Abstract
Melatonin is a neurohormone produced in both animals and plants. It binds at least three G-protein-coupled receptors: MT1 and MT2, and Mel1cGPR. Mammalian GPR50 evolved from the reptilian/avian Mel1c and lost its capacity to bind melatonin in all the therian mammal species that have been tested. In order to determine if binding is lost in the oldest surviving mammalian lineage of monotremes we investigated whether the melatonin receptor has the ability to bind melatonin in the platypus (Ornithorhynchus anatinus), and evaluated its pharmacological profile. Sequence and phylogenetic analysis showed that platypus has in fact retained the ancestral Mel1c and has the capacity to bind melatonin similar to other mammalian melatonin receptors (MT1 and MT2), with an affinity in the 1 nM range. We also investigated the binding of a set of melatoninergic ligands used previously to characterize the molecular pharmacology of the melatonin receptors from sheep, rats, mice, and humans and found that the general profiles of these compounds make Mel1c resemble human MT1 more than MT2. This work shows that the loss of GPR50 binding evolved after the divergence of monotremes less than 190MYA in therian mammals.
Introduction
Melatonin is a neurohormone produced by the pineal gland and maintains the circadian information through diffusion in the blood circulation to peripheral organs. Melatonin exists in almost all living organisms, and its actions are mediated by binding to two membrane receptors (MT1 and MT2) [1], a nuclear receptor [2] (although this observation has been challenged [3]), or a protein (quinone reductase 2; QR2) [4]. If the MT1 and MT2 receptors bind melatonin with an affinity of 1 nM and below, QR2 has an affinity for melatonin in the 10 μM range. In mammals, a third receptor exists: the melatonin-related receptor (MRR), recently renamed GPR50 [5,6], that does not bind melatonin but could regulate the activity, specificity, and signaling pathways of MT1 via dimerization [1]. GPR50 is thus an orphan receptor because its ligand, if any, is unknown. Mel1c has been described as a melatonin-binding receptor in amphibians, birds, and fish [5]. Dufourny et al. [7] demonstrated that the melatonin-related receptor or GPR50 in mammals is likely the protein that evolved from Mel1c on the basis of phylogenetic and sequence analyses. However, this discovery raised the question of why this receptor no longer binds melatonin in mammals.
Although the melatonin binding capacity of Mel1c and pharmacological profiles (e.g., in chicken) have been known for quite a long time [8–10], in depth comparative molecular pharmacological evidence is lacking. In fact, the few data available on the activity of the Mel1c receptor, particularly from Xenopus laevis, are functional ones, with some compounds using a cell-based assay, pigment aggregation in melanophores [9,11–14]. Further to this, some data is available on binding recorded using chicken brain membranes (see for example, Sugden [15] or Pegurier et al. [16]).
Genes involved in reproduction undergo accelerated evolution [17–19], and it seems that the evolution of genes, when fast, is often linked to reproduction. In this sense, we assumed that the passage from eggs to viviparity could be accompanied by many different changes, including from a melatonin-binding Mel1c (in amphibians and birds) to a sequence-related non-melatonin binding GPR50. In that sense, we wanted to better understand the evolution of Mel1c to GPR50 binding in mammals by investigating the oldest surviving monotreme lineage. Monotremes (platypus, echidna) are the oldest surviving mammals and provide unprecedented insight into mammalian evolution regarding, for example, the evolution of sex chromosomes [20,21].
Here we analyzed the evolution and function of the melatonin receptor Mel1c/GPR50 in the iconic egg-laying platypus. Interestingly, we found that the platypus Mel1c receptor shares high similarity with bird and other vertebrate Mel1c, while the therian ortholog GPR50 is more diverged. Furthermore, we found that platypus Mel1c binds melatonin while this binding has been lost in other mammals. Further, we analyzed if this change resulted in other pharmacological responses, which is not the case. Overall this work provides novel information about the functional evolution of this enigmatic receptor.
Materials and methods
Reagents and ligands
The radioligand 2-[125I]-iodomelatonin (specific activity: 2200 Ci/mmol) was purchased from Perkin Elmer (Courtaboeuf, France). Melatonin was obtained from Sigma (St Louis, MO) and 4-phenyl-2-propionamidotetraline (4P-PDOT) and 2-benzyl-N-acetyltryptamine (luzindole) from Tocris (Bristol, UK). Compounds were dissolved in dimethylsulfoxide (DMSO) at a stock concentration of 10 mM and stored at –20°C.
RNA extraction and cDNA synthesis
Platypus specimens were killed with an intraperitoneal injection of pentobarbitone sodium (Nembutal) or pentobarbital (Lethabarb) at a dose of 0.1mg g-1. Tissue samples were snap-frozen. RNA was extracted from whole brain tissue using TRIzol (Invitrogen, USA) according to the manufacturer’s instructions. The RNA was resuspended in nuclease-free water and stored at -80°C. The cDNA was synthesized from 3 μg RNA using Superscript III Reverse Transcriptase (Invitrogen) following the manufacturer’s instructions. Briefly, RNA was treated with DNase I (Roche) to remove genomic DNA, and then incubated with 50 ng of random hexamers and 0.5 μl of 10 mM dNTPs for 5 min at 65°C. After incubation, 2 μl of 5× First-strand RT buffer, 0.5 μl of 0.1 M dithiothreitol (DTT), 0.5 μl of RNaseOUT™ (40 U/μl), and 0.5 μl of SuperScript III Reverse Transcriptase (200 U/μl) were added and the mixture incubated at 25°C for 10 min. This was followed by incubation at 50°C for 50 min and the final termination at 85°C for 5 min. Finally, 0.2 μl of RNase H (Biolabs, 5 U/μl) was added to each tube and incubated at 37°C for 20 min. The cDNA was stored at -20°C. All experimental approaches were performed according to the University of Adelaide biosafety and ethics committee regulations (Institutional Biosafety Committee, Dealing ID 12713) concerning this and the previous experiments of Dr. Grützner’s laboratory. Platypus samples were collected in 2002 (AEEC permit R.CG.07.03, Environment ACT permit LI 2002 270, NPWS permit A193) and 2008 (AEC permit N°. S-49-2006) at the Upper Barnard River (New South Wales, Australia).
Cloning of platypus Mel1c receptor
PCR reactions were performed in a final volume of 50 μl using Q5 High-Fidelity DNA Polymerase (NEB, Ipswich, MA, USA) following the manufacturer’s protocol with 2 μl of platypus brain cDNA using the primers described in Table 1. The PCR conditions were as follows: 98°C for 3 min, followed by 10 cycles of 98°C for 10s, 65°C−1°C/cycle for 30s, 72°C for 30 s, and then 30 cycles of 98°C for 10s, X°C for 30s, 72°C for 30 s, and finally 2 min at 72°C.
Table 1. Primer sequences used in this study.
| Amplicons | Forward primer sequences | Reverse primer sequences | Annealing temperature |
|---|---|---|---|
| (5’–3’) | (3’–5’) | (°C) | |
| Exon 1 |
ATGCCGGGAGCCGGGAAC GGGACCTGCCCGGGCT |
CGTTGCGGAGCTTTCTGTT CCTCAGC |
60 |
| Exon 2 |
GCAACGCCGGGAACATCTT TGTGATCAG |
TTACACATGGATTTCAGCTT GATTTTTCTTTG |
58 |
Amplicons were separated on 1% agarose gels stained with ethidium bromide and the gel bands revealed with U Genius (Syngene, Frederick, USA). PCR products were purified using the High Pure Purification kit (Roche Mannhein Germany). Eluted DNA were inserted into the blunt pJET vector using the CloneJET PCR Cloning Kit (Thermo Fisher Scientific, Waltham, USA) and transformed into DH10β chemically competent Escherichia coli cells (NEB, Ipswich, MA, USA). Forward and reverse sequencing reactions were performed using the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA) and pJET1.2 Sequencing Primers. Sequencing products were purified using the BigDye XTerminator® Purification Kit (Thermo Fisher Scientific, Waltham, USA) and run on an ABI 3730 XL automated sequencer (Applied Biosystems). Data were analyzed by Sequencher® version 5.4.6 DNA sequence analysis software (Gene Codes Corporation, Ann Arbor, MI USA).
Phylogenetic analysis
All phylogenetic analyses were undertaken using Geneious V10.0.2 (http://www.geneious.com, [22]). Platypus (O. anatinus), chicken (G. gallus) and the clawed frog (X. laevis) nucleotide sequences were imported into Geneious and translated to protein sequences. Nucleotide sequences for a range of therian mammals (group consisting of eutherians and marsupials) were downloaded from NCBI (National Center for Biotechnology Information); names and GenBank accession numbers are shown in Table 2. Introns were removed and coding regions were translated according to their individual start and stop positions. DNA and protein alignments were undergone using ClustalW algorithm. Phylogenetic tree was built using Neighbour-Joining method, with Jukes-Cantor substitution model, 1000 bootstrap replicates and no outgroup was selected.
