Abstract
Modular polyketide synthases (PKSs) produce numerous structurally complex natural products with diverse applications in medicine and agriculture. They typically consist of several multienzyme subunits that utilize structurally-defined docking domains (DDs) at their N- and C-termini to ensure correct assembly into functional multi-protein complexes. Here we report a fundamentally different mechanism for subunit assembly in trans-AT modular PKSs at the junction between ketosynthase (KS) and dehydratase (DH) domains. This involves direct interaction of a largely unstructured docking domain (DD) at the C-terminus of the KS with the surface of the downstream DH. Acyl transfer assays and mechanism-based crosslinking established that the DD is required for the KS to communicate with the acyl carrier protein appended to the DH. Two distinct regions for binding of the DD to the DH were identified using NMR spectroscopy, carbene footprinting and mutagenesis, providing a foundation for future elucidation of the molecular basis for interaction specificity.
Introduction
Modular PKSs assemble structurally diverse carbon frameworks via a succession of covalently tethered intermediates1. Due to their modular architecture, which is responsible for a logical series of enzymatic chain elongation and modification reactions, such PKSs are frequently likened to molecular production lines2. These fascinating molecular machines usually consist of several multi-enzyme subunits, which must undergo non-covalent self-assembly to produce functional megasynthases with molecular weights typically in the MDa range3,4.
Three domains are required for chain elongation in a PKS module: an acyl carrier protein (ACP) domain, which is post-translationally modified via attachment of a coenzyme A-derived phosphopantetheine (ppant) “arm” to a conserved Ser residue; an acyltransferase (AT) domain, which loads an (alkyl)malonyl extender unit onto the ACP domain’s ppant thiol; and a ketosynthase (KS) domain, which utilizes a conserved active site Cys residue to receive the growing polyketide chain from the ppant thiol of the ACP domain in the upstream module and joins it to the extender unit via a decarboxylative Claisen condensation5. The α- and β-carbon atoms in the resulting β-ketothioester can be modified by optional catalytic domains, including ketoreductases (KRs), dehydratases (DHs), enoylreductases, and C- and O-methyltransferases (MTs)6,7.
Two phylogenetically distinct classes of modular PKSs, known as cis-AT and trans-AT, have been identified8. In the former an AT domain is typically found in every module, whereas in the latter the modules lack such domains and a single standalone AT loads a malonyl extender unit onto each ACP domain8. The archetypal example of a cis-AT PKS is the 6-deoxyerthronolide B synthase (DEBS), which assembles the polyketide precursor of erythromycin A9. The three subunits of this well-studied system self-assemble via N- and/or C-terminal docking domains (DDs)10–12, which are portable and can be exploited to produce engineered systems with unnatural subunit combinations13.
In contrast to their cis-AT counterparts, the mechanism(s) by which subunits self-assemble in trans-AT PKSs are less well understood. A key difference between these two types of assembly line is that, unlike cis-AT PKSs, the junctions between subunits in trans-AT systems often occur within a module. This gives rise to various types of split module (e.g. with junctions between KS/KR, KS/DH, KR/MT and DH/KR domains)14. Recently, DDs in the trans-AT PKSs responsible for the biosynthesis of virginiamycin and macrolactin have been reported15,16. These consist of approximately 25 residue appendages at the N- and C-termini of subunits, which form a four-helix bundle upon docking with a cognate partner and have been found at ACP/KS, DH/KR and KS/KR junctions. The complexes formed by such DDs were found to have dissociation constants in 0.8-10 μM range and a high degree of selectivity for cognate over non-cognate partners was observed.
Despite these advances, the mechanism of communication across KS/DH junctions in trans-AT PKSs remains obscure. To address this problem, we elected to study the GbnD4 and GbnD5 subunits of the gladiolin PKS, which have KS and DH domains at their C- and N-termini, respectively. Gladiolin is a novel macrolide antibiotic with promising activity against drug resistant strains Mycobacterium tuberculosis and negligible toxicity towards mammalian cells that we recently discovered as a metabolite of Burkholderia gladioli BCC0238, a clinical isolate from a cystic fibrosis patient17. Here we show that a largely unstructured DD at the C-terminus of the GbnD4 KS domain interacts directly with the GbnD5 DH domain, revealing a new paradigm for subunit communication across KS/DH junctions in PKS assembly lines.
Results
Identifying putative docking domains at KS/DH junctions
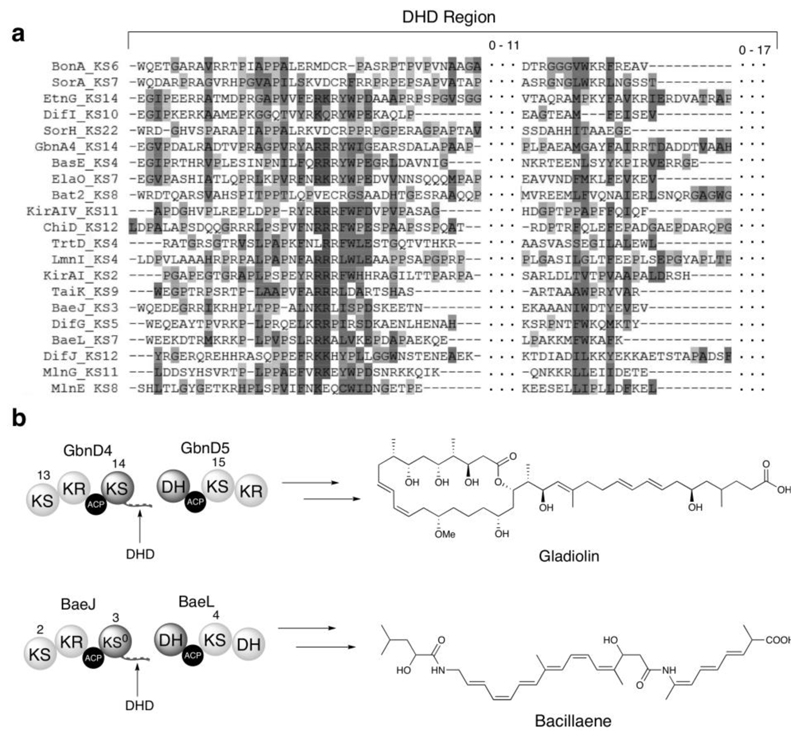
Inspection of KS/DH junctions in trans-AT PKSs suggested that, unlike other types of catalytic domain at subunit interfaces, the DH domains lack an N-terminal DD. On the other hand, the KS domains have an approximately 40-80 residue unannotated region fused to their C-termini, which could function as DDs that interact directly with the downstream DH domain (Fig. 1). These putative DH docking (DHD) domains were found to be present at most, but not all KS/DH junctions in trans-AT PKSs for which the metabolic product is known (Supplementary Fig. 1). A profile hidden Markov model based on the sequences of the putative DHD domains in these systems was developed,18 but was found to perform poorly in the identification of additional examples, consistent with the low level of conservation observed for these regions.
Figure 1. Bioinformatics analysis of DHD domains and examples from the gladiolin and bacillaene PKSs.
(a) Multiple sequence alignment of DHD domains identified by manual inspection of KS/DH boundaries in trans-AT PKSs for which the metabolic product is known. Localized conservation is observed in the N-terminal region, but the C-terminal region is poorly conserved. Dotted sections indicate residues between the conserved regions of the DHD domain, and the number of residues located in these regions is indicated above. Sequences from the following pathways were used: Bae, bacillaene (B. amyloliquefaciens); Bas, basiliskamide (B. laterosporus PE36); Bat, batumin/kalimantacin (P. fluorescens); Dfn, difficidin (B. amyloliquefaciens); Etn, etnangien (S. cellulosum So ce56); Ela, elansolid (Chitinophaga pinensis); Gbn, gladiolin (B. gladioli); Kir, kirromycin (S. collinus); Lnm, leinamycin (S. atroolivaceus); Mln, macrolactin (B. amyloliquefaciens); Mmp, mupirocin (Pseudomonas fluorescens); Sor, sorangicin (Sorangium cellulosum); Tai, thailandamide (B. thailandensis E264); Trt, tartrolon (T. turnerae T7901).
