Abstract
The tight correlation between mRNA distribution and subsequent protein localization and function indicate a major role for mRNA localization within the cell. RNA localization, followed by local translation, presents a mechanism for spatial and temporal gene expression regulation utilized by various cell types. However, little is known about mRNA localization and translation in the mammalian oocyte and early embryo. Importantly, fully-grown oocyte becomes transcriptionally inactive and only utilizes transcripts previously synthesized and stored during earlier development. We discovered an abundant RNA population in the oocyte and early embryo nucleus together with RNA binding proteins. We also characterized specific ribosomal proteins, which contribute to translation in the oocyte and embryo. By applying selected markers to mouse and human oocytes, we found that there might be a similar mechanism of RNA metabolism in both species. In conclusion, we visualized the localization of RNAs and translation machinery in the oocyte, that could shed light on this terra incognita of these unique cell types in mouse and human.
Introduction
Meiotic maturation of mammalian oocytes and oocyte-to-zygote transition proceed without transcription and depend entirely on the post-transcriptional regulation of maternal mRNAs. The overall translation gradually decreases during oocyte meiotic maturation [1], but the activators of cap-dependent translation become more active during this period, implying a role for translation of specific mRNAs in the regulation of meiosis [2,3]. These mRNAs need to be recruited to translational machinery and subsequently degraded in a tightly controlled temporal manner [4]. Spatial segregation of protein synthesis in cells involves the positioning of mRNAs according to where their protein products are required, and results in local or compartmentalized gene expression. mRNA localization can occur during different stages of development. While these processes were studied in other cell types [5–7], little is known about mRNA localization and translation in the mammalian oocyte or early embryo.
Our previous study indicated that the oocyte nucleus contains an RNA population in the fully grown oocyte that most likely contributes to translation in the vicinity of chromosomes after nuclear envelope break down (NEBD) [8,9]. Localization of mRNAs at the spindle is evolutionarily conserved in mammals and has also been seen in mitotic cells [10,11]. The differences in enrichment of 384 mRNAs between the meiotic spindle area and cortical regions have been analyzed by microarrays [12]. Investigations of mRNA localization have been conducted in the oocytes of Drosophila [13,14], Xenopus [15,16] and mouse [4]. The localization of mRNA molecules within the cytoplasm provides a basis for cell polarization, underlying developmental processes such as asymmetric cell division during meiosis, and embryonic patterning [17].
Endogenous mRNAs do not exist alone; they bind to a number of proteins to form mRNA-protein complexes [18]. RNA-binding proteins (RBPs) and molecular motors mediate the transport of mRNAs along the cytoskeleton of cells, resulting in an asymmetric distribution of RNAs. RBPs are capable of regulating mRNA stability and translation. Some RBPs, such as CPE-binding protein (CPEB), Deleted in azoospermia-like (DAZL), post-transcriptionally regulate mRNA by binding to its 7-methylguanosine cap structure at the 5′-untranslated end (5′UTR) or to 3′-+ regions (3′UTRs). Regulation of the initiation of cap dependent translation is controlled via the translational repressor 4E-binding protein 1 (4E-BP1). Hierarchical phosphorylation of 4E-BP1 leads to its dissociation from eIF4E. In a previous study [19], we detected enriched localization of 4E-BP1 in the nucleus of fully grown oocytes. Another common mechanism regulating the recruitment and stability of dormant maternal mRNAs is reversible polyadenylation, which is controlled by cytoplasmic polyadenylation elements (CPEs) [20]. CPEs are specific sequences in 3′UTRs of dormant maternal mRNAs that serve as a binding platform for the CPE-binding proteins (CPEBs), which control polyadenylation-induced translation. The CPEBs family has four members. The most studied is CPE-binding protein 1 (CPEB1), which functions as a translational activator or repressor according to its phosphorylation state [21]. Other RBPs that regulate RNA processing are known as heterogeneous nuclear ribonucleoproteins (hnRNPs) [22]. This family contains more than 20 members. The key characteristic of the hnRNPs is their nucleocytoplasmic shuttling [23]. The hnRNP proteins A0, A1, A2/B1 and A3 were considered to be the prime constituents of 40S heterogeneous nuclear ribonucleoprotein particles, which bind to and stabilize nascent pre-mRNA [24,25]. The exon junction complex consists of RNA binding proteins and contains Eukaryotic initiation factor 4A-III (eIF4A3), a DEAD-box RNA helicase–a member of the eIF4A family of translation initiation factors [26]. Exon junction complex proteins play important roles in post-splicing events–including mRNA export, cytoplasmic localization, and nonsense-mediated decay [26,27,28]. The presence of assembled ribosomes is directly linked to protein synthesis during crucial periods of development and is tightly associated with the developmental competence of oocytes. It has been proposed that the mRNAs for 27 ribosomal proteins are expressed at higher levels in developmentally competent oocytes compared to non-competent ones [29].
A remarkable feature of mammalian oocyte maturation is the significant elimination of rRNA and ribosomes [30]. Moreover, genome-wide transcriptome analysis has shown that mRNAs coding for ribosomal proteins are degraded during oocyte maturation and after fertilization [31,32]. The eukaryotic ribosome contains 4 RNAs and ~80 ribosomal proteins (RPs), forming its two subunits: small 40S and large 60S. RPL24 is positioned between the two subunits with an N-terminal domain in 60S [33], while its C-terminal part interacts with RPS6 [34]. Mitogens and growth factors are responsible for RPS6 phosphorylation on five residues [35,36]. It was shown that RPS6 phosphorylation has an important function in the translational control of the subclass of mRNAs that harbor the 5′ tract oligopyrimidine (5′ TOP) sequence and this level of regulation may imbue the ribosome with greater specificity [37]. Similar to RPS6, RPS14 also plays a role in the regulation of the MDM2-p53 pathway [38,39].
In this study we focus on the visualization of transcriptome in association with the regulation of translation and visualization of translation in the mammalian oocyte and early embryo. Extensive analyses of the transcriptionally inactive germinal vesicle (GV) oocytes and the 2-cell embryos during transcriptional reprograming show substantial measurements of both similarities and differences between the two cell types, as well as between mouse and human. Here, we also present the distribution and expression of specific components of translational machinery in the mammalian oocyte and early embryo.
Methods
Oocyte isolation and cultivation
Mouse ovaries were obtained from CD1 mice which were at least 6 weeks old and had been stimulated to superovulate by intraperitoneal injection of 5 UI pregnant mare serum gonadotropin (PMSG; Folligon, Merck Animal Health) 46 h prior to collection. Growing and fully grown GV oocytes were isolated subsequently into M2 medium (Millipore) supplemented with 100 μM of 3-isobutyl-1-methylxanthine (IBMX, Sigma Aldrich) to prevent resumption of meiosis. Selected oocytes were stripped of cumulus cells and cultured in M16 medium (Millipore) without IBMX at 37°C, 5% CO2. Zygotes were obtained by mating females with males after injections of 5 UI PMSG and then (after 46 h) 5 UI of human chorionic gonadotropin (hCG; Merck Animal Health). Zygotes were isolated 17 h after mating and cultured in vitro in M16 under mineral oil for 20 h, then 2-cell embryos were collected. All animal work was conducted according to Act No 246/1992 on the protection of animals against cruelty. Human oocytes, not used in human reproduction, were obtained from the Obstetrics and Gynecology Clinic of the General University Hospital in Prague. The project was accredited (#30/12) by the Ethical Committee of the General Hospital, Prague.
Immunocytochemistry
Oocytes were fixed 15 min in 4% paraformaldehyde (PFA, Sigma Aldrich) in PBS and permeabilized 10 min in 0.1% Triton X-100 in PBS with one drop of ActinGreen probe Phalloidin488 (Thermo Fisher). Then the oocytes were incubated overnight at 4°C with primary antibodies diluted in PBS/0.2% normal bovine serum. The following antibodies were used in 1:150 dilution: rabbit anti-4E-BP1 (CST); rabbit anti-Ribosomal S14 (Santa Cruz); rabbit anti-Ribosomal S3 (CST); rabbit anti-RPL24 (Thermo Fisher); mouse anti-RPS6 (Santa Cruz); rabbit anti-CPEB4 (Thermo Fisher); mouse anti-hnRNPA1 (Sigma Aldrich); mouse anti-eIF4A3 (Abcam), rabbit anti-RPL7 (Abcam); mouse anti-m3G cap/m7G cap (Thermo Fisher). Mouse anti-5.8S rRNA antibody (Abcam), was also diluted 1:150 and incubated at room temperature for 2 h. After washing in PBS for 2x15 min, detection of the primary antibodies was performed by cultivation of the oocytes with relevant Alexa Fluor 488/594 conjugates (diluted 1:250) for 1 h at room temperature. Oocytes were then washed 2x15 min in PBS and mounted using Vectashield Mounting Medium with DAPI (Vector Laboratories). For nascent protein synthesis oocytes and embryos were incubated 1 h with 1 μgml-1 puromycin (Sigma Aldrich) or 1 μM cycloheximide, specific stage (GV, 2-cell embryo). For measuring nascent translation oocytes were cultured in methionine-free medium (Gibco) supplemented with 1% dialyzed fetal bovine serum (10,000MW; Sigma) and 50 mM HPG for 30 min. HPG was detected by using Click-iT Cell Reaction Kit (Life Technologies) according manufacture instruction. Samples were visualized using Leica SP5 inverted confocal microscope in 16 bit depth. Images were assembled in Photoshop CS3 and quantified by Image J software (http://rsbweb.nih.gov/ij/). Experiments were performed three times with 25 oocytes/embryos per experiment.
Preparation of slides for imaging
Nunc Lab-Tek II Chamber Slide System (Thermo Fisher) glass coverslips were coated at 37°C for 2 h with poly-L-ornithine (Sigma Aldrich) diluted in RNase-free water 1:250 and then overnight with laminin (Sigma Aldrich) diluted in PBS 1:1000 also at 37°C.
