Abstract
Purpose
IκB kinase epsilon (IKBKE; IKKε), a member of the nuclear factor-κB kinase inhibitor family, is upregulated in several human cancers, including breast cancer, prostate cancer, and ovarian cancer. Esophageal squamous cell carcinoma (ESCC) is one of the most common and most aggressively malignant cancers with dismal prognosis. However, the state of IKBKE expression in ESCC is still unknown and its potential value remains unexplored.
Patients and methods
IKBKE protein expression was evaluated by immunohistochemistry in 118 paraffin specimens of ESCC treated by curative surgery. All patients were regularly followed up by telephone over 3 years after surgery. The chi-square test, Kaplan–Meier method, and Cox proportional hazard regression model were used to analyze the relationship of IKBKE expression, clinicopathological characteristics, and prognostic value for ESCC.
Results
IKBKE expression was 61.9% (73/118) in paraffin-embedded archived ESCC. Its expression was significantly associated with tumor differentiation grade (p=0.045) and advanced TNM (pathologic tumor node metastasis) stages (p=0.023). In univariate analysis, IKBKE expression was closely associated with decreased 3-year disease-free survival (HR 1.804, 95% CI 1.076–3.027; p=0.023) and overall survival (HR 2.118, 95% CI 1.189–3.773; p=0.009). Meanwhile, in multivariate analysis it was identified as an independent prognostic factor for 3-year disease-free survival (HR 1.777, 95% CI 1.034–3.054; p=0.037) and overall survival (HR 2.078, 95% CI 1.138–3.796; p=0.017).
Conclusion
Our data indicated for the first time that IKKε expression is a highly recurrent event in ESCC and could play a pivotal role in the evaluation of prognosis. IKBKE upregulation is negatively associated with disease-free survival and overall survival. Therefore, IKBKE could serve as a prognostic variable and potential therapeutic target for this malignancy.
Keywords: IKBKE, esophageal squamous cell carcinoma, biomarker, prognosis, immunohistochemistry
Introduction
Esophageal carcinoma is the eighth most common cancer and the sixth leading cause of cancer mortality worldwide.1 It consists of various histological types with different incidence patterns. Among them, two histopathological types – esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC) – are the most common. Even so, they still have different cells of origin, risk factors, and causality. In the United States, 15,590 deaths and 16,980 new esophageal cancer cases were estimated to occur in 2015.2 In some high-risk regions, especially in developing countries, including People’s Republic of China and other East Asian countries, ESCC is responsible for 90% of all cases with an increasing incidence rate.3 Initially, barium esophagography is widely used for assessment in patients who present with progressive dysphagia symptoms of esophageal carcinoma. Currently, more examination techniques such as computed tomography (CT), magnetic resonance imaging, positron emission tomography-CT, endoscopic ultrasonography, and minimally invasive staging biopsy have been used for its diagnosis. Also, some new methods such as chemotherapy, radiotherapy, and minimally invasive esophagectomy have been used to treat esophageal cancer;1 nevertheless, the prognosis of patients remains poor and the 5-year overall survival (OS) is only 26.2%–49.4% after surgery.4
So it is urgent to identify effective and independent markers for early diagnosis and clinical prognostic prediction. During the past years, researchers worked on in-depth analysis of biological factors associated with malignant behavior and the signaling pathway outcome for certain molecules. Significantly, some new therapies to target tissue polypeptide antigen have been applied. An increasing number of related schemes are becoming available. Nuclear factor-κB (NF-κB) has already been identified as a transcription factor, which combines with a DAN element in B cell kappa immunoglobulin light-chain enhancer.5 It is characterized by a conserved 300 amino acid Rel homology domain located in the N terminus of the protein, and is used to bind DNA. NF-κB could dimerize DNA and cause the cells to become cancerous. Meanwhile, it also interacts with specific inhibitory factors known as IκB proteins.6 NF-κB mediates apoptotic signaling pathways in different types of human cancers.7,8 Some studies have shown that antiapoptotic function of activated NF-κB was an obstacle to cancer therapy.9,10 Thus, we consider whether suppressing NF-κB signaling will show an important step in the process of optimal antiesophageal cancer therapies. Normally, NF-κB complexes are sequestered in unstimulated cytoplasm as homo-heterodimers bound to IκB proteins. In stimulated cells, with tumor necrosis factor, interleukin-1, or protein kinase cascades, IκB can be phosphorylated, ubiquitinated, and subsequently degraded.11 This will result in NF-κB translocation to the nucleus and increased expression of NF-κB-regulated genes. Of course, there are a large number of active complexes to phosphorylate IκB. For example, inhibitor of NF-κB kinase epsilon (IKBKE, IKKε) is a serine/threonine kinase that belongs to the IKK family, which includes IKKα, IKKβ, IKKε, and TBK1.12,13 Comparing the amino acid sequences of IKKα, IKKβ, and IKKε showed significant sequence similarities. They have similar molecular weights of about 85 kDa, and their functional domains are approximately organized. IKKε together with IKKα and IKKβ activate the NF-κB pathway by phosphorylation and degradation of IκB.14,15 Among these IKK proteins, IKBKE protein exhibits 31% and 33% identities at the amino acid level.16 Also, only IKBKE has been proven to be overexpressed in cancers, including upregulation in ovarian cancer, prostate cancer, breast cancer, and nonsmall-cell lung carcinoma (NSCLC).17,18 However, two studies have focused on the role of IKKε in immunity and inflammation.19,20 The molecular basis of IKKε overex-pression in tumors is still unknown.