(b) Examples of DHD domains in the gladiolin (top) and bacillaene (bottom) PKSs and structures of the metabolites produced by these assembly lines.
Secondary structure analysis of the aligned DHD domain sequences using the PsiPred and IUpred servers indicated a high degree of disorder with the propensity for β-strand formation in two distinct regions19,20. The DHD domains are predicted to have flexible or context dependent dynamics, with most order parameters, S2, below 0.8 (S2=1 indicates complete rigidity, S2=0 indicates unrestricted motion), in both the presence and the absence of the corresponding KS domain (Supplementary Fig. 2). Regions of the DHD domains with higher order parameters appear to coincide with amino acids predicted to be involved in binding events by the web server ANCHOR21. Despite the lack of sequence conservation, the predicted binding sites seemingly occur in similar two site patterns for all DHD domains (Supplementary Fig. 3). In addition, DHD domains possess a notably low pI, which may facilitate association with the downstream DH domain via electrostatic interactions. Interestingly, phylogenetic analysis of all trans-AT PKS DH domains revealed that those predicted to be interaction partners for DHD domains belong to a discrete clade (Supplementary Fig. 4).
Acyl transfer across a trans-AT PKS KS/DH junction
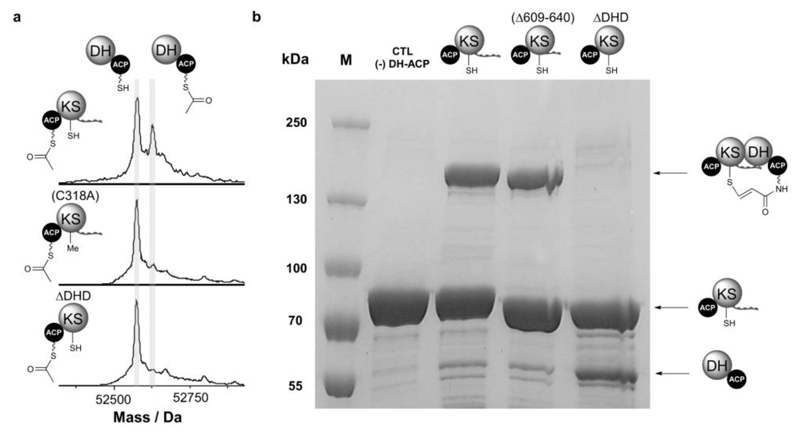
To investigate the role played by DHD domains in communication across KS/DH junctions the C-terminal ACP-KS-DHD tri-domain of GbnD4 and the N-terminal DH-ACP di-domain of GbnD5 (Fig. 1) were overproduced in E. coli as N-terminal His6 fusion proteins, which were purified to near-homogeneity using immobilized metal-ion affinity chromatography (IMAC) (Supplementary Fig. 5). The identity of the purified proteins was confirmed by ESI-Q-TOF-MS analyses (Supplementary Fig. 6). The gladiolin PKS contains 20 KS domains, 17 of which are predicted to catalyze chain elongation and 3 of which appear to function as transacylases that shuttle particular chain elongation intermediates between ACP domains.17 In most other trans-AT PKSs, a conserved active site His residue, which plays an essential role in chain elongation, is lacking from transacylating KS domains. However, this is not the case for the gladiolin PKS and it is unclear whether the KS domain at the C-terminus of GbnD4 catalyzes chain elongation or transacylation. We thus sought to establish whether the KS domain in the ACP-KS-DHD tri-domain construct is active by investigating whether it can catalyze transfer of an acetyl group from the upstream ACP domain to the corresponding domain in the DH-ACP construct. To do this we exploited the inability of E. coli to convert ACP domains of PKSs into their phosphopantetheinylated holo form. Sfp, a substrate tolerant phosphopanthetheinyl transferase, was used to transfer the S-acetyl-ppant moiety of acetyl-CoA onto the ACP domain of the ACP-KS-DHD construct (Supplementary Fig. 7). The DH-ACP di-domain was converted to its holo form by treatment with Sfp and coenzyme A (Supplementary Fig. 7), then incubated with a twofold excess of the acetylated holo-ACP-KS-DHD tri-domain. Separation and analysis of the protein mixture using UHPLC-ESI-Q-TOF-MS showed that 39.9% (σ2=1.34) of the DH-ACP di-domain was acetylated (Supplementary Fig. 8 and Fig. 2a). The level of DH-ACP di-domain acetylation was much lower (5.9%, σ2=0.39) in an analogous experiment employing an ACP-KS-DHD variant in which the active site Cys residue of the KS domain has been mutated to Ala, confirming that the KS domain catalyzes the acetyl transfer reaction (Fig. 2a).
Figure 2. Acyl transfer and protein crosslinking assays demonstrate that DHD domains play a key role in communication across KS/DH interfaces.
(a) Deconvoluted mass spectra of GbnD5 holo-DH-ACP, following incubation with GbnD4 Ac-ACP-KS-DHD (top), Ac-ACP-KS(C318A)-DHD (middle) and Ac-ACP-KS(ΔDHD) (bottom). Transfer of the acetyl group onto the holo-DH-ACP di-domain results in a +42 Da mass shift. The data show that an acyl group can be transferred across the subunit interface (top), and that the KS active site Cys residue plays an important role in this process (middle). No acetylation of the holo- DH-ACP di-domain is observed when the DHD domain is removed (bottom).
(b) SDS-PAGE (6%) analysis of crosslinking reactions between the GbnD5 DH-ACP di-domain loaded with the chloroacrylamide-terminated ppant analogue and the GbnD4 ACP-KS-DHD tri-domain (left), GbnD4 ACP-KS-DHD(Δ609-640) truncated tri-domain (middle) and the ACP-KS(ΔDHD) di-domain (right). Efficient formation of a crosslinked complex (˜130 kDa) is observed for the ACP-KS-DHD tri-domain (left) and GbnD4 ACP-KS-DHD(Δ609-640) truncated tri-domain (middle), but not the ACP-KS(ΔDHD) di-domain (right).
We next investigated whether the C-terminal DHD domain of GbnD4 and the N-terminal DH domain of GbnD5 play a role in the KS-catalyzed transacylation reaction. A GbnD4 apo-ACP-KS(ΔDHD) di-domain, lacking the C-terminal DHD domain, was overproduced, purified and converted to its acetylated holo form using Sfp and acetyl-CoA (Supplementary Figs 5-7). Negligible acetylation of the holo-ACP-DH di-domain was observed when it was incubated with the acetylated holo-ACP-KS(ΔDHD) di-domain (Fig. 2a). Similarly, the GbnD5 ACP domain, lacking the upstream DH domain, was overproduced, purified and phosphopantetheinylated using Sfp and coenzyme A (Supplementary Figs 5-7). A small amount of acetyl transfer was observed (6.6%, σ2=0.63) when the GbnD5 ACP domain was incubated with the acetylated GbnD4 holo-ACP-KS-DHD tri-domain (Supplementary Fig. 9). These data demonstrate that both the DHD domain and the DH are required for efficient acyl transfer across the GbnD4-GbnD5 interface.
Trapping the ACP-KS-DHD/DH-ACP complex via crosslinking
Type II fatty acid synthase ACPs post-translationally modified with an analogue of ppant in which the terminal thiol has been replaced by trans-β-chloroacrylamide have been shown to react efficiently with the active site Cys residue of cognate KSs to generate covalently crosslinked complexes22. Such an approach has also been shown to be effective for crosslinking ACP and KS domains from the DEBS cis-AT PKS23. The extent of crosslinking in these experiments correlates with the strength of the interaction between the proteins. We exploited this mechanism-based crosslinker to further explore the interaction between the GbnD4 ACP-KS-DHD tri-domain and the GbnD5 DH-ACP di-domain. The trans-β-chloroacrylamide-containing pantetheine analogue was synthesized, then enzymatically converted to the corresponding coenzyme A analogue, which was used by Sfp to post-translationally modify the ACP domain of the apo-DH-ACP construct (Supplementary Fig. 10), using an established procedure23. Incubation of the resulting crosslinker-modified DH-ACP di-domain with the GbnD4 apo-ACP-KS-DHD tri-domain resulted in efficient formation of a covalent complex (Fig. 2b). No such complex was observed when the crosslinker-modified DH-ACP di-domain was omitted from the reaction, or when the wild type apo-ACP-KS-DHD tri-domain was replaced by the C318A mutant (Fig. 2b and Supplementary Fig. 11). Similarly, incubation of the crosslinker-modified GbnD5 DH-ACP di-domain, with the ACP-KS(ΔDHD) di-domain yielded no covalent complex (Fig. 2b), confirming the important role played by the DHD domain in communication across the GbnD4/GbnD5 interface.
