMPR1A protein followed by activation of the Smad pathway, indicating that the BMP-BMPR1A signaling axis is critical for the pathogenesis of AS. Moreover, a point mutation in UBE3A has also been identified in patients with autism spectrum disorders (ASD) [129,130], suggesting that the UBE3A-BMPR1A-BMP signaling pathway might be associated with the pathogenesis of a broad range of neurodevelopmental disorders, including ASD.
Mowat-Wilson syndrome
Mowat-Wilson syndrome (MWS) (OMIM No. 235730) is a mental retardation multiple congenital anomaly syndrome characterized by typical fancies, severe mental retardation, epilepsy, and variable congenital malformations, including Hirschsprung disease (HSCR), genital anomalies, congenital heart disease (CHD), and agenesis of the corpus callosum (ACC) [131]. MWS is a very rare disease first described in 1998 [132], and the prevalence is between 1 in 50,000–70,000 [133]. Heterozygous mutation in the Smad-interacting protein-1 (SIP1 also known as ZEB2 or ZFHX1B) gene causes MWS [131]. SIP1 represses Smad signaling in response to activation by BMPs and leads to the induction of neural fate [134–136] (Table 2). The BMP-Smad pathway is crucial for the generation of neural crest cells [137,138], which gives rise to a variety of cell populations in the PNS and contributes to the formation of forebrain and midbrain [139]. In zebrafish, KD of two Sip1 ortholog results in a loss of vagal/postotic neural crest cell derivatives [53]. In Sip1-KO mouse embryos, depletion of neural crest cells and inhibition in the neural crest of Msh homeobox 1 (Msx1), a BMP target gene, can be observed, suggesting that functional BMP signaling is impaired in these mice [54]. Sip1 also acts as an essential modulator of CNS myelination by inducing Smad7, an antagonist of the BMP-Smad pathway, specifically in oligodendrocytes, of which it promotes the differentiation [12]. However, the molecular mechanisms by which the SIP1 mutation affects BMP signaling in MWS patients and how SIP1-BMP signaling leads to the MWS pathogenesis remain unclear.
Hereditary spastic paraplegia
Hereditary spastic paraplegia (HSP), also known as familial spastic paraplegia, is a group of neurodegenerative disorders that leads to spastic weakness of the lower extremities [140]. HSP is classified into two groups, ‘pure HSP’ and ‘complex HSP’, according to whether the HSP occurs alone or is accompanied by additional neurological syndromes, such as cognitive impairment, dementia, epilepsy, and polyneuropathy, respectively [141]. HSP is rare and its prevalence is estimated from 1.27 to 9.6 per 100,000 [140]. Mutations in more than 40 distinct loci and 21 spastic paraplegia (SPG) genes have been associated with HSP. SPG genes encode proteins that are involved in the maintenance of corticospinal tract neurons [142] including Spartin (SPG20), Atlastin-1 (SPG3A), Spastin (SPG4), and non-imprinted in Prader-Willi/AS1 (NIPA1, also known as SPG6), some of which are inhibitory to BMP signaling [55,57]. The characteristics of different classes of HSP and their associated genes, such as Troyer syndrome (SPG20), SPG3A, SPG4, and SPG6, are discussed below.
Troyer syndrome (SPG20)
Troyer syndrome (OMIM No. 275,900) is a child-onset autosomal recessive complex HSP caused by mutation in SPART, which encodes Spartin [143]. Troyer syndrome is characterized by a progressive spastic partial paralysis of the lower limbs, motor speech disorder, and pseudobulbar palsy (inability to control facial movements), distal muscular atrophy, motor and cognitive delay, short stature, and subtle skeletal abnormalities, with both developmental and degenerative features [144]. Only 21 patients in the USA [143,144], 6 in Oman [145], and 3 in the Philippines [9] have been reported. Spartin is known to have multiple functions, including cytokinesis, endosomal trafficking and degradation of the epidermal growth factor receptor (EGFR), lipid/cholesterol metabolism, and mitochondrial function [147–152]. Interestingly, Spartin inhibits the BMP signaling pathway [55,56]. Spartin-KO mice develop progressive impairments in motor function similar to Troyer syndrome [153]. The cerebral cortical neurons from Spartin-KO mice exhibit increased branching, density of dendrites, and elongated axon length in cerebral cortical neurons [153]. In the Drosophila NMJ, Spartin localizes presynaptically [56]. Spartin homozygous mutants in Drosophila show an increase of the BMP signaling, overgrowth of synapses, and progressive neurodegeneration [56]. Mutation in Wit or the BMP inhibitor Daughters against dpp (Dad) (Table 1), an ortholog of Smad6, rescues the synapse number and neurodegeneration phenotype, suggesting that Spartin negatively controls BMP signaling by promoting endocytic degradation of Wit [56]. Therefore, these observations suggest that abnormal activation of BMP signaling is linked to the pathogenesis of Troyer syndrome (Table 2).
Spastic paraplegia-3A (SPG3A)
Mutations in the Atlastin-1 (ATL1) gene are the most common cause of pure and complex HSP with early onset at around 4 years old [141]. The ATL1 is a GTPase of the dynamin superfamily implicated in facilitating membrane interactions, fission, and fusion [154,155]. The mutants exhibit reduced GTPase activity and a prominently disrupted morphology of the endoplasmic reticulum (ER) network [156]. Knockdown of ATL1 increases (while overexpression represses) BMP signaling in zebrafish [157]. ATL1 co-localizes with the Type I BMP receptor [157] and associates with BMPR2 [57]. HSP-related mutations in ATL1 interfere with BMPR2 trafficking to the cell surface and attenuate BMP signaling [57]. On the contrary, null mutations of Drosophila dATL1 [58] and of ATL1 in mammalian cells [59] show augmented BMP activities, indicating that ATL1 may also modulate trafficking, and similarly inhibit BMP signaling in mammals by interfering with microtubule dynamics and axon guidance [58,59] (Table 2).
Spastic paraplegia-4 (SPG4)
Mutations in the Spastin (SPAST) gene are commonly found in autosomal dominant pure and complex HSP. SPG4 patients show sensory disturbances, sphincter dysfunction, tremor, cognitive impairment, dementia, and ataxia. SPG4 is age-dependent, and the onset peak is 10 and 30 years old [141]. SPAST interacts with microtubules and promotes microtubule disassembly, which is essential for axon growth, branching, and neuronal plasticity [158]. Mutations in SPAST cause abnormal microtubule disassembly [159]. The dSpastin mutant Drosophila shows disorganized microtubules and disrupted proximal-distal transmission gradient along axon branches, which causes increased BMP signaling at the NMJ [57,60] (Table 2).
Spastic paraplegia-6 (SPG6)
SPG6 is a teenage-onset form of pure HSP, with some patients with complex HSP showing memory deficit and epilepsy [141]. All NIPA1 mutations in SPG6 are missense mutations that affect trafficking of proteins and trapping in the ER [160]. Patients lacking one copy of the NIPA1 gene seldom develop clinical HSP, thus it is unlikely that SPG6 is caused by haploinsufficiency of NIPA1, but rather by a ‘toxic gain of function’. NIPA1 binds ATL1 and promotes efficient cell surface trafficking of ATL1 [160]. NIPA1 also directly binds BMPR2 [55]. NIPA1 mutants, which are prone to ER trapping, reduce endocytosis, lysosomal degradation, and recycling of BMPR2, and thus augment the amount of BMPR2 on the cell surface [55] (Table 2). Transgenic rats expressing a NIPA1 mutant in neurons show an increase of BMPR2 in the spinal cord [61]. Interestingly, a null mutation of Spichthyin (Spict), a Drosophila ortholog of NIPA1, causes a distal axonal abnormality with synaptic overgrowth at the NMJ, due to the attenuation of Wit endocytosis [16]. Spict preferentially localizes in early endosomes and presynaptically regulates not only synaptic bouton formation at the NMJ but also microtubule maintenance and axonal transport, by inhibiting BMP signaling [16].
Taken together, abnormal BMPR2 endocytosis and trafficking, followed by atypical activation of BMP signaling, is closely linked to the pathology of several subtypes of HSP.
TGF-β/BMP Signaling in Neurodegenerative Disorders
The TGF-β family plays an extensive role in the survival of neurons [161,162]. Levels of the ligands and the receptors of TGF-β family are regulated following neural injury and repair for proper function of CNS [163–165]. Hippocampal astrocytes from old rats secrete higher amounts of TGF-βs compared with postnatal or young rats [166]. The level of BMP4 also increases in an age-dependent manner in both human and mouse hippocampus [167], suggesting that the changes of activities of the TGF-β family of signaling affect the homeostasis of aging brains by altering the function and the niches of neurons. Thus, it is not surprising that the TGF-β family is implicated in the pathogenesis of age-related diseases, such as Alzheimer’s and Parkinson’s disease, as described below.
Alzheimer’s disease
Alzheimer’s disease (AD) (OMIM No. 104300) is an age-related, progressive neurodegenerative disease characterized by progressive cognitive impairment and pathological abnormalities, such as loss of neurons, amyloid plaque, and hyperphosphorylated tau in intracellular neurofibrillary tangles in the brain [168]. In addition to the neurodegenerative hallmarks, synaptic plasticity and neuronal integrity are also impaired in AD brains [169]. AD affects approximately 29.8 million people and is a major health concern worldwide [170]. Although pathological mutations in the amyloid precursor protein (APP), presenilin-1, and presenilin-2 are found in familial AD, more than 95% of AD patients do not carry these gene mutations [171]. The molecular mechanisms of pathogenesis of AD remain largely unclear.