Table 2. Therian mammals used for phylogenetic analysis.
| Scientific Name | Common Name | GenBank Accession Number |
|---|---|---|
| Pan troglodytes | Chimpanzee | 736267 |
| Bos Taurus | Cow | 530065 |
| Sus scrofa | Pig | 100620104 |
| Canis lupus familiaris | Dog | 492213 |
| Dipodomys ordii | Ord's kangaroo rat | 105992838 |
| Monodelphis domestica | Gray short-tailed opossum | 100025945 |
| Phascolarctos cinereus | Koala | 110199994 |
| Sarcophilus harrisii | Tasmanian devil | 100913368 |
Establishment of the transient COS7/3HA-Mel1c cell line
Coding sequences for the platypus (XM 001512887), X. laevis (U67880), and G. gallus (NM 205361.1) Mel1c melatonin receptors were synthesized by Genecust (Dudelange, Luxemburg) and subcloned into the pCIneo expression vector (Promega Madison, Wisconsin, USA). COS7 cells obtained from the American Type Culture Collection were maintained in DMEM GlutaMAX (Thermo Fisher Scientific, Waltham, USA) supplemented with 10% (v/v) fetal calf serum. Nucleofection of COS7 cells was performed according to the manufacturer's instructions using the Nucleofector machine (Amaxa, Cologne, Germany). Adherent COS7 cells were washed in phosphate-buffered saline (PBS), trypsinized, and gently resuspended in V nucleofection solution to a final concentration of 7 × 106 cells/100 μl. Next, 5 μg of pCIneo/Mel1c vector was mixed with 100 μl of the COS7 cell suspension, transferred to a 2.0 ml electroporation cuvette, and nucleofected using program A024 and an Amaxa Nucleofector apparatus. Just after the nucleofection, 500 μl of media supplemented with 20% (v/v) fetal calf serum was added to the cell suspension and cultured in a humidified 37°C, 5% CO2 incubator for 20 minutes. Fourteen million cells were transferred to a T225 flask containing 50 ml of media supplemented with 20% (v/v) fetal calf serum and cultured in a humidified 37°C, 5% CO2 incubator. Two days after transfection, the cells were harvested in PBS, pelleted, and stored at -80°C until use.
Indirect immunofluorescence
COS7 cells transiently expressing platypus (O. anatinus), X. laevis, or G. gallus Mel1c receptors were seeded at 50,000 cells per well in eight-well Lab-Tek chamber slides (Nunc, Naperville, IL) in 0.5 ml of medium. Cells were fixed by treatment with 4% formaldehyde in PBS for 15 min then incubated with glycine (100 mM) in PBS for 10 min at room temperature (RT). The cells were blocked with 0.2% bovine serum albumin (BSA) in PBS for 5 min at RT. For visualization of HA epitope-tagged Mel1c receptors, cell surface receptors were stained using a mouse monoclonal anti-HA antibody (0.25 μg/ml) for 30 min at 37°C in PBS containing 0.2% BSA. After washing in PBS supplemented with 0.2% BSA, cells were blocked with a solution of 0.2% BSA and 10% fetal calf serum. The cells were then incubated in a humidified chamber for 60 min at 37°C with secondary antibody (1 μg/ml Alexa fluor 488-conjugated goat anti-mouse antibody; St Louis, MO). The cells were washed twice with PBS/0.2% BSA and coverslips applied using Vectashield® Mounting medium containing 4′, 6′-diamidino-2-phenylindole to stain nuclei (Vector Lab, Burlingame, CA). Fluorescence microscopy was performed using a Zeiss Axiovert 200M microscope (Zeiss, Oberkochen, Germany). Immunofluorescent images were captured by an AxioCam MR3 camera and AxioVision software (Zeiss, Oberkochen, Germany).
Establishment of stable CHO-FlpIn Mel1c cell lines
CHO-Flp-In cells (Thermo Fisher Scientific, Waltham, USA) were cultivated in the presence of zeocin (0.1 mg/ml). The cells were cotransfected with either platypus (cloned sequence that will be submitted to GenBank) Mel1c/pcDNA5-FRT (platypus Mel1c) or X. laevis (NM 001087919.1) Mel1c/pcDNA5-FRT (clawed frog Mel1c) and Flp recombinase expression pOG44 plasmid using PEIpro (Invitrogen, Carlsbad, CA). CHO cells stably expressing platypus Mel1c or clawed frog Mel1c were selected using hygromycin (0.6 mg/ml). CHO-Flp-In-platypus Mel1c cells were grown in HAM F12 with 2 mM glutamine and supplemented with 10% fetal calf serum. Cells were selected using antibiotic pressure (hygromycin) for 72 h. The resistant cells were pooled and the presence of the transgene confirmed by RT-PCR (not shown) using 500,000 cells. After confirmation of the presence of the transgene, the cells were amplified up to 2 x 109 cells. This material was used to prepare the cell membranes used in further pharmacological experiments.
Membrane preparation
This standard procedure was described in a previous experiment with CHO cells expressing the sheep MT2 receptor [23]. Briefly, CHO cells expressing either the platypus or clawed frog Mel1c melatonin receptor were grown to confluence, harvested in phosphate buffer containing 2 mM EDTA, and centrifuged at 1000g for 5 min at 4°C. The resulting pellet was suspended in 5 mM Tris/HCl (pH 7.4) containing 2 mM EDTA and homogenized using a Kinematica polytron. The homogenate was then centrifuged (20,000g, 30 min, 4°C) and the resulting pellet suspended in 75 mM Tris/HCl (pH 7.4) containing 2 mM EDTA and 12.5 mM MgCl2. The protein content was determined according to the method of Lowry et al. [24] using the Bio-Rad kit (Bio-Rad SA, Ivry-sur-Seine, France). Aliquots of membrane preparations were stored in binding buffer (50 mM Tris/HCl, pH 7.4 containing 5 mM MgCl2 and 1 mM EDTA) at –80°C until use.
2-[125I]-melatonin binding assay
This approach has been described very often by our laboratory and others. Briefly, membranes were incubated for 2 hours at 37°C in a final volume of 250 μl of binding buffer containing 50 pM 2-[125I]-melatonin for competition experiments. The results were expressed as Ki, taking into account the concentration of radioligand used in each experiment. Non-specific binding was defined with 10 μM melatonin. The reaction was stopped by rapid filtration through GF/B Unifilters, followed by three successive washes with ice-cold buffer. Data were analyzed by using the program PRISM (GraphPad Software Inc., San Diego, CA). For saturation assays, the density of binding sites (Bmax) and the dissociation constant of the radioligand (KD) were calculated according to the Scatchard method. For competition experiments, inhibition constants (Ki) were calculated according to the Cheng-Prusoff equation: Ki = IC50/[1+(L/KD)], where IC50 is the measured 50% inhibitory concentration, L is the concentration of 2-[125I]-iodomelatonin, and KD is the dissociation constant. A similar protocol was used when the binding was performed on whole cells, as described by Legros et al. [25]. As per Plos One policy, all the raw data–listings from radioactivity counter are attached as raw files (Supplementary Tables A to M in S1 Data).
Results
Cloning of platypus Mel1c receptor
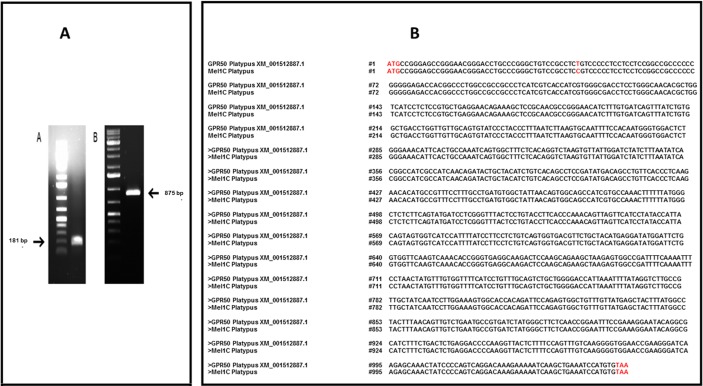
The two amplicons isolated from exons 1 and 2 are depicted in Fig 1A. They were fully sequenced. Fig 1B shows the complete nucleotide sequence of the gene. The alignment between the predicted and experimental sequences is shown. The platypus Mel1c sequence fits the predicted sequence XM 001512887 (www.ncbi.nlm.nih.gov/nuccore/XM/001512887) except for a single mutation in position 44 (T>C). This mutation changes the nature of the amino acid encoded by this triplet at position 15 (L>P).
Fig 1. Cloning of Mel1c from platypus brain and comparison of the actual and predicted sequences.
The cloning was performed using the predicted sequence XM_001512887. A, lane 1: Exon 1 amplicon, 181 bp; lane 2: Exon 2 amplicon, 875 bp. B, Comparison of the obtained sequence. In red: ATG starting codon and TAA stop codon. At position 44, a single mutation from the predicted sequence changes one amino acid in the protein sequence at position 15 (L>P).
Gene organization and phylogenetic analysis
Platypus Mel1c is organized in a similar way as any melatonin receptor described in other vertebrates. Platypus Mel1c contains two exons [separated by a long intron (~30,000 bp) (Panel A in S1 Fig)] that encode the transmembrane domain 1 and the other six transmembrane domains. Protein alignment with chicken and the clawed frog shows that Mel1c in platypus is well conserved at the amino acid level. From Thr44 to Ala352, amino acid sequence identity was comprised between 72 and 78% for the 3 receptors. (see Table 3 and Panel B in S1 Fig). Interestingly, the Mel1c family has much more variation at the nucleotide level throughout these species (Panel C in S1 Fig).