Bioinformatics analyses indicate that DHD domains contain two separate DH-binding regions (Supplementary Fig. 3). To explore the contribution made by each of these to communication across the junction between KS and DH domains, a GbnD4 ACP-KS-DHD(Δ609-640) construct was produced. In this construct, the C-terminal DH-binding region of the DHD domain has been removed, but the N-terminal DH-binding region remains intact. Incubation of the crosslinker-modified DH-ACP di-domain with the ACP-KS-DHD(Δ609-640) construct resulted in a similar amount of covalent complex formation to that observed with the ACP-KS-DHD tri-domain (Fig. 2b). This indicates that the N-terminal DH-binding region plays a key role in the formation of a complex between the DHD and DH domains. A modest reduction in the amount of acetyl transfer was also observed (31.1%, σ2=0.16), providing additional support for this hypothesis. Further experiments will be required to clarify the role played by the C-terminal DH-binding region, which shows a lower degree of conservation than the N-terminal region (Fig. 1a).
Characterization of the DHD domain by NMR spectroscopy
The GbnD4 DHD domain was overproduced as an N-terminal His6 fusion and purified via IMAC, allowing its structural and dynamic properties to be investigated using NMR spectroscopy, which is an ideal tool for atomic resolution characterization of largely unstructured proteins24–26. Sharp signals with narrow dispersion in the 1H dimension of 1H-15N HSQC spectra indicated that the DHD domain adopts a mostly disordered structure. Signals due to 83% of the backbone heavy atoms were assigned using a standard set of triple resonance experiments on a [U-13C-15N]-labelled sample. Some of the residues were found to give rise to a second, weaker set of signals suggesting the presence of multiple conformations that slowly interconvert (Fig. 3a). The slow rate of exchange is possibly due, at least in part, to the relatively low temperature at which the spectra were acquired. The chemical shifts of signals due to the major conformational isomer were used to calculate secondary structure propensity scores, allowing regions of the DHD domain with a tendency to form α-helices or β-strands to be detected. No propensity for secondary structure formation was found for the minor conformer. Only very small propensities, which barely reached the threshold for any type of secondary structure element, were identified for the major conformer (Supplementary Fig. 12). Consistent with this, the experimental chemical shift values were in good agreement with those calculated for a random coil conformation of the DHD domain (Supplementary Fig. 12). Moreover, a 1H-1H NOESY experiment indicated a lack of long range contacts, suggesting that the DHD domain does not adopt a defined tertiary structure. Finally, the circular dichroism spectrum of the DHD domain was consistent with a largely unstructured protein, indicating only a modest propensity for β-strand formation (Supplementary Fig. 13).
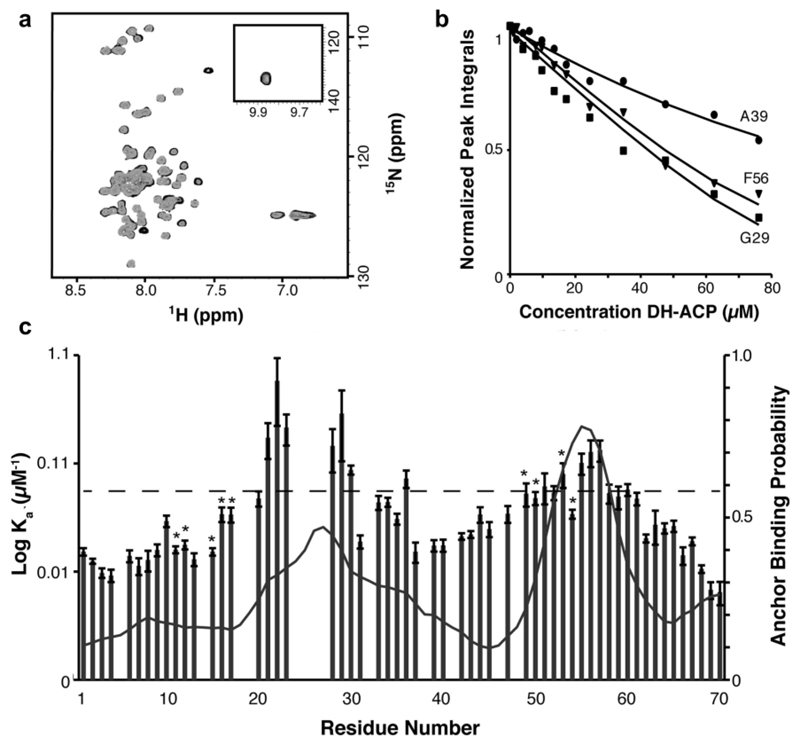
Figure 3. NMR titrations reveal two regions of DHD domains involved in the interaction with DH-ACP di-domains.
(a) Overlaid 2D BEST-TROSY-15N-HSQC spectra of [U-15N]-labeled GbnD4 DHD domain titrated with unlabeled GbnD5 apo-DH-ACP di-domain. The spectrum in black shows the DHD domain alone. The grey spectrum show the DHD domain in a presence of an equivalent amount of the GbnD5 DH-ACP di-domain. As the concentration of the GbnD5 DH-ACP di-domian was increased, several signals were observed to decrease in intensity, suggestive of slow to intermediate exchange with the binding affinity in the lower μM range.
(b) Experimental data points and individual fits of normalized signal integrals for selected residues located in the first (Gly29, squares) and second (Phe56, triangles) DH-ACP-interacting regions of the DHD domain, and the region between them (Ala39, circles).
(c) LogKa values calculated from signal integral values for individual residues of the DHD domain. Residues with values above the threshold (dotted line – defined in the online methods) are involved in interaction with the DH-ACP di-domain. These data agree well with the binding site probabilities predicted by the ANCHOR web server (solid line). Asterisks indicate residues for which a second set of weaker resonances was observed in the spectra. The error bars correspond to two standard deviations of 1000 iterations of fits using Monte Carlo error analysis.
Additional insight into the structural properties of the Gbn4 DHD domain was provided by backbone dynamics measurements. As is typical for an intrinsically disordered protein, steady-state {1H}-15N NOE, 15N spin-lattice (R1) and spin-spin (R2) relaxation rates revealed that the DHD domain has high local mobility on the fast picosecond to nanosecond timescale (Supplementary Fig. 14). While the low overall {1H}-15N NOE values for the DHD domain show that it is generally very flexible, Val19 to Glu30 and, to a lesser extent, Ala49 to Arg60 gave higher {1H}-15N NOE values, indicative of reduced mobility on the fast time scale in these regions. A similar trend, manifested by higher R2 rates in the Val19 to Glu30 and Ala49 to Arg 60 regions, was also observed in the 15N relaxation data (Supplementary Fig. 14).
Probing the interaction between the DHD and DH domains
To investigate the nature of the interaction between the DHD and DH domains, we examined how the 2-D BEST-TROSY-15N-HSQC spectrum of the [U-15N]-labeled GbnD4 DHD domain changes when increasing concentrations of the GbnD5 DH-ACP di-domain are added. Several signals were observed to decrease in intensity without any significant change in chemical shift and no new signals appeared in the spectrum (Fig. 3a), suggesting a slow to intermediate exchange process and a dissociation constant in the low μM range. Interestingly, the residues of the DHD domain for which the strongest decrease in signal intensity as a function of DH-ACP concentration was observed were localized in the Val19 to Glu30 and Ala49 to Arg60 regions (Fig. 3b and c). This suggests that two distinct sites of the DHD domain bind to the DH-ACP di-domain. Unfortunately, detection of any structural or conformational changes in the DHD domain upon binding to the DH-ACP di-domain was precluded by the large size of the complex, which caused broadening of the signals in the NMR spectrum beyond the limit of detection. Due to the slow/intermediate rate of exchange and the two-site interaction of the DHD domain with the DH-ACP di-domain, it was only possible to estimate the dissociation constant of the complex. The rate of signal intensity decay as a function of DH-ACP di-domain concentration yields apparent dissociation constants on the residue level (Fig. 3b). Compared to the average of these apparent dissociation constants for the full DHD domain (approximately 40 μM) the average values for the Val19-Glu30 and Ala49-Arg60 regions suggest that these bind to the DH-ACP di-domain with significantly higher affinities (average apparent dissociation constants in the 7-15 μM range).