It is likely that growth factors, such as TGF-βs and BMPs, are involved in the pathogenesis of AD [172] (Table 2). The phosphorylation of Smad2/3 is decreased in the AD brain, indicative of impaired TGF-β signaling [62]. Nuclear Smad2, Smad3, and Smad4 are also decreased in the temporal cortex of AD patients [63]. TGF-β receptor Type 2 (TβR2) expression is decreased in neurons in AD patients [64]. It has been proposed that impaired TGF-β signaling in neurons contributes to β amyloid (Aβ) accumulation, microglia activation [65], and neurodegeneration [64]. Accordingly, exogenous TGF-β1 induces activation of microglia and clearance of Aβ and protects against Aβ-induced synapse loss, neurodegeneration, and apoptosis [66–69]. However, conflicting results have been also reported. In the brain of a mouse model of familial AD, there is an increase of TGF-β1 and Smad7, an antagonist of TGF-β signal, which are thought to mediate neural apoptosis [70] (Table 2). Additionally, TGF-β1 also affects the microglia and ameliorates neuroinflammation in AD [71]. Because TGF-β1 seems to act as both survival and apoptotic factor depending on the context, the exact role of TGF-β1 in AD is still unclear.
The BMP signaling pathway is also involved in AD-related neurodegeneration. In the hippocampus of AD patients, BMP6, but not BMP2 or BMP7, is augmented [72]. Aβ exposure induces BMP6, which then inhibits proliferation of NPCs [72]. Increase of BMP4 is also observed in AD mouse brains [73]. These observations suggest that augmented BMP6 is involved in AD-related altered neurogenesis. BMP4 is increased and noggin, an antagonist of BMPs, is decreased in the dentate gyrus of the AD mouse model and the apolipoprotein E (ApoE)-KO mice [74,75]. Several observations suggest the implication of TGF-β/BMP signaling in the development and progression of AD. To further understand the molecular mechanism of the pathogenesis of AD and to develop therapeutics for AD, a more precise role of TGF-β/BMP signaling in AD brains should be clarified in the future.
Parkinson’s disease
Parkinson’s disease (PD) is the second most common neurodegenerative disorder after AD, affecting 6.2 million people globally [170]. The clinical symptoms of PD include tremor at rest, rigidity, bradykinesia, postural abnormalities and a freezing phenomenon [173]. The pathological findings in PD include a loss of nigrostriatal dopaminergic (DA) neurons with a subsequent loss of the neurotransmitter dopamine in the corpus striatum, an area of the brain which is critical for the control of movement [174]. One of the pathological hallmarks of PD is the presence of intracellular protein aggregates called Lewy bodies [174]. Approximately 5% of PD patients carry a familial form of PD and several causal genes have been identified, including leucine rich repeat kinase 2 (LRRK2), Parkinsonism associated deglycase (PARK7), PTEN-induced putative kinase 1 (PINK1), parkin RBR E3 ubiquitin protein ligase (PRKN), and synuclein alpha (SNCA), but the genetic cause of 95% of PD is unknown [175].
It has been reported that the TGF-β superfamily signaling pathway controls DA neuron development and survival [76] (Table 2). Genetic studies suggest an association of single nucleotide polymorphisms (SNPs) in the TGFB2 gene with PD [77]. Tgfb2+/−;Tgfb3−/− and Tgfb2−/−;Tgfb3+/− mice show a significant reduction of DA neurons at E14.5 [76]. Tgfb2+/−, Tgfb3−/−, and Smad3−/− mice show postnatal or age-dependent loss of DA neurons [76]. Adult Tgfb2+/− mice show more significant loss of striatal dopamine compared with young mice [176]. These data suggest that TGF-βs play more critical role in adult brain function and homeostasis. Neuron-specific expression of a kinase-inactive mutant TβR2 in mice displays age-dependent neurodegeneration in the nigrostriatal system [78]. TGF-β activation by overexpressing constitutively active TβR1 aborts degeneration of DA neurons in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced PD mouse model [78]. In the cerebrospinal fluid of PD patients, the amount of TGF-β1 and TGF-β2 is increased [79].
BMP signaling is also involved in DA neuron differentiation and protection from neurodegeneration [80–84]. Catecholaminergic neuron-specific knock-in of a dominant negative form of BMPR2 (a truncation mutant) results in a decrease of tyrosine hydroxylase (TH), which catalyzes the formation of L-dihydroxyphenylalanine (L-DOPA), the rate-limiting step in the biosynthesis of DA, and a number of neurons in substantia nigra compacta [80]. Pretreatment with BMP7 reduces neurodegeneration in 6-hydroxydopamine (6-OHDA)-induced PD [81]. BMP7 heterozygous KO (Bmp7+/−) rats exhibit an increased sensitivity of adult DA neurons to methamphetamine [84]. BMP7 induces DA neuron differentiation from mesencephalic precursor cells in vitro [82]. BMP2 induces neurite outgrowth through Smad activation via BMPR1B in SH-SY5Y cells, which are capable of differentiating to DA neurons [83].
Finally, glial-derived neurotrophic factor (GDNF) heterozygous KO (Gdnf+/−) mice show an accelerated decline of DA neurons during aging [85]. Based on these observations, GDNF was used in clinical trials for PD [86]. GDF5 and GDF15 also play important roles in DA neuron development and survival [76]. Transplantation of GDF5-expressing CHO cells into the striatum exhibits neuroprotective and neurorestorative effects on DA neurons in a 6-OHDA-induced PD rat [87]. Thus, impairment of TGF-β superfamily signaling is closely associated with the pathogenesis of PD.
Huntington’s disease
Huntington’s disease (HD) (OMIM No. 143100) is the most common inherited neurodegenerative disorder caused by the expansion of a polyglutamine (polyQ) stretch within the coding sequence of Huntingtin (HTT) [177]. HD is characterized by motor, cognitive, and emotional defects [178]. The incidence is 0.38 per 100,000 persons per year [179]. The expansion of a polyQ repeat causes HTT protein aggregation [180]. There is conflicting evidence on how aggregated HTT causes the neurotoxicity [181], but, in general, the polyQ repeat mediates neurotoxicity through impaired vesicle trafficking and axonal transport, altered proteasomal degradation, mitochondrial dysfunction, and transcriptional deregulation [182]. Recently it has been found that an accumulation of pathogenic HTT protein in nerve terminals interferes with endosomal recycling and leads to buildup of early endosomal signaling compartments, such as BMP signaling molecules in Drosophila [93]. The augmented BMP signaling molecules trigger a robust overgrowth of synaptic boutons at NMJ [93] (Table 2). Disruption of BMP signaling rescues abnormal synapse formation and neurotoxicity of the pathogenic HTT, suggesting that the aberrant activation of BMP signaling is involved in the neuronal dysfunction in HD [93]. Indeed, extensive dendritic branching with increased number and size of spines are observed in striatal spiny neurons in HD patients [183].
Deregulation of TGF-β signaling appears to be involved in the pathogenesis of HD [88–92]. The amount of circulating TGF-β1 in asymptomatic HD patients is decreased [88], while it is increased in symptomatic HD patients [88] (Table 2). The amount of TGF-β1 in cortical neurons is also reduced in post-mortem brain samples from both asymptomatic and symptomatic HD patients as well as HD model mice [89]. Transcriptome analysis using iPSCs and neural stem cells (NSCs) from HD patients reveals that TGF-β signaling molecules are increased in HD [90]. An increase of the TGF-β signaling pathway is also observed in striatal cell lines expressing HTT mutant and iPSC-derived neural progenitor cells (NPCs) [92]. Smad7, an antagonist of TGF-β signaling, is significantly decreased in these cells, further supporting the increase of TGF-β signaling in HD neurons. It is possible that the boost in TGF-β signaling might be a compensatory response to neurodegeneration [92], or that it predisposes the NSCs toward quiescence during the neurodegeneration process [91]. It has also been reported that Smad3 binds to the promoter region of the HTT gene and activates transcription [92]. Further studies are required to clarify molecular link between the TGF-β family signaling to the pathogenesis of HD.
Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS) (OMIM No. 105400) is a progressive neurodegenerative disease which affects the upper and lower motor neurons [184]. ALS is characterized by muscle stiffness, twitching, weakness, and atrophy throughout the body [184]. ALS affects about 2 in 100,000 individuals [184]. Mutations in >25 genes, including superoxide dismutase 1 (SOD1), alsin Rho guanine nucleotide exchange factor (ALS2), fused in sarcoma (FUS), TAR DNA-binding protein 43 (TDP43, also known as TARDBP), and chromosome 9 open reading frame 72 (C9orf72), have been identified in association with ALS, but for 90%–95% of the patients with ALS, the causal genes are unknown [185]. Irregular TGF-β signaling has been implicated in ALS pathogenesis [94–97] (Table 2). TGF-β1 is elevated in astrocytes in the spinal cord of Sod1 mutant mice and sporadic ALS patients [94]. TGF-β1−3 are also augmented in the muscle of ALS patients and Sod1 mutant mice [95]. More TGF-β1 is also detected in the serum and plasma of ALS patients [96]. Nuclear phosopho-Smad2/3 (P-Smad2/3), readout of the TGF-β activity, is increased in neurons and glial cells in the spinal cord of Sod1 mutant mice as well as both familial and sporadic ALS patients [97]. Administration of TGF-β inhibitor ameliorates ALS progression in Sod1 mutant mice [94]. These reports suggest that astrocyte-derived TGF-βs inhibit neuroprotective responses, promote motor neuron axon degeneration, and contribute to ALS.