RNA fluorescent in situ hybridization (FISH)
RNA FISH was performed with small changes according to [40]. Oocytes were fixed for 10 min in 4% paraformaldehyde and permeabilized in 0.1% Triton X-100 in PBS with 40 units/20 μl of RNAseOut (Invitrogen), then mounted to pre-coated Lab-Tek II Chamber Slide System slides using 80% methanol pre-frozen to -20°C. Oocytes were washed in Wash Buffer A (Biosearch Technologies) and incubated overnight at 30°C in hybridization buffer (Biosearch Technologies) with 75 nM oligo-d(T) probe (Biotech Generi); Neat2 CalFluorRed610 (Biosearch Technologies); Dazl (Biosearch Technologies) and β-actin labelled with Cy5 (Biotech Generi); GFP CalFluorRed635 (Biosearch Technologies) with 75 nM (protected from light). Oocytes were then washed 3x in Wash Buffer A and 2x in 2xSSC (Sigma Aldrich). For visualization of chromatin structure the oocytes were incubated 1 min with 10 nM DAPI (Sigma Aldrich) in 2xSSC; then washed 1x with 2xSSC and scanned in 2xSSC. For negative control RNase A (Ambion) was used for 2 h at 37°C after the permeabilization step. Forty-five oocytes and embryos was analyzed using ImageJ/FIJI (http://rsbweb.nih.gov/ij/) for quantification of fluorescence intensity in the cytoplasm and the nucleus.
Rolling circle amplification (RCA) FISH
RCA FISH was performed according to [41] protocol with the following changes: oocytes were fixed 10 min in 4% PFA (Sigma Aldrich) and permeabilized in 0.1% Triton X-100 in PBS for 10 min and subsequently in 70% ethanol, pre-frozen to -20°C, for 10 seconds. The whole transcriptome was converted into cDNA by M-MuLV reverse transcriptase (Enzymatics) and the reaction mix was prepared according to the mentioned protocol. The cDNA fragments were fixed to the cellular protein matrix using a nonreversible amine cross-linker BS(PEG)9 (Sigma Aldrich) and circularized after degrading the RNA residues. The circular templates were amplified using RCA primers 100 μM (TCTTCAGCGTTCCCGA*G*A; where * is phosphorothioate, Generi Biotech) complementary to the adapter sequence in the presence of aminoallyl-dUTP and stably cross-linked. For visualization of endogenous cDNA we used fluorescently labeled random octamers, which were labeled by two fluorophore (green-488 nm) or (red-561 nm). Chromatin was visualized by incubation (1min) with 10 nM DAPI (Sigma Aldrich) in 2xSSC; then washed 1x with 2xSSC. We scanned the samples in 2xSSC. Quantification of fluorescent intensity between cytoplasm and nucleus was used ImageJ/FIJI (http://rsbweb.nih.gov/ij/). Per one experiment was used 12 oocyte and 10 embryos, we quantified 3 experiments.
In situ proximity ligation assay (PLA)
Proximity ligation assay was performed according to manual instructions of PLA Duolink kit (Sigma Aldrich). Oocytes and embryos were incubated 1 h with 1 μgml-1 puromycin (Sigma Aldrich) to decrease translation, then fixed 15 min in 4% paraformaldehyde in PBS and permeabilized 10 min in 0.1% Triton X-100 in PBS. We added PLA Duolink kit blocking solution to each sample. Oocytes were incubated with primary antibodies rabbit anti-RPL24 (Thermo Fisher) and mouse anti-RPS6 (Santa Cruz) at 4°C overnight. The samples were washed in PBS and then in Wash Buffer A (Sigma Aldrich). The samples were incubated with 40 μl reaction mixture (8 μl PLA probe MINUS stock, 8 μl PLA probe PLUS stock and 24 μl PBS) in a chamber for 1 h at 37°C. The slides were washed in 1x Wash Buffer A for 6x2 min and ligation was performed in 40 μl reaction: 1 μl of ligase to 39 μl of ligation solution. Samples were incubated in ligation reaction mixture for 30 min at 37°C then washed 6x2 min in Wash Buffer A. 40 μl of amplification reaction (0.5 μl polymerase and 39.5 μl amplification solution) was added to each sample before incubation at 37°C for 100 min. Next, the samples were washed in Wash Buffer B (Sigma Aldrich) for 3x5 min and in 0.01 Wash Buffer B for 2 min. The samples were mounted by Vectashield Mounting Medium with DAPI (Vector Laboratories). Quantification of interaction foci was performed using ImageJ/FIJI. We performed 5 experiments with 150 oocytes/embryos.
Live cell imaging of nascent translation (ReAsH method)
The ReAsH method [42] was used to detect in situ translation. For the ReAsH we used plasmid #27123 (Adgene;[43]). The growing oocytes were injected according to the protocol by [44] with a plasmid diluted to ~40 ng/μl into nuclei. Oocytes were incubated overnight in 1μM cycloheximide (CHX; Sigma Aldrich) in M16 medium to prevent translation. After CHX wash oocytes were incubated for 30 min in M16 supplemented with ReAsH dye (final concentration 20 μM, Thermo Fisher) and then transferred into 250 μM 2,3-dimercaptopropanol (BAL buffer, Thermo Fisher) in M16 and immediately scanned on confocal microscope Leica TCS SP5. Thirty oocytes was quantified from three independent experiments.
Western blot (WB)
Lysates of embryos and oocytes was analysed on 4–12% gradient acrylamide gel. Samples were transferred to polyvinyliden-fluoride membrane (Immobilon P; Merckmillipore) using blotting system (Biometra GmbH) at 5 mA/cm2 during 25 minute. Membranes were blocked for 1 h, at room temperature and then they were incubated at 4 ᵒC overnight with the following primary antibodies: rabbit anti-4E-BP1 (CST); rabbit anti-Ribosomal S14 (Santa Cruz); rabbit anti-Ribosomal S3 (CST); rabbit anti-RPL24 (Thermo Fisher); mouse anti-RPS6 S235/236 (Santa Cruz); rabbit anti-CPEB4 (Thermo Fisher); mouse anti-hnRNPA1 (Sigma Aldrich); mouse anti-eIF4A3 (Abcam), rabbit anti-RPL7 (Abcam), with 1% milk/TTBS (Tween-Tris-buffer saline; NaCl, Tween 20,2M; Tris pH 7,6; dH2O). After 3 cycles of 10 minute washing in 0,05% TTBS membrane was incubated at room temperature for 1 h in 3% milk with secondary antibody conjugated with peroxidase (Jackson Immunoresearch). Following washing step with 0,05% TTBS. Immunodetected proteins were visualized by ECL (Amersham, GE Healthecare Life Science) according instruction from supplier. Films were scanned using a GS-800 calibrated densitometer (Bio-Rad) and quantified 3 independent experiment for each protein for one experiment was used 100 oocytes/embryos, we normalized quantification to GAPDH, using ImageJ/FIJI (http://rsbweb.nih.gov/ij/).
Statistical analysis
Mean and SD values were calculated using MS Excel, statistical significance of the differences between the groups was tested using Student’s t-test (PrismaGraph5) and P<0.05 was considered as statistically significant (marked by asterisk) *p<0.05; **p<0.01; ***p<0.001.
Results
Detection of global transcriptome in the oocyte and embryo
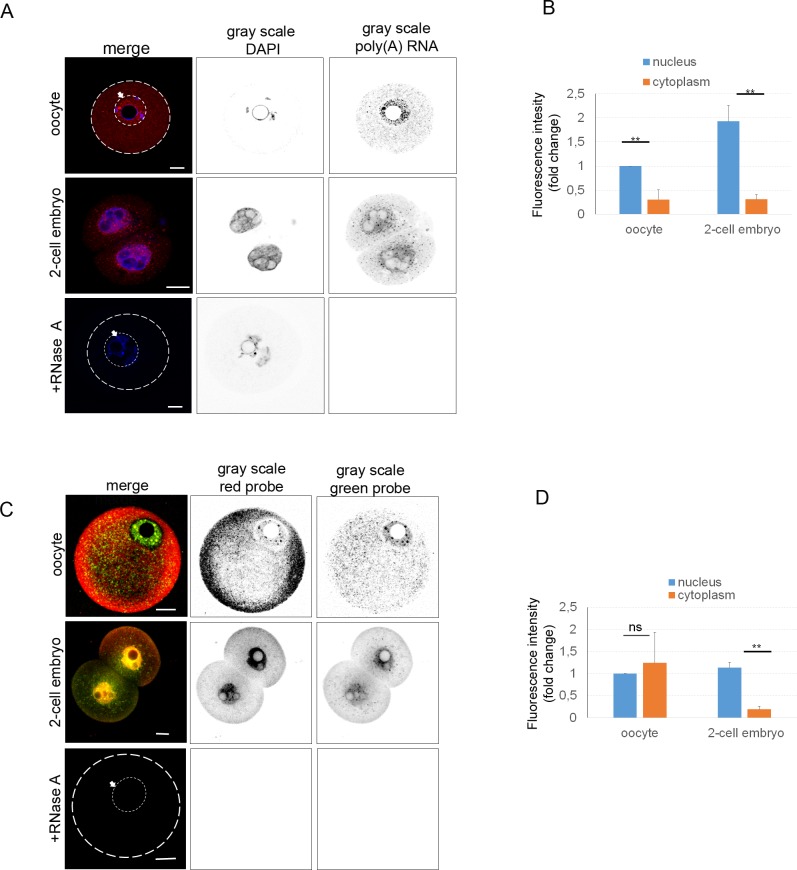
mRNA localization generally leads to targeted translation [45]. We have asked how the transcriptome is distributed in the mammalian oocyte and 2-cell embryo. It is known that 70% of RNAs are polyadenylated [46]. To detect this subpopulation, i.e. to detect RNA localization in the oocyte and 2-cell embryo, fluorescently (CY5) labeled oligo(dT) probes were used (Fig 1A). Treatment by RNase A eliminated the fluorescence signal, thus confirming its RNA origin in the rest of the experiments (Fig 1A). PolyA RNA was detected in the cytoplasm with a significant decrease (0.7-fold) compared to the nucleus of the oocyte. The nucleus of the 2-cell embryo shows higher (2-fold) presence of polyA RNA in the comparison with the oocyte. However, cytoplasmic signal intensity of poly(A) RNA is similar in both cell types (Fig 1B).
Fig 1. Localization of transcriptome in oocyte and embryo.
A) Single Z-stack from confocal images of GV (germinal vesicle) oocyte stage and 2-cell embryo. RNA FISH detecting poly(A) RNA subpopulation (red; oligo(dT) probe), and the gray scale shows separated light channels (DAPI and oligo(dT) probe). The arrow with the white line indicates the nucleus of the oocyte. As a negative control RNA was digested by RNase A after the cell permeabilization step. Scale bars 20 μm. The cortex of the oocyte is indicated by the white line. B) Quantification of fluorescence intensity of poly(A) RNA of equatorial Z-stack, in the nucleus and cytoplasm of oocyte and embryo, relatively compared to the nucleus of the oocyte. The experiment was repeated 3 times, with 15 oocytes and embryos per experiment. Data are represented as mean ± s.d.; the values bars with ns are not significant, and the asterisk denotes statistically significant differences *p<0.05; **p<0.01; ***p<0.001. C) Rolling circle amplification FISH using random hexamers probes showing distribution of global RNA. The arrow with the white line indicates the nucleus of the oocyte. The gray scale shows separated channels (red and green probe). The cortex of the oocyte is indicated by the white line. As a negative control RNA was digested by RNase A after cell permeabilization step. Scale bars 20 μm. D) Quantification of fluorescence intensity of whole transcriptome in the nucleus and cytoplasm. The value of the nucleus was set as 1. The experiment was repeated 3 times, with 12 oocytes and 10 embryos per experiment. Data are represented as mean ± s.d.; the values bars with ns are not significant, and the asterisk denotes statistically significant differences *p<0.05; **p<0.01; ***p<0.001.