Mapping of interaction sites by carbene footprinting
To establish the site(s) of interaction of the GbnD4 DHD domain with the GbnD5 DH domain, recently developed carbene footprinting methodology was employed27. This involves labeling of the solvent-exposed surface of a protein with a highly reactive carbene, followed by proteolytic digestion and LC-MS/MS analysis. When this experiment is carried out in the presence and absence of an interaction partner, differential labeling is observed allowing the site of interaction to be identified.
LC-MS analysis of tryptic digests of the DHD domain and DH-ACP di-domain resulted in 70% and 52% sequence coverage, respectively (Supplementary Fig. 15). Differential labeling of five tryptic fragments from the DH domain was observed when carbene footprinting was carried out in the presence and absence of the DHD domain (Supplementary Fig. 16). MS/MS analysis of these differentially labeled tryptic fragments allowed near-residue level identification of the DHD domain’s binding site (Supplementary Fig. 17). On the other hand, differential labeling of only a single tryptic fragment of the DHD domain was observed (Supplementary Fig. 16) and MS/MS analysis showed that the masked region overlaps with the C-terminal DH-interacting region of the DHD domain (Ala49-Arg60) identified by NMR spectroscopic analysis (Supplementary Fig. 17). Unfortunately, no labeling data was available for the other DH-interacting region identified by the NMR experiments, (Val19-Glu30), due to the presence of three contiguous Arg residues in the sequence.
We also sought to map the interaction of the ACP domain with the DH domain in the GbnD5 DH-ACP di-domain. To do this, the DH domain was overproduced as N-terminal His6 fusion protein in E. coli and purified using IMAC (Supplementary Fig. 5). 39% sequence coverage was observed in LC-MS analyses of tryptic digests of the ACP domain (Supplementary Fig. 15). Three tryptic fragments from the DH domain and one fragment from the ACP domain were found to be differentially labeled when the data from carbene footprinting of the DH-ACP di-domain, and the standalone DH and ACP domains were compared (Supplementary Fig. 18). As before, the interaction interface between the DH and the ACP was mapped to near-residue level using MS/MS (Supplementary Fig. 19).
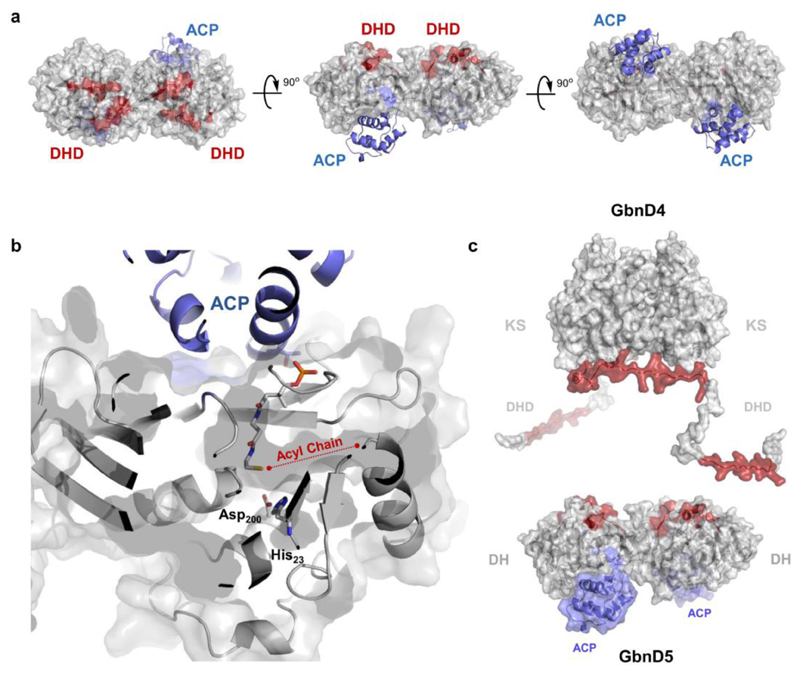
The results of these experiments were visualized on a homology model of the GbnD5 DH domain, revealing distinct binding sites for the DHD and ACP domains. These are located adjacent to each other, allowing simultaneous interaction of the DHD and ACP domains with the DH domain (Fig. 4a). This brings the GbnD4 KS domain into close proximity to the GbnD5 ACP domain, facilitating acyl transfer or chain elongation across the subunit interface. The GbnD5 ACP domain also needs to be capable of delivering the acyl group attached to its ppant thiol into the active site of the DH domain. Docking simulations using residue constraints derived from the results of the carbene footprinting experiments place the conserved Ser residue that undergoes phosphopantetheinylation in the ACP domain at the entrance to the DH active site (Fig. 4b). Taken together, these data suggest that a single ACP binding site is sufficient to allow the ppant arm to access the active sites of both the GbnD4 KS domain and the GbnD5 DH domain (Fig. 4c).
Figure 4. Mapping DHD and ACP domain interaction sites on the DH domain.
(a) Identification of the interaction sites for the GbnD4 DHD domain and GbnD5 ACP domain on the dimeric GbnD5 DH domain using carbene foot-printing. Regions masked by the DHD and ACP domains are highlighted in red and blue, respectively, on a homology model of GbnD5 DH domain (grey). Residue constraints derived from the foot-printing experiments were used to dock the ACP domain (blue) onto the DH domain. Residues used to constrain the docking calculations are as follows; DH: Leu31, Arg61, Leu298, Val299, Asp300; ACP: Leu394, Ala395, Leu396. The model shows adjacent but distinct binding sites for the DHD and ACP domains. These place the GbnD4 KS domain and the GbnD5 ACP domain in close proximity, facilitating efficient communication across the subunit interface.
(b) Predicted location of the ppant arm in the model of the GdnD5 DH-ACP di-domain complex based on the X-ray crystal structure of a crosslinked complex of the ACP and DH from the E. coli fatty acid synthase (PDB accession no: 4KEH). The docking site for the ACP domain is situated near the entrance to the active site of the DH domain. This allows the thiol of the ppant arm to sit adjacent to the catalytic Asp and His residues in the active site of the DH domain. A putative binding pocket for the acyl chain of the substrate sits to the right of the ppant thiol.
(c) Overall model for the interaction between the GbnD4 KS-DHD (red) and GbnD5 DH-ACP (grey/blue) di-domains, both of which form homodimers. The red patches on the surface of the DH and DHD domains indicate mutual interaction sites.
Probing the specificity of the DHD/DH interaction
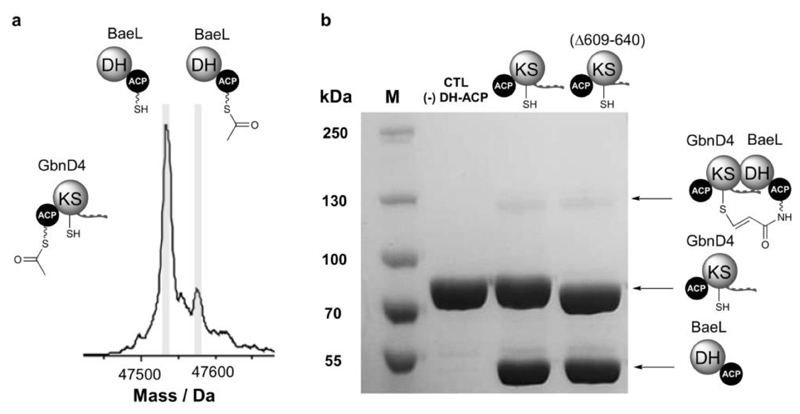
To investigate whether the GbnD4 DHD domain is able to interact with DH domains from other systems, we overproduced and purified the N-terminal DH-ACP di-domain from the BaeL subunit of the bacillaene PKS (Supplementary Fig. 5) and examined its ability to communicate with the GbnD4 ACP-KS-DHD tri-domain using the acyl transfer and crosslinking assays (Fig. 5). The extent of acyl transfer and crosslinking for the non-cognate proteins was substantially reduced in comparison with that observed for the cognate pair (Fig. 5a), congruent with the notion that a degree of specificity in the interaction between DHD/DH domain pairs is required for the correct assembly of trans-AT PKS subunits, especially in systems that contain multiple KS-DHD/DH-ACP junctions within a single assembly line (Supplementary Fig. 20). In addition, we examined whether the GbnD4 ACP-KS-DHD(Δ609-640) construct can interact with the DH-ACP di-domain from BaeL. However, a covalent complex was not formed when the chloroacrylamide-modified DH-ACP di-domain was incubated with this construct (Fig. 5b), indicating that the N-terminal DH-binding region of the DHD domain and/or the KS domain are important determinants of interaction specificity.