Table 3. Comparison of Mel1c sequences in platypus, xenopus and chicken to the human GPR50.
| Gallus gallus Mel1c | Homo sapiens GPR50 | Ornithorynchus anatinus Mel1c | Xenopus laevis Mel1c | |
|---|---|---|---|---|
| Gallus gallus Mel1c | 46.10% | 78.40% | 78.70% | |
| Homo sapiens GPR50 | 46.10% | 48.60% | 39.10% | |
| Ornithorynchus anatinus Mel1c | 78.40% | 48.60% | 72.30% | |
| Xenopus laevis Mel1c | 78.70% | 39.10% | 72.30% |
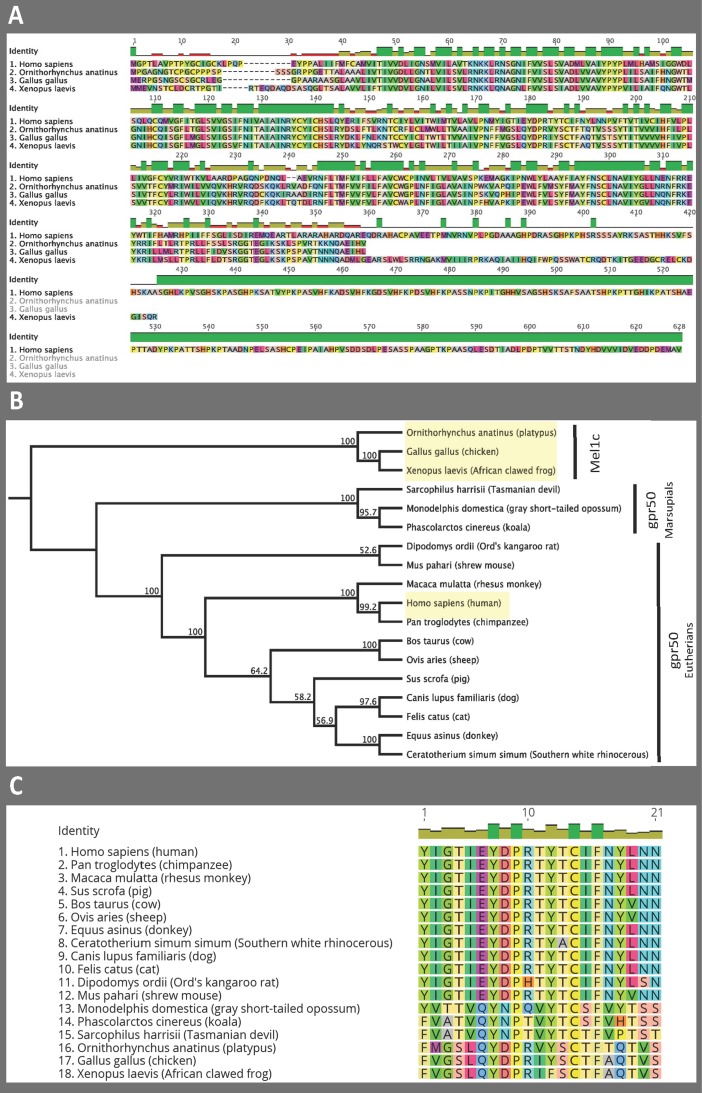
When Mel1c is aligned with the human sequence of GPR50 (the therian ortholog of Mel1c), the divergence between the two is apparent (Fig 2A). Phylogenetic analysis shows platypus clustering with chicken and the clawed frog Mel1c to the exclusion of an array of therian mammal GPR50 (Fig 2B), showing here that platypus has retained the ancestral Mel1c gene and its ortholog GPR50 evolved after the divergence of monotremes from therian mammals. Although GPR50 has lost its capacity to bind melatonin, the sequence has been conserved in mammals (Table 3), with a major difference being that the sequence of human GPR50 has 270 additional amino acids compared to the platypus (Fig 2A), rendering the C-terminus of GPR50 one of the longest known G-protein-coupled receptors (GPCRs).
Fig 2. Platypus has retained ancestral Mel1c showing that the ortholog GPR50 evolved after the divergence of monotremes from therian mammals.
A) Protein alignment of human (Homo sapiens) GPR50 with platypus (Ornithorhynchus anatinus), chicken (Gallus gallus) and the clawed frog (Xenopus laevis) Mel1c. Each amino acid is depicted by its single letter symbol and an associated colour. Identity bar above alignment denotes the similarity at each amino acid position between the four species: green = all homologous; yellow = three species homologous; red = two species homologous; no bar = no homology. Every 50 amino acid positions are labeled. B) Neighbour-Joining phylogenetic tree showing platypus clustering together with chicken and the clawed frog Mel1c while marsupial GPR50 and eutherian GPR50 form their own distinct clusters. Bootstrap support values are shown for each node. The four species from protein alignment in panel A are highlighted in yellow. C) Amino acid sequence of the second extracellular loop (E2) of Mel1c and GPR50 for all species used in the phylogenetic tree from panel B. Each amino acid is depicted by its single letter symbol and an associated colour. Identity bar above alignment denotes the similarity at each amino acid position between all species: green = all homologous; yellow = some species homologous. Every 10 amino acid positions are labeled.
A recent study has highlighted the importance of the second extracellular domain (E2) of melatonin receptors in their ability to bind to melatonin [26]. Therefore, we also looked at this region between Mel1c and GPR50 in all species used for phylogenetic analyses. Here we found that platypus, chicken and the clawed frog contained a Gln (Q) at position 18 of E2 (Fig 2C), which was shown by Clement et al. [26] to be important for the binding function of melatonin receptors. However, all eutherian species contained a Thr (Y) at this position instead, while the marsupials had Thr (Y; opossum), His (H; koala) or Pro (P; Tasmanian devil) (Fig 2C). If this Gln in E2 is in fact important for binding in melatonin receptors, then the lack of it in all therian mammals tested here supports that hypothesis and also suggests the transition from Mel1c to GPR50 occurred between the divergence of monotremes and therian mammals– 190 to 160 million years ago (MYA).
Expression and subcellular localization of recombinant platypus, clawed frog and chicken Mel1c receptors in COS7 cells
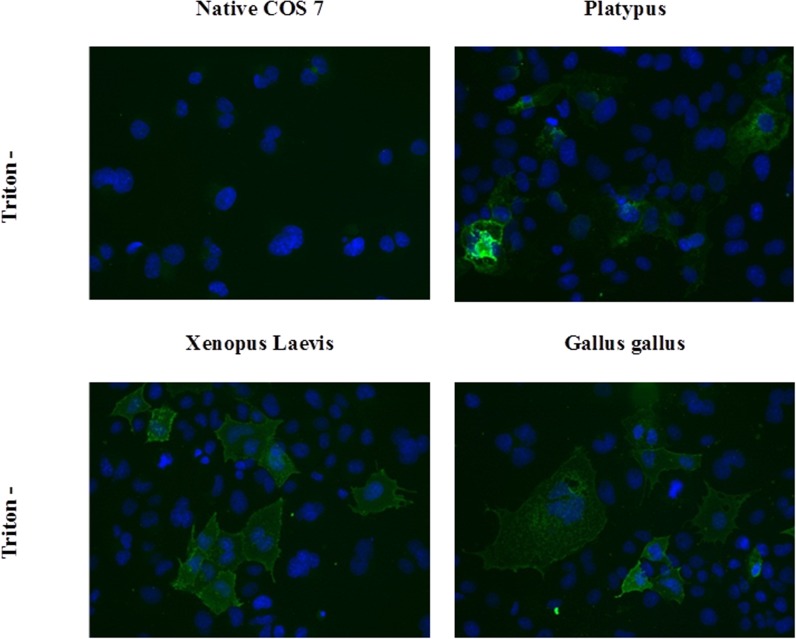
Mel1c from G. gallus and X. laevis was synthesized according to the reported predicted sequences and built in the same expression vector as platypus Mel1c. The first step was to determine whether they are expressed and active in a cellular model. We started by fusing the N-terminus of the three sequences to a 3HA flag, for which potent antibodies exist. For this particular purpose, the transition expression of the receptors into COS7 cells led to the images in Fig 3. Immune staining of non-permeabilized COS7/3HA-Mel1c cells with a fluorescent anti-HA antibody revealed major expression of platypus, X. laevis, and G. gallus Mel1c receptors at the plasma membrane. No fluorescent signal was detected from native COS7 cells.
Fig 3. Expression of Mel1c receptors in COS7 cells.
Subcellular localization of HA epitope-tagged Mel1c receptors. Immunofluorescence studies were performed with transfected COS7cells. COS7 cells and COS7 expressing 3HA-platypus Mel1C or 3HA-X. laevis Mel1C or 3HA-G. gallus Mel1C receptors were probed with mouse monoclonal anti-HA antibody directed against the N-terminal epitope tag present on recombinant receptor. Experiments were carried out with paraformaldehyde-fixed and non-permeabilized cells. The fluorescence images were obtained by using Alexa 488-conjugated goat anti- mouse IgG secondary antibody. COS7 cell nuclei were stained with 4′, 6′-diamidino-2-phenylindole. Native COS7 cells were used as a negative control.