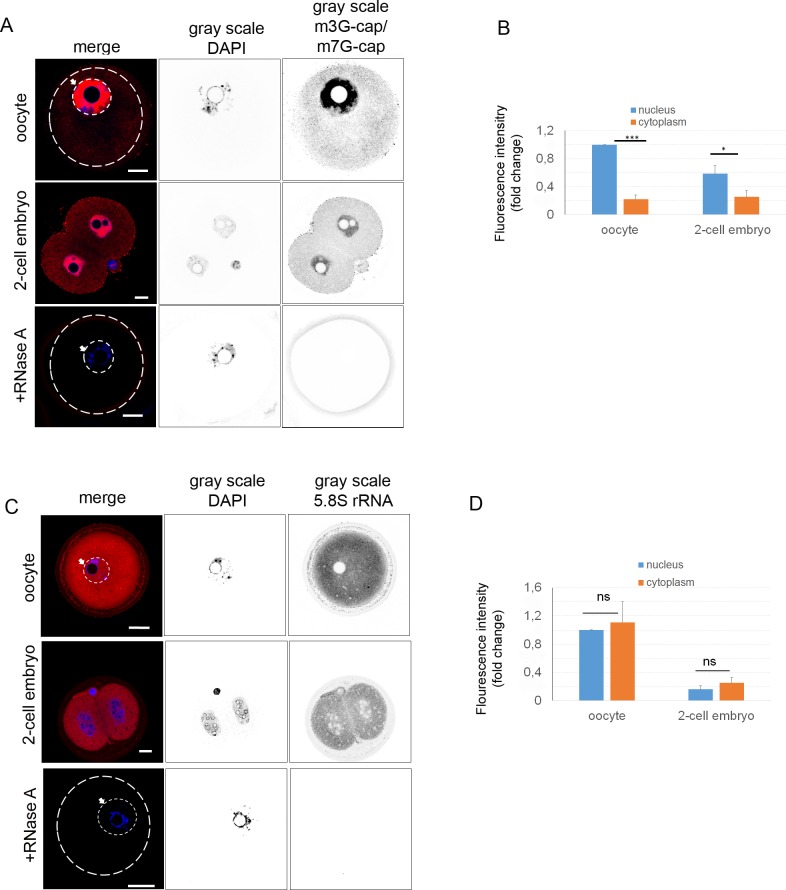
Rolling circle amplification (RCA) [41,47] was used to visualize the whole cellular transcriptome, the method comprising reverse in situ transcription followed by hybridization of fluorescently labeled random octamers, which were labeled by fluorophore, green (488 nm) and red (561 nm). By RCA, we observed a similar localization of RNA as with RNA FISH (Fig 1A and 1C). The different color pattern might be the result of a different affinity of the labeled probes. Similarly, the degradation of RNA by RNase A treatment significantly decreased the fluorescence signal (Fig 1C). Fluorescence intensity of global transcriptome was at the same level in both compartments of the oocyte (Fig 1D). However, in the cytoplasm of the 2-cell embryo RNA intensity was significantly lower (about 0.8-fold; Fig 1D). In addition to the previous methods for visualization of the global transcriptome, we used an antibody detecting the 5‘ UTR cap both m7G-cap (present in most RNAs) and m3G-cap (present in small nuclear RNAs, snRNAs) structures [48]. We were able to detect these cap structures (Fig 2A) with similar RNA distribution in both oocytes and 2-cell embryos, again comparable to RNA FISH and RCA methods (Fig 2A). Similar to RNA FISH and RCA results, we observed significantly higher fluorescence intensity of the m3G/m7G-cap in the nuclei of the oocytes and embryos (about 0.7-fold; Fig 2B).
Fig 2. Localization of rRNA and RNA in oocyte and embryo.
A) Antibody detecting m3G-cap and m7G-cap indicates cap-structure at the 5’UTR of mRNA (red). DNA stained with DAPI (blue). The gray scale shows separated light channels. The arrow with the white line indicates the nucleus of the oocyte. As a negative control RNA was digested by RNase A after the cell permeabilization step. The cortex of the oocyte is indicated by the white line. Scale bars 20 μm. The experiments were repeated 3 times, with 25 oocytes/embryos per experiment. B) Quantification of fluorescence intensity of 5’UTR cap-structure of equatorial Z-stacks, in the nucleus and cytoplasm of oocyte and embryo, relatively compared to nucleus of oocyte. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. Data are represented as mean ± s.d.; the value bars with ns are not significant, and the asterisk denotes statistically significant differences * p<0.05; **p<0.01; ***p<0.001. C) Distribution of 5.8S rRNA in the oocyte and early embryo (red). DNA stained with DAPI (blue). The gray scale shows separated light channels. The arrow with the white line indicates the nucleus of the oocyte. As a negative control RNase A digestion was used after the cell permeabilization step. The white line indicates the oocyte cortex. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. D) Quantification of fluorescence intensity of 5.8S rRNA in the nucleus and cytoplasm from equatorial Z-stacks. The value of the oocyte nucleus was set as 1; other values are represented as a ratio to the intensity of the oocyte nucleus. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. Data are represented as mean ± s.d.; the value bars with ns are not significant, and the asterisk denotes statistically significant differences * p<0.05; **p<0.01; ***p<0.001.
Using RNA FISH, RCA and an antibody against RNA cap structure produced results showing a high uniformity concerning the localization of global transcriptome. We found the nuclei significantly enriched with RNA in both cell types.
In addition, using immunocytochemistry (ICC) with antibody against 5.8S ribosomal RNA, which is a structural component of 40S subunit of ribosome [49,50], we found a strong fluorescence signal in the cytoplasm of the oocyte and the embryo, with a decrease in the nucleoplasm of both stages (Fig 2C). Treatment with RNase A led to the loss of fluorescent signal. Quantification of 5.8S rRNA signal showed similar intensities in both cellular compartments in the oocyte/embryo. We observed a significant decrease of global 5.8S ribosomal RNA (about 0.8-fold) in the 2-cell embryo compared to the oocyte (Fig 2D).
Using direct detection of whole transcriptome by RCA, RNA FISH and ICC, we detected the localization of various RNA types and ribosomal rRNA in the mammalian oocyte and early embryo.
Identification of specific RNAs in the oocyte and embryo
Pinpointing the subcellular localization of specific RNA species might lead to the unveiling of their potential molecular role in such large cells. Single molecule RNA FISH (smRNA FISH) was used to visualize specific RNAs. First, we determined the localization of Deleted in azoospermia-like RNA (Dazl)–a germ cell specific transcript. smRNA FISH showed an even distribution of the mRNA in the cytoplasm and a weak signal in the nucleoplasm of both, the oocyte and the early embryo (Fig 3A and 3D) with increased levels in the cytoplasm of the 2-cell embryo (Fig 3A and 3D), suggesting early transcription of Dazl mRNA in the embryo. Cumulus cells (depicted by asterisk, Fig 3A) and mouse fibroblasts NIH3T3 (Fig 3A) were negative and RNase A treatment of the oocyte and embryo resulted in the absence of a fluorescent signal (S1B Fig), supporting the specificity of Dazl mRNA detection in the oocyte and early embryo. Another detected mRNA was β-actin, which was present in both subcellular compartments, nucleus and cytoplasm of the oocyte, and as dot-like structures in the cytoplasm of the early embryo (Fig 3B and 3E). The localization in the NIH3T3 was mostly at the leading edges (Fig 3B). Next, we also detected long noncoding RNA Neat2/Malat1 (Nuclear-Enriched Abundant Transcript 2/Metastasis Associated Lung Adenocarcinoma Transcript 1) which is known to be localized in the nuclear speckles of HeLa cells [51]. Neat2/Malat1 was exclusively localized in the nucleus of the GV oocyte and NIH3T3 cells. However, localized in the cytoplasm of the embryo (Fig 3C and 3F). RNase A treatment eliminated the fluorescent signal (S1B Fig) which corroborates the specificity of our RNA detection. The quantification of the intensity of the specific RNA signal shows that Dazl mRNA is distributed in both compartments, i.e. the nucleus and cytoplasm, of the oocyte (Fig 3D). However, in the embryo, this mRNA is elevated and localized predominantly in the cytoplasm (1.3-fold; Fig 3D). β-actin mRNA shows equal localization in the nucleus and embryo in both cell types (Fig 3E). Neat2 lncRNA shows significant presence in the nucleus (about 0.85-fold) of the oocyte. In contrast, in the embryo, the fluorescence intensity was significantly higher (about 0.7-fold) in the cytoplasm (Fig 3F).
Fig 3. Localization of specific RNAs in GV oocyte, 2-cell embryo and NIH3T3 cells.
A) Confocal images of smRNA FISH of Dazl (green) in GV oocyte, 2-cell embryo and NIH3T3 cells. DNA stained with DAPI (blue). The gray scale shows separated channels. The arrow with the white line indicates the nucleus of the oocyte. The asterisk indicates cumulus cells. The white line indicates the cortex of the oocyte. Scale bars 20 μm. The experiment was repeated 3 times, with 15 oocytes/embryos per experiment. B) Confocal images of smRNA FISH of β-actin (green) in GV oocyte, 2-cell embryo and NIH3T3 cells (DNA stained with DAPI (blue)). The gray scale shows separated light channels. Scale bars 20 μm. The experiment was repeated 3 times, with 15 oocytes and embryos per experiment. C) Confocal images of smRNA FISH of Neat2 lncRNA (green) in GV oocyte, 2-cell embryo and NIH3T3 cells. DNA stained with DAPI (blue). The gray scale shows separated channels. The arrow with the white line indicates the nucleus of the oocyte. The white line indicates the cortex of the oocyte. Scale bars 20 μm. The experiment was repeated 3 times, with 15 oocytes/embryos per experiment. D) Quantification of fluorescence intensity of Dazl mRNA in the nucleus and cytoplasm from equatorial Z-stacks. The value of the oocyte nucleus was set as 1. The experiment was repeated 3 times, with 15 oocytes/embryos per experiment. Data are represented as mean ± s.d.; the values bars with ns are not significant, and the asterisk denotes statistically significant differences * p<0.05; **p<0.01; ***p<0.001. E) Quantification of fluorescence intensity of β-actin mRNA in the nucleus and cytoplasm from equatorial Z-stacks. Value of oocyte nucleus was set as 1. The experiment was repeated 3 times, with 15 oocytes and embryos per experiment. Data are represented as mean ± s.d.; the value bars with ns are not significant, asterisk denotes statistically significant differences * p<0.05; **p<0.01; ***p<0.001. F) Quantification of fluorescence intensity of Neat2 lncRNA in the nucleus and cytoplasm from equatorial Z-stacks. The value of the oocyte nucleus was set as 1. The experiment was repeated 3 times, with 15 oocytes and embryos per experiment. Data are represented as mean ± s.d.; the value bars with ns are not significant, and the asterisk denotes statistically significant differences * p<0.05; **p<0.01; ***p<0.001.