Figure 5. DHD domains interact selectively with their cognate DH domain partners.
(a) Deconvoluted mass spectrum of BaeL holo-DH-ACP di-domain following incubation with GbnD4 Ac-ACP-KS-DHD tri-domain. In comparison with the GbnD4 holo-DH-ACP di-domain, only low levels of acetylation can be observed, suggesting that the GbnD4 DHD and BaeL DH domains interact weakly.
(b) SDS-PAGE (6%) analysis of the crosslinking reaction between GbnD4 ACP-KS-DHD tri-domain and GbnD4 ACP-KS-DHD(Δ609-640) truncated tri-domain with BaeL DH-ACP di-domain loaded with the β-chloroacrylamido ppant analogue. Only trace amounts of the crosslinked complex (˜130 kDa) can be observed, providing further evidence for a weak interaction between non-cognate DHD and DH domains.
Discussion
The mechanism illuminated here for inter-subunit communication across KS/DH junctions differs markedly from that reported for other types of junction in trans-AT PKSs,15,16 which involve mutually compatible DDs at both the N- and C-termini of the interacting subunits. Indeed, to the best of our knowledge, the utilization of only a single DD at the C-terminus of the upstream subunit to interact directly with a catalytic domain at the N-terminus of the downstream subunit represents a fundamentally new mechanism for ensuring productive subunit interaction in assembly line multienzymes.
Unlike other types of DD in trans-AT (and other) PKSs, the DHD domain is largely unstructured (note, however, that the four-helix bundle DD at the N-terminus of VirG appears to show elements of disorder15). Despite this, the dissociation constant estimated for the DHD domain from its DH domain partner is in the low μM range, which is similar to that reported for other types of DD in both cis- and trans-AT PKSs10,15,16. The occurrence of intrinsically disordered regions in prokaryotic proteins is not uncommon, but is more common in eukaryotes, 28,29 where they play important roles in intracellular signaling and the ordered assembly of macromolecular complexes30. Such regions evolve faster than globular regions31, and are not only subject to enhanced Darwinian selection32, but tolerate a greater range of mutations, allowing faster adjustments to specificity33. The short and disordered nature of the two DH-binding regions of the DHD domain suggests that they should be classified as ‘short linear motifs’ (SLiMs)34. The utilization of SLiMs to mediate inter-subunit communication in trans-AT PKSs may facilitate the evolution of novel biosynthetic pathways35.
The carbene footprinting analyses show that the GbnD4 DHD domain interacts with two distinct regions on the surface of the GbnD5 DH domain. These lie on opposite sides of a hydrophobic cavity, which in conventional DH domains (i.e. those internal to a subunit), in both cis- and trans-AT PKSs, is occupied by an a ~20 residue N-terminal region that is lacking in DH domains from KS/DH junctions36. A conserved HPLL motif in the N-terminal appendage of conventional DH domains occludes the region corresponding to the binding site for the DHD domain on the surface of DH domains associated with KS/DH junctions. On the basis of structural studies of a DH domain from a KS/DH junction in another trans-AT PKS system, this very recently led others to speculate that this region may provide a binding site for a putative DD at the C-terminus of the upstream subunit Supplementary Fig. 21)37.
Bioinformatics analyses suggest that the two regions in the GbnD4 DHD domain that have been identified to interact with the GbnD5 DH domain are likely to be a general feature of all DHD domains (Supplementary Fig. 3). Higher levels of conservation are observed for the N-terminal region, consistent with the observation that it is critical for complex formation (Fig. 2b).
In conclusion, complementary data from a range of structural and biochemical techniques has afforded unparalleled insights into the mechanism of communication across a ubiquitous type of subunit interface in trans-AT PKSs. These insights could ultimately facilitate future efforts to create hybrid assembly lines incorporating subunits from different trans-AT PKS systems that are capable of producing radically new chemotypes.
Online Methods
Profile Hidden Markov Model
Amino acid sequences corresponding to putative DHD domains lying downstream of trans-AT KS domains at KS/DH junctions were excised manually and aligned using ClustalX38. Using this alignment file, a profile hidden Markov model (pHMM) was built from these sequences using the hmmbuild function of HMMER 3.1b218, with default parameters.
Gene Synthesis, Cloning and Mutagenesis
The following constructs were purchased from Epoch Life Sciences: pET24a_ACP-KS-DHD, pET24a_ACP-KS(Δ609-640) (residues 1-608 of the tri-domain construct), pET24a_ACP-KS(ΔDHD) (residues 1-588 of the tri-domain construct) and pET24a_DHD (residues 571-640 of the tri-domain construct). The amplification of GbnD5 DH-ACP, GbnD5 DH and GbnD5 ACP from B. gladioli gDNA was performed with Phusion DNA polymerase (NEB). The primers used in each instance are as follows; DH-ACP_For(5’-CACCATGACTCATCGCCATGCA-3’), DH-ACP_Rev-(5’-TCATGCAGCCACCGATTCGCT-3’); DH_For-(5’-CACCATGACTCATCGCCATGCA-3’), DH_Rev-(5’-TCAGACGTCGGCGCCAGCGGA-3’); ACPDH_For(5’-CACCGTGGCGGCCGGGTACGA-3’), ACPDH_Rev(5’-TCATGCAGCCACCGATTCGCT-3’). PCR products were separated on a 1% agarose gel and bands were excised and purified with a Gene Jet gel extraction kit (Thermo Scientific). The insert was ligated with pET151 (Invitrogen) following the manufacturer’s instructions. The resulting vector was used to transform E. coli TOP10 cells (Invitrogen), which were plated on LB agar containing ampicillin (100 μg/mL). Colonies were picked and grown overnight in LB media. Plasmids were isolated from the culture using a miniprep kit (Thermo), and the inserts were sequenced to verify their integrity. The ACP-KS(C318A)-DHD mutant was constructed using the Q5 site-directed mutagenesis kit (NEB), with the following primers: ACP-KS(C318A)-DHD_For(5’-CGATACGGCCGCCGCCAGCGCGC-3’), ACP-KS(C318A)-DHD_Rev(5’-ACCACCACGTTCGGGCCG-3’).