Melatonin binding
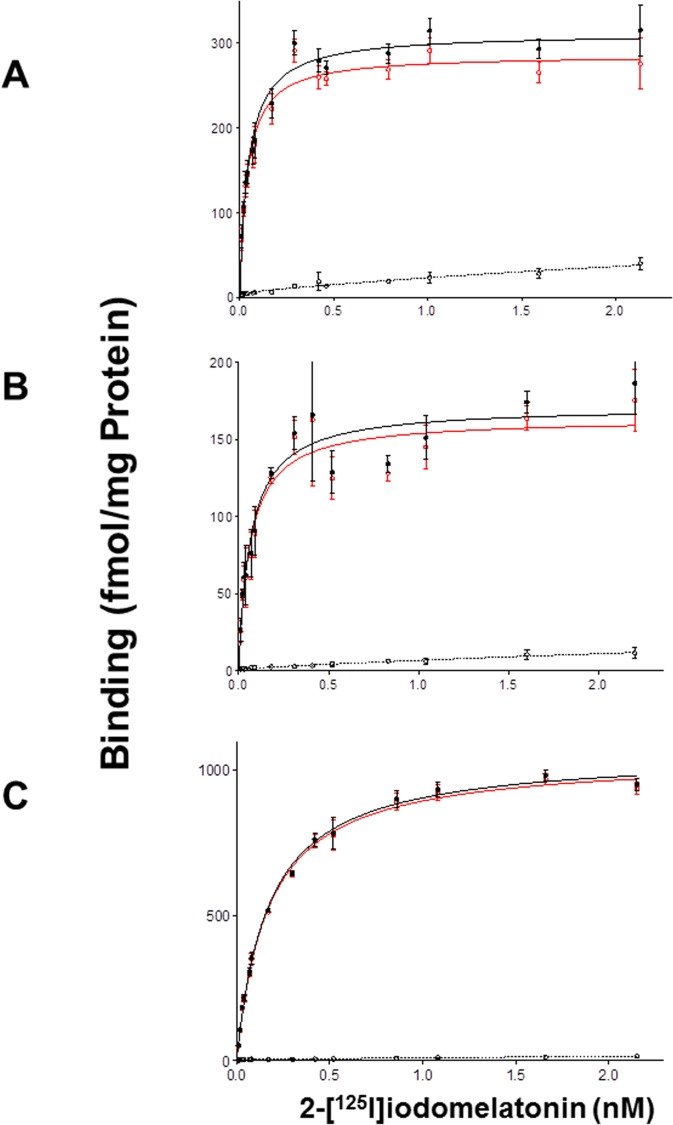
Next, we prepared membranes for clawed frog, platypus, and chicken from cells overexpressing their respective Mel1c receptors and attempted to measure the binding of 2[125I]-iodomelatonin. Saturations were obtained and showed specific bindings for all species tested. This was the first time that the Mel1c receptor from a mammal was shown to bind 2[125I]-iodomelatonin. The saturation curves (Fig 4) for the three species were similar to those obtained under similar conditions for the melatonin receptors that have been cloned, expressed, and characterized in our laboratory. The expression levels of the Mel1c receptors from these species stably expressed in CHO cells were 18 fmol/mg of protein for clawed frog and 36 fmol/mg of protein for the platypus. These levels are in the lower range of expression for melatonin receptors (MT1 and MT2 receptors) from more classical species we generated previously, with expression levels ranging from 80 to 2650 fmol/mg of protein in rats and humans [27] and 300 to 700 fmol/mg of protein for mice [28]. In contrast, the KD of the two Mel1c receptors for melatonin was 133 pM for clawed frog and 67 pM for platypus.
Fig 4. Saturation binding experiments for 2-[125I]-iodomelatonin.
Membranes from COS7 cells transfected with platypus Mel1c (A), clawed frog Mel1c (B), and chicken Mel1c (C) were used to measure the binding at Mel1c receptors. Red line represents specific binding, black line represents total binding and dotted line represents non-specific binding.
Molecular pharmacology
To obtain stable expression of the receptor for future studies, a preliminary step involving transient expression of the receptor was performed. The Mel1c receptors from the three species were expressed in transiently transfected COS7 cells. A limited number of compounds were tested in order to gain a preliminary perception of the characteristics of Mel1c from these species. The profiles for chicken, clawed frog, and platypus are reported here for the first time. The comparative data are given in Table 4. The profiles comprise the binding affinities of a dozen compounds for the receptors of the three species. These profiles were quite similar, which was not surprising for the chicken and clawed frog, despite their evolutionary distance. However, the similarity with pharmacological profiles of the monotreme Mel1c was somewhat unexpected. Simple linear correlations between the sets of data (taken 2 by 2) led to R2 > 0.95. These correlations translate a view of the proximity of the receptor affinities to some compounds from a pure biochemical point of view.
Table 4. Comparison of the molecular pharmacology of platypus, xenopus and chicken melatonin Mel1c receptors.
| pKi Ornithorhyncus anatinus | pKi Xenopus laevis | pKi Gallus gallus | |
|---|---|---|---|
| Melatonin | 8.6 | 8.8 | 8.6 |
| 2-Iodomelatonin | 10.2 | 10.0 | 9.8 |
| S 70254 | 6.6 | 6.8 | 6.6 |
| 4P-P-DOT | 6.7 | 6.7 | 6.4 |
| Agolmelatin/S 20098 | 9.6 | 9.2 | 8.9 |
| S 22153 | 7.6 | 7.0 | 6.7 |
| Ramelteon/ FLN68 | 10.1 | 10.1 | 9.8 |
| Luzindole | 5.9 | 5.7 | 5.4 |
Binding experiments were performed with 2-[125I]-iodomelatonin as the radioligand using COS7 cells transiently transfected with the respective Mel1c gene. Experiments using two different transfections were performed separately. Each point was the mean of a triplicate determination. Concentration isotherm curves were obtained using 10 concentrations of each product from 10−13 to 10−4 M. S70254, 2-iodo-N-2-[5-methoxy-2-(naphthalen-1-yl)-1H-pyrrolo[3,2-b]pyridine-3-yl]) acetamide; 4P-P-DOT, N-[(2S,4S)-4-phenyl-1,2,3,4-tetrahydronaphthalen-2-yl]propanamide; Agomelatin® (S20098), N-(2-(7-methoxynaphthalen-1-yl)ethyl)acetamide; S22153, N-[2-(5-ethylbenzothiophen-3-yl)ethyl]acetamide; Ramelteon® (FLN68), (S)-N-(2-(1,6,7,8-tetrahydro-2H-indeno-(5,4)furan-8-yl)ethyl)propionamide; Luzindole, N-acetyl-2-benzyl-tryptamine.
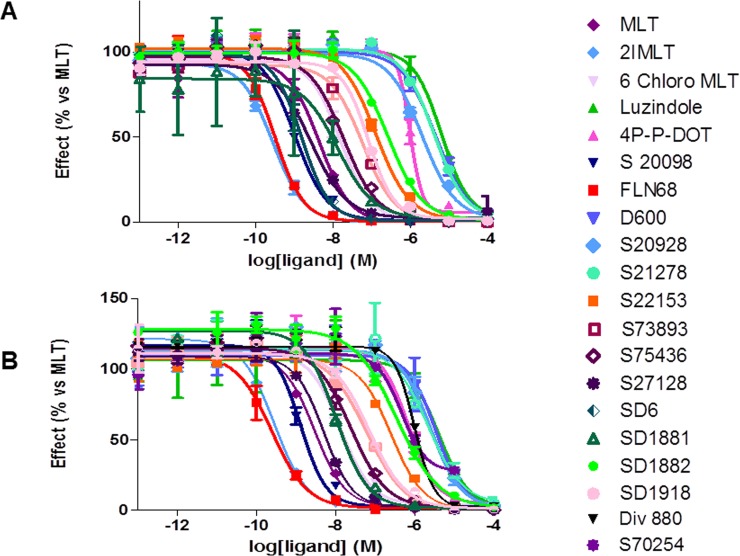
Because the profiles of the Mel1c bindings are similar between birds (chicken) and amphibians (clawed frog), it seemed important to proceed to studies using only the Mel1c from clawed frog. Thus, we focused on the stable expression in CHO cells of the platypus and clawed frog Mel1c receptors. These two receptors were expressed at workable levels, although other melatonin receptors were sometimes expressed at up to 2 pmol/mg of protein [27] depending on the method chosen for establishing the stable cell line. The last step of this characterization was to obtain on this biological material, a pharmacological profile for a series of 19 ligands for these two species compared to the human melatonin receptors MT1 and MT2. The curves obtained for platypus Mel1c with the products are given in Fig 5A and for the clawed frog Mel1c in Fig 5B as examples.
Fig 5.