TDP43 is one of the ALS-related genes [186,187]. Pathological TDP43 can lead to deregulation of the TGF-β and BMP signaling pathways in ALS [98,99]. Mutant TDP43 protein found in ALS is prone to aggregate [188]. Interestingly, in some sporadic ALS patients, wild-type TDP43 protein is also aggregated [189,190]. The intracytoplasmic inclusion bodies containing TDP43 are associated with frontotemporal dementia and cognitive impairment in ALS [191,192]. It has also been reported that Smurf2, an E3 ubiquitin ligase that promotes ubiquitin-dependent degradation of Smad2/3 proteins, and phosphorylated Smad2/3 proteins are colocalized with TDP43 and ubiquitin within neuronal intracytoplasmic inclusions in the spinal cord and medulla oblongata of sporadic ALS patients, suggesting that the TGF-β signaling pathway is decreased in neuron [98] (Table 2). In Drosophila model of ALS, BMP signaling pathway genes (Smox, SkpA, and Sax) (Table 1) are elevated in CNS of dTDP43 mutant Drosophila according to the genome-wide transcriptome analysis [99]. Aberrant expression of TDP43 in Drosophila motor neurons mediates defective endosomal trafficking of Tkv and reduced synaptic BMP signal, leading to the impaired synaptic growth at NMJ accompanied by the abnormal larval crawling [100]. These reports suggest that deregulation of TDP43 can cause the neurological impairment in ALS pathogenesis through the impairment of the TGF-β/BMP signaling.
Mutations in the Vesicle-associated membrane protein-associated protein B (VAPB) gene are found in patients with familial ALS [193]. VAPB protein is reduced in the spinal cord of sporadic ALS patients [194]. VAPB is a Type II integral membrane protein that mainly localizes at the ER and implicated in a variety of cellular processes, including ER stress response and the unfolded protein response (UPR). A Drosophila expressing the mutant form of dVAP33A, a Drosophila ortholog of VAPB, shows reduced phospho-Mad both at NMJ and in CNS, indicative of reduced BMP signaling [101]. While it is unclear how dVAP33A modulates the BMP signaling, it is proposed that VAPB mutant might be involved in the abnormal UPR [102].
It is essential to elucidate the molecular mechanisms by which the deregulation of TGF-β signaling/BMP signaling pathway promoting ALS pathogenesis in order to develop a treatment for ALS.
Multiple sclerosis
Multiple sclerosis (MS) is an autoimmune disease with demyelination in CNS neurons [195] which affects over 2.3 million people[170]. Typically, MS patients are diagnosed in young adulthood with a higher incidence in women [195]. Clinical symptoms include blurred vision, muscle weakness and spasms as well as motor problems [195]. There are two major clinical courses of MS: (i) relapsing-remitting MS and (ii) progressive MS [195]. Despite these subtypes, all patients with MS have progressive and irreversible neurological disabilities [195]. Genome-wide association studies demonstrate that SNPs in the major histocompatibility complex (MHC) class II and DR beta 1 (HLA-DRB1) are highly associated with MS [196,197], suggesting that chronic neuroinflammation and failure of the myelin-producing cells, followed by neurodegeneration, are involved in the pathogenesis of MS [198]. Myelin is critical for the propagation of nervous impulses and axonal maintenance and is synthesized as the plasma membrane of the oligodendrocytes in the CNS [199]. During neural development, myelin and oligodendrocytes are generated from oligodendrocyte progenitors under the control of various growth factors [200]. MS plaques are characterized by the presence of immune cell infiltration, demyelination, death of mature oligodendrocytes, axonal damage and neurodegeneration [195,201]. NPCs and oligodendrocyte precursor cells (OPCs) are present in the MS lesions [202–204], suggesting that the failure of maturation of NPCs and OPCs is involved in MS pathogenesis.
The BMP signaling pathway is involved in NPC differentiation into astrocytes with concurrent suppression of oligodendroglial differentiation in adult brains, thus, deregulation of BMP signaling can contribute to demyelination in MS [106] (Table 2). Both BMPs and BMP antagonist noggin are potentially involved in MS pathology through the functions in neuronal differentiation, myelination, and immune system regulation [107]. It is thought that noggin, which is in a niche of NPCs, is associated with neurogenesis [205,206]. NPCs treated with or overexpressing noggin can differentiate into astrocytes, oligodendrocytes, and mature neurons [205,206]. Noggin is highly expressed in T cells, and the amount is reduced in MS patients [108], suggesting that the reduced production of noggin by T cells might contribute to demyelination in MS. It has also been reported that BMP4 is increased in the caudal cerebellar peduncle of rats in ethidium bromide-induced demyelinated lesions [109]. BMP4 and BMP5 are expressed at the lesions in post-mortem brain tissues from MS patients, and BMP5 expression is augmented in MS patients compared to healthy controls [110] (Table 2). T cell-derived BMP2, BMP4, and BMP5 are increased in peripheral blood mononuclear cells from MS patients [111]. The BMP signaling regulates myelination and demyelination through oligodendrogenesis [11,12]. Additionally, abnormal trafficking of BMP signaling molecules may also contribute to MS pathology [112]. Recent genome-wide association studies demonstrate that SNPs at the 16p13 locus containing C-type lectin domain family 16, member A (CLEC16A) increase the risk of MS as well as other autoimmune diseases [196,197,207,208]. CLEC16A protein in the white matter and peripheral blood mononuclear cells is increased in MS patients [209]. It has been suggested that CLEC16A promotes late endosomal maturation to disrupt the HLA-II antigen presentation pathway in MS [209]. In Drosophila, a mutation in Ema, an ortholog of CLEC16A, causes abnormal synaptic growth and defective protein trafficking [112]. Ema is an endosomal membrane protein that interacts with the class C Vps-HOPS complex to promote endosomal maturation [112]. The Ema mutant fails to form mature late endosomes and lysosomes. In the Ema mutant, Tkv, phospho-Mad, and synaptic bouton number are all increased, but they can all be reversed by overexpression of human CLEC16A [112]. These results suggest that Ema and CLE16A inhibit BMP signaling through endolysosomal trafficking and degradation of the signaling components [112]. Compared with the BMP pathway, studies linking the TGF-β pathway to MS are limited. TGF-β is augmented in peripheral blood and cerebrospinal fluid of MS patients [103]. The amount of TGF-β and the activity of the disease are linearly correlated [104,105], suggesting that the amount of circulating TGF-β can be used as a biomarker for MS. MS is yet another example of neurodegenerative disorder associated with the deregulation of both TGF-β and BMP signaling pathways.
Anxiety, Depression, and Dementia
Anxiety, depression, and dementia are common neurological disorders that are also linked to the deregulation of TGF-β signaling [2–218]. Forebrain-specific Bmpr2-KO mice exhibit reduced anxiety-related behavior [2]. Forebrain-specific activin transgenic mice also show decreased anxiety-related behavior [211]. Major depressive disorder patients show a reduction in TGF-β in the serum or a polymorphism in the TGF-β gene [212–214]. Antidepressant treatment increases TGF-β [215]. These reports suggest that TGF-β superfamily signaling pathways modulate psychiatric disorders. Blocking BMP signaling by either transgenic or pharmacological methods significantly enhances hippocampus-dependent learning memory behaviors [216], suggesting that BMP signaling is involved in learning and memory processes. Charged multivesicular body protein 2B (CHMP2B) mutations are related with frontotemporal dementia [217]. Overexpression of Rab8, a negative regulator of TGF-β signaling, rescued synapse overgrowth phenotype in Chmp2b mutant Drosophila [218], implying that an overactive TGF-β signaling pathway is involved in frontotemporal dementia pathogenesis.
Conclusion
The TGF-β family of growth factors plays essential roles during embryonic development and in the regulation of tissue homeostasis. Here we summarized studies describing the association of deregulation of TGF-β signaling with neuronal development and neurological disorders. Abundant evidence in both invertebrates and vertebrates indicates that the TGF-β pathways play important roles in the maintenance of neuron and spine homeostasis. Causal links between deregulation of TGF-β signaling pathway and human disorders such as cancer and cardiovascular or bone diseases have been well documented, and a number of therapeutic molecules have been generated. Compared to other human diseases, the current knowledge of how TGF-β pathways lead to various neurological abnormalities is limited. Many studies are rather descriptive than mechanistic. We hope that this article will provide a basis for future research aimed at providing more mechanistic insights into neurological abnormalities stemming from deregulation of TGF-β signaling, which are essential for the future development of targeted therapies.
Funding
This work was supported by the grants from the NIH (Nos. HL093154 and HL108317) and the Catalyst Award from UCSF Clinical and Translational Science Institute (CTSI).
Acknowledgements
We thank Dr. G. Lagna (Cardiovascular Research Institute, University of California, San Francisco, USA) for critical reading of the review. We also thank all current and past members of the Hata lab for their contributions to the scientific work performed from our laboratory.