Protein Overproduction and Purification
A single colony of E. coli BL21(DE3) that had been transformed with the appropriate expression vector was picked and used to inoculate LB medium (10 mL) containing kanamycin (50 μg/mL) or ampicillin (100 μg/mL). The resulting culture was incubated overnight at 37 °C and 180 rpm then used to inoculate LB medium (1 L) containing kanamycin (50 μg/mL) or ampicillin (100 μg/mL). The resulting culture was incubated at 37 °C and 180 rpm until the optical density of the culture at 595 nm reached 0.6, then IPTG (1 mM) was added and growth was continued overnight at 15 °C and 180 rpm. The cells were harvested by centrifugation (4,000 x g, 15 min, 4 °C) and re-suspended in buffer (20 mM Tris-HCl, 100 mM NaCl, 20 mM Imidazole, pH 7.4) at 10 mL/L of growth medium then lysed using a Constant Systems cell disruptor. The lysate was centrifuged (37,000 x g, 30 min, 4 °C) and the resulting supernatant was loaded onto a HiTrap Chelating Column (GE Healthcare), which had been pre-loaded with 100 mM NiSO4 and equilibrated in re-suspension buffer (20 mM Tris-HCl, 100 mM NaCl, 20 mM Imidazole, pH 7.4). Proteins were eluted in a stepwise manner using re-suspension buffer containing increasing concentrations of imidazole – 50 mM (5 mL), 100 mM (3 mL), 200 mM (3 mL) and 300 mM (3 mL). The presence of the protein of interest in fractions was confirmed by SDS-PAGE, and an additional gel filtration step (Superdex 75/200, GE Healthcare) was used to further purify proteins where necessary. Fractions containing the protein of interest were pooled and concentrated to 130 - 300 μM using a Viva-Spin centrifugal concentrator at an appropriate MWCO (5 kDa for ACP/DHD domains, 30 kDa for ACP-KS-DHD/DH-ACP/DH constructs). Glycerol was added to the concentrated protein samples to a final concentration of 10% (v/v) and they were snap-frozen in liquid N2 and stored at -80 °C.
Preparation of [U-13C, 15N]-Labelled DHD domain
The [U-13C, 15N]-labelled DHD domain was obtained from E. coli BL21(DE3) cells that were first grown in 1 L of LB medium at 37 °C to an optical density (595 nm) >1.0. The cells were pelleted by centrifugation at 4000 x g for 15 min and washed with PBS. After centrifuging again, the cells were re-suspended in 500 mL of M9 medium supplemented with 15NH4Cl and [U-13C] glucose. After incubation for 2 h at 37 °C, expression was induced with 1 mM IPTG and the culture was incubated at 18 °C overnight. [U-15N] labelled DHD was produced analogously using unlabeled glucose instead of [U-13C] glucose in 1 L of M9 medium. Subsequent purification steps were conducted as described above, except that the protein was lyophilized for storage at -20 °C
Acyl Transfer Assays
GbnD4 ACP-KS-DHD/ACP-KS(C318A)-DHD/ACP-KS(ΔDHD) (˜200 μM) and GbnD5 DH-ACP/BaeL DH-ACP (~130 μM) proteins were converted to their acetylated and holo- forms, respectively, by incubation in 20 mM Tris, 100 mM NaCl, 10% glycerol, 2 μM Sfp, 400 μM acetyl-CoA or CoA and 10 mM MgCl2 in a total volume of 50 μL. Excess acetyl-CoA/CoA-SH was removed by two successive 10-fold dilutions with 20 mM Tris, 100 mM NaCl, followed by concentration to 50 μL using a Viva-Spin centrifugal concentrator with a 30 kDa MWCO. Acyl transfer reactions were performed in 50 μL total volume of 20 mM Tris, 100 mM NaCl. Acetyl-ACP-KS (100 μM) was incubated with holo-DH-ACP (50 μM) at room temperature for 6 h, then analyzed by UHPLC-ESI-Q-TOF-MS.
UHPLC-ESI-Q-TOF-MS Analysis of Intact Proteins
All acyl-transfer assays were analyzed on a Bruker MaXis II ESI-Q-TOF-MS connected to a Dionex 3000 RS UHPLC fitted with an ACE C4-300 RP column (100 x 2.1 mm, 5 μm, 30 °C). The column was eluted with a linear gradient of 5–100% MeCN containing 0.1% formic acid over 30 min. The mass spectrometer was operated in positive ion mode with a scan range of 200–3000 m/z. Source conditions were: end plate offset at −500 V; capillary at −4500 V; nebulizer gas (N2) at 1.8 bar; dry gas (N2) at 9.0 L min−1; dry temperature at 200 °C. Ion transfer conditions were: ion funnel RF at 400 Vpp; multiple RF at 200 Vpp; quadrupole low mass at 300 m/z; collision energy at 8.0 eV; collision RF at 2000 Vpp; transfer time at 110.0 μs; pre-pulse storage time at 10.0 μs.
Protein Crosslinking
The chloroacrylamido-pantetheine crosslinker was synthesized according to literature procedures39,40, and attached to GbnD5 DH-ACP and BaeL DH-ACP using previously described methodology22,23. Typically, loading reactions were carried out using either DH-ACP (130 μM) or ACP (150 μM) in 20 mM Tris, 100 mM NaCl, 10 mM MgCl2 and 10% glycerol (total volume: 50 μL). The loading reaction was initiated by addition of 2 μM Sfp, 1 μM CoaA, 1 μM CoaD, 1 μM CoaE and 500 μM crosslinker, and was allowed to proceed for 2 h at 25 °C. Removal of excess crosslinker was achieved by two successive 10-fold dilutions with 20 mM Tris, 100 mM NaCl, followed by concentration using Viva-Spin centrifugal concentrators with 30 kDa (DH-ACP) or 5 kDa (ACP) MWCOs. Loading reactions were monitored by UHPLC-ESI-Q-TOF-MS. Crosslinking reactions were performed in a total volume of 25 μL of 20 mM Tris, 100 mM NaCl. Reactions were initiated by addition of apo-ACP-KS-DHD/ACP-KS(C318A)-DHD/ACP-KS(ΔDHD) (100 μM) to GbnD5 DH-ACP/BaeL DH-ACP that had been loaded with the crosslinker (200 μM). The reactions were allowed to proceed for 16 h at 25 °C before analysis by SDS-PAGE (Supplementary Fig. S12).
NMR Experiments
[U-13C,15N]-labelled DHD domain was dissolved in 200 μL of measurement buffer (50 mM potassium phosphate, 150 mM NaCl, 10% D2O, pH 6.5) to obtain a protein concentration of 450 μM. Spectra were acquired in a 3 mm tube at 280 K on a Bruker Avance 700 MHz spectrometer equipped with a TCI cryoprobe. TopSpin 3.2 was used to process the spectra and the data were analyzed with CCPNMR41. Triple resonance 3D BEST-HNCACB, BEST-HNCA, BEST-HN(CO)CACB, BEST-HNCO, and BEST-HN(CA)CO,42 were used to assign 1H, 15N and 13C resonances (deposited at BMRB, accession code 27016). The obtained chemical shifts were compared to predicted random coil chemical shifts and used for neighbor-corrected secondary structure propensity calculations using the ncSSP server. To detect long range 1H-1H contacts, a 1H-1H NOESY spectrum with a mixing time of 100 ms was acquired. 15N longitudinal (R1), 15N transverse (R2) and {1H}–15N heteronuclear NOE relaxation data were obtained using standard methods43 and fitted using the CCPNMR rate analysis tool41. Relaxation delays from 0 to 1000 ms and 316.8 ms were employed for R1 and R2 measurements. For relaxation rates, the in CCPNMR implemented covariance method was chosen to estimate the standard deviation. The {1H}–15N heteronuclear NOEs were obtained for 1H-saturated and-unsaturated spectra (6 s saturation time). For binding studies, [U-15N]-labelled DHD domain was dissolved in 200 μL of buffer (50 mM potassium phosphate, 150 mM NaCl, 10% D2O, pH 7.4) to a final concentration of 80 μM. DH-ACP (415 μM) was added in 13 steps to obtain final concentrations of 1.0, 2.0, 4.0, 5.9, 7.8, 9.8, 13.5, 17.2, 24.4, 34,7, 47.6, 62.4, and 76.1 μM. A 2-D 1H-15N BEST-TROSY-HSQC spectrum was acquired at each titration step. The data were analyzed using the CCPNMR rate analysis tool41 and Statdisk (Triola Statistics) for statistical evaluation. Equation 1 defines the dissociation of the DHD domain (P) from DHD/DH-ACP complex (PL) at a single site on the DH-ACP di-domain (L). This is a valid description because the affinity of each residue in the DHD domain for the DH-ACP di-domain is examined individually. The sum of the concentrations of the bound and unbound states gives the total concentrations of L and P (Equation 2)44. By combining Equations 1 and 2, and including a multiplier constant, A, which takes into account error in the integral of the first data point, Equation 3 can be used to determine an approximate value of the dissociation constant (Kd) for each residue of the DHD domain in the DHD/DH-ACP complex. The decay in integral value for each signal as a function of DH-ACP concentration was fitted to Equation 3. Regions affected by the interaction were defined as above the threshold, which was calculated as previously described.45 Briefly the average Ka (Ka = 1/Kd) and standard deviation were calculated for all residues. Subsequently, residues with Ka values larger than the average plus 3 standard deviations were removed and the average and standard deviations were recalculated. This process was repeated iteratively until no additional residues with Ka values larger than the average plus 3 standard deviations remained in the set. The threshold was set to the average plus the standard deviation for the final set of residues. Errors were propagates using Monte Carlo error analysis. The signal decay curves were back-predicted from the best fit parameters, random noise within the estimated range of integral errors was added to the back-predicted points and data refitted. Error bars reflect two standard deviations over 1000 iterations of the described procedure.