Molecular pharmacology of the Mel1c receptors from platypus (A) and clawed frog (B). The ligand was 2-[125I]iodomelatonin. Independent experiments were performed at least twice using different batches of membranes from stably transfected CHO cells and each point was obtained in triplicate. SD6, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]iodoacetamide; 2IMLT, 2-iodomelatonin; 6-Cl- Chloro MLT, 6-chloromelatonin; Luzindole, N-acetyl-2-Br-MLT; 2-bromomelatonin, benzyltryptamine; 4P-P-DOT, N-[(2S,4S)-4-phenyl-1,2,3,4-tetrahydronaphthalen-2-yl]propanamide; Agomelatin® (S20098), N-(2-(7-methoxynaphthalen-1-yl)ethyl)acetamide; Ramelteon® (FLN68), (S)-N-(2-(1,6,7,8-tetrahydro-2H-indeno-(5,4)furan-8-yl)ethyl)propionamide; D600, methoxyverapamil; 5HT, 5-hydroxytryptamine; S20928, (N-[2-(1-naphthyl)ethyl] cyclobutanecarboxamide); S21278, N-[2-(6-methoxybenzimidazol-1-yl)ethyl]acetamide; S22153, N-[2-(5-ethylbenzothiophen-3-yl)ethyl]acetamide; S27128-1, N-[2-(2-iodo-5-methoxy-6-nitro-1H-indol-3-yl)ethyl]acetamide; S73893, N-[3-methoxy-2-(7-methoxy-1-naphthyl)propyl]acetamide; S75436, 2-fluoro-N-[3-hydroxy-2-(7-methoxy-1-naphthyl) propyl]acetamide; S77834S27128, N-[(8-[2-(2-iodo-5-methoxy-10,11-dihydro-5H-dibenzo[a,d][7] annulen-106-nitro-1H-indol-3-yl)methylethyl]acetamide; S77840, 1-[(8-methoxy-10,11-dihydro-5H-dibenzo[a,d][7]annulen-101H-indol-3-yl)methyl]urea ethyl]iodoacetamide; SD1881, N-[2-(6-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamideiodomelatonin; SD1882, N-[2-(4-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide; SD1918, N-[7-iodomelatonin; Div 880, 2-(7-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide2-[(2-iodo-4,5-dimethoxyphenyl)methyl]-4,5-dimethoxy phenyl; S70254, 2-iodo-N-2-[5-methoxy-2-(naphthalen-1-yl)-1H-pyrrolo[3,2-b]pyridine-3-yl])acetamide. Concentration isotherms were obtained using 10 concentrations of each product from 10−13 to 10−4 M.
Nineteen compounds were tested, from the classic melatonin ligands to the more obscure D600, a specific MT1 ligand [25], and the marketed drugs Ramelteon® and Agomelatin®, as well as the iodinated derivatives of melatonin (e.g., 4-iodo, 2-iodo, or 6-iodo melatonin). The data were compared to that of the human MT1 and MT2 (Table 5).
Table 5. Molecular pharmacology of Mel1c receptors in platypus and xenopus compared to the human MT1 and MT2 melatonin receptors.
| Compounds | Platypus Mel1C | Xenopus Mel1C | hMT1 | hMT2 |
|---|---|---|---|---|
| Melatonin | 8.7 | 8.7 | 9.6 | 9.3 |
| 2-iodomelatonin | 9.9 | 9.7 | 10.7 | 9.8 |
| 6-chloromelatonin | 8.1 | 8.1 | 8.7 | 9.6 |
| Luzindole | 5.6 | 5.6 | 6.6 | 7.6 |
| 4P-PDOT | 6.2 | 6.1 | 6.8 | 8.9 |
| Agomelatin ® | 9.4 | 9.1 | 9.9 | 9.9 |
| Ramelteon ® | 9.9 | 9.9 | 10.1 | 10.3 |
| D600 | 5.7 | 5.7 | 7.0 | <5 |
| S20928 | 6.1 | 5.9 | 7.1 | 7.1 |
| S21278 | 5.7 | 5.8 | 6.2 | 6.2 |
| S22153 | 7.3 | 6.9 | 8.2 | 8.0 |
| S73893 | 7.7 | 7.4 | 8.4 | 8.1 |
| S75436 | 8.2 | 7.8 | 7.9 | 8.9 |
| S27128 | 9.0 | 8.6 | 8.9 | 9.2 |
| DIV880 | 6.2 | 5.9 | 6.1 | 8.0 |
| SD6 | 9.4 | 9.3 | 9.9 | 9.9 |
| 6-Iodomelatonin | 8.4 | 8.1 | 8.8 | 8.6 |
| 4-Iodomelatonin | 7.0 | 6.6 | 7.8 | 7.9 |
| 7-Iodomelatonin | 7.5 | 7.3 | 7.3 | 7.3 |
| S70254 | ND | 6.3 | 7.0 | 9.0 |
ND: not determined. Luzindole, N-acetyl-2-benzyltryptamine; 4P-PDOT, N-[(2S,4S)-4-phenyl-1,2,3,4-tetrahydronaphthalen-2-yl]propanamide; D600, methoxyverapamil; S20928, (N-[2-(1-naphthyl)ethyl] cyclobutanecarboxamide); S21278, N-[2-(6-methoxybenzimidazol-1-yl)ethyl]acetamide; S22153, N-[2-(5-ethylbenzothiophen-3-yl)ethyl]acetamide; S73893, N-[3-methoxy-2-(7-methoxy-1-naphthyl)propyl]acetamide; S75436, 2-fluoro-N-[3-hydroxy-2-(7-methoxy-1-naphthyl)propyl]acetamide; S27128-1, N-[(8-methoxy-6-nitro-1H-indol-3-yl)ethyl]acetamide; DIV880, 2-(2-[(2-iodo-4,5-dimethoxyphenyl)methyl]-4,5-dimethoxy phenyl; SD6, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]iodoacetamide; S70254, 2-iodo-N-2-[5-methoxy-2-(naphthalen-1-yl)-1H-pyrrolo[3,2-b]pyridine-3-yl])acetamide. Binding experiments were performed with 2-[125I]-iodomelatonin as the radioligand, Experiments were run in triplicate, at least twice using different batches of membranes from stably transfected CHO cells. Concentration isotherms were obtained using 10 concentrations of each product from 10−13 to 10−4 M.
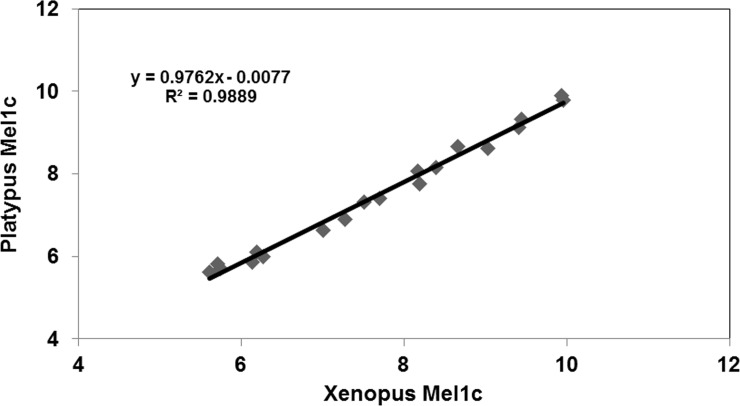
We obtained an almost perfect match between the results obtained with chicken Mel1c, which is a reference in this domain, and those obtained with the platypus Mel1c that was cloned and characterized here for the first time. The simplest correlation coefficient (R2) was 0.98 (Fig 6).
Fig 6. Correlation between the data obtained from platypus Mel1c and those obtained with clawed frog Mel1c.
Data were obtained in a binding assay with 2-[125I]-iodomelatonin as the radioligand. See Fig 5.
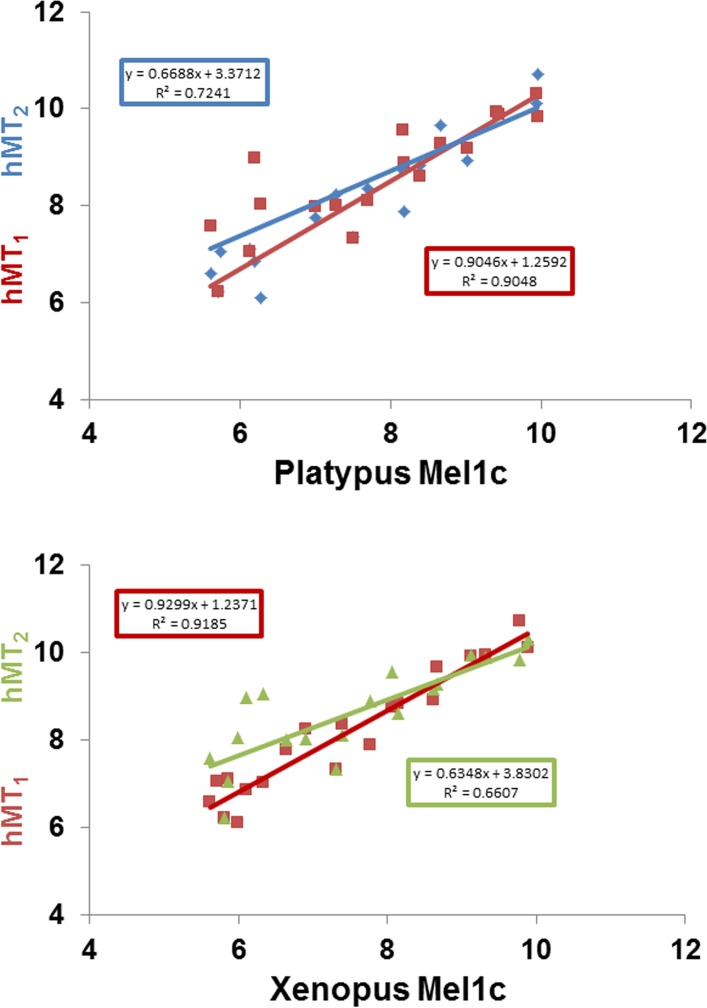
At first glance, the data do not seem to differ from one species to another, with either the Mel1c receptor, as shown above with transiently expressed receptor (Table 4), or the human MT1 and MT2 receptors. In order to validate this observation, the data obtained with the clawed frog Mel1c receptor and platypus Mel1c receptor were plotted against the data obtained for MT1 and MT2 (Fig 7).