References
- 1. Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol 2007, 8: 970–982. [DOI] [PubMed] [Google Scholar]
- 2. Heldin CH, Moustakas A. Signaling receptors for TGF-beta family members. Cold Spring Harb Perspect Biol 2016, 8: a022053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol 2012, 13: 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang YE. Non-Smad signaling pathways of the TGF-beta family. Cold Spring Harb Perspect Biol 2017, 9: a022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meyers EA, Kessler JA. TGF-beta family signaling in neural and neuronal differentiation, development, and function. Cold Spring Harb Perspect Biol 2017, 9: a022244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hegarty SV, O’Keeffe GW, Sullivan AM. BMP-Smad 1/5/8 signalling in the development of the nervous system. Prog Neurobiol 2013, 109: 28–41. [DOI] [PubMed] [Google Scholar]
- 7. Bond AM, Bhalala OG, Kessler JA. The dynamic role of bone morphogenetic proteins in neural stem cell fate and maturation. Dev Neurobiol 2012, 72: 1068–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kalcheim C, Kumar D. Cell fate decisions during neural crest ontogeny. Int J Dev Biol 2017, 61: 195–203. [DOI] [PubMed] [Google Scholar]
- 9. Le Dreau G, Marti E. The multiple activities of BMPs during spinal cord development. Cell Mol Life Sci 2013, 70: 4293–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu A, Niswander LA. Bone morphogenetic protein signalling and vertebrate nervous system development. Nat Rev Neurosci 2005, 6: 945–954. [DOI] [PubMed] [Google Scholar]
- 11. See JM, Grinspan JB. Sending mixed signals: bone morphogenetic protein in myelination and demyelination. J Neuropathol Exp Neurol 2009, 68: 595–604. [DOI] [PubMed] [Google Scholar]
- 12. Weng Q, Chen Y, Wang H, Xu X, Yang B, He Q, Shou W, et al. . Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron 2012, 73: 713–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dutta DJ, Zameer A, Mariani JN, Zhang J, Asp L, Huynh J, Mahase S, et al. . Combinatorial actions of Tgfbeta and Activin ligands promote oligodendrocyte development and CNS myelination. Development 2014, 141: 2414–2428. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 14. Lee-Hoeflich ST, Causing CG, Podkowa M, Zhao X, Wrana JL, Attisano L. Activation of LIMK1 by binding to the BMP receptor, BMPRII, regulates BMP-dependent dendritogenesis. EMBO J 2004, 23: 4792–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhong J, Zou H. BMP signaling in axon regeneration. Curr Opin Neurobiol 2014, 27: 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Shaw WR, Tsang HT, Reid E, O’Kane CJ. Drosophila spichthyin inhibits BMP signaling and regulates synaptic growth and axonal microtubules. Nat Neurosci 2007, 10: 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith RB, Machamer JB, Kim NC, Hays TS, Marques G. Relay of retrograde synaptogenic signals through axonal transport of BMP receptors. J Cell Sci 2012, 125: 3752–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Unsicker K, Meier C, Krieglstein K, Sartor BM, Flanders KC. Expression, localization, and function of transforming growth factor-beta s in embryonic chick spinal cord, hindbrain, and dorsal root ganglia. J Neurobiol 1996, 29: 262–276. [DOI] [PubMed] [Google Scholar]
- 19. Poon VY, Choi S, Park M. Growth factors in synaptic function. Front Synaptic Neurosci 2013, 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shoji-Kasai Y, Ageta H, Hasegawa Y, Tsuchida K, Sugino H, Inokuchi K. Activin increases the number of synaptic contacts and the length of dendritic spine necks by modulating spinal actin dynamics. J Cell Sci 2007, 120: 3830–3837. [DOI] [PubMed] [Google Scholar]
- 21. Xiao L, Michalski N, Kronander E, Gjoni E, Genoud C, Knott G, Schneggenburger R. BMP signaling specifies the development of a large and fast CNS synapse. Nat Neurosci 2013, 16: 856–864. [DOI] [PubMed] [Google Scholar]
- 22. Diniz LP, Matias IC, Garcia MN, Gomes FC. Astrocytic control of neural circuit formation: highlights on TGF-beta signaling. Neurochem Int 2014, 78: 18–27. [DOI] [PubMed] [Google Scholar]
- 23. Upadhyay A, Moss-Taylor L, Kim MJ, Ghosh AC, O’Connor MB. TGF-beta family signaling in Drosophila. Cold Spring Harb Perspect Biol 2017, 9: a031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fuentes-Medel Y, Ashley J, Barria R, Maloney R, Freeman M, Budnik V. Integration of a retrograde signal during synapse formation by glia-secreted TGF-beta ligand. Curr Biol 2012, 22: 1831–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fukushima T, Liu RY, Byrne JH. Transforming growth factor-beta2 modulates synaptic efficacy and plasticity and induces phosphorylation of CREB in hippocampal neurons. Hippocampus 2007, 17: 5–9. [DOI] [PubMed] [Google Scholar]
- 26. Koeglsperger T, Li S, Brenneis C, Saulnier JL, Mayo L, Carrier Y, Selkoe DJ, et al. . Impaired glutamate recycling and GluN2B-mediated neuronal calcium overload in mice lacking TGF-beta1 in the CNS. Glia 2013, 61: 985–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aberle H, Haghighi AP, Fetter RD, McCabe BD, Magalhaes TR, Goodman CS. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron 2002, 33: 545–558. [DOI] [PubMed] [Google Scholar]
- 28. McCabe BD, Marques G, Haghighi AP, Fetter RD, Crotty ML, Haerry TE, Goodman CS, et al. . The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction. Neuron 2003, 39: 241–254. [DOI] [PubMed] [Google Scholar]
- 29. Krieglstein K, Zheng F, Unsicker K, Alzheimer C. More than being protective: functional roles for TGF-beta/activin signaling pathways at central synapses. Trends Neurosci 2011, 34: 421–429. [DOI] [PubMed] [Google Scholar]
- 30. Zheng F, Adelsberger H, Muller MR, Fritschy JM, Werner S, Alzheimer C. Activin tunes GABAergic neurotransmission and modulates anxiety-like behavior. Mol Psychiatry 2009, 14: 332–346. [DOI] [PubMed] [Google Scholar]
- 31. Zheng F, Puppel A, Huber SE, Link AS, Eulenburg V, van Brederode JF, Muller CP, et al. . Activin controls ethanol potentiation of inhibitory synaptic transmission through GABAA receptors and concomitant behavioral sedation. Neuropsychopharmacology 2016, 41: 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saldarriaga W, Tassone F, Gonzalez-Teshima LY, Forero-Forero JV, Ayala-Zapata S, Hagerman R. Fragile X syndrome. Colomb Med (Cali) 2014, 45: 190–198. [PMC free article] [PubMed] [Google Scholar]
- 33. Santos AR, Kanellopoulos AK, Bagni C. Learning and behavioral deficits associated with the absence of the fragile X mental retardation protein: what a fly and mouse model can teach us. Learn Mem 2014, 21: 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Richter JD, Bassell GJ, Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci 2015, 16: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 2001, 107: 489–499. [DOI] [PubMed] [Google Scholar]
- 36. Ascano M Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, et al. . FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 2012, 492: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kashima R, Roy S, Ascano M, Martinez-Cerdeno V, Ariza-Torres J, Kim S, Louie J, et al. . Augmented noncanonical BMP type II receptor signaling mediates the synaptic abnormality of fragile X syndrome. Sci Signal 2016, 9: ra58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamauchi K, Varadarajan SG, Li JE, Butler SJ. Type Ib BMP receptors mediate the rate of commissural axon extension through inhibition of cofilin activity. Development 2013, 140: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosso S, Bollati F, Bisbal M, Peretti D, Sumi T, Nakamura T, Quiroga S, et al. . LIMK1 regulates Golgi dynamics, traffic of Golgi-derived vesicles, and process extension in primary cultured neurons. Mol Biol Cell 2004, 15: 3433–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xie J, Li X, Zhang W, Chai X, Huang Y, Li K, Cheng X, et al. . Aberrant expression of LIMK1 impairs neuronal migration during neocortex development. Histochem Cell Biol 2017, 147: 471–479. [DOI] [PubMed] [Google Scholar]
- 41. Grossman AW, Elisseou NM, McKinney BC, Greenough WT. Hippocampal pyramidal cells in adult Fmr1 knockout mice exhibit an immature-appearing profile of dendritic spines. Brain Res 2006, 1084: 158–164. [DOI] [PubMed] [Google Scholar]
- 42. McKinney BC, Grossman AW, Elisseou NM, Greenough WT. Dendritic spine abnormalities in the occipital cortex of C57BL/6 Fmr1 knockout mice. Am J Med Genet B Neuropsychiatr Genet 2005, 136B: 98–102. [DOI] [PubMed] [Google Scholar]
- 43. Irwin SA, Idupulapati M, Gilbert ME, Harris JB, Chakravarti AB, Rogers EJ, Crisostomo RA, et al. . Dendritic spine and dendritic field characteristics of layer V pyramidal neurons in the visual cortex of fragile-X knockout mice. Am J Med Genet 2002, 111: 140–146. [DOI] [PubMed] [Google Scholar]
- 44. Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex 2000, 10: 1038–1044. [DOI] [PubMed] [Google Scholar]
- 45. Kashima R, Redmond PL, Ghatpande P, Roy S, Kornberg TB, Hanke T, Knapp S, et al. . Hyperactive locomotion in a Drosophila model is a functional readout for the synaptic abnormalities underlying fragile X syndrome. Sci Signal 2017, 10: eaai8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Piccioli ZD, Littleton JT. Retrograde BMP signaling modulates rapid activity-dependent synaptic growth via presynaptic LIM kinase regulation of cofilin. J Neurosci 2014, 34: 4371–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eaton BA, Davis GW. LIM Kinase1 controls synaptic stability downstream of the type II BMP receptor. Neuron 2005, 47: 695–708. [DOI] [PubMed] [Google Scholar]
- 48. Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, et al. . Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell 2001, 107: 591–603. [DOI] [PubMed] [Google Scholar]