Despite these previous research studies, little is known about IKBKE expression and its exact role in esophageal carcinoma. Therefore, in this report, through clinicopatho-logical analysis of esophagus lesions sampled from esophageal cancer patients, we investigate the relationship between IKBKE expression and tumor malignant level and prognosis. These findings suggest that IKKε could be a therapeutic target and prognostic marker for this malignancy.
Materials and methods
Patients and specimens
Between January 1st and December 31st 2013, 118 patients with ESCC who received radical surgery in the Department of Thoracic Surgery at Qilu Hospital of Shandong University, Jinan, People’s Republic of China were admitted into our retrospective study. The patients had the same surgical procedure – transthoracic esophageal resection. All patients had undergone left or right thoracotomy for curative resection by subtotal or total thoracic esophagectomy, as well as at least a two-field lymphadenectomy. No patient underwent transhiatal esophagectomy. The final day guaranteed a minimum follow-up of 36 months for all included patients. Patients did not receive radiotherapy and chemotherapy before surgery. Patients were already excluded because of stage 0 disease, distant metastasis, resection not for curative intent, lack of tumor tissue, and loss to follow up. Both pathological reports and hematoxylin and eosin (H&E)-stained tissue from the primary tumor were collected from the Department of Pathology at Qilu Hospital of Shandong University. The tumor stage was divided according to the American Joint Committee on Cancer 7th edition staging manual. All of the data, including age, gender, drinking and smoking habit, histologic grade, invasion depth, lymph node metastasis, distant metastasis (pTNM stage), and number of dissected lymph nodes, was abstracted from the clinical or pathologic records. The study was approved by the Ethics Committee of Qilu Hospital of Shandong University.
Follow-up
All patients were regularly followed up by telephone every 3 months during the first 2 years after surgery. Subsequently, their information was acquired every 6 months to the end of the study (December 2016) or death. Additional interim visits were scheduled if complaints such as unexplained weight loss (more than 10% of the original body weight), pain, or renewed dysphagia arose before the next scheduled follow-up. Follow-up items included a clinical history, a physical examination, laboratory blood or urine tests, barium meal fluoroscopy, and CT. The endpoints of study were 3-year disease-free survival (DFS) and 3-year OS. DFS was measured as the time interval following surgery that patients lived free of tumor or the earliest advancement that occurred including locoregional recurrence, distant dissemination, or death from any cause. OS was the time interval from the surgery date to the last day of follow-up or the date of death.
Immunohistochemical analysis
IKBKE protein expression was examined in the 118 ESCC surgical specimens using immunohistochemistry (IHC). Briefly, 4-μm-thick formalin-fixed paraffin-embedded sections were baked at 70°C for 1 h. The sections were deparaffinized with xylenes, and rehydrated through a graded ethanol series. Then, we placed specimens in EDTA antigen retrieval buffer and microwaved them. Endogenous peroxidase activity was blocked using a solution of 3% hydrogen peroxide in methanol. Nonspecific binding sites were blocked through 20-min incubation in 1% bovine serum albumin at room temperature. The sections were incubated with anti-IKBKE rabbit polyclonal antibody (1:100, Anti-IKKi/IKKe antibody [EPR4904]; Abcam, Shanghai, People’s Republic of China) by the manufacturer’s protocol with some modification at 4°C overnight.