Fig 7.
Comparison of the correlations between the molecular pharmacology of platypus Mel1c (A) or clawed frog Mel1c (B) and human melatonin receptors MT1 and MT2. The correlation in red is between hMT1 and platypus Mel1c; in blue, between hMT2 and Platypus Mel1c (upper panel). The correlation in red is between hMT1 and clawed frog Mel1c; and in green between hMT2 and clawed frog Mel1c (lower panel.) See Fig 5 for data.
At first glance, Mel1c seems to behave identically towards this selection of compounds among the two species (human versus platypus), but it is quite different when the correlation coefficients are examined. Surprisingly, the Mel1c data correlate highly (R2 = 0.9) with MT1 and more poorly with MT2 (R2 = 0.7 for platypus and 0.6 for Clawed frog).
Discussion
Platypus and echidna are the oldest surviving mammalian lineage, having diverged from other mammals approximately 190 MYA [29]. These species have an astonishing track record of informing us about the evolution of the mammalian genome [30] and individual genes (see for example Grutzner et al. [17], Cortez et al. [20], Tsend-Ayush et al. [31] and Hu et al. [32]). Comparing the GPCRs from different species can help us understand the structure of these receptors, as well as their binding characteristics [33]. To prepare for the expression, purification, crystallization, and characterization of receptors, a preliminary approach is to closely study the various receptors from diverse species. Natural sequences have evolved throughout these species; though maintaining specificity towards their natural ligand (see Li et al. for complete melatonin receptor evolutionary divergences [34]). They show that it is slightly different for the melatonin-related receptor (GPR50) and Mel1c.
The seminal work of Dufourny et al. [7] demonstrated from minute genetic analyses that the Mel1c receptor from amphibians and birds gave rise to GPR50 in mammals, which is still categorized as an orphan receptor because it does not bind melatonin or analogs, confirmed independently by us and others (C Ouvry & JA Boutin, unpublished). It has been a challenge for a long time to understand when GPR50 lost its binding during mammalian evolution and what this means for the function and evolution of melatonin and GPR50.
Studies of molecular evolution have been reported frequently in the literature. The trees of phylogenetic affiliation has been used to better understand the passage from one species to another, especially by studying species that seem far away from each other, as well as those that are closely related. Monotremes occupy a key position in mammalian phylogeny and the present paper attempted to determine the evolution and binding of melatonin. Therefore, whether the melatonin receptor cloned from platypus is in fact Mel1c or GPR50 was of interest.
The predicted sequence was very close to the actual sequence with a single mutation at position 44. The structure of the gene was as expected because of the common nature of all of the melatonin receptors throughout living species, two exons separated by a long intron of ~30,000 bp (Panel A in S1 Fig). Alignment of the platypus receptor with chicken and frog Mel1c shows very high similarity at the amino acid level (Panel B in S1 Fig). However, when including human GPR50 in the protein alignment, it is apparent that the platypus is much more similar to chicken and from than to human GPR50 (Fig 2A). Furthermore, phylogenetic analysis (including a range of eutherian and marsupial species) shows the platypus grouping with chicken and frog to the exclusion of all therian species (Fig 2B). This suggests that platypus does in fact have the ancestral Mel1c receptor and that the “mammalian” GPR50 evolved after monotremes diverged from therian mammals ~190 MYA. Of interest in the present study is the similarity of the melatonin binding Mel1c sequence throughout the millions of years of evolution between amphibians, birds and even monotremes (Panel B in S1 Fig). In stark contrast is the highly variable GPR50 in therian mammals that have lost their ability to bind melatonin. This strongly suggests that the initial loss of binding allowed GPR50 to then continue to diverge in therian mammals.
There is still a lack of consensus on which specific amino acid changes in GPR50 have caused the lack of binding of this receptor to melatonin, despite many mutation and modeling studies on functional melatonin receptors (see Pala et al., for review [35]). The majority of studies have focused on the transmembrane domains. However, Clement et al. proposed that the second extracellular loop (E2) in MT1 is instead important for melatonin binding [26]. They found that Gln181 in hMT1 was highly important for melatonin binding through a stabilizing interaction with Phe179. All melatonin-binding receptors investigated had Gln at this position, while human GPR50 instead had a Thr. Upon examination of the same E2 region of Mel1c in the platypus, chicken and frog we found there also to be a Gln at this same position (Fig 2C), while all eutherian species at this position contained Thr and the three marsupial species analysed had Thr, His or Pro (Fig 2C). This supports the proposal from Clement et al [26] that if Gln in the E2 region of melatonin receptors is important for binding melatonin, then the change of this amino acid to Thr (or His/Pro) may be responsible for the inability of GPR50 to be a functional melatonin receptor. As the marsupial species also lacked Gln, this further suggests that Mel1c changing to GPR50 occurred after the divergence of monotremes and before the therian split, giving a tighter timeframe of 190–160MYA. Further support for this hypothesis is that the marsupial GPR50 also has the characteristic elongated C-tail that eutherian GPR50 has [7]. Functional studies using marsupial GPR50 would need to be undertaken to verify its inability to bind melatonin before committing to this timeframe.
In order to confirm the observations we found in the amino acid sequence for platypus melatonin receptor, we cloned platypus Mel1c for the first time and expressed it to study its binding capacity and function. The three Mel1c receptors studied here (from frog, chicken and platypus) were expressed at the cell surface. Considering the binding aspects, few data have been reported in the literature on the pharmacological profiles of receptors from uncommon species. Chicken and clawed frog Mel1c have been known for some time to bind melatonin [8,36]. To the best of our knowledge, few data have been published on the binding characteristics of Mel1c receptors from other species, and even less has been published regarding various molecules in a binding assay [9,10]. Pharmacological profiles have been published, but using a pigment aggregation X. laevis melanophore-based assay (i.e., a functional assay) rather than a molecular one [9,11]. Overall, with the few compounds that have been described in clawed frog Mel1c [36] and in chicken Mel1c [9,11], no major discrepancies were recorded here. Though the present work provides new information on the molecular pharmacology of platypus Mel1c, the ancestor of the GPR50 orphan receptor in mammals, the available data on the roles of those two receptors remains scarce. Interestingly, GPR50 is expressed strongly in specific brain areas (hypothalamo-pituitary regions [37–40] and ependymal cell layer of the third ventricle [41]) in many mammalian species, and Mel1c is widely expressed in the chicken brain [8]. Because we cloned the receptor from whole brain, one can conclude that Mel1c is also expressed broadly in this organ in the platypus. Furthermore, from the molecular pharmacology point of view, Mel1c is closer to MT1 than to MT2; its profile based on a set of 19 compounds as diverse as possible from the melatoninergic prospect is similar to human MT1.
Complementing the initial work of Dufourny et al. [7], mammalian Mel1c from the platypus fills a gap in terms of sequence and gene structure, as well as molecular pharmacology characteristics, between the melatoninergic systems of amphibians and birds and that of mammals. This work also complements nicely our current efforts to clone melatonin receptors from various species to better understand their sequence/pharmacology relationships [27,28,42,43].
In conclusion, the present work establishes for the first time that melatonin binding to GPR50 was lost after the divergence of monotremes and that this even likely sparked rapid divergence of GPR50 in therian mammals
Supporting information
Structure of Mel1c in platypus. The entire gene is 34,831 bp in length, where ~30,000 bp intron separates two exons: exon 1 is 184 bp long and exon 2 is 866 bp long. Every 5000 bp are labeled. B) Protein alignment of Mel1c from platypus (O. anatinus), chicken (G. gallus) and the clawed frog (X. laevis), showing that Mel1c is very conserved at the amino acid level. Each amino acid is depicted by its single letter symbol and an associated colour. Identity bar above alignment denotes the similarity at each amino acid position between the four species: green = all homologous; yellow = two species homologous; no bar = no homology. Every 10 amino acid positions are labeled. C) DNA alignment of Mel1c exons from platypus (O. anatinus), chicken (G. gallus) and the clawed frog (X. laevis), showing that at a nucleotide level Mel1c has more variation compared to at the amino acid level. Grey bars indicate homology between all three species while colours indicate SNPs (Single Nucleotide Polymorphisms); red = A; green = T; yellow = G; purple = C. Identity bar above alignment denotes the similarity at each nucleotide position between the three species: green = all homologous; yellow = two species homologous; no bar = no homology. Every 20 bp are labeled.