By detection of specific RNAs, we determined the localization of specific mRNAs and lncRNA in the oocytes, 2-cell embryos and mouse embryonic fibroblasts.
Expression and localization of RNA binding proteins in the oocyte and 2-cell embryo
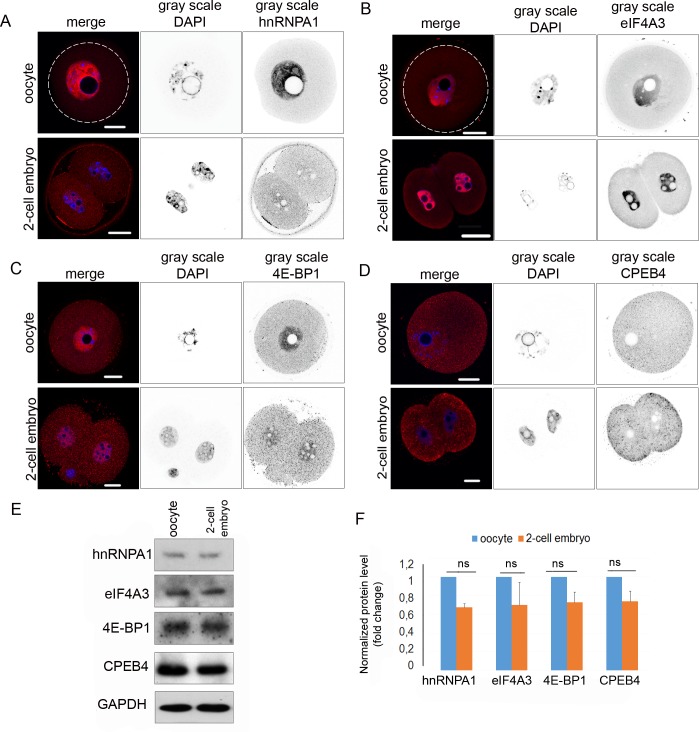
RBPs are essential for RNA metabolism and localization [52–54], and so we accordingly examined the localization of a number of these proteins in the oocytes and early embryos. First, we studied the ubiquitously binding RBPs, heterogeneous nuclear ribonucleoproteins (hnRNP), which participate in pre-mRNA processing and are important determinants of mRNA export localization, translation, and stability [55]. Heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) was localized mostly in the nucleus of the oocyte (Fig 4A). However, in the embryo, the protein was present throughout the whole volume of the cell (Fig 4A). Next, we analyzed the distribution of the exon junction complex protein eIF4A3, which is deposited onto mRNA during splicing and is released during the first round of translation [26,56]. ICC showed a high abundance of eIF4A3 in the nuclei of both cell types, with a weak presence in the cytoplasm (Fig 4B). Another protein, 5’UTR cap-binding protein 4E-BP1, functions as a repressor of cap dependent translation [19,57,58]. In our experiments, 4E-BP1 showed a granular structure in the cytoplasm with a significant increase of signal in the nucleoplasm (Fig 4C and S2 Fig). RNase A treatment disrupted the granular pattern in the oocyte (S2 Fig). CPEB4 is responsible for meiotic progression between MI and MII and for the regulation of cytostatic factors in the frog oocyte [59]. CPEB4 analysis showed a granular distribution in the whole volume of the cells (Fig 4D). Next, WB was used to validate the antibodies (Fig 4E and 4F and S3 Fig). Expression levels of the hnRNPA1, eIF4A3, 4E-BP1 and CPEB4 showed no significant decrease in the early embryo (Fig 4F). The GAPDH protein was used as a loading control for WB.
Fig 4. Localization and expression levels of RNA binding proteins in GV oocyte and 2-cell embryo.
A) Representative confocal images of GV oocyte and 2-cell stage embryo stained with hnRNPA1 antibody (red), DNA stained with DAPI (blue); the gray scale images show separated light channels. The white line indicates the cortex of the oocyte. Scale bars 20 μm. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. B) Representative confocal image of GV oocyte and 2-cell embryo stained with eIF4A3 antibody (red), DNA stained with DAPI (blue); the gray scale images show separated light channels. The white line indicates the cortex of the oocyte. Scale bars 20 μm. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. C) Representative confocal image of GV oocyte and 2-cell embryo stained with 4E-BP1 antibody (red), DNA stained with DAPI (blue); the gray scale images show separated light channels. White line indicate cortex of oocyte. Scale bars 20 μm. The experiment was repeated 3 times, with 25 oocytes per experiment. D) Representative confocal image of GV oocytes and 2-cell embryo stained with CPEB4 antibody (red), DNA stained with DAPI (blue); the gray scale images show separated light channels. The white line indicates the cortex of the oocyte. Scale bars 20 μm. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. E) Representative images from WB probed by antibodies for hnRNPA1, eIF4A3, 4E-BP1, and CPEB4 proteins in the GV oocytes and 2-cell stage embryos. GAPDH was used as a loading control. WBs were performed 3 times, with 100 cells per experiment. F) Quantification of hnRNPA1, eIF4A3, 4E-BP1 and CPEB4 expression in the oocytes and embryos. Data were normalized to GAPDH. The values of oocytes were set as 1. Data are represented as mean ± s.d.; the value bars with ns are not significant, and the asterisk denotes statistically significant differences * p<0.05; **p<0.01; ***p<0.001.
Localization of translational machinery
To characterize the localization and expression of ribosomal proteins in the GV oocyte and 2-cell embryo, we selected several components of the 40S ribosomal subunit–ribosomal proteins S14 (RPS14) and S3 (RPS3) and RPS6 phosphorylated on S235/236 –as well as of the 60S subunit, ribosomal protein L7 (RPL7) and L24 (RPL24). These subunits combine to form the eukaryotic (80S) ribosome [33], a large molecular machine that catalyzes the synthesis of proteins. ICC analysis showed that RPS14 was localized at the cortex of the oocyte and embryo and in the nucleoplasm of the oocyte (Fig 5A). However, in the 2-cell embryo we found a significant decrease of fluorescence in the nucleoplasm and cytoplasm, which was also confirmed by WB analysis (Fig 5A, 5F and 5G). The RPS3 protein was localized evenly in both compartments–nucleus and cytoplasm in the oocyte and the embryo (Fig 5B). The protein level of RPS3 was stable in the oocyte and 2-cell embryo (Fig 5F and 5G). It is known that phosphorylation of RPS6 on S235/236 increases its affinity to the cap structure of RNA, which strongly implies that RPS6 phosphorylation enhances mRNA translation initiation [60]. RPS6 protein phosphorylated on S235/236 was distributed throughout the whole cytoplasm with a decreased signal in the nucleoplasm in both the oocyte and the embryo (Fig 5C). RPS6(S235/236) significantly decreased its presence in the 2-cell embryo (Fig 5C, 5F and 5G). Protein RPL7 was distributed in the cytoplasm of the oocyte and embryo and with higher intensity in the nucleus of the oocyte (Fig 5D) compared to the embryo. Protein levels of RPL7 were stable in the oocyte and 2-cell embryo (Fig 5F and 5G). RPL24 showed more abundant distribution in the cytoplasm than in the nucleus in both cell types (Fig 5E). The quantification of the expression of ribosome components by WB in the oocyte and the embryo showed a significant decrease only for RPS14 (0.67-fold) and RPS6 phosphorylated on S235/236 (0.85-fold) in the 2-cell embryos (Fig 5F and 5G). Other components of ribosome, RPS3, RPL7 and RPL24 showed no significant decrease in the embryo (Fig 5F and 5G). The GAPDH protein was used as a loading control.
Fig 5. Localization and expression levels of selected ribosomal proteins.
A) Confocal images of GV oocyte and 2-cell stage embryo probed with RPS14 antibody (red), DNA stained with DAPI (blue); the gray scale shows separated light channels. Scale bars 20 μm. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. B) Confocal images of GV oocyte and 2-cell stage embryo probed with RPS3 antibody (red), DNA stained with DAPI (blue); the gray scale shows separated light channels. The asterisk indicates cumulus cells. Scale bars 20 μm. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. C) Confocal images of GV oocyte and 2-cell stage embryo probed with RPS6(S235/236) antibody (red), DNA stained with DAPI (blue); the gray scale shows separated light channels. Scale bars 20 μm. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. D) Confocal images of GV oocyte and 2-cell stage embryo probed with RPL7 antibody (red); DNA stained with DAPI (blue); the gray scale shows separated light channels. Scale bars 20 μm. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. E) Confocal images of GV oocyte and 2-cell stage embryo probed with RPL24 antibody (red). DNA stained with DAPI (blue); the gray scale images shows separated light channels. Scale bars 20 μm. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. F) Representative images from WBs for RPS14, RPS3, RPS6 (S235/236), RPL7, RPL24 protein expression in GV oocytes and 2-cell stage embryos, and the loading control (GAPDH); WB was repeated 3 times, with 100 oocytes/embryos per experiment. G) Quantification of expression of RPS14, RPS3, RPS6 (S235/236), RPL7 and RPL24 proteins in the oocyte and embryo. Data were normalized to GAPDH. Data are represented as the mean ± s.d.; the values obtained from oocytes were set as 1. The value bars with ns are not significant, the asterisk denotes statistically significant differences *p<0.05; **p<0.01; ***p<0.001.
We showed different localization and expression of components of 40S and 60S subunits in the mouse oocyte and early embryo.