The anti-IKBKE antibody was performed for the specimen for at 37°C for 1 hour. Then the PBS was utilized to wash the section three times for 5 minutes at a time. Normal goat serum was applied as a negative control replacing the primary antibody. After incubation with biotinylated anti-rabbit secondary antibody for 30 min, the specimen was incubated with streptavidin alkaline peroxidase complex (Biotin-Streptavidin HRP Detection Systems SP9000; ZSGB-Bio, Beijing, People’s Republic of China). Then, the sections were color developed in AEC reagent and counterstained using 10% Mayer’s hematoxylin, dehydrated. Finally, the sections were mounted with an aqueous mounting medium provided with the kit.
Evaluation of immunostaining
The degree of immunostaining was counted independently by two observers (Wenjing Yang and Yan Qu). IKBKE expression was considered positive when yellow particles were observed in cytoplasm. The two observers recorded the average number of immunopositive cytoplasms. The recorded cytoplasms were from 400 tumor cells of about 5–10 scientifically randomized fields throughout the sections. The tumor sections were selected by observing at low power magnification (×100), with the highest number of positive cell infiltration in intratumoral areas. It is worth noting that intratumoral tissue has the highest density of tumor cell infiltration. Whenever the observers encountered heterogeneous staining, counting was carried out in the areas of highest density of tumor cells and identified at medium magnification (×200), because those tumor areas are likely to be of biologic significance. No lymphoid cells were covered in the counts although they expressed IKBKE. The Axio Vision Rel. 4.6 (Carl Zeiss, Meditec AG, Jena, Germany) computerized image analysis system was used to analyze the difference in IHC stain intensities in tumors.
Statistical analysis
All data analyses were conducted using the SPSS 16.0 (SPSS Inc., Chicago, IL, USA) statistical software package. Statistical analysis was shown by paired-sample t-test to contrast quantitative variables between groups. The associations between IKBKE expression and clinicopathological features, such as tumor T categorization, clinical stage, and lymph node metastasis status, were verified using Fisher exact tests or Pearson’s χ2 tests. Spearman’s rank correlation coefficient was used to determine the associations between numerical variables. Survival curves were plotted by the Kaplan–Meier method and compared using the log rank test. Also, in order to identify independent prognostic factors, we used Cox’s proportional hazards model through multivariate analysis. Two-sided p-values <0.05 were regarded as significant.
Results
Clinicopathological characteristics of patients
The clinicopathologic characteristics of our 118 patients are shown in Table 1. This study included 23 (19.5%) females and 95 (80.5%) males. The median age of all patients was 60.5 years, ranging from 42 to 78 years at the date of surgery. Tumor location was the cervical esophagus in 3 patients; 66 (55.9%) patients’ tumor location was in the middle third of the thoracic esophagus while 37 (31.4%) patients’ tumor occurred in the thoracic lower, and only about 12 (10.2%) patients’ tumor location was located in the upper third. The median length of the primary tumor was 3.5 cm (range, 0.5–8.5 cm). Forty-three patients (36.4%) had T1/T2 tumors, but others were T3/T4. When it came to the histopathological differentiation, 32 cases were well and 51 cases were moderate. There were 47 (39.8%) patients with positive lymph nodes, and the number of lymphatic metastases exceeded 2 in 26 (22.0%) patients. The pathological stages in all patients were 18 (15.3%) stage I, 57 (48.3%) stage II, and stage III in 43 (36.4%) patients. The median follow-up was 26.5 months (range, 3–50 months). Of the 68 (57.6%) patients who suffered recurrence during the follow-up, 4 cases were surgical anastomosis recurrences, 18 cases had lymph node metastasis, 19 were local recurrences, and 27 were distant recurrences. Median DFS was 38.7 months (range, 1.5–71.5 months). Median OS was 43.5 months (range, 2.6–71.5 months). We differentiated between IKBKE expression and no expression, depending on evaluation of immunostaining. A proportion of IKBKE-expressing cells exceeding 25% was defined as high IKBKE expression and if not, then low expression. There were 73 patients with high expression, accounting for 61.9%.
Table 1.