(TIF)
They are counter listings, formatted to give the individual numbers used to calculate all the saturation curves and affinities reported in the present paper. The plates are all arranged the same ways: Saturation: The 3 first columns are used for increasing low concentrations (nM): A: 0.01; B: 0.02; C: 0.04; D: 0.05; E: 0.08; F: 0.1; G: 0.2; in triplicate. The 3 next columns (4 to 6) were used for the nonspecific binding. The 3 next columns (7 to 9) were used for higher concentrations: A: 0.3; B: 0.4; C: 0.5; D: 0.8; E: 1; F: 1.5 and G: 2. The last 3 columns, same concentrations, nonspecific binding. Nonspecific binding was done in the presence of 10 μM of cold melatonin. The H line was not used. R: 11 concentrations of each product. The concentrations of the products were from 10-14M to 10-4M (from column 1 to 11). Colum 12 is for unspecific binding. Two lines (A&B; C&D, etc.) were used per compounds. For DR in COS7 cell membranes, only 8 compounds were tested in that order from top to bottom: melatonin, 2-iodomelatonin, S 70254, 4P-P-DOT, S 20098/agomelatonin, S 22153, FLN68/ramelteon and Luzindole. For DR in CHO cell membranes in that order from top to bottom: melatonin, 2-iodomelatonin, 6-chlmromeltonin, Luzindole, 4PPDOT, S 20098/agomelatin, FLN68/ramelteon, D600, S20928, S21278, S22153, S70254, S73893, S75436, S27128, DIV880, SD6, SD1881, SD1882 and SD1918. If needed, more information can be obtained from the corresponding author upon request. Table A. Raw data for calculation of COS7 Xenopus Mel1c (n = 1 & 2) saturations. Table B. Raw data for calculation of COS7 Platypus Mel1c (n = 1) saturation. Table C. Raw data for calculation of COS7 Platypus (n = 2) & Xenopus (n = 3) Mel1c saturations. Table D. Raw data for calculation of COS7 Mel1c Platypus (n = 3) and naïve cells saturation. Table E. Raw data for calculation of CO7 Mel1c Chicken (n = 1 & 2) and naïve cells saturations. Table F. Raw data for calculation of CHO Mel1c Xenopus (n = 1) saturation. Table G. Raw data for calculation of CHO Mel1c Platypus (n = 1 & 2) saturation. Table H. Raw data for calculation of CHO Mel1c Platypus (n = 3), Human MT1 and MT2 & Xenopus Mel1c (n = 2) saturation. Table I. Raw data for calculation of COS7 Mel1c Xenopus saturation (n = 4 & 5) and dose-responses (n = 1 & 2). Table J. Raw data for calculation of COS7 Mel1c Platypus (n = 1 & 2) dose-responses. Table K. Raw data for calculation of COS7 Chicken Mel1c (n = 1 & 2) dose-responses. Table L. Raw data for calculation of CHO Mel1C Platypus (n = 2) & Xenopus (n = 2) dose-responses. Table M. Raw data for calculation CHO Mel1C Platypus (n = 1) & Xenopus (n = 1) dose-responses.
(ZIP)
Acknowledgments
JAB is thankful to Dr. Laurent Ripoll for numerous helpful discussions and M. Xavier Deguercy for his help. Frank Grützner is supported by an Australian Research Council Fellowship. Tahlia J. Perry is supported by an Australian Postgraduate Award.
Data Availability
All relevant data are within the paper and its Supporting Information files. Additional data have been uploaded to the NCBI GenBank and are accessible using the following link: https://www.ncbi.nlm.nih.gov/nuccore/XM_001512887.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Levoye A, Dam J, Ayoub MA, Guillaume JL, Couturier C, Delagrange P, et al. The orphan GPR50 receptor specifically inhibits MT1 melatonin receptor function through heterodimerization. EMBO J. 2006; 25: 3012–3023. doi: 10.1038/sj.emboj.7601193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker-Andre M, Wiesenberg I, Schaeren-Wiemers N, Andre E, Missbach M, Saurat JH, et al. Pineal gland hormone melatonin binds and activates an orphan of the nuclear receptor superfamily. J Biol. Chem. 1994; 269: 28531–28534. [PubMed] [Google Scholar]
- 3.Jetten AM, Joo JH. Retinoid-related Orphan Receptors (RORs). Roles in Cellular Differentiation and Development. Adv. Dev. Biol. 2006; 16: 313–355. doi: 10.1016/S1574-3349(06)16010-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nosjean O, Ferro M, Coge F, Beauverger P, Henlin JM, Lefoulon F, et al. Identification of the melatonin-binding site MT3 as the quinone reductase 2. J Biol. Chem. 2000; 275: 31311–31317. doi: 10.1074/jbc.M005141200 [DOI] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR, Ebisawa T, Mahle CD, Kolakowski LFJ. Cloning of a melatonin-related receptor from human pituitary. FEBS Lett. 1996; 386: 219–224. [DOI] [PubMed] [Google Scholar]
- 6.Gubitz AK, Reppert SM. Assignment of the melatonin-related receptor to human chromosome X (GPR50) and mouse chromosome X (Gpr50). Genomics. 1999; 55: 248–251. doi: 10.1006/geno.1998.5661 [DOI] [PubMed] [Google Scholar]
- 7.Dufourny L, Levasseur A, Migaud M, Callebaut I, Pontarotti P, Malpaux B, et al. GPR50 is the mammalian ortholog of Mel1c. Evidence of rapid evolution in mammals. BMC. Evol. Biol. 2008; 8: 105 doi: 10.1186/1471-2148-8-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reppert SM, Weaver DR, Cassone VM, Godson C, Kolakowski LFJ. Melatonin receptors are for the birds. Molecular analysis of two receptor subtypes differentially expressed in chick brain. Neuron. 1995; 15: 1003–1015. [DOI] [PubMed] [Google Scholar]
- 9.Pickering H, Sword S, Vonhoff S, Jones R, Sugden D. Analogues of diverse structure are unable to differentiate native melatonin receptors in the chicken retina, sheep pars tuberalis and Xenopus melanophores. Br. J. Pharmacol. 1996; 119: 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jockers R, Petit L, Lacroix I, de CP, Barrett P, Morgan PJ, et al. Novel isoforms of Mel1c melatonin receptors modulating intracellular cyclic guanosine 3',5'-monophosphate levels. Mol. Endocrinol. 1997; 11: 1070–1081. doi: 10.1210/mend.11.8.9964 [DOI] [PubMed] [Google Scholar]
- 11.Teh MT, Sugden D. Comparison of the structure-activity relationships of melatonin receptor agonists and antagonists. Lengthening the N-acyl side-chain has differing effects on potency on Xenopus melanophores. Naunyn Schmiedebergs Arch. Pharmacol. 1998; 358: 522–528. [DOI] [PubMed] [Google Scholar]
- 12.Garratt PJ, Jones R, Tocher DA, Sugden D. Mapping the melatonin receptor. 3. Design and synthesis of melatonin agonists and antagonists derived from 2-phenyltryptamines. J Med. Chem. 1995; 38: 1132–1139. [DOI] [PubMed] [Google Scholar]
- 13.Sugden D, Yeh LK, Teh MT. Design of subtype selective melatonin receptor agonists and antagonists. Reprod Nutr Dev. 1999; 39: 335–344. [DOI] [PubMed] [Google Scholar]
- 14.Faust R, Garratt PJ, Jones R, Yeh LK, Tsotinis A, Panoussopoulou M, et al. Mapping the melatonin receptor. 6. Melatonin agonists and antagonists derived from 6H-isoindolo2,1-aindoles, 5,6-dihydroindolo2,1-aisoquinolines, and 6,7-dihydro-5H-benzocazepino2,1-aindoles. J Med. Chem. 2000; 43: 1050–1061. [DOI] [PubMed] [Google Scholar]
- 15.Sugden D. N-acyl-3-amino-5-methoxychromans. A new series of non-indolic melatonin analogues. Eur. J. Pharmacol. 1994; 254: 271–275. [DOI] [PubMed] [Google Scholar]
- 16.Pégurier C, Morellato L, Chahed E, Andrieux J, Nicolas JP, Boutin JA, et al. Synthesis of new arylalkoxy amido derivatives as melatoninergic ligands. Bioorg Med Chem. 2003; 11: 789–800. [DOI] [PubMed] [Google Scholar]
- 17.Grutzner F, Rens W, Tsend-Ayush E, El-Mogharbel N, O'Brien PC, Jones RC, et al. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature. 2004; 432: 913–917. doi: 10.1038/nature03021 [DOI] [PubMed] [Google Scholar]
- 18.Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008; 18: 965–973. doi: 10.1101/gr.7101908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002; 3: 137–144. doi: 10.1038/nrg733 [DOI] [PubMed] [Google Scholar]
- 20.Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, et al. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014; 508: 488–493. doi: 10.1038/nature13151 [DOI] [PubMed] [Google Scholar]
- 21.Daish TJ, Casey AE, Grutzner F. Lack of sex chromosome specific meiotic silencing in platypus reveals origin of MSCI in therian mammals. BMC. Biol. 2015; 13: 106 doi: 10.1186/s12915-015-0215-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic. An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012; 28: 1647–1649. doi: 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coge F, Guenin SP, Fery I, Migaud M, Devavry S, Slugocki C, et al. The end of a myth. Cloning and characterization of the ovine melatonin MT(2) receptor. Br. J. Pharmacol. 2009; 158: 1248–1262. doi: 10.1111/j.1476-5381.2009.00453.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowry OH, Rosenbrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol. Chem. 1951; 193: 265–275. [PubMed] [Google Scholar]