Nascent translation in oocyte and 2-cell embryo
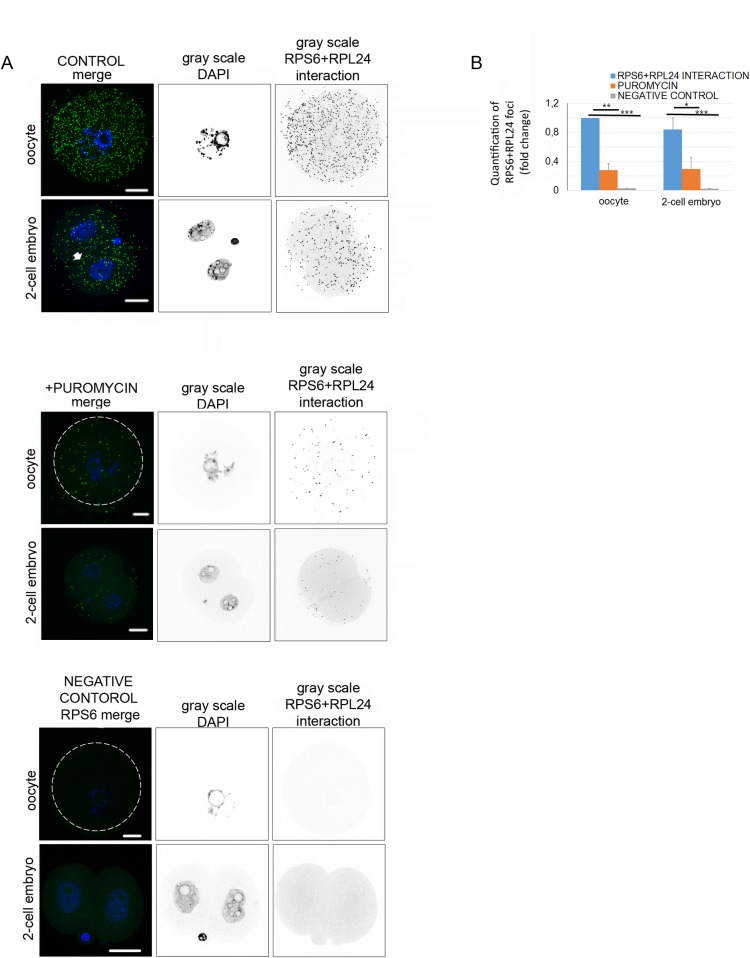
In order to detect ongoing protein synthesis, we used the Duolink in situ proximity ligation assay (PLA) [61,62] with RPL24 and RPS6 specific antibodies to detect assembled 80S ribosomes in the oocyte and embryo. The positive interaction of the RPL24 and RPS6 ribosomal proteins suggests completion of 80S ribosome and ongoing translation. Through PLA, we detected significant interactions of these two proteins in the cytoplasm of the oocyte and 2-cell embryo (Fig 6A). The quantification of the RPL24 and RPS6 interaction foci showed a similar number of RPL24 and RPS6 interactions in the oocyte and the embryo (Fig 6B). The distribution of interactions was even, with increased intensity at the cortex of blastomeres. The cultivation of oocytes or embryos in the presence of translational inhibitor puromycin (which disrupts translating ribosomes) showed a significant decrease (0.7-fold) in the number of interactions in both cell types (Fig 6A and 6B). Moreover, using RPS6 antibody alone as a negative control of PLA showed 0.98-fold significant decrease (Fig 6A and 6B). In order to detect nascent translation in situ, the methionine analog L-homoproparglycine (HPG) was incorporated into the translated proteins during a short 30-minute cultivation period followed by the Click-IT protocol to fluorescently label HPG in the cell [63]. The fluorescent signals of HPG were observed in the whole oocyte, with an increase in the perinuclear area (S4A Fig), while in the 2-cell embryo we detected strong signals at the dividing ridge of the blastomeres. As expected, the disruption of the ribosomes by puromycin decreased the intensity of the HPG signal in the oocyte by about 95%, and in the 2-cell embryo by about 80% (S4A and S4B Fig). In the presence of cycloheximide (CHX; protein elongation inhibitor) the intensity of the HPG signal in the oocyte and the embryo decreased to 15% and 13% respectively (S4A and S4B Fig).
Fig 6. Detection of in situ translation in oocyte and embryo.
A) Proximity ligation assay (PLA) detects interaction of two translational components RPL24 and RPS6 in the oocyte and 2-cell embryo. Fluorescent foci indicate RPL24 and RPS6 interactions (green) in the oocyte and embryo. As negative controls, oocytes and 2-cell embryos were treated with the translational inhibitor puromycin, or a single RPS6 antibody was used. DNA stained with DAPI (blue); the gray scale shows separated channels. Scale bar 20μm. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. B) Graph shows quantification of RPL24 and RPS6 interactions in the whole cell volume. The values obtained from the oocyte were set as 1. The experiment was repeated 3 times, with 25 oocytes/embryos per experiment. Data are represented as mean ± s.d.; the value bars with ns are not significant, and the asterisk denotes statistically significant differences *p<0.05; **p<0.01; ***p<0.001.
The combination of PLA and HPG methods provided consistent visualization of the in situ translation in the oocyte and 2-cell embryo.
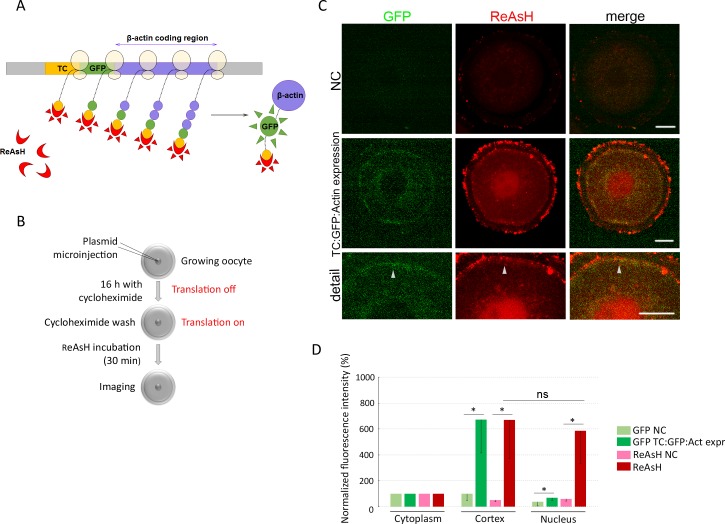
Visualization of translation of endogenous β-actin mRNA in live oocyte
The ReAsH method [42] was used to detect in situ translation of the specific transcript β-actin (Fig 7A). The plasmid coding the sequence of the β-actin ORF with tetracysteine (TC) and GFP domain [43] was injected into the nucleus of transcriptionally active oocytes in the presence of cycloheximide (CHX, protein synthesis inhibitor) (Fig 7B). Overnight cultivation of oocytes led to the transcription of the construct, which mimicked the endogenous β-actin mRNA. We found translation of β-actin in patches at the cortex of the oocyte, where ReAsH and GFP fluorescence were localized. GFP visualized mature protein on the cortex and ReAsH showed spots of in situ translation of β-actin (Fig 7C). A quantification of the images showed a sevenfold increase of the fluorescent intensity in ReAsH and GFP in the patches in the cortex (Fig 7D) compared with other areas of the cell or with negative control (non-injected oocytes). However, we detected a high presence of the ReAsH dye in the nucleus as well, where GFP was not detected (Fig 7C), which might suggest non-specific incorporation or an unknown process in the oocyte [64].
Fig 7. Visualization of translation of endogenous β-actin mRNA in live oocyte.
A) Scheme of the TC GFP β-actin construct and detection of nascent translation. TC—tetracystein sequence; GFP—green fluorescent protein sequence; β-actin—open reading frame; ReAsH—resorufin arsenical hairpin binder. B) Scheme of the experimental procedures for translational detection of the β-actin RNA in the live oocyte. C) Representative confocal images of the oocyte microinjected with TC GFP β-actin plasmid and of non-injected control. ReAsH dye labels TC tag (red) of newly translated β-actin protein. GFP (green) was used as a marker of fully translated β-actin. The arrowheads depict nascent translation of β-actin RNA. Scale bars 10 µm. D) Quantification of the fluorescence intensity in the cytoplasm, cortex and nucleus. The values of the cytoplasm were set as 100%. The experiment was repeated 3 times, with 10 oocytes per experiment. Data are represented as mean ± s.d.; the value bars with ns are not significant; and the asterisk denotes statistically significant differences *p<0.05; **p<0.01; ***p<0.001.
We were able to detect translation of the endogenous β-actin mRNA in the living oocyte, which resembled the known localization of filamentous actin [65,66], shown also in S5 Fig.
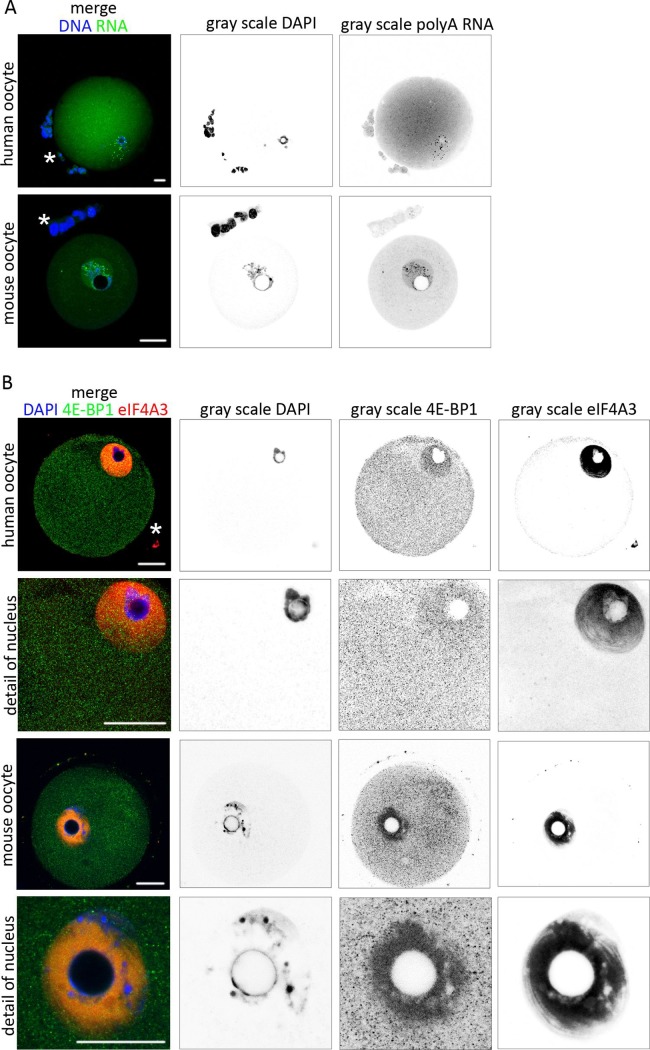
Localization of global RNA and RBPs in the human oocyte
In order to study the similarity between mouse and human oocytes, a localization of the global transcriptome was determined through the visualization of the poly(A) RNA population. We found that the poly(A) RNA fluorescence signal was distributed evenly throughout the cytoplasm and nucleoplasm with the presence of the abundant poly(A) RNA foci in the cytoplasm and nucleus of the human oocyte (Fig 8A). Next, the ICC labeling of RBPs, 4E-BP1 (green) and eIF4A3 (red) was performed, uncovering a similar localization of these proteins in human oocytes to our previously reported findings in the mouse oocyte [19] (Fig 8B). 4E-BP1 was distributed in both, the cytoplasm and the nucleoplasm. Contrastingly, eIF4A3 was localized in the nucleoplasm only (Fig 8B). Through analyses of the poly(A) RNA population and RBPs, we found similar localization of the selected transcriptome markers in both mammalian species.