Clinical and histopathological characteristics of 118 esophageal squamous cell carcinoma patients
| Characteristic | Patients, n | Percentage |
|---|---|---|
| Gender | ||
| Female | 23 | 19.5 |
| Male | 95 | 80.5 |
| Age (years) | ||
| Mean ± SD | 61.1±8.2 | |
| Median | 60.5 | |
| Tumor location | ||
| Cervical | 3 | 2.5 |
| Upper | 12 | 10.2 |
| Middle | 66 | 55.9 |
| Lower | 37 | 31.4 |
| Tumor length (cm) | ||
| Mean ± SD | 3.84±1.59 | |
| Median (range) | 3.5 (0.5–8.5) | |
| T stage | ||
| T1 | 17 | 14.4 |
| T2 | 26 | 22.0 |
| T3 | 67 | 56.8 |
| T4 | 8 | 6.8 |
| Differentiation grade | ||
| Well | 32 | 27.1 |
| Middle | 51 | 43.2 |
| Poor | 35 | 29.7 |
| Lymphatic metastasis | ||
| Yes | 47 | 39.8 |
| No | 71 | 60.2 |
| Lymphatic metastasis number | ||
| <2 | 21 | 17.8 |
| ≥2 | 26 | 22.0 |
| Mean ± SD | 2.55±2.28 | |
| Median (range) | 2 (1–10) | |
| Tumor thrombus | ||
| Yes | 5 | 4.2 |
| No | 113 | 95.8 |
| TNM stage | ||
| I | 18 | 15.3 |
| II | 57 | 48.3 |
| III | 43 | 36.4 |
| IKBKE expressiona | ||
| High | 73 | 61.9 |
| Low | 45 | 38.1 |
Note:
The proportion of IKBKE-expressing cells that exceeded 25% was grouped into high expression, and if not, then low expression.
Abbreviations: T, tumor; TNM, pathological tumor node metastasis; IKBKE, inhibitor of NF-κB kinase epsilon.
IKBKE expression in ESCC and adjacent tissue
IKBKE protein expression was mainly localized in the cytoplasm. In order to examine the difference of IKBKE expression in nontumor and tumor cells, we compared its expression in adjacent normal tissues and ESCC tumor. Representative images of IKBKE positive cell infiltration are shown in Figure 1. The median proportion of IKBKE-expression cells in tumors and nontumors was 57.2% (range, 0–90.0%) and 26.1% (range, 0–53.2%), respectively (Figure 2). IKBKE expression had a 1.3-fold increase in tumors. In conclusion, IKBKE was more highly expressed in ESCC tumor cells as compared to adjacent nontumor tissues (p=0.03), and showed a positive correlation of IKBKE expression and ESCC tumors.
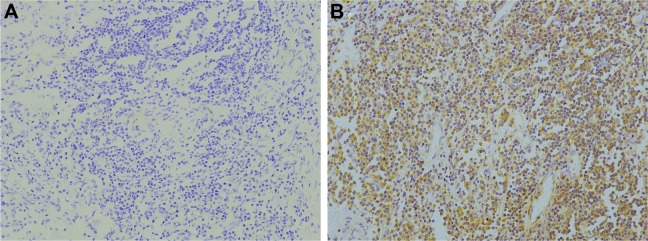
Figure 1.
Inhibitor of NF-κB kinase epsilon (IKBKE, IKKε) was significantly upregulated in esophageal squamous cell carcinoma (ESCC). Immunohistochemistry assays of IKBKE expression in 118 ESCC patients. (A) Tumor tissues with low IKKε expression. (B) High IKBKE expression in ESCC tissues. The panels were observed at medium magnification (×200). Scale bars, 1:100 μm.
Figure 2.
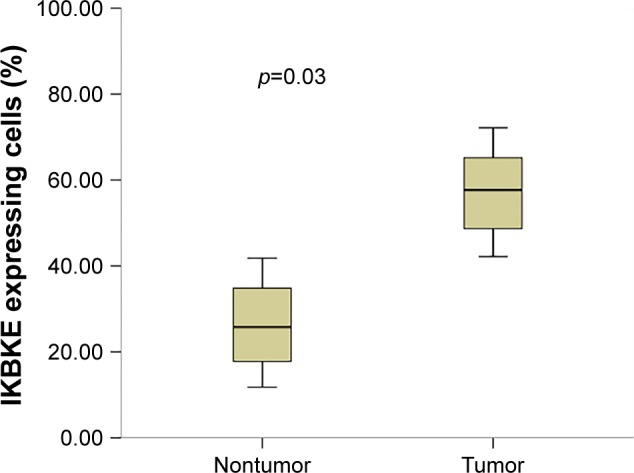
Inhibitor of NF-κB kinase epsilon (IKBKE) expression compared in tumor and adjacent nontumor tissues.