- 25.Legros C, Devavry S, Caignard S, Tessier C, Delagrange P, Ouvry C, et al. Melatonin MT(1) and MT(2) receptors display different molecular pharmacologies only in the G-protein coupled state. Br. J. Pharmacol. 2014; 171: 186–201. doi: 10.1111/bph.12457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clement N, Renault N, Guillaume J- L, Cecon E, Journé A-S, Laurent X, et al. Importance of the second extracellular loop for melatonin MT 1 receptor function and absence of melatonin binding in GPR50. Br. J. Pharmacol. 2017; 16: 168 doi: 10.1111/bph.14029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Audinot V, Bonnaud A, Grandcolas L, Rodriguez M, Nagel N, Galizzi JP, et al. Molecular cloning and pharmacological characterization of rat melatonin MT1 and MT2 receptors. Biochem. Pharmacol. 2008; 75: 2007–2019. doi: 10.1016/j.bcp.2008.02.022 [DOI] [PubMed] [Google Scholar]
- 28.Devavry S, Legros C, Brasseur C, Cohen W, Guenin SP, Delagrange P, et al. Molecular pharmacology of the mouse melatonin receptors MT(1) and MT(2). Eur. J. Pharmacol. 2012; 677: 15–21. doi: 10.1016/j.ejphar.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 29.Phillips MJ, Bennett TH, Lee MSY. Molecules, morphology, and ecology indicate a recent, amphibious ancestry for echidnas. Proc Natl Acad Sci U S A. 2009; 106: 17089–17094. doi: 10.1073/pnas.0904649106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grutzner F, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008; 453: 175–183. doi: 10.1038/nature06936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsend-Ayush E, He C, Myers MA, Andrikopoulos S, Wong N, Sexton PM, et al. Monotreme glucagon-like peptide-1 in venom and gut. One gene—two very different functions. Sci. Rep. 2016; 6: 37744 doi: 10.1038/srep37744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Q, Tan H, Irwin DM. Evolution of the Vertebrate Resistin Gene Family. PLoS. One. 2015; 10: e0130188 doi: 10.1371/journal.pone.0130188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He C, Tsend-Ayush E, Myers MA, Forbes BE, Grutzner F. Changes in the ghrelin hormone pathway maybe part of an unusual gastric system in monotremes. Gen. Comp Endocrinol. 2013; 191: 74–82. doi: 10.1016/j.ygcen.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 34.Li DY, Smith DG, Hardeland R, Yang MY, Xu HL, Zhang L, et al. Melatonin receptor genes in vertebrates. Int. J Mol. Sci. 2013; 14: 11208–11223. doi: 10.3390/ijms140611208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pala D, Lodola A, Bedini A, Spadoni G, Rivara S. Homology models of melatonin receptors. Challenges and recent advances. Int. J Mol. Sci. 2013; 14: 8093–8121. doi: 10.3390/ijms14048093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebisawa T, Karne S, Lerner MR, Reppert SM. Expression cloning of a high-affinity melatonin receptor from Xenopus dermal melanophores. Proc. Natl. Acad. Sci. U. S. A. 1994; 91: 6133–6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanova EA, Bechtold DA, Dupre SM, Brennand J, Barrett P, Luckman SM, et al. Altered metabolism in the melatonin-related receptor (GPR50) knockout mouse. Am. J Physiol Endocrinol. Metab. 2008; 294: E176–E182. doi: 10.1152/ajpendo.00199.2007 [DOI] [PubMed] [Google Scholar]
- 38.Drew JE, Barrett P, Mercer JG, Moar KM, Canet E, Delagrange P, et al. Localization of the melatonin-related receptor in the rodent brain and peripheral tissues. J Neuroendocrinol. 2001; 13: 453–458. [DOI] [PubMed] [Google Scholar]
- 39.Batailler M, Mullier A, Sidibe A, Delagrange P, Prevot V, Jockers R, et al. Neuroanatomical distribution of the orphan GPR50 receptor in adult sheep and rodent brains. J Neuroendocrinol. 2012; 24: 798–808. doi: 10.1111/j.1365-2826.2012.02274.x [DOI] [PubMed] [Google Scholar]
- 40.Sidibe A, Mullier A, Chen P, Baroncini M, Boutin JA, Delagrange P, et al. Expression of the orphan GPR50 protein in rodent and human dorsomedial hypothalamus, tanycytes and median eminence. J Pineal Res. 2010; 48: 263–269. doi: 10.1111/j.1600-079X.2010.00750.x [DOI] [PubMed] [Google Scholar]
- 41.Barrett P, Ivanova E, Graham ES, Ross AW, Wilson D, Ple H, et al. Photoperiodic regulation of cellular retinol binding protein, CRBP1 [corrected] and nestin in tanycytes of the third ventricle ependymal layer of the Siberian hamster. J Endocrinol. 2006; 191: 687–698. doi: 10.1677/joe.1.06929 [DOI] [PubMed] [Google Scholar]
- 42.Audinot V, Mailliet F, Lahaye-Brasseur C, Bonnaud A, Le GA, Amosse C, et al. New selective ligands of human cloned melatonin MT1 and MT2 receptors. Naunyn Schmiedebergs Arch. Pharmacol. 2003; 367: 553–561. doi: 10.1007/s00210-003-0751-2 [DOI] [PubMed] [Google Scholar]
- 43.Mailliet F, Audinot V, Malpaux B, Bonnaud A, Delagrange P, Migaud M, et al. Molecular pharmacology of the ovine melatonin receptor. Comparison with recombinant human MT1 and MT2 receptors. Biochem. Pharmacol. 2004; 67: 667–677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure of Mel1c in platypus. The entire gene is 34,831 bp in length, where ~30,000 bp intron separates two exons: exon 1 is 184 bp long and exon 2 is 866 bp long. Every 5000 bp are labeled. B) Protein alignment of Mel1c from platypus (O. anatinus), chicken (G. gallus) and the clawed frog (X. laevis), showing that Mel1c is very conserved at the amino acid level. Each amino acid is depicted by its single letter symbol and an associated colour. Identity bar above alignment denotes the similarity at each amino acid position between the four species: green = all homologous; yellow = two species homologous; no bar = no homology. Every 10 amino acid positions are labeled. C) DNA alignment of Mel1c exons from platypus (O. anatinus), chicken (G. gallus) and the clawed frog (X. laevis), showing that at a nucleotide level Mel1c has more variation compared to at the amino acid level. Grey bars indicate homology between all three species while colours indicate SNPs (Single Nucleotide Polymorphisms); red = A; green = T; yellow = G; purple = C. Identity bar above alignment denotes the similarity at each nucleotide position between the three species: green = all homologous; yellow = two species homologous; no bar = no homology. Every 20 bp are labeled.
(TIF)
They are counter listings, formatted to give the individual numbers used to calculate all the saturation curves and affinities reported in the present paper. The plates are all arranged the same ways: Saturation: The 3 first columns are used for increasing low concentrations (nM): A: 0.01; B: 0.02; C: 0.04; D: 0.05; E: 0.08; F: 0.1; G: 0.2; in triplicate. The 3 next columns (4 to 6) were used for the nonspecific binding. The 3 next columns (7 to 9) were used for higher concentrations: A: 0.3; B: 0.4; C: 0.5; D: 0.8; E: 1; F: 1.5 and G: 2. The last 3 columns, same concentrations, nonspecific binding. Nonspecific binding was done in the presence of 10 μM of cold melatonin. The H line was not used. R: 11 concentrations of each product. The concentrations of the products were from 10-14M to 10-4M (from column 1 to 11). Colum 12 is for unspecific binding. Two lines (A&B; C&D, etc.) were used per compounds. For DR in COS7 cell membranes, only 8 compounds were tested in that order from top to bottom: melatonin, 2-iodomelatonin, S 70254, 4P-P-DOT, S 20098/agomelatonin, S 22153, FLN68/ramelteon and Luzindole. For DR in CHO cell membranes in that order from top to bottom: melatonin, 2-iodomelatonin, 6-chlmromeltonin, Luzindole, 4PPDOT, S 20098/agomelatin, FLN68/ramelteon, D600, S20928, S21278, S22153, S70254, S73893, S75436, S27128, DIV880, SD6, SD1881, SD1882 and SD1918. If needed, more information can be obtained from the corresponding author upon request. Table A. Raw data for calculation of COS7 Xenopus Mel1c (n = 1 & 2) saturations. Table B. Raw data for calculation of COS7 Platypus Mel1c (n = 1) saturation. Table C. Raw data for calculation of COS7 Platypus (n = 2) & Xenopus (n = 3) Mel1c saturations. Table D. Raw data for calculation of COS7 Mel1c Platypus (n = 3) and naïve cells saturation. Table E. Raw data for calculation of CO7 Mel1c Chicken (n = 1 & 2) and naïve cells saturations. Table F. Raw data for calculation of CHO Mel1c Xenopus (n = 1) saturation. Table G. Raw data for calculation of CHO Mel1c Platypus (n = 1 & 2) saturation. Table H. Raw data for calculation of CHO Mel1c Platypus (n = 3), Human MT1 and MT2 & Xenopus Mel1c (n = 2) saturation. Table I. Raw data for calculation of COS7 Mel1c Xenopus saturation (n = 4 & 5) and dose-responses (n = 1 & 2). Table J. Raw data for calculation of COS7 Mel1c Platypus (n = 1 & 2) dose-responses. Table K. Raw data for calculation of COS7 Chicken Mel1c (n = 1 & 2) dose-responses. Table L. Raw data for calculation of CHO Mel1C Platypus (n = 2) & Xenopus (n = 2) dose-responses. Table M. Raw data for calculation CHO Mel1C Platypus (n = 1) & Xenopus (n = 1) dose-responses.
(ZIP)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Additional data have been uploaded to the NCBI GenBank and are accessible using the following link: https://www.ncbi.nlm.nih.gov/nuccore/XM_001512887.