Fig 8. Localization of poly(A) RNA and RNA binding proteins in human oocyte.
A) Single Z-stack from confocal images of human and mouse GV oocytes. RNA FISH detecting poly(A) RNA subpopulation (green) and DNA (blue). The gray scale shows separated light channels. The asterisk indicates cumulus cells. The experiment was repeated 3 times, with 15 mouse oocytes per experiment and 3 human oocytes per experiment. Scale bars 20 μm. B) Single Z-stack of human and mouse GV oocytes and 2-cell stage mouse embryos probed with 4E-BP1 (green) and eIF4A3 (red) and DAPI (blue). The gray scale shows separated channels. The experiment was repeated 3 times, with 15 mouse oocytes and 3 human oocytes. Scale bars 20 μm. The asterisk indicates cumulus cells.
Discussion
Post-transcriptional control of gene expression at the level of translation has been shown to be essential for the regulation of a number of cellular processes during development [19,67]. Mammalian oocytes are transcriptionally active during their growth period. However, after resumption of meiosis, the oocytes reach a phase of transcriptional quiescence and utilize a store of maternally synthesized RNAs. The progression through meiosis and early embryogenesis is therefore regulated in the oocyte at the level of mRNA stabilization, translation and post-translational modifications. The aim of this study was to detect the global transcriptome present in the mammalian GV oocyte and the 2-cell embryo as a prerequisite for protein synthesis. Using RCA, poly(A) and RNA antibody detection techniques, we observed RNA distribution in both the cytoplasm and the nucleus of transcriptionally inactive oocytes, as well as in active 2-cell stage embryos, consistent with previous analyses [1,8]. Our results lead us to the conclusion that the transcriptionally inactive nucleus might serve as a repository for translationally dormant mRNAs, which may later be translated when the nuclear envelope breaks down after the resumption of meiosis. However, the presence of RNAs in the nucleus of the 2-cell embryo might also be the result of transcription. Our results from RNA FISH and immunocytochemistry analyses positively correlate with previously published results [8,19]. In addition to global RNA found in the nucleus, we detected the partial presence of specific mRNAs Dazl and β-actin in the nucleoplasm. Neat2 lncRNA was present exclusively in the nucleus of the oocyte. In contrast, in the 2–cell embryo, Neat2 appeared also in the cytoplasm. Neat2 lncRNA and other RNAs of this class are known to be localized in the nuclei of mouse embryonic fibroblasts and HeLa cells [7,51].
Although findings on translation in the nuclei of cells have been published [68,69], and we detected a number of ribosome components (5.8S rRNA RPS14, RPS3, RPL7) in the nuclei, our other results (absence of phosphorylated RPS6 in the nucleus of the both cell types, and presence of the translational inhibitor 4E-BP1 in the nucleoplasm) indicate that the mRNAs present in the nucleus are translationally dormant [70]. In addition, the translational repressor 4E-BP1 was active in both stages, the oocyte and the embryo (consistently with [19]). The localization of 4E-BP1 in the 2-cell embryo might have a role in the post-transcriptional regulation [71] of the newly synthetized RNA upon embryonic gene activation [72]. The localization of ribosomal components in the nucleoplasm suggests a known mechanism of ribosome biogenesis in the nucleus [73,74]. eIF4A3 is the component of translation initiation complex and also of the exon junction complex, and has a dominant nuclear localization signal sequence [75] which might be responsible for the localization of the protein after the pioneer round of translation [76]. CPEB proteins are key players in the polyadenylation of maternal transcripts [11,20,77] and thus ensure their translation. Our previous work on the mammalian oocyte [21] revealed that CPEB1 was degraded after the resumption of meiosis. Igea and Mendez (2010) proposed that, in the frog oocyte, CPEB4 is accumulated during the second meiotic division and thus can substitute for degraded CPEB1. These authors further suggest that a mechanism is present in Xenopus which enables CPEB4 to replace later polyadenylation events in the oocyte. However, we found that CPEB4 was also abundant in the oocyte prior to the resumption of meiosis when CPEB1 was also abundant and functional, suggesting a different mechanism of CPEB regulation in the mammalian oocyte.
We found that selected RBPs, hnRNPA1, eIF4A3, CPEB4 and translational repressor 4E-BP1 were expressed at the same level in the oocyte and in the embryo. However, the ribosomal components RPS14 and phosphorylated RPS6(S235/236) showed significant reduction in the 2-cell embryos. On the other hand, ribosomal components RPS3, RPL7 and RPL24 did not show significant changes in the embryos. We observed different localization of components of ribosomal subunits. During ribosome biogenesis, proteins change their localization, and they also have different localization if they are unbound from ribosomal subunits. We assume that localization of RPS3, RPS14 and RPL7 in the nucleus is necessary for ribosome biogenesis [76, 77, 78, 79] prior to nuclear envelope breakdown to efficiently localize translational machinery to the newly developing spindle [8,9,58].
It is accepted that maternal components (RNA, proteins, organelles) are eliminated after fertilization [32,80]. It is possible that mammalian oocytes and embryos adjust their pool of rRNA and ribosomes to match the mRNA pool and protein requirements, with high levels during oocyte growth, and a relative quiescence in MII stage is accompanied by global reduction of protein synthesis [81–83]. The presence or absence of specific ribosomal proteins in the ribosome is known to control translation of specific subsets of mRNAs [84,85]. This suggests that the elimination of maternal ribosomes might lead to changes in the RNA metabolism between oocyte to embryo transition. Ellederova et al. (2006) and Susor et al. (2008) observed a decrease in global translation during early embryo development, which supports our results generated by PLA showing that 2-cell embryos display a 16% significant decrease in ribosome assembly. In addition, we found that the interaction between RPL24 and RPS6 decreased after the disruption of active ribosomes by puromycin [86], indicating that the interaction foci were active translational sites. We propose that the studied components of the translational machinery are essential for oocyte and embryo development, consistent with [8,19,58]. Monti et al. (2013) discovered that RPL24 and RPS6 are transcribed at the end of the transcriptional activity of the oocyte and are essential for the oocyte to support early embryo development. Ellederova et al. (2006) and Tomek et al. (2002) also observed that despite a decrease in overall protein synthesis in the mammalian oocyte during meiosis, there is a regulatory program that ensures temporal and spatial synthesis of specific proteins that are essential for meiotic progression and embryo development. Similarly with this basic principle we have confirmed the decrease in translation in 2-cell embryos by PLA and HPG methods.
Moreover, we showed in situ translation of the well-known cytoskeletal protein β-actin in the live oocyte. A quantification of the images showed a sevenfold increase of fluorescent intensity in ReAsH and GFP in the patches within the cortex. In vivo transcription of the construct DNA showed the translation of β-actin RNA in the area of protein localization in the cell.
Although the human oocyte is extremely valuable as the gold standard for assessing clinical relevance, using this cell type is limited in several ways. By starting with the identification of the localization of transcriptome and RBPs in the mouse oocyte and the application of selected markers to the human oocyte, we have found a similar localization of poly(A) RNA and proteins 4E-BP1 and eIF4A3 in both mammalian species, which suggests similar RNA metabolism in human and mouse GV oocytes.
Our findings provide a fundamental insight into the cellular architecture of maternal RNAs in oocytes and embryos from two different mammalian species, mouse and human. We provide a view of the localization of ribosomal proteins, revealing unique and unexpected roles for the translation machinery itself in directing essential aspects of oocyte and early embryo development.
Supporting information
A) Detection of RNA coding GFP in the oocyte. Detection of non-endogenous RNA coding eGFP in the oocyte. Non-endogenous RNA detection was used as a negative control. GFP (green) and DAPI (blue). The gray scale shows separated light channels. The white line indicates the cortex of the oocyte and the arrow with the white line indicates the nucleus of the oocyte. The experiment was repeated 3 times, with 15 mouse oocytes per experiment. Scale bars 20 μm. B) As negative controls for smRNA FISH, RNA was digested by RNase A treatment in the fixed oocytes. Oocytes were probed for Dazl, β-actin and Neat2 RNAs. The gray scale shows separated light channels: RNA (green) and DNA (blue). The white line indicates the cortex of the oocyte. The arrow with the white line indicates the nucleus of the oocyte. The asterisk indicates cumulus cells. Scale bars 20 μm.
(TIF)
Treatment with RNase A leads to decrease of signal intensity and disruption of granular structure of 4E-BP1. Single Z-stack from confocal images of mouse GV oocytes and detail of nuclei. We used for 4E-BP1 (green) and eIF4A3 (red) antibodies and stained DNA with DAPI (blue). The gray scale shows different light channels. The experiment was repeated 3 times, with 25 oocytes per experiment. Scale bars represent 10 μm.
(TIF)
Images of WBs probed for specific proteins with depicted molecular size.
(TIF)
A) Single Z-stack of confocal image shows nascent translation (red) in GV oocytes and 2-cell embryos. Cells were cultured for 30 minutes in the presence of methionine analog HPG and nascent translation was visualized by Click-IT chemistry. The suppression of translation by potent inhibitors puromycin and cycloheximide (CHX) inhibits the incorporation of HPG to the newly synthetized proteins. The white line indicates the cortex of the oocyte. Representative images of at least three independent experiments are shown. DNA stained with DAPI (blue). The gray scale shows light channel for HPG. The arrow indicates the ridge of blastomeres. The asterisk indicates cumulus cells. Scale bars 20μm.