- 49. Pan L, Zhang YQ, Woodruff E, Broadie K. The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr Biol 2004, 14: 1863–1870. [DOI] [PubMed] [Google Scholar]
- 50. Myrick LK, Deng PY, Hashimoto H, Oh YM, Cho Y, Poidevin MJ, Suhl JA, et al. . Independent role for presynaptic FMRP revealed by an FMR1 missense mutation associated with intellectual disability and seizures. Proc Natl Acad Sci USA 2015, 112: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hoogenraad CC, Akhmanova A, Galjart N, De Zeeuw CI. LIMK1 and CLIP-115: linking cytoskeletal defects to Williams syndrome. Bioessays 2004, 26: 141–150. [DOI] [PubMed] [Google Scholar]
- 52. Li W, Yao A, Zhi H, Kaur K, Zhu YC, Jia M, Zhao H, et al. . Angelman syndrome protein Ube3a regulates synaptic growth and endocytosis by inhibiting BMP signaling in Drosophila. PLoS Genet 2016, 12: e1006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Delalande JM, Guyote ME, Smith CM, Shepherd IT. Zebrafish sip1a and sip1b are essential for normal axial and neural patterning. Dev Dyn 2008, 237: 1060–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D, Higashi Y. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am J Hum Genet 2003, 72: 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tsang HT, Edwards TL, Wang X, Connell JW, Davies RJ, Durrington HJ, O’Kane CJ, et al. . The hereditary spastic paraplegia proteins NIPA1, spastin and spartin are inhibitors of mammalian BMP signalling. Hum Mol Genet 2009, 18: 3805–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nahm M, Lee MJ, Parkinson W, Lee M, Kim H, Kim YJ, Kim S, et al. . Spartin regulates synaptic growth and neuronal survival by inhibiting BMP-mediated microtubule stabilization. Neuron 2013, 77: 680–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhao J, Hedera P. Hereditary spastic paraplegia-causing mutations in atlastin-1 interfere with BMPRII trafficking. Mol Cell Neurosci 2013, 52: 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Summerville JB, Faust JF, Fan E, Pendin D, Daga A, Formella J, Stern M, et al. . The effects of ER morphology on synaptic structure and function in Drosophila melanogaster. J Cell Sci 2016, 129: 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhao G, Zhu PP, Renvoise B, Maldonado-Baez L, Park SH, Blackstone C. Mammalian knock out cells reveal prominent roles for atlastin GTPases in ER network morphology. Exp Cell Res 2016, 349: 32–44. [DOI] [PubMed] [Google Scholar]
- 60. Ball RW, Peled ES, Guerrero G, Isacoff EY. BMP signaling and microtubule organization regulate synaptic strength. Neuroscience 2015, 291: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Watanabe F, Arnold WD, Hammer RE, Ghodsizadeh O, Moti H, Schumer M, Hashmi A, et al. . Pathogenesis of autosomal dominant hereditary spastic paraplegia (SPG6) revealed by a rat model. J Neuropathol Exp Neurol 2013, 72: 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ueberham U, Ueberham E, Gruschka H, Arendt T. Altered subcellular location of phosphorylated Smads in Alzheimer’s disease. Eur J Neurosci 2006, 24: 2327–2334. [DOI] [PubMed] [Google Scholar]
- 63. Ueberham U, Hilbrich I, Ueberham E, Rohn S, Glockner P, Dietrich K, Bruckner MK, et al. . Transcriptional control of cell cycle-dependent kinase 4 by Smad proteins—implications for Alzheimer’s disease. Neurobiol Aging 2012, 33: 2827–2840. [DOI] [PubMed] [Google Scholar]
- 64. Tesseur I, Zou K, Esposito L, Bard F, Berber E, Can JV, Lin AH, et al. . Deficiency in neuronal TGF-beta signaling promotes neurodegeneration and Alzheimer’s pathology. J Clin Invest 2006, 116: 3060–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tichauer JE, von Bernhardi R. Transforming growth factor-beta stimulates beta amyloid uptake by microglia through Smad3-dependent mechanisms. J Neurosci Res 2012, 90: 1970–1980. [DOI] [PubMed] [Google Scholar]
- 66. Wyss-Coray T, Lin C, Yan F, Yu GQ, Rohde M, McConlogue L, Masliah E, et al. . TGF-beta1 promotes microglial amyloid-beta clearance and reduces plaque burden in transgenic mice. Nat Med 2001, 7: 612–618. [DOI] [PubMed] [Google Scholar]
- 67. Caraci F, Battaglia G, Bruno V, Bosco P, Carbonaro V, Giuffrida ML, Drago F, et al. . TGF-beta1 pathway as a new target for neuroprotection in Alzheimer’s disease. CNS Neurosci Ther 2011, 17: 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fisichella V, Giurdanella G, Platania CB, Romano GL, Leggio GM, Salomone S, Drago F, et al. . TGF-beta1 prevents rat retinal insult induced by amyloid-beta (1-42) oligomers. Eur J Pharmacol 2016, 787: 72–77. [DOI] [PubMed] [Google Scholar]
- 69. Diniz LP, Tortelli V, Matias I, Morgado J, Bergamo Araujo AP, Melo HM, Seixas da Silva GS, et al. . Astrocyte transforming growth factor beta 1 protects synapses against Abeta oligomers in Alzheimer’s disease model. J Neurosci 2017, 37: 6797–6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Salins P, He Y, Olson K, Glazner G, Kashour T, Amara F. TGF-beta1 is increased in a transgenic mouse model of familial Alzheimer’s disease and causes neuronal apoptosis. Neurosci Lett 2008, 430: 81–86. [DOI] [PubMed] [Google Scholar]
- 71. Huang WC, Yen FC, Shie FS, Pan CM, Shiao YJ, Yang CN, Huang FL, et al. . TGF-beta1 blockade of microglial chemotaxis toward Abeta aggregates involves SMAD signaling and down-regulation of CCL5. J Neuroinflammation 2010, 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Crews L, Adame A, Patrick C, Delaney A, Pham E, Rockenstein E, Hansen L, et al. . Increased BMP6 levels in the brains of Alzheimer’s disease patients and APP transgenic mice are accompanied by impaired neurogenesis. J Neurosci 2010, 30: 12252–12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kwak YD, Hendrix BJ, Sugaya K. Secreted type of amyloid precursor protein induces glial differentiation by stimulating the BMP/Smad signaling pathway. Biochem Biophys Res Commun 2014, 447: 394–399. [DOI] [PubMed] [Google Scholar]
- 74. Tang J, Song M, Wang Y, Fan X, Xu H, Bai Y. Noggin and BMP4 co-modulate adult hippocampal neurogenesis in the APP(swe)/PS1(DeltaE9) transgenic mouse model of Alzheimer’s disease. Biochem Biophys Res Commun 2009, 385: 341–345. [DOI] [PubMed] [Google Scholar]
- 75. Li G, Bien-Ly N, Andrews-Zwilling Y, Xu Q, Bernardo A, Ring K, Halabisky B, et al. . GABAergic interneuron dysfunction impairs hippocampal neurogenesis in adult apolipoprotein E4 knockin mice. Cell Stem Cell 2009, 5: 634–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hegarty SV, Sullivan AM, O’Keeffe GW. Roles for the TGFbeta superfamily in the development and survival of midbrain dopaminergic neurons. Mol Neurobiol 2014, 50: 559–573. [DOI] [PubMed] [Google Scholar]
- 77. Goris A, Williams-Gray CH, Foltynie T, Brown J, Maranian M, Walton A, Compston DA, et al. . Investigation of TGFB2 as a candidate gene in multiple sclerosis and Parkinson’s disease. J Neurol 2007, 254: 846–848. [DOI] [PubMed] [Google Scholar]
- 78. Tesseur I, Nguyen A, Chang B, Li L, Woodling NS, Wyss-Coray T, Luo J. Deficiency in neuronal TGF-beta signaling leads to nigrostriatal degeneration and activation of TGF-beta signaling protects against MPTP neurotoxicity in mice. J Neurosci 2017, 37: 4584–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vawter MP, Dillon-Carter O, Tourtellotte WW, Carvey P, Freed WJ. TGFbeta1 and TGFbeta2 concentrations are elevated in Parkinson’s disease in ventricular cerebrospinal fluid. Exp Neurol 1996, 142: 313–322. [DOI] [PubMed] [Google Scholar]
- 80. Chou J, Harvey BK, Ebendal T, Hoffer B, Wang Y. Nigrostriatal alterations in bone morphogenetic protein receptor II dominant negative mice. Acta Neurochir Suppl 2008, 101: 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Harvey BK, Mark A, Chou J, Chen GJ, Hoffer BJ, Wang Y. Neurotrophic effects of bone morphogenetic protein-7 in a rat model of Parkinson’s disease. Brain Res 2004, 1022: 88–95. [DOI] [PubMed] [Google Scholar]
- 82. Lee JY, Koh HC, Chang MY, Park CH, Lee YS, Lee SH. Erythropoietin and bone morphogenetic protein 7 mediate ascorbate-induced dopaminergic differentiation from embryonic mesencephalic precursors. Neuroreport 2003, 14: 1401–1404. [DOI] [PubMed] [Google Scholar]
- 83. Hegarty SV, Sullivan AM, O’Keeffe GW. BMP2 and GDF5 induce neuronal differentiation through a Smad dependant pathway in a model of human midbrain dopaminergic neurons. Mol Cell Neurosci 2013, 56: 263–271. [DOI] [PubMed] [Google Scholar]
- 84. Chou J, Luo Y, Kuo CC, Powers K, Shen H, Harvey BK, Hoffer BJ, et al. . Bone morphogenetic protein-7 reduces toxicity induced by high doses of methamphetamine in rodents. Neuroscience 2008, 151: 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Boger HA, Middaugh LD, Huang P, Zaman V, Smith AC, Hoffer BJ, Tomac AC, et al. . A partial GDNF depletion leads to earlier age-related deterioration of motor function and tyrosine hydroxylase expression in the substantia nigra. Exp Neurol 2006, 202: 336–347. [DOI] [PubMed] [Google Scholar]
- 86. Sullivan AM, Toulouse A. Neurotrophic factors for the treatment of Parkinson’s disease. Cytokine Growth Factor Rev 2011, 22: 157–165. [DOI] [PubMed] [Google Scholar]
- 87. Costello DJ, O’Keeffe GW, Hurley FM, Sullivan AM. Transplantation of novel human GDF5-expressing CHO cells is neuroprotective in models of Parkinson’s disease. J Cell Mol Med 2012, 16: 2451–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chang KH, Wu YR, Chen YC, Chen CM. Plasma inflammatory biomarkers for Huntington’s disease patients and mouse model. Brain Behav Immun 2015, 44: 121–127. [DOI] [PubMed] [Google Scholar]