Correlation of IKBKE expression with clinicopathological parameters
The clinical characteristics of 118 ESCC patients were grouped by IKBKE low and high expression, as presented in Table 2. Based on the proportion of expression, 73 (61.9%) cases were assigned into the IKBKE group whereas the other 45 (38.1%) cases were assigned into the no expression group. No significant difference was discovered regarding gender, age, tumor location, tumor length, tumor thrombus, pathological tumor status, lymphatic metastasis number, and recurrence between the two groups. However, increased IKBKE expression was significantly associated with differentiation grade (p=0.045) and advanced pTNM stages (p=0.023). Furthermore, IKBKE expression was more frequently observed in poor pathologic differentiation and advanced cancer.
Table 2.
Correlation of IKBKE expression with clinicopathological parameters
| Characteristic | IKBKE low expressiona | IKBKE high expressiona | p-value |
|---|---|---|---|
| Gender | 0.122 | ||
| Male | 33 | 62 | |
| Female | 12 | 11 | |
| Age | 0.57 | ||
| <60.5 years | 21 | 38 | |
| ≥60.5 years | 24 | 35 | |
| Tumor location | 0.601 | ||
| Cervical/upper | 7 | 8 | |
| Middle | 26 | 40 | |
| Lower | 12 | 25 | |
| Tumor length | 0.544 | ||
| <3.84 cm | 26 | 38 | |
| ≥3.84 cm | 19 | 35 | |
| T stage | 0.071 | ||
| 1 and 2 | 21 | 22 | |
| 3 and 4 | 24 | 51 | |
| Differentiation grade | 0.045 | ||
| Well | 16 | 16 | |
| Middle | 13 | 38 | |
| Poor | 16 | 19 | |
| LN metastasis | 0.129 | ||
| Negative | 31 | 40 | |
| Positive | 14 | 33 | |
| Positive LN | 0.313 | ||
| 0 | 31 | 40 | |
| 1 | 8 | 13 | |
| 2 | 3 | 8 | |
| ≥3 | 3 | 12 | |
| pTNM stage | 0.023 | ||
| I | 12 | 6 | |
| II | 20 | 37 | |
| III | 13 | 30 | |
| Recurrence | 0.059 | ||
| No | 24 | 26 | |
| Yes | 21 | 47 | |
| Tumor thrombus | 0.648 | ||
| No | 44 | 69 | |
| Yes | 1 | 4 |
Notes: Values in bold signify p<0.05, considered significant.
The proportion of IKBKE-expressing cells that exceeded 25% was grouped into high expression, and if not, then low expression.
Abbreviations: IKBKE, inhibitor of NF-κB kinase epsilon; T, tumor; LN, lymph node; pTNM, pathological tumor node metastasis.
Correlation of IKBKE with other prognostic factors
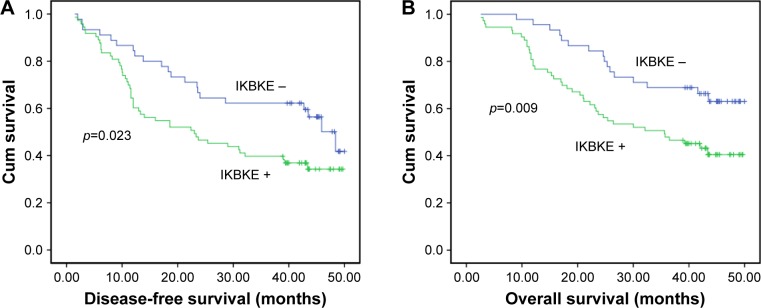
We used Kaplan–Meier curves for DFS (p<0.05) and OS (p<0.05) based on preoperative IKBKE (Figure 3). The 3-year DFS and OS for patients with IKBKE low expression were 67.1% and 71.2%, respectively, compared with 40.7% and 49.6% for patients with high expression. Patients with high preoperative IKBKE had a significant worse prognosis for DFS (p=0.023) and OS (p=0.009) than those with no IKBKE. The difference of curves between the two control groups remained statistically significant.
Figure 3.