B) Quantification of fluorescence intensity of HPG signal after treatment by puromycin or CHX in GV oocytes and 2-cell stage embryos. The experiment was repeated 3 times, with 25 oocytes per experiment. Data are represented as mean ± s.d.; the values bars with ns are not significant, and the asterisk denotes statistically significant differences *p<0.05; **p<0.01; ***p<0.001.
(TIF)
Single Z-stack of confocal images. Actin filaments visualized by phalloidin (green) and DNA stained with DAPI (blue). Representative images of at least three independent experiments are shown. Scale bars represent 10 μm.
(TIF)
Acknowledgments
We thank Jaroslava Supolikova and Marketa Hancova for their exceptional technical assistance with experiments and also to Lenka Gahurova, PhD for valuable advices and comments. We are also grateful to the members of the Obstetrics and Gynecology Clinic of the General University Hospital in Prague–Mgr. Martina Moosova, Dr. Karel Rezabek and Dr. Jana Pavelkova–for obtaining human oocytes.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Charles University in Prague, Faculty of Cell Biology-GAUK 243-227026 to DJ. The Academy of Sciences of the Czech Republic, http://www.cas.cz/, GACR13–12291S to AS, GACR15–22765S to MK, EXCELLENCECZ.02.1.01/0.0/0.0/15_003/0000460 OP RDE and Institutional Research Concept RVO67985904.
References
- 1.Schultz RM. Regulation of zygotic gene activation in the mouse. BioEssays News Rev Mol Cell Dev Biol. 1993;15: 531–538. doi: 10.1002/bies.950150806 [DOI] [PubMed] [Google Scholar]
- 2.Ellederova Z, Kovarova H, Melo-Sterza F, Livingstone M, Tomek W, Kubelka M. Suppression of translation during in vitro maturation of pig oocytes despite enhanced formation of cap-binding protein complex eIF4F and 4E-BP1 hyperphosphorylation. Mol Reprod Dev. 2006;73: 68–76. doi: 10.1002/mrd.20368 [DOI] [PubMed] [Google Scholar]
- 3.Tomek W, Melo Sterza FA, Kubelka M, Wollenhaupt K, Torner H, Anger M, et al. Regulation of translation during in vitro maturation of bovine oocytes: the role of MAP kinase, eIF4E (cap binding protein) phosphorylation, and eIF4E-BP1. Biol Reprod. 2002;66: 1274–1282. [DOI] [PubMed] [Google Scholar]
- 4.Flemr M, Ma J, Schultz RM, Svoboda P. P-body loss is concomitant with formation of a messenger RNA storage domain in mouse oocytes. Biol Reprod. 2010;82: 1008–1017. doi: 10.1095/biolreprod.109.082057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280: 585–590. [DOI] [PubMed] [Google Scholar]
- 6.Trcek T, Chao JA, Larson DR, Park HY, Zenklusen D, Shenoy SM, et al. Single-mRNA counting using fluorescent in situ hybridization in budding yeast. Nat Protoc. 2012;7: 408–419. doi: 10.1038/nprot.2011.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16: 20 doi: 10.1186/s13059-015-0586-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Susor A, Jansova D, Cerna R, Danylevska A, Anger M, Toralova T, et al. Temporal and spatial regulation of translation in the mammalian oocyte via the mTOR-eIF4F pathway. Nat Commun. 2015;6: 6078 doi: 10.1038/ncomms7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Susor A, Kubelka M. Translational Regulation in the Mammalian Oocyte Oocytes: Springer, Cham; 2017. pp. 257–295. doi: 10.1007/978-3-319-60855-6_12 [DOI] [PubMed] [Google Scholar]
- 10.Blower MD, Feric E, Weis K, Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol. 2007;179: 1365–1373. doi: 10.1083/jcb.200705163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groisman I, Huang Y-S, Mendez R, Cao Q, Theurkauf W, Richter JD. CPEB, Maskin, and Cyclin B1 mRNA at the Mitotic Apparatus: Implications for Local Translational Control of Cell Division. Cell. 2000;103: 435–447. doi: 10.1016/S0092-8674(00)00135-5 [DOI] [PubMed] [Google Scholar]
- 12.VerMilyea MD, Maneck M, Yoshida N, Blochberger I, Suzuki E, Suzuki T, et al. Transcriptome asymmetry within mouse zygotes but not between early embryonic sister blastomeres. EMBO J. 2011;30: 1841–1851. doi: 10.1038/emboj.2011.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becalska AN, Gavis ER. Lighting up mRNA localization in Drosophila oogenesis. Dev Camb Engl. 2009;136: 2493–2503. doi: 10.1242/dev.032391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnstone O, Lasko and P. Translational Regulation and RNA Localization in Drosophila Oocytes and Embryos. Annu Rev Genet. 2001;35: 365–406. doi: 10.1146/annurev.genet.35.102401.090756 [DOI] [PubMed] [Google Scholar]
- 15.King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97: 19–33. doi: 10.1042/BC20040067 [DOI] [PubMed] [Google Scholar]
- 16.Kloc M, Etkin LD. RNA localization mechanisms in oocytes. J Cell Sci. 2005;118: 269–282. doi: 10.1242/jcs.01637 [DOI] [PubMed] [Google Scholar]
- 17.Medioni C, Mowry K, Besse F. Principles and roles of mRNA localization in animal development. Dev Camb Engl. 2012;139: 3263–3276. doi: 10.1242/dev.078626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Müller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet. 2013;14: 275–287. doi: 10.1038/nrg3434 [DOI] [PubMed] [Google Scholar]
- 19.Jansova D, Koncicka M, Tetkova A, Cerna R, Malik R, Del Llano E, et al. Regulation of 4E-BP1 activity in the mammalian oocyte. Cell Cycle Georget Tex. 2017; 1–13. doi: 10.1080/15384101.2017.1295178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32: 279–285. doi: 10.1016/j.tibs.2007.04.004 [DOI] [PubMed] [Google Scholar]
- 21.Komrskova P, Susor A, Malik R, Prochazkova B, Liskova L, Supolikova J, et al. Aurora kinase A is not involved in CPEB1 phosphorylation and cyclin B1 mRNA polyadenylation during meiotic maturation of porcine oocytes. PloS One. 2014;9: e101222 doi: 10.1371/journal.pone.0101222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Görlach M, Burd CG, Portman DS, Dreyfuss G. The hnRNP proteins. Mol Biol Rep. 1993;18: 73–78. [DOI] [PubMed] [Google Scholar]
- 23.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355: 730–732. doi: 10.1038/355730a0 [DOI] [PubMed] [Google Scholar]
- 24.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13: 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jean-Philippe J, Paz S, Caputi M. hnRNP A1: The Swiss Army Knife of Gene Expression. Int J Mol Sci. 2013;14: 18999–19024. doi: 10.3390/ijms140918999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, et al. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10: 200–209. doi: 10.1261/rna.5230104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Murigneux V, Le Hir H. Transcriptome-wide modulation of splicing by the exon junction complex. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0551-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue K, Ohno M, Sakamoto H, Shimura Y. Effect of the cap structure on pre-mRNA splicing in Xenopus oocyte nuclei. Genes Dev. 1989;3: 1472–1479. [DOI] [PubMed] [Google Scholar]
- 29.Monti M, Zanoni M, Calligaro A, Ko MSH, Mauri P, Redi CA. Developmental Arrest and Mouse Antral Not-Surrounded Nucleolus Oocytes. Biol Reprod. 2013;88: 2 doi: 10.1095/biolreprod.112.103887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clegg KB, Pikó L. RNA synthesis and cytoplasmic polyadenylation in the one-cell mouse embryo. Nature. 1982;295: 342–345. doi: 10.1038/295342a0 [DOI] [PubMed] [Google Scholar]
- 31.Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272: 483–496. doi: 10.1016/j.ydbio.2004.05.018 [DOI] [PubMed] [Google Scholar]
- 32.Su Y-Q, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, et al. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302: 104–117. doi: 10.1016/j.ydbio.2006.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334: 1524–1529. doi: 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- 34.Krieg J, Hofsteenge J, Thomas G. Identification of the 40 S ribosomal protein S6 phosphorylation sites induced by cycloheximide. J Biol Chem. 1988;263: 11473–11477. [PubMed] [Google Scholar]
- 35.Rosner M, Fuchs C, Dolznig H, Hengstschläger M. Different cytoplasmic/nuclear distribution of S6 protein phosphorylated at S240/244 and S235/236. Amino Acids. 2011;40: 595–600. doi: 10.1007/s00726-010-0684-2 [DOI] [PubMed] [Google Scholar]
- 36.Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 2006;31: 342–348. doi: 10.1016/j.tibs.2006.04.003 [DOI] [PubMed] [Google Scholar]
- 37.Meyuhas O, Dreazen A. Ribosomal protein S6 kinase from TOP mRNAs to cell size. Prog Mol Biol Transl Sci. 2009;90: 109–153. doi: 10.1016/S1877-1173(09)90003-5 [DOI] [PubMed] [Google Scholar]
- 38.Kim T-H, Leslie P, Zhang Y. Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. Oncotarget. 2014;5: 860–871. doi: 10.18632/oncotarget.1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal Protein S14 Unties the MDM2-p53 Loop Upon Ribosomal Stress. Oncogene. 2013;32: 388–396. doi: 10.1038/onc.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jansova D. Single Molecule RNA FISH in the Mammalian Oocyte. Bio-protocol. 2015;2015: e1666 https://doi.org/10.21769/BioProtoc.1666 [Google Scholar]
- 41.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10: 442–458. doi: 10.1038/nprot.2014.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Machleidt T, Robers M, Hanson GT. Protein labeling with FlAsH and ReAsH. Methods Mol Biol Clifton NJ. 2007;356: 209–220. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. Visualization of mRNA translation in living cells. J Cell Biol. 2006;175: 67–76. doi: 10.1083/jcb.200512137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tetkova A, Hancova M. Mouse Oocyte Isolation, Cultivation and RNA Microinjection. BIO-Protoc. 2016;6 doi: 10.21769/BioProtoc.1729 [Google Scholar]
- 45.Jambor H, Surendranath V, Kalinka AT, Mejstrik P, Saalfeld S, Tomancak P. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. eLife. 2015;4 doi: 10.7554/eLife.05003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clegg KB, Pikó L. Poly(A) length, cytoplasmic adenylation and synthesis of poly(A)+ RNA in early mouse embryos. Dev Biol. 1983;95: 331–341. [DOI] [PubMed] [Google Scholar]
- 47.Larsson C, Grundberg I, Söderberg O, Nilsson M. In situ detection and genotyping of individual mRNA molecules. Nat Methods. 2010;7: 395–397. doi: 10.1038/nmeth.1448 [DOI] [PubMed] [Google Scholar]
- 48.Bochnig P, Reuter R, Bringmann P, Lührmann R. A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. Eur J Biochem. 1987;168: 461–467. [DOI] [PubMed] [Google Scholar]