- 89. Battaglia G, Cannella M, Riozzi B, Orobello S, Maat-Schieman ML, Aronica E, Busceti CL, et al. . Early defect of transforming growth factor beta1 formation in Huntington’s disease. J Cell Mol Med 2011, 15: 555–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ring KL, An MC, Zhang N, O’Brien RN, Ramos EM, Gao F, Atwood R, et al. . Genomic analysis reveals disruption of striatal neuronal development and therapeutic targets in human Huntington’s disease neural stem cells. Stem Cell Rep 2015, 5: 1023–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kandasamy M, Couillard-Despres S, Raber KA, Stephan M, Lehner B, Winner B, Kohl Z, et al. . Stem cell quiescence in the hippocampal neurogenic niche is associated with elevated transforming growth factor-beta signaling in an animal model of Huntington disease. J Neuropathol Exp Neurol 2010, 69: 717–728. [DOI] [PubMed] [Google Scholar]
- 92. Bowles KR, Stone T, Holmans P, Allen ND, Dunnett SB, Jones L. SMAD transcription factors are altered in cell models of HD and regulate HTT expression. Cell Signal 2017, 31: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Akbergenova Y, Littleton JT. Pathogenic Huntington Alters BMP Signaling and synaptic growth through local disruptions of endosomal compartments. J Neurosci 2017, 37: 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Endo F, Komine O, Fujimori-Tonou N, Katsuno M, Jin S, Watanabe S, Sobue G, et al. . Astrocyte-derived TGF-beta1 accelerates disease progression in ALS mice by interfering with the neuroprotective functions of microglia and T cells. Cell Rep 2015, 11: 592–604. [DOI] [PubMed] [Google Scholar]
- 95. Si Y, Kim S, Cui X, Zheng L, Oh SJ, Anderson T, AlSharabati M, et al. . Transforming growth factor beta (TGF-beta) Is a muscle biomarker of disease progression in ALS and correlates with Smad expression. PLoS One 2015, 10: e0138425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ilzecka J, Stelmasiak Z, Dobosz B. Transforming growth factor-Beta 1 (tgf-Beta 1) in patients with amyotrophic lateral sclerosis. Cytokine 2002, 20: 239–243. [DOI] [PubMed] [Google Scholar]
- 97. Nakamura M, Ito H, Wate R, Nakano S, Hirano A, Kusaka H. Phosphorylated Smad2/3 immunoreactivity in sporadic and familial amyotrophic lateral sclerosis and its mouse model. Acta Neuropathol 2008, 115: 327–334. [DOI] [PubMed] [Google Scholar]
- 98. Nakamura M, Kaneko S, Wate R, Asayama S, Nakamura Y, Fujita K, Ito H, et al. . Regionally different immunoreactivity for Smurf2 and pSmad2/3 in TDP-43-positive inclusions of amyotrophic lateral sclerosis. Neuropathol Appl Neurobiol 2013, 39: 144–156. [DOI] [PubMed] [Google Scholar]
- 99. Hazelett DJ, Chang JC, Lakeland DL, Morton DB. Comparison of parallel high-throughput RNA sequencing between knockout of TDP-43 and its overexpression reveals primarily nonreciprocal and nonoverlapping gene expression changes in the central nervous system of Drosophila. G3 (Bethesda) 2012, 2: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Deshpande M, Feiger Z, Shilton AK, Luo CC, Silverman E, Rodal AA. Role of BMP receptor traffic in synaptic growth defects in an ALS model. Mol Biol Cell 2016, 27: 2898–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ratnaparkhi A, Lawless GM, Schweizer FE, Golshani P, Jackson GR. A Drosophila model of ALS: human ALS-associated mutation in VAP33A suggests a dominant negative mechanism. PLoS One 2008, 3: e2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kanekura K, Nishimoto I, Aiso S, Matsuoka M. Characterization of amyotrophic lateral sclerosis-linked P56S mutation of vesicle-associated membrane protein-associated protein B (VAPB/ALS8). J Biol Chem 2006, 281: 30223–30233. [DOI] [PubMed] [Google Scholar]
- 103. Link J, Soderstrom M, Olsson T, Hojeberg B, Ljungdahl A, Link H. Increased transforming growth factor-beta, interleukin-4, and interferon-gamma in multiple sclerosis. Ann Neurol 1994, 36: 379–386. [DOI] [PubMed] [Google Scholar]
- 104. Carrieri PB, Provitera V, Bruno R, Perrella M, Tartaglia G, Busto A, Perrella O. Possible role of transforming growth factor-beta in relapsing-remitting multiple sclerosis. Neurol Res 1997, 19: 599–600. [DOI] [PubMed] [Google Scholar]
- 105. Bertolotto A, Capobianco M, Malucchi S, Manzardo E, Audano L, Bergui M, Bradac GB, et al. . Transforming growth factor beta1 (TGFbeta1) mRNA level correlates with magnetic resonance imaging disease activity in multiple sclerosis patients. Neurosci Lett 1999, 263: 21–24. [DOI] [PubMed] [Google Scholar]
- 106. Grinspan JB. Bone morphogenetic proteins: inhibitors of myelination in development and disease. Vitam Horm 2015, 99: 195–222. [DOI] [PubMed] [Google Scholar]
- 107. Eixarch H, Calvo-Barreiro L, Montalban X, Espejo C. Bone morphogenetic proteins in multiple sclerosis: role in neuroinflammation. Brain Behav Immun 2017, doi:10.1016/j.bbi.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 108. Urshansky N, Mausner-Fainberg K, Auriel E, Regev K, Bornstein NM, Karni A. Reduced production of noggin by immune cells of patients with relapsing-remitting multiple sclerosis. J Neuroimmunol 2011, 232: 171–178. [DOI] [PubMed] [Google Scholar]
- 109. Zhao C, Fancy SP, Magy L, Urwin JE, Franklin RJ. Stem cells, progenitors and myelin repair. J Anat 2005, 207: 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Deininger M, Meyermann R, Schluesener H. Detection of two transforming growth factor-beta-related morphogens, bone morphogenetic proteins-4 and -5, in RNA of multiple sclerosis and Creutzfeldt-Jakob disease lesions. Acta Neuropathol 1995, 90: 76–79. [DOI] [PubMed] [Google Scholar]
- 111. Mausner-Fainberg K, Urshansky N, Regev K, Auriel E, Karni A. Elevated and dysregulated bone morphogenic proteins in immune cells of patients with relapsing-remitting multiple sclerosis. J Neuroimmunol 2013, 264: 91–99. [DOI] [PubMed] [Google Scholar]
- 112. Kim S, Wairkar YP, Daniels RW, DiAntonio A. The novel endosomal membrane protein Ema interacts with the class C Vps-HOPS complex to promote endosomal maturation. J Cell Biol 2010, 188: 717–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mervis CB, Klein-Tasman BP. Williams syndrome: cognition, personality, and adaptive behavior. Ment Retard Dev Disabil Res Rev 2000, 6: 148–158. [DOI] [PubMed] [Google Scholar]
- 114. Morris CA, Demsey SA, Leonard CO, Dilts C, Blackburn BL. Natural history of Williams syndrome: physical characteristics. J Pediatr 1988, 113: 318–326. [DOI] [PubMed] [Google Scholar]
- 115. Chailangkarn T, Trujillo CA, Freitas BC, Hrvoj-Mihic B, Herai RH, Yu DX, Brown TT, et al. . A human neurodevelopmental model for Williams syndrome. Nature 2016, 536: 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Meyer-Lindenberg A, Mervis CB, Berman KF. Neural mechanisms in Williams syndrome: a unique window to genetic influences on cognition and behaviour. Nat Rev Neurosci 2006, 7: 380–393. [DOI] [PubMed] [Google Scholar]
- 117. Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu WY, et al. . Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron 2002, 35: 121–133. [DOI] [PubMed] [Google Scholar]
- 118. Frangiskakis JM, Ewart AK, Morris CA, Mervis CB, Bertrand J, Robinson BF, Klein BP, et al. . LIM-kinase1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell 1996, 86: 59–69. [DOI] [PubMed] [Google Scholar]
- 119. Tassabehji M, Metcalfe K, Fergusson WD, Carette MJ, Dore JK, Donnai D, Read AP, et al. . LIM-kinase deleted in Williams syndrome. Nat Genet 1996, 13: 272–273. [DOI] [PubMed] [Google Scholar]
- 120. Fiumara A, Pittala A, Cocuzza M, Sorge G. Epilepsy in patients with Angelman syndrome. Ital J Pediatr 2010, 36: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Buiting K, Williams C, Horsthemke B. Angelman syndrome—insights into a rare neurogenetic disorder. Nat Rev Neurol 2016, 12: 584–593. [DOI] [PubMed] [Google Scholar]
- 122. Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993, 75: 495–505. [DOI] [PubMed] [Google Scholar]
- 123. Sato M. Early origin and evolution of the Angelman syndrome ubiquitin ligase Gene Ube3a. Front Cell Neurosci 2017, 11: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, et al. . The Angelman syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell 2010, 140: 704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet 2008, 17: 111–118. [DOI] [PubMed] [Google Scholar]
- 126. Margolis SS, Salogiannis J, Lipton DM, Mandel-Brehm C, Wills ZP, Mardinly AR, Hu L, et al. . EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell 2010, 143: 442–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, et al. . Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 1998, 21: 799–811. [DOI] [PubMed] [Google Scholar]
- 128. Lu Y, Wang F, Li Y, Ferris J, Lee JA, Gao FB. The Drosophila homologue of the Angelman syndrome ubiquitin ligase regulates the formation of terminal dendritic branches. Hum Mol Genet 2009, 18: 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Yi JJ, Berrios J, Newbern JM, Snider WD, Philpot BD, Hahn KM, Zylka MJ. An Autism-linked mutation disables phosphorylation control of UBE3A. Cell 2015, 162: 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, et al. . The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014, 515: 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zweier C, Albrecht B, Mitulla B, Behrens R, Beese M, Gillessen-Kaesbach G, Rott HD, et al. . ‘Mowat-Wilson’ syndrome with and without Hirschsprung disease is a distinct, recognizable multiple congenital anomalies-mental retardation syndrome caused by mutations in the zinc finger homeo box 1B gene. Am J Med Genet 2002, 108: 177–181. [PubMed] [Google Scholar]
- 132. Mowat DR, Croaker GD, Cass DT, Kerr BA, Chaitow J, Ades LC, Chia NL, et al. . Hirschsprung disease, microcephaly, mental retardation, and characteristic facial features: delineation of a new syndrome and identification of a locus at chromosome 2q22-q23. J Med Genet 1998, 35: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Adam MP, Conta J, Bean LJH Mowat-Wilson Syndrome. 1993.