| Equation 1 |
| Equation 2 |
| Equation 3 |
CD Spectroscopic Analysis of the DHD domain
CD spectra of the DHD domain (0.2 mg/mL dissolved in 50 mM potassium phosphate, 50 mM sodium chloride, pH 7.4) were recorded between 190 and 260 nm at room temperature on a JASCO J-1500 using a 1 mm path length quartz cuvette. The spectra were analyzed using DichroWeb46 and fitted with the CDSSTR method using the reference data set SP 17547.
Carbene Footprinting
Interaction of GbnD5 DH and ACP domains
A 60 μM solution of DH, ACP or DH-ACP in 10 μL of 20 mM Tris, 150 mM NaCl was mixed with an equal volume of a 10 mM solution of aryldiazirine photoreagent in the same buffer. The mixture was left to equilibrate for 5 minutes at room temperature before 6 μL aliquots were placed in crystal clear vials (Fisher Scientific UK) and snap-frozen in liquid nitrogen. The labelling reaction was initiated by photolysis of the mixture using the third harmonic of a Nd:YLF laser (Spectra Physics, repetition frequency 1000 Hz, pulse energy 125 μJ) at a wavelength of 347 nm. The frozen samples were irradiated for 16 s. All experiments were performed in triplicate.
Interaction of the GbnD5 DH-ACP di-domain with the GbnD4 DHD domain
A solution of 20 μM DH-ACP in 20 mM Tris, 150 mM NaCl (10 μL total volume) was mixed with an equal volume of a 40 μM DHD domain solution (or buffer for unbound DH-ACP experiments). A 100 mM solution of photoreagent in the same buffer was added to the mixture to a final concentration of 5 mM. The same labelling protocol described above was used to start the reaction. All experiments were performed in triplicate.
Following irradiation, samples were reduced (DTT, 10 mM in 10 mM ammonium bicarbonate), alkylated (Iodoacetamide, 55 mM in 10 mM ammonium bicarbonate) and incubated at 37 °C with trypsin overnight (1:20 protease/protein ratio in 10 mM ammonium bicarbonate) in the presence of Protease MAX (Promega). The digestion was quenched by adjusting the pH to below 6 by adding formic acid to 2% final concentration.
LC-MS Analysis of Tryptic Digests from Carbene Footprinting
The analysis of the digests was carried out in load-trapping mode on a Dionex Ultimate 3000 Nano LC system using a C18 trapping column and a Pepmap C18 analytical column (Thermo Scientific, 75 μm, 150 mm, 5 μm particle size, 300 Å pore size). Peptides were eluted using a 30 min linear gradient of solvent B (water/acetonitrile 5/95 with 0.1% formic acid) from 0% to 55%, followed by 5 minutes at 90% B and a 20 minutes re-equilibration segment to 100% A (water/acetonitrile 95/5 with 0.1 % formic acid) at a flow rate of 0.3 μL/min. The HPLC system was coupled to a Thermo Scientific LTQ FT Ultra mass spectrometer equipped with a nanoelectrospray ionization source. A 1.7 kV voltage was applied to a coated PicoTip emitter (New Objective). The capillary temperature was set at 275 °C, with inner capillary voltage value set on 37 V and tube lens value of 145 V. Spectra were acquired in positive ion mode for a 400-2000 m/z range at a nominal resolution of 100,000. Identification of peptides was conducted in data-dependent mode. The 5 most intense ions for each scan were isolated and subjected to CID in the linear ion trap using a nominal energy of 35.0. Signals with +1 charge state were rejected. The data were searched against a custom FASTA database including the DH, DHD and ACP sequences using Bioworks software (Thermo Fisher Scientific). Peptide tolerance was set at 2.0 Dalton, fragment ion tolerance at 0.8 and 2 missed cleavage sites were allowed. Addition of the photo-reagent was included as a variable modification on all residues and carbamoylation of cysteine was set as fixed modification. Targeted CID experiments to locate and quantitate the modification at residue level were performed at a nominal energy of 15.0. The precursors were isolated within a window of 8 Th and the activation time was set at 30 ms.
Analysis of LC-MS data from Carbene Footprinting
Peptide level analysis
A previously described method was used to quantitate the fraction of each peptide modified27. Briefly, the chromatograms for each singly-labeled and unlabeled peptide were extracted within a range of ±0.1 m/z and the spectrum for each peak was manually inspected to ensure the sampling of the correct ion only. The peptide fractional modification was calculated using Equation 4.
| Equation 4 |
Where Alabelled and Aunlabelled correspond respectively to the peak area of each labeled and unlabeled peptide. Differences in the extent of labeling between peptides were considered significant when the p-value obtained from a Student t-test was <0.05. Peptides that showed significant differences between the different states of the proteins were subjected to MS/MS experiments in order to locate and quantitate the modification at the residue level.
Residue level analysis
Modification sites on the peptides that showed significant differences at the peptide level were investigated using tandem MS. The fractional modification on the ni residue was calculated using Equation 5.
| Equation 5 |
In which I(ni labelled) and I(ni unlabelled) correspond to the measured intensity of the fragment ni in its labeled and unlabeled version. The difference in fractional modification between two consecutive fragments, multiplied by the fractional amount of labeling on the peptide gives the absolute level of modification on the ni residue using Equation 6.
| Equation 6 |
In the case of the impossibility of detection of a ni ion, its fractional modification was grouped together with its subsequent fragment. The few fractional modification values of residues that showed an anomalous increase or decrease in respect to the fractional modification curve trend were excluded from the dataset and were grouped together with the value corresponding to the adjacent residue resulting in only a small loss of resolution. Per-residue modification values from each repeat were averaged and the averages compared between the two states of the protein. Differences in per-residue modification were considered significant when the p-value obtained from a Student t-test was <0.05.
Homology Modelling and Docking
Homology modelling of GbnD5 DH (using entry 4LN9 from the PDB as a template) and ACP domains was conducted using the I-TASSER server48, and further refinement of the model was achieved using the MolProbity server49. Docking of the ACP domain was carried out using the ZDOCK server50 with residue constraints on the DH and ACP domains obtained from the carbene footprinting analysis. The residue constraints are detailed in the legend of Fig. 4.
Supplementary Material
Acknowledgements
This research was supported by grants from the BBSRC (BB/L021692/1 to G.L.C. and BB/L022761/1 to J.R.L.). The Bruker MaXis II instrument used in this study was funded by the BBSRC (BB/M017982/1). N.J.O, J.E.M, L.M. and A.S.B. thank the University of Nottingham for funding. The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013) / ERC Grant Agreement 639907 (to J.R.L.). G.L.C. is the recipient of a Wolfson Research Merit Award from the Royal Society (WM130033).
Footnotes
Author Contributions
MJ and GLC designed the study and all authors contributed to performing the research as follows: MJ and DG carried out bioinformatics analyses; MJ overproduced and purified all recombinant proteins, conducted acyl transfer assays and LC-MS analyses of intact proteins; PP synthesised the chloroacrylamido pantetheine cross linker, and PP and MJ carried out the crosslinking experiments; SK and JRL conducted the NMR and CD experiments, and analyzed the data; ASB and JEM synthesized the diazirine reagent used in the footprinting experiments; LM and NJO carried out the footprinting experiments and analyzed the data; MJ, SK, JRL and GLC wrote the paper and all authors contributed to revision of the manuscript.
Competeing Financial Interests Statement
The authors declare no competing financial interests.