Kaplan–Meier analysis of inhibitor of NF-κB kinase epsilon (IKBKE) expression in 118 cases with esophageal squamous cell carcinoma. IKBKE expression was significantly associated with decreased disease-free survival (A) and overall survival (B).
Abbreviation: Cum, cumulative.
In order to identify variables that affect the prognosis in ESCC patients, univariate and multivariate analyses were applied to investigate the influence between IKBKE expression and other clinicopathological parameters on the prognosis for the 118 ESCC patients. In the univariate analysis of 3-year DFS and OS, the HR of IKBKE was 1.804 (95% CI 1.076–3.027; p=0.023) and 2.118 (95% CI 1.189–3.773; p=0.009), respectively (Table 3). In the univariate model, variables closely associated with poor DFS and OS were lymphatic metastasis number (≥2), advanced tumor stages, and advanced pTNM stages (p<0.05). No prognostic importance was found for tumor location, tumor length, and differentiation grade. The multivariate analysis showed increased IKBKE expression was independently associated with decreased DFS and poor OS. The HRs for IKBKE were 1.777 (95% CI 1.034–3.054; p=0.037) and 2.078 (95% CI 1.138–3.796; p=0.017), respectively (Table 4). Therefore, IKBKE was an independent prognostic variable for 3-year DFS and OS.
Table 3.
Univariate analysis of prognostic factors in ESCC with respect to DFS and OS
| Univariate analysis | Disease-free survival
|
Overall survival
|
||
|---|---|---|---|---|
| HR (95% CI) | p-value | OS 95% CI | p-value | |
| Gender | 1.270 (0.715–2.257) | 0.415 | 1.311 (0.718–2.395) | 0.378 |
| Age | 0.868 (0.539–1.399) | 0.562 | 1.032 (0.616–1.727) | 0.906 |
| Location | ||||
| Upper | Ref | 0.148 | Ref | 0.278 |
| Middle | 1.992 (0.963–4.118) | 0.063 | 1.863 (0.845–4.107) | 0.123 |
| Low | 1.128 (0.647–1.966) | 0.671 | 1.136 (0.623–2.07) | 0.678 |
| Length | 1.196 (0.743–1.928) | 0.461 | 1.484 (0.886–2.486) | 0.133 |
| Differentiation grade | ||||
| High | Ref | 0.396 | Ref | 0.333 |
| Middle | 0.662 (0.341–1.284) | 0.222 | 0.637 (0.328–1.236) | 0.182 |
| Low | 0.974 (0.562–1.690) | 0.926 | 0.971 (0.560–1.684) | 0.916 |
| T stage (1, 2 vs 3, 4) | 1.736 (1.034–2.914) | 0.037 | 2.526 (1.380–4.622) | 0.003 |
| Lymph nodes | 2.438 (1.509–3.938) | <0.001 | 2.861 (1.697–4.823) | <0.001 |
| pTNM (I, II vs III, IV) | 2.288 (1.416–3.697) | 0.001 | 2.650 (1.576–4.456) | <0.001 |
| IKBKE expression | 1.804 (1.076–3.027) | 0.023 | 2.118 (1.189–3.773) | 0.009 |
Note: Values in bold signify p<0.05, considered significant.
Abbreviations: CI, confidence interval; ESCC, esophageal squamous cell carcinoma; DFS, disease-free survival; HR, hazard ratio; OS, overall survival; Ref, reference; T, tumor; pTNM, pathological tumor node metastasis; IKBKE, inhibitor of NF-κB kinase epsilon.
Table 4.
Multivariate analysis of prognostic factors in ESCC with respect to DFS and OS
| Multivariate analysis | Disease-free survival
|
Overall survival
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95%CI | p-value | |
| Gender | ||||||
| Male vs female | 1.562 | 0.827–2.953 | 0.17 | 1.701 | 0.874–3.311 | 0.118 |
| Age | ||||||
| <61.1 vs ≥61.1 | 0.963 | 0.559–1.660 | 0.893 | 1.366 | 0.765–2.438 | 0.292 |
| Tumor location | ||||||
| Upper | Ref | Ref | 0.243 | 0.337 | ||
| Middle | 1.939 | 0.891–4.216 | 0.095 | 1.909 | 0.808–4.511 | 0.141 |
| Lower | 1.358 | 0.741–2.940 | 0.322 | 1.282 | 0.652–2.522 | 0.472 |
| T | ||||||
| 1, 2 vs 3, 4 | 1.362 | 0.753–2.462 | 0.307 | 2.05 | 0.998–4.215 | 0.051 |
| Differentiation grade | ||||||
| Well | Ref | Ref | 0.869 | 0.573 | ||
| Middle | 0.943 | 0.447–1.990 | 0.879 | 0.653 | 0.279–1.525 | 0.325 |
| Poor | 0.855 | 0.470–1.554 | 0.607 | 0.777 | 0.415–1.454 | 0.43 |
| LN-M | ||||||
| Negative vs positive | 1.533 | 0.686–3.429 | 0.289 | 1.635 | 0.644–4.153 | 0.301 |
| pTNM | ||||||
| I, II vs III, IV | 1.576 | 0.686–3.622 | 0.283 | 1.467 | 0.551–3.905 | 0.443 |
| IKBKE | ||||||
| Low vs high | 1.777 | 1.034–3.054 | 0.037 | 2.078 | 1.138–3.796 | 0.017 |
Note: Values in bold signify p<0.05, considered significant.