- 134. Lerchner W, Latinkic BV, Remacle JE, Huylebroeck D, Smith JC. Region-specific activation of the Xenopus brachyury promoter involves active repression in ectoderm and endoderm: a study using transgenic frog embryos. Development 2000, 127: 2729–2739. [DOI] [PubMed] [Google Scholar]
- 135. Nitta KR, Tanegashima K, Takahashi S, Asashima M. XSIP1 is essential for early neural gene expression and neural differentiation by suppression of BMP signaling. Dev Biol 2004, 275: 258–267. [DOI] [PubMed] [Google Scholar]
- 136. van Grunsven LA, Taelman V, Michiels C, Verstappen G, Souopgui J, Nichane M, Moens E, et al. . XSip1 neuralizing activity involves the co-repressor CtBP and occurs through BMP dependent and independent mechanisms. Dev Biol 2007, 306: 34–49. [DOI] [PubMed] [Google Scholar]
- 137. Liem KF Jr, Tremml G, Roelink H, Jessell TM. Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell 1995, 82: 969–979. [DOI] [PubMed] [Google Scholar]
- 138. Wilson PA, Hemmati-Brivanlou A. Induction of epidermis and inhibition of neural fate by Bmp-4. Nature 1995, 376: 331–333. [DOI] [PubMed] [Google Scholar]
- 139. Le Douarin NM, Brito JM, Creuzet S. Role of the neural crest in face and brain development. Brain Res Rev 2007, 55: 237–247. [DOI] [PubMed] [Google Scholar]
- 140. Fink JK. Hereditary spastic paraplegia. Curr Neurol Neurosci Rep 2006, 6: 65–76. [DOI] [PubMed] [Google Scholar]
- 141. Finsterer J, Loscher W, Quasthoff S, Wanschitz J, Auer-Grumbach M, Stevanin G. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance. J Neurol Sci 2012, 318: 1–18. [DOI] [PubMed] [Google Scholar]
- 142. Harding AE. Classification of the hereditary ataxias and paraplegias. Lancet 1983, 1: 1151–1155. [DOI] [PubMed] [Google Scholar]
- 143. Patel H, Cross H, Proukakis C, Hershberger R, Bork P, Ciccarelli FD, Patton MA, et al. . SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat Genet 2002, 31: 347–348. [DOI] [PubMed] [Google Scholar]
- 144. Cross HE, McKusick VA. The Troyer syndrome. A recessive form of spastic paraplegia with distal muscle wasting. Arch Neurol 1967, 16: 473–485. [DOI] [PubMed] [Google Scholar]
- 145. Manzini MC, Rajab A, Maynard TM, Mochida GH, Tan WH, Nasir R, Hill RS, et al. . Developmental and degenerative features in a complicated spastic paraplegia. Ann Neurol 2010, 67: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Butler S, Helbig KL, Alcaraz W, Seaver LH, Hsieh DT, Rohena L. Three cases of Troyer syndrome in two families of Filipino descent. Am J Med Genet A 2016, 170: 1780–1785. [DOI] [PubMed] [Google Scholar]
- 147. Lu J, Rashid F, Byrne PC. The hereditary spastic paraplegia protein spartin localises to mitochondria. J Neurochem 2006, 98: 1908–1919. [DOI] [PubMed] [Google Scholar]
- 148. Bakowska JC, Jupille H, Fatheddin P, Puertollano R, Blackstone C. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Mol Biol Cell 2007, 18: 1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Eastman SW, Yassaee M, Bieniasz PD. A role for ubiquitin ligases and Spartin/SPG20 in lipid droplet turnover. J Cell Biol 2009, 184: 881–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Hooper C, Puttamadappa SS, Loring Z, Shekhtman A, Bakowska JC. Spartin activates atrophin-1-interacting protein 4 (AIP4) E3 ubiquitin ligase and promotes ubiquitination of adipophilin on lipid droplets. BMC Biol 2010, 8: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Renvoise B, Parker RL, Yang D, Bakowska JC, Hurley JH, Blackstone C. SPG20 protein spartin is recruited to midbodies by ESCRT-III protein Ist1 and participates in cytokinesis. Mol Biol Cell 2010, 21: 3293–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Spiegel R, Soiferman D, Shaag A, Shalev S, Elpeleg O, Saada A. Novel homozygous missense mutation in SPG20 gene results in Troyer syndrome associated with mitochondrial cytochrome c oxidase deficiency. JIMD Rep 2017, 33: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Renvoise B, Stadler J, Singh R, Bakowska JC, Blackstone C. Spg20-/- mice reveal multimodal functions for Troyer syndrome protein spartin in lipid droplet maintenance, cytokinesis and BMP signaling. Hum Mol Genet 2012, 21: 3604–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, Tukel T, et al. . Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet 2001, 29: 326–331. [DOI] [PubMed] [Google Scholar]
- 155. McNew JA, Sondermann H, Lee T, Stern M, Brandizzi F. GTP-dependent membrane fusion. Annu Rev Cell Dev Biol 2013, 29: 529–550. [DOI] [PubMed] [Google Scholar]
- 156. Guelly C, Zhu PP, Leonardis L, Papic L, Zidar J, Schabhuttl M, Strohmaier H, et al. . Targeted high-throughput sequencing identifies mutations in atlastin-1 as a cause of hereditary sensory neuropathy type I. Am J Hum Genet 2011, 88: 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Fassier C, Hutt JA, Scholpp S, Lumsden A, Giros B, Nothias F, Schneider-Maunoury S, et al. . Zebrafish atlastin controls motility and spinal motor axon architecture via inhibition of the BMP pathway. Nat Neurosci 2010, 13: 1380–1387. [DOI] [PubMed] [Google Scholar]
- 158. Canbaz D, Kirimtay K, Karaca E, Karabay A. SPG4 gene promoter regulation via Elk1 transcription factor. J Neurochem 2011, 117: 724–734. [DOI] [PubMed] [Google Scholar]
- 159. Errico A, Ballabio A, Rugarli EI. Spastin, the protein mutated in autosomal dominant hereditary spastic paraplegia, is involved in microtubule dynamics. Hum Mol Genet 2002, 11: 153–163. [DOI] [PubMed] [Google Scholar]
- 160. Botzolakis EJ, Zhao J, Gurba KN, Macdonald RL, Hedera P. The effect of HSP-causing mutations in SPG3A and NIPA1 on the assembly, trafficking, and interaction between atlastin-1 and NIPA1. Mol Cell Neurosci 2011, 46: 122–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Roussa E, Farkas LM, Krieglstein K. TGF-beta promotes survival on mesencephalic dopaminergic neurons in cooperation with Shh and FGF-8. Neurobiol Dis 2004, 16: 300–310. [DOI] [PubMed] [Google Scholar]
- 162. Krieglstein K, Unsicker K. Distinct modulatory actions of TGF-beta and LIF on neurotrophin-mediated survival of developing sensory neurons. Neurochem Res 1996, 21: 843–850. [DOI] [PubMed] [Google Scholar]
- 163. Lindholm D, Castren E, Kiefer R, Zafra F, Thoenen H. Transforming growth factor-beta 1 in the rat brain: increase after injury and inhibition of astrocyte proliferation. J Cell Biol 1992, 117: 395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Tomoda T, Shirasawa T, Yahagi YI, Ishii K, Takagi H, Furiya Y, Arai KI, et al. . Transforming growth factor-beta is a survival factor for neonate cortical neurons: coincident expression of type I receptors in developing cerebral cortices. Dev Biol 1996, 179: 79–90. [DOI] [PubMed] [Google Scholar]
- 165. Sometani A, Kataoka H, Nitta A, Fukumitsu H, Nomoto H, Furukawa S. Transforming growth factor-beta1 enhances expression of brain-derived neurotrophic factor and its receptor, TrkB, in neurons cultured from rat cerebral cortex. J Neurosci Res 2001, 66: 369–376. [DOI] [PubMed] [Google Scholar]
- 166. Bellaver B, Souza DG, Souza DO, Quincozes-Santos A. Hippocampal astrocyte cultures from adult and aged rats reproduce changes in glial functionality observed in the aging brain. Mol Neurobiol 2017, 54: 2969–2985. [DOI] [PubMed] [Google Scholar]
- 167. Meyers EA, Gobeske KT, Bond AM, Jarrett JC, Peng CY, Kessler JA. Increased bone morphogenetic protein signaling contributes to age-related declines in neurogenesis and cognition. Neurobiol Aging 2016, 38: 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Burns A, Iliffe S. Alzheimer’s disease. Br Med J 2009, 338: b158. [DOI] [PubMed] [Google Scholar]
- 169. Ondrejcak T, Klyubin I, Hu NW, Barry AE, Cullen WK, Rowan MJ. Alzheimer’s disease amyloid beta-protein and synaptic function. Neuromolecular Med 2010, 12: 13–26. [DOI] [PubMed] [Google Scholar]
- 170. GBD 2015 Disease and Injury Incidence and Prevalence Collaborators Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388: 1545–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Cacace R, Sleegers K, Van Broeckhoven C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimers Dement 2016, 12: 733–748. [DOI] [PubMed] [Google Scholar]
- 172. Lauzon MA, Daviau A, Marcos B, Faucheux N. Growth factor treatment to overcome Alzheimer’s dysfunctional signaling. Cell Signal 2015, 27: 1025–1038. [DOI] [PubMed] [Google Scholar]
- 173. Sveinbjornsdottir S. The clinical symptoms of Parkinson’s disease. J Neurochem 2016, 139(Suppl 1): 318–324. [DOI] [PubMed] [Google Scholar]
- 174. Lotharius J, Brundin P. Pathogenesis of Parkinson’s disease: dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci 2002, 3: 932–942. [DOI] [PubMed] [Google Scholar]