- 49.Elela SA, Nazar RN. Role of the 5.8S rRNA in ribosome translocation. Nucleic Acids Res. 1997;25: 1788–1794. doi: 10.1093/nar/25.9.1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lerner EA, Lerner MR, Janeway CA, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A. 1981;78: 2737–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18: 738–751. doi: 10.1261/rna.028639.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deshler JO, Highett MI, Abramson T, Schnapp BJ. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr Biol. 1998;8: 489–496. doi: 10.1016/S0960-9822(98)70200-3 [DOI] [PubMed] [Google Scholar]
- 53.Tolino M, Köhrmann M, Kiebler MA. RNA-binding proteins involved in RNA localization and their implications in neuronal diseases. Eur J Neurosci. 2012;35: 1818–1836. doi: 10.1111/j.1460-9568.2012.08160.x [DOI] [PubMed] [Google Scholar]
- 54.Zhang Z, Wang Y, Jiang Y, Lin P, Jia X, Zou Z. Ribosomal protein L24 is differentially expressed in ovary and testis of the marine shrimp Marsupenaeus japonicus. Comp Biochem Physiol B Biochem Mol Biol. 2007;147: 466–474. doi: 10.1016/j.cbpb.2007.02.013 [DOI] [PubMed] [Google Scholar]
- 55.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3: 195–205. doi: 10.1038/nrm760 [DOI] [PubMed] [Google Scholar]
- 56.Shibuya T, Tange TØ, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11: 346–351. doi: 10.1038/nsmb750 [DOI] [PubMed] [Google Scholar]
- 57.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13: 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romasko EJ, Amarnath D, Midic U, Latham KE. Association of maternal mRNA and phosphorylated EIF4EBP1 variants with the spindle in mouse oocytes: localized translational control supporting female meiosis in mammals. Genetics. 2013;195: 349–358. doi: 10.1534/genetics.113.154005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Igea A, Méndez R. Meiosis requires a translational positive loop where CPEB1 ensues its replacement by CPEB4. EMBO J. 2010;29: 2182–2193. doi: 10.1038/emboj.2010.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, et al. RAS/ERK Signaling Promotes Site-specific Ribosomal Protein S6 Phosphorylation via RSK and Stimulates Cap-dependent Translation. J Biol Chem. 2007;282: 14056–14064. doi: 10.1074/jbc.M700906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benesova V, Kinterova V, Kanka J, Toralova T. Characterization of SCF-Complex during Bovine Preimplantation Development. PloS One. 2016;11: e0147096 doi: 10.1371/journal.pone.0147096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Söderberg O, Leuchowius K-J, Gullberg M, Jarvius M, Weibrecht I, Larsson L-G, et al. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods San Diego Calif. 2008;45: 227–232. doi: 10.1016/j.ymeth.2008.06.014 [DOI] [PubMed] [Google Scholar]
- 63.Dieterich DC, Hodas JJL, Gouzer G, Shadrin IY, Ngo JT, Triller A, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13: 897–905. doi: 10.1038/nn.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reid DW, Nicchitta CV. The enduring enigma of nuclear translation. J Cell Biol. 2012;197: 7–9. doi: 10.1083/jcb.201202140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Azoury J, Verlhac M-H, Dumont J. Actin filaments: key players in the control of asymmetric divisions in mouse oocytes. Biol Cell Auspices Eur Cell Biol Organ. 2009;101: 69–76. [DOI] [PubMed] [Google Scholar]
- 66.Schuh M, Ellenberg J. Self-Organization of MTOCs Replaces Centrosome Function during Acentrosomal Spindle Assembly in Live Mouse Oocytes. Cell. 2007;130: 484–498. doi: 10.1016/j.cell.2007.06.025 [DOI] [PubMed] [Google Scholar]
- 67.Curtis D, Lehmann R, Zamore PD. Translational regulation in development. Cell. 1995;81: 171–178. [DOI] [PubMed] [Google Scholar]
- 68.Belgrader P, Cheng J, Maquat LE. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci U S A. 1993;90: 482–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287: 5518–5527. doi: 10.1074/jbc.M111.312280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ford CL, Randal-Whitis L, Ellis SR. Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Res. 1999;59: 704–710. [PubMed] [Google Scholar]
- 71.Siemer C, Smiljakovic T, Bhojwani M, Leiding C, Kanitz W, Kubelka M, et al. Analysis of mRNA associated factors during bovine oocyte maturation and early embryonic development. Mol Reprod Dev. 2009;76: 1208–1219. doi: 10.1002/mrd.21096 [DOI] [PubMed] [Google Scholar]
- 72.Hamatani T, Carter MG, Sharov AA, Ko MSH. Dynamics of Global Gene Expression Changes during Mouse Preimplantation Development. Dev Cell. 2004;6: 117–131. doi: 10.1016/S1534-5807(03)00373-3 [DOI] [PubMed] [Google Scholar]
- 73.David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, et al. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197: 45–57. doi: 10.1083/jcb.201112145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13: 355–369. doi: 10.1038/nrm3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.SHIBUYA T, TANGE TØ, STROUPE ME, MOORE MJ. Mutational analysis of human eIF4AIII identifies regions necessary for exon junction complex formation and nonsense-mediated mRNA decay. RNA. 2006;12: 360–374. doi: 10.1261/rna.2190706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maquat LE, Tarn W-Y, Isken O. The Pioneer Round of Translation: Features and Functions. Cell. 2010;142: 368–374. doi: 10.1016/j.cell.2010.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eliscovich C, Peset I, Vernos I, Méndez R. Spindle-localized CPE-mediated translation controls meiotic chromosome segregation. Nat Cell Biol. 2008;10: 858–865. doi: 10.1038/ncb1746 [DOI] [PubMed] [Google Scholar]
- 78.Meyuhas O, Klein A. The mouse ribosomal protein L7 gene. Its primary structure and functional analysis of the promoter region. J Biol Chem. 1990;265: 11465–11473. [PubMed] [Google Scholar]
- 79.Hemmerich P, von Mikecz A, Neumann F, Sözeri O, Wolff-Vorbeck G, Zoebelein R, et al. Structural and functional properties of ribosomal protein L7 from humans and rodents. Nucleic Acids Res. 1993;21: 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alizadeh Z, Kageyama S-I, Aoki F. Degradation of maternal mRNA in mouse embryos: selective degradation of specific mRNAs after fertilization. Mol Reprod Dev. 2005;72: 281–290. doi: 10.1002/mrd.20340 [DOI] [PubMed] [Google Scholar]
- 81.Schultz RM, Wassarman PM. Biochemical studies of mammalian oogenesis: Protein synthesis during oocyte growth and meiotic maturation in the mouse. J Cell Sci. 1977;24: 167–194. [DOI] [PubMed] [Google Scholar]
- 82.Susor A, Jelínková L, Karabínová P, Torner H, Tomek W, Kovárová H, et al. Regulation of cap-dependent translation initiation in the early stage porcine parthenotes. Mol Reprod Dev. 2008;75: 1716–1725. doi: 10.1002/mrd.20913 [DOI] [PubMed] [Google Scholar]
- 83.Ellederová Z, Cais O, Susor A, Uhlírová K, Kovárová H, Jelínková L, et al. ERK1/2 map kinase metabolic pathway is responsible for phosphorylation of translation initiation factor eIF4E during in vitro maturation of pig oocytes. Mol Reprod Dev. 2008;75: 309–317. doi: 10.1002/mrd.20690 [DOI] [PubMed] [Google Scholar]
- 84.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145: 383–397. doi: 10.1016/j.cell.2011.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, et al. mTORC1 Phosphorylation Sites Encode Their Sensitivity to Starvation and Rapamycin. Science. 2013;341: 1236566 doi: 10.1126/science.1236566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Azzam ME, Algranati ID. Mechanism of Puromycin Action: Fate of Ribosomes after Release of Nascent Protein Chains from Polysomes. Proc Natl Acad Sci U S A. 1973;70: 3866–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Detection of RNA coding GFP in the oocyte. Detection of non-endogenous RNA coding eGFP in the oocyte. Non-endogenous RNA detection was used as a negative control. GFP (green) and DAPI (blue). The gray scale shows separated light channels. The white line indicates the cortex of the oocyte and the arrow with the white line indicates the nucleus of the oocyte. The experiment was repeated 3 times, with 15 mouse oocytes per experiment. Scale bars 20 μm. B) As negative controls for smRNA FISH, RNA was digested by RNase A treatment in the fixed oocytes. Oocytes were probed for Dazl, β-actin and Neat2 RNAs. The gray scale shows separated light channels: RNA (green) and DNA (blue). The white line indicates the cortex of the oocyte. The arrow with the white line indicates the nucleus of the oocyte. The asterisk indicates cumulus cells. Scale bars 20 μm.
(TIF)
Treatment with RNase A leads to decrease of signal intensity and disruption of granular structure of 4E-BP1. Single Z-stack from confocal images of mouse GV oocytes and detail of nuclei. We used for 4E-BP1 (green) and eIF4A3 (red) antibodies and stained DNA with DAPI (blue). The gray scale shows different light channels. The experiment was repeated 3 times, with 25 oocytes per experiment. Scale bars represent 10 μm.
(TIF)
Images of WBs probed for specific proteins with depicted molecular size.
(TIF)
A) Single Z-stack of confocal image shows nascent translation (red) in GV oocytes and 2-cell embryos. Cells were cultured for 30 minutes in the presence of methionine analog HPG and nascent translation was visualized by Click-IT chemistry. The suppression of translation by potent inhibitors puromycin and cycloheximide (CHX) inhibits the incorporation of HPG to the newly synthetized proteins. The white line indicates the cortex of the oocyte. Representative images of at least three independent experiments are shown. DNA stained with DAPI (blue). The gray scale shows light channel for HPG. The arrow indicates the ridge of blastomeres. The asterisk indicates cumulus cells. Scale bars 20μm.
B) Quantification of fluorescence intensity of HPG signal after treatment by puromycin or CHX in GV oocytes and 2-cell stage embryos. The experiment was repeated 3 times, with 25 oocytes per experiment. Data are represented as mean ± s.d.; the values bars with ns are not significant, and the asterisk denotes statistically significant differences *p<0.05; **p<0.01; ***p<0.001.
(TIF)
Single Z-stack of confocal images. Actin filaments visualized by phalloidin (green) and DNA stained with DAPI (blue). Representative images of at least three independent experiments are shown. Scale bars represent 10 μm.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.