Abbreviations: CI, confidence interval; HR, hazard ratio; ESCC, esophageal squamous cell carcinoma; DFS, disease-free survival; OS, overall survival; Ref, reference; T, tumor; LN-M, lymph node metastasis; pTNM, pathological tumor node metastasis; IKBKE, inhibitor of NF-κB kinase epsilon.
Discussion
Tumor suppressor genes and mutational proto-oncogenes permit incipient cancer cells to escape the physical normal regulation maintaining cell and tissue homeostasis. Especially, kinases play important roles in many aspects of human cancers and also are particularly attractive therapeutic targets. Therefore, several kinase inhibitors are already used in clinical treatment. The transcription factor NF-κB participates in numerous physiological processes such as immunity, inflammation, cell proliferation, and control of cell death.21–23 The five members of the NF-κB family, p50 (NF-κB1), p52 (NF-κB2), p65 (RelA), cRel, and RelB, can make up different dimer combinations, but a heterodimer is the most frequently detected form between p50 and the transactivating p65 subunit.24 RelA is the most abundantly and ubiquitously expressed.25 Moreover, RelA is the transcriptionally active subunit of the predominant RelA–p50 dimer. Usually, p50 (NF-κB1) and p65 (RelA), the two members of the NF-κB family, are kept inactive through complex formation with inhibitory kappa B proteins (IκBs) in cytoplasm.26 Optimal NF-κB activation is initiated by viral or bacterial infections through Toll-like receptors, phorbol 12-myristate 13-acetate, or interferon. This process involves the signal-induced phosphorylation and subsequent ubiquitin-mediated degradation of IκB by the classical IκB kinase (IKK)-dependent pathway.25,27–29 Studies have confirmed that although some dimers play a regulatory role, NF-κB proteins form dimers which canonically activate gene expression. The most prominent NF-κB dimer is present in an inactive state bound to a natural inhibitor termed IκBα. NF-κB activation is dependent on polyubiquitination that targets IκBα for degradation, releasing NF-κB dimers from the NF-κB–IκBα complex, followed by translocation to the nucleus and binding to κB enhancer elements of target genes.30 In all members of the IKK kinase family, IκB kinase (IKK) epsilon (IKKε) is different from the others in identity and similarity at the amino acid level. It is a noncanonical IKK homologue, which shares 33% and 31% amino acid identity with the corresponding domains in IKKα and IKKβ, and plays an important role in activating NF-κB signaling by phosphorylating IκBα.12,31,32 It has been well demonstrated that IKKα and IKKβ form a complex with scaffold protein IKKγ (also called NEMO) and activate NF-κB.19,33 We want to know whether IKKε has a similar function in ESCC. Gene deletion experiments suggested that IKKε was dispensable for the canonical NF-κB activation pathway.34 However, although some late NF-κB target genes could not be upregulated in IKKε cells,20 it has been presented that IKKε might regulate NF-κB at some later step. The kinase was first identified as an IKK kinase family member of the NF-κB activation by lipopolysaccharides and phorbolesters.12,15 IKKε mainly mediates NF-κB activation induced by the T cell receptor, phorbol 12-myristate 13-acetate, or interferon, but not by interleukin-1 and tumor necrosis factor, which activate IKBKA and IKBKB. Mattioli et al32 indicated that IKKε mediated inducible phosphorylation of NF-κB p65 at serine 468 in response to T cell costimulation. This phosphorylation affected RelA/p65-dependent transactivation. Adi Mazhar et al35 showed that ectopically expressed IKKε induces Ser-536 p65/RelA phosphorylation, NF-κB activation, and NF-κB-dependent gene expression. They also raised that IKKε worked in an autoregulatory loop, leading to its own expression. However, using transfection and Western blots, Koop et al36 showed that phosphorylation of p65/RelA even at both sites was insufficient to activate NF-κB-driven gene expression, indicating that most likely several mechanisms were involved in IKKε-mediated NF-κB activation. Subsequently, studies37 found that, independently of the classical IκB/IKK pathway, IKKε could directly phosphorylate