- 175. Houlden H, Singleton AB. The genetics and neuropathology of Parkinson’s disease. Acta Neuropathol 2012, 124: 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Andrews ZB, Zhao H, Frugier T, Meguro R, Grattan DR, Koishi K, McLennan IS. Transforming growth factor beta2 haploinsufficient mice develop age-related nigrostriatal dopamine deficits. Neurobiol Dis 2006, 21: 568–575. [DOI] [PubMed] [Google Scholar]
- 177. The Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72: 971–983. [DOI] [PubMed] [Google Scholar]
- 178. Mendez MF. Huntington’s disease: update and review of neuropsychiatric aspects. Int J Psychiatry Med 1994, 24: 189–208. [DOI] [PubMed] [Google Scholar]
- 179. Pringsheim T, Wiltshire K, Day L, Dykeman J, Steeves T, Jette N. The incidence and prevalence of Huntington’s disease: a systematic review and meta-analysis. Mov Disord 2012, 27: 1083–1091. [DOI] [PubMed] [Google Scholar]
- 180. Hoffner G, Djian P. Polyglutamine aggregation in Huntington disease: does structure determine toxicity? Mol Neurobiol 2015, 52: 1297–1314. [DOI] [PubMed] [Google Scholar]
- 181. Jimenez-Sanchez M, Licitra F, Underwood BR, Rubinsztein DC. Huntington’s disease: mechanisms of pathogenesis and therapeutic strategies. Cold Spring Harb Perspect Med 2017, 7: a024240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Takahashi T, Katada S, Onodera O. Polyglutamine diseases: where does toxicity come from? what is toxicity? where are we going? J Mol Cell Biol 2010, 2: 180–191. [DOI] [PubMed] [Google Scholar]
- 183. Ferrante RJ, Kowall NW, Richardson EP Jr. Proliferative and degenerative changes in striatal spiny neurons in Huntington’s disease: a combined study using the section-Golgi method and calbindin D28k immunocytochemistry. J Neurosci 1991, 11: 3877–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, Burrell JR, et al. . Amyotrophic lateral sclerosis. Lancet 2011, 377: 942–955. [DOI] [PubMed] [Google Scholar]
- 185. Marangi G, Traynor BJ. Genetic causes of amyotrophic lateral sclerosis: new genetic analysis methodologies entailing new opportunities and challenges. Brain Res 2015, 1607: 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, et al. . TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 2008, 319: 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, et al. . TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 2008, 40: 572–574. [DOI] [PubMed] [Google Scholar]
- 188. Kabashi E, Lin L, Tradewell ML, Dion PA, Bercier V, Bourgouin P, Rochefort D, et al. . Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum Mol Genet 2010, 19: 671–683. [DOI] [PubMed] [Google Scholar]
- 189. Deng HX, Zhai H, Bigio EH, Yan J, Fecto F, Ajroud K, Mishra M, et al. . FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann Neurol 2010, 67: 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, et al. . Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314: 130–133. [DOI] [PubMed] [Google Scholar]
- 191. Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol 2010, 9: 995–1007. [DOI] [PubMed] [Google Scholar]
- 192. Chio A, Calvo A, Moglia C, Restagno G, Ossola I, Brunetti M, Montuschi A, et al. . Amyotrophic lateral sclerosis-frontotemporal lobar dementia in 3 families with p.Ala382Thr TARDBP mutations. Arch Neurol 2010, 67: 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, et al. . A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet 2004, 75: 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Anagnostou G, Akbar MT, Paul P, Angelinetta C, Steiner TJ, de Belleroche J. Vesicle associated membrane protein B (VAPB) is decreased in ALS spinal cord. Neurobiol Aging 2010, 31: 969–985. [DOI] [PubMed] [Google Scholar]
- 195. Huang WJ, Chen WW, Zhang X. Multiple sclerosis: pathology, diagnosis and treatments. Exp Ther Med 2017, 13: 3163–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, Shah TS, et al. . Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 2013, 45: 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, Dilthey A, et al. . Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011, 476: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Dendrou CA, Fugger L, Friese MA. Immunopathology of multiple sclerosis. Nat Rev Immunol 2015, 15: 545–558. [DOI] [PubMed] [Google Scholar]
- 199. Osso LA, Chan JR. Architecting the myelin landscape. Curr Opin Neurobiol 2017, 47: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Bergles DE, Richardson WD. Oligodendrocyte development and plasticity. Cold Spring Harb Perspect Biol 2015, 8: a020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Plemel JR, Liu WQ, Yong VW. Remyelination therapies: a new direction and challenge in multiple sclerosis. Nat Rev Drug Discov 2017, 16: 617–634. [DOI] [PubMed] [Google Scholar]
- 202. Wolswijk G. Chronic stage multiple sclerosis lesions contain a relatively quiescent population of oligodendrocyte precursor cells. J Neurosci 1998, 18: 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Chang A, Tourtellotte WW, Rudick R, Trapp BD. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med 2002, 346: 165–173. [DOI] [PubMed] [Google Scholar]
- 204. Snethen H, Love S, Scolding N. Disease-responsive neural precursor cells are present in multiple sclerosis lesions. Regen Med 2008, 3: 835–847. [DOI] [PubMed] [Google Scholar]
- 205. John GR, Shankar SL, Shafit-Zagardo B, Massimi A, Lee SC, Raine CS, Brosnan CF. Multiple sclerosis: re-expression of a developmental pathway that restricts oligodendrocyte maturation. Nat Med 2002, 8: 1115–1121. [DOI] [PubMed] [Google Scholar]
- 206. Setoguchi T, Nakashima K, Takizawa T, Yanagisawa M, Ochiai W, Okabe M, Yone K, et al. . Treatment of spinal cord injury by transplantation of fetal neural precursor cells engineered to express BMP inhibitor. Exp Neurol 2004, 189: 33–44. [DOI] [PubMed] [Google Scholar]
- 207. Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, et al. . Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med 2007, 357: 851–862. [DOI] [PubMed] [Google Scholar]
- 208. Hoppenbrouwers IA, Aulchenko YS, Janssens AC, Ramagopalan SV, Broer L, Kayser M, Ebers GC, et al. . Replication of CD58 and CLEC16A as genome-wide significant risk genes for multiple sclerosis. J Hum Genet 2009, 54: 676–680. [DOI] [PubMed] [Google Scholar]
- 209. van Luijn MM, Kreft KL, Jongsma ML, Mes SW, Wierenga-Wolf AF, van Meurs M, Melief MJ, et al. . Multiple sclerosis-associated CLEC16A controls HLA class II expression via late endosome biogenesis. Brain 2015, 138: 1531–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. McBrayer ZL, Dimova J, Pisansky MT, Sun M, Beppu H, Gewirtz JC, O’Connor MB. Forebrain-specific loss of BMPRII in mice reduces anxiety and increases object exploration. PLoS One 2015, 10: e0139860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Ageta H, Murayama A, Migishima R, Kida S, Tsuchida K, Yokoyama M, Inokuchi K. Activin in the brain modulates anxiety-related behavior and adult neurogenesis. PLoS One 2008, 3: e1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Sutcigil L, Oktenli C, Musabak U, Bozkurt A, Cansever A, Uzun O, Sanisoglu SY, et al. . Pro- and anti-inflammatory cytokine balance in major depression: effect of sertraline therapy. Clin Dev Immunol 2007, 2007: 76396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Musil R, Schwarz MJ, Riedel M, Dehning S, Cerovecki A, Spellmann I, Arolt V, et al. . Elevated macrophage migration inhibitory factor and decreased transforming growth factor-beta levels in major depression—no influence of celecoxib treatment. J Affect Disord 2011, 134: 217–225. [DOI] [PubMed] [Google Scholar]
- 214. Mihailova S, Ivanova-Genova E, Lukanov T, Stoyanova V, Milanova V, Naumova E. A study of TNF-alpha, TGF-beta, IL-10, IL-6, and IFN-gamma gene polymorphisms in patients with depression. J Neuroimmunol 2016, 293: 123–128. [DOI] [PubMed] [Google Scholar]
- 215. Lee KM, Kim YK. The role of IL-12 and TGF-beta1 in the pathophysiology of major depressive disorder. Int Immunopharmacol 2006, 6: 1298–1304. [DOI] [PubMed] [Google Scholar]
- 216. Gobeske KT, Das S, Bonaguidi MA, Weiss C, Radulovic J, Disterhoft JF, Kessler JA. BMP signaling mediates effects of exercise on hippocampal neurogenesis and cognition in mice. PLoS One 2009, 4: e7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, Nielsen JE, et al. . Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet 2005, 37: 806–808. [DOI] [PubMed] [Google Scholar]
- 218. West RJ, Lu Y, Marie B, Gao FB, Sweeney ST. Rab8, POSH, and TAK1 regulate synaptic growth in a Drosophila model of frontotemporal dementia. J Cell Biol 2015, 208: 931–947. [DOI] [PMC free article] [PubMed] [Google Scholar]