Data Availability Statement
NMR data from this study are deposited at BMRB (accession code 27016). All other data are available from the authors upon request.
References
- 1.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 2009;48:4688–716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 2.Keatinge-Clay AT. The structures of type I polyketide synthases. Nat Prod Rep. 2012;29:1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 3.Richter CD, Nietlispach D, Broadhurst RW, Weissman KJ. Multienzyme docking in hybrid megasynthetases. Nat Chem Biol. 2008;4:75–81. doi: 10.1038/nchembio.2007.61. [DOI] [PubMed] [Google Scholar]
- 4.Weissman KJ, Muller R. Protein-protein interactions in multienzyme megasynthetases. Chembiochem. 2008;9:826–848. doi: 10.1002/cbic.200700751. [DOI] [PubMed] [Google Scholar]
- 5.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 6.Keatinge-Clay AT. Stereocontrol within polyketide assembly lines. Nat Prod Rep. 2016;33:141–149. doi: 10.1039/c5np00092k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner DT, et al. alpha-Methylation follows condensation in the gephyronic acid modular polyketide synthase. Chem Commun. 2016;52:8822–8825. doi: 10.1039/c6cc04418b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helfrich EJN, Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2016;33:231–316. doi: 10.1039/c5np00125k. [DOI] [PubMed] [Google Scholar]
- 9.Katz L. Chapter 6 The DEBS Paradigm for Type I Modular Polyketide Synthases and Beyond. Methods Enzymol. 2009;459:113–142. doi: 10.1016/S0076-6879(09)04606-0. [DOI] [PubMed] [Google Scholar]
- 10.Broadhurst RW, Nietlispach D, Wheatcroft MP, Leadlay PF, Weissman KJ. The structure of docking domains in modular polyketide synthases. Chem Biol. 2003;10:723–731. doi: 10.1016/s1074-5521(03)00156-x. [DOI] [PubMed] [Google Scholar]
- 11.Gokhale RS, Khosla C. Role of linkers in communication between protein modules. Curr Opin Chem Biol. 2000;4:22–27. doi: 10.1016/s1367-5931(99)00046-0. [DOI] [PubMed] [Google Scholar]
- 12.Buchholz TJ, et al. Structural Basis for Binding Specificity between Subclasses of Modular Polyketide Synthase Docking Domains. ACS Chem Biol. 2009;4:41–52. doi: 10.1021/cb8002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo J, Lynch SR, Liu CW, Xiao X, Khosla C. Partial In Vitro Reconstitution of an Orphan Polyketide Synthase Associated with Clinical Cases of Nocardiosis. ACS Chem Biol. 2016;11:2636–2641. doi: 10.1021/acschembio.6b00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulder T, Freeman M, Piel J. Top Curr Chem. 2011:1–53. doi: 10.1007/128_2010_113. [DOI] [PubMed] [Google Scholar]
- 15.Dorival J, et al. Characterization of Intersubunit Communication in the Virginiamycin trans-Acyl Transferase Polyketide Synthase. J Am Chem Soc. 2016;138:4155–4167. doi: 10.1021/jacs.5b13372. [DOI] [PubMed] [Google Scholar]
- 16.Zeng J, et al. Portability and Structure of the Four-Helix Bundle Docking Domains oftrans-Acyltransferase Modular Polyketide Synthases. ACS Chem Biol. 2016;11:2466–2474. doi: 10.1021/acschembio.6b00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song L, et al. Discovery and biosynthesis of gladiolin: A novel macrolide antibiotic with promising activity against Mycobacterium tuberculosis. J Am Chem Soc. 2017;139:7974–7981. doi: 10.1021/jacs.7b03382. [DOI] [PubMed] [Google Scholar]
- 18.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 19.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 20.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 21.Dosztanyi Z, Meszaros B, Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worthington AS, Rivera H, Torpey JW, Alexander MD, Burkart MD. Mechanism-based protein cross-linking probes to investigate carrier protein-mediated biosynthesis. ACS Chem Biol. 2006;1:687–691. doi: 10.1021/cb6003965. [DOI] [PubMed] [Google Scholar]
- 23.Kapur S, et al. Mechanism based protein crosslinking of domains from the 6-deoxyerythronolide B synthase. Bioorg Med Chem Lett. 2008;18:3034–3038. doi: 10.1016/j.bmcl.2008.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosol S, Contreras-Martos S, Cedeño C, Tompa P. Structural Characterization of Intrinsically Disordered Proteins by NMR Spectroscopy. Molecules. 2013;18:10802–10828. doi: 10.3390/molecules180910802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen MR, Ruigrok RW, Blackledge M. Describing intrinsically disordered proteins at atomic resolution by NMR. Curr Opin Struct Biol. 2013;23:426–435. doi: 10.1016/j.sbi.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Uversky VN. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015;282:1182–1189. doi: 10.1111/febs.13202. [DOI] [PubMed] [Google Scholar]
- 27.Manzi L, et al. Carbene footprinting accurately maps binding sites in protein-ligand and protein-protein interactions. Nat Commun. 2016 doi: 10.1038/ncomms13288. E13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlovic-Lazetic GM, et al. Bioinformatics analysis of disordered proteins in prokaryotes. BMC Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vucetic S, et al. DisProt: a database of protein disorder. Bioinformatics. 2005;21:137–140. doi: 10.1093/bioinformatics/bth476. [DOI] [PubMed] [Google Scholar]
- 30.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin YS, Hsu WL, Hwang JK, Li WH. Proportion of solvent-exposed amino acids in a protein and rate of protein evolution. Mol Biol Evol. 2007;24:1005–1011. doi: 10.1093/molbev/msm019. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson J, Grahn M, Wright APH. Proteome-wide evidence for enhanced positive Darwinian selection within intrinsically disordered regions in proteins. Genome Biol. 2011;12:R65. doi: 10.1186/gb-2011-12-7-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellay J, et al. Bringing order to protein disorder through comparative genomics and genetic interactions. Genome Biol. 2011;12:R14. doi: 10.1186/gb-2011-12-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neduva V, Russell RB. Peptides mediating interaction networks: new leads at last. Curr Opin Biotechnol. 2006;17:465–471. doi: 10.1016/j.copbio.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Gitlin L, Hagai T, LaBarbera A, Solovey M, Andino R. Rapid Evolution of Virus Sequences in Intrinsically Disordered Protein Regions. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004529. E1004529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keatinge-Clay A. Crystal Structure of the Erythromycin Polyketide Synthase Dehydratase. J Mol Biol. 2008;384:941–953. doi: 10.1016/j.jmb.2008.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner DT, et al. Structural and Functional Trends in Dehydrating Bimodules from trans -Acyltransferase Polyketide Synthases. Structure. 2017;25:1045–1055. doi: 10.1016/j.str.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 39.Worthington AS, Burkart MD. One-pot chemo-enzymatic synthesis of reporter-modified proteins. Org Biomol Chem. 2006;4:44–6. doi: 10.1039/b512735a. [DOI] [PubMed] [Google Scholar]
- 40.Meier JL, Mercer AC, Rivera H, Burkart MD. Synthesis and Evaluation of Bioorthogonal Pantetheine Analogues for in Vivo Protein Modification. J Am Chem Soc. 2006;128:12174–12184. doi: 10.1021/ja063217n. [DOI] [PubMed] [Google Scholar]
- 41.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins-Structure Funct Bioinforma. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 42.Lescop E, Schanda P, Brutscher B. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J Magn Reson. 2007;187:163–169. doi: 10.1016/j.jmr.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Palmer AG. NMR characterization of the dynamics of biomacromolecules. Chem Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 44.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Schumann FH, et al. Combined chemical shift changes and amino acid specific chemical shift mapping of protein–protein interactions. J Biomol NMR. 2007;39:275–289. doi: 10.1007/s10858-007-9197-z. [DOI] [PubMed] [Google Scholar]
- 46.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lees JG, Miles AJ, Wien F, Wallace BA. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics. 2006;22:1955–1962. doi: 10.1093/bioinformatics/btl327. [DOI] [PubMed] [Google Scholar]
- 48.Yang JY, et al. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierce BG, et al. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.