the C-terminal domain of cRel in vivo and in vitro within two regions of the transactivation domain and regulated nuclear accumulation of cRel. IκBα degradation was not affected, but IKKε-mediated phosphorylation of cRel would result in dissociation of the IκBα–cRel complex, in a manner which was independent of IκBα Ser32 and Ser36 phosphorylation. Nevertheless, nuclear retention of phosphorylated cRel was not enough to induce strong cRel transactivation. This activation step could involve additional cRel posttranslational modification, association with chromatin modification, or coactivators.37 Meanwhile, chromatin immunoprecipitation experiments were used to reveal the inducible association of IKKε to the control regions of several NF-κB target genes and nuclear NF-κB activity.38 Chen et al30 discovered by IHC staining and Western blot analysis that deregulation of NF-κB activity is the result of altered translation, stability, and/or phosphorylation of proteins rather than upregulation of the genes that encode proteins such as IKKε, IκBα, or NF-κB p65. However, its precise mechanism of action remains under study.
With regard to a role in cancer, IKKε has been shown to be upregulated in breast, ovarian, and endometrial cancers as well as gliomas.39–41 Recently, many important functions of IKBKE were discovered. Research revealed that IKBKE can inhibit the function of cell growth inhibitor FOXO3a in NSCLC and breast cancer cells through SerS644 phosphorylation.22 In breast cancer, Colas et al,42 moreover, using fluorescence in situ hybridization for IKKε amplification, showed that 10 of 21 breast cancers and 13 of 17 cell lines founded IKKε protein overexpression in the absence of 1q32 copy number changes. RAS can stimulate many pathways, such as the mitogen-activated protein kinase, phosphoinositide 3 kinase, and Ral guanine nucleotide dissociation stimulator signaling pathways.43 IKBKE could work as the RAS oncogene downstream effector in NSCLC cells and meanwhile mediates Ras-based active IKKεation of NF-κB and tumorigenic activity.44 Beyond that, Boehm et al,39 using complementary genetic approaches, identified IKBKE as a breast cancer oncogene by analyzing the downstream pathways for RAS to induce tumor transformation. In order to seek kinases that took the place of AKT in the RAS–AKT pathway to promote cellular transformation, they filtrated a myristoylated kinase expression library and identified that IKKε could replace AKT in transformation and that AKT-driven cell transformation requires NF-κB activity and IKBKE. Furthermore, researchers also found that IKKε can functionally replace the AKT oncogene to promote malignant transformation by phosphorylating Akt-Thr308 and Ser473, with AKT activation.23 In this study, we did not investigate the mechanism underlying IKKε significant upregulation in human esophageal squamous carcinoma cells, but molecular studies on the precise signal pathways are still needed.
In order to identify kinase-specific expression patterns for tumor tissues, we analyzed 118 ESCC patients who received radical surgery. We utilized IHC methods to investigate the prognostic value of tumor-infiltrating IKBKE and to analyze the relationship between IKBKE expression and clinicopathological parameters in ESCC. It indicated the expression level of IKBKE in patients of the Discovery Group (all study subjects) (61.9% vs 38.1%). IKKε overexpression was positively correlated with poor prognosis, increased progression, and resistance to therapy. That was similar to our current observations. But our study firstly established that increased IKBKE expression in ESCC was significantly associated with advanced pTNM stages, lymph node metastasis, and a trend toward advanced pathological stages. Moreover, in Kaplan–Meier analysis, we presented that IKBKE expression had statistical relation with OS and DFS in 118 patients and increased IKKε expression was a poor prognosis with poor DFS and OS. It was independent of well-established clinical features, including lymphatic metastasis number (≥2), pTNM stages, and depth of invasion. Most importantly, we could indicate that IKBKE was overexpressed in ESCC tissue and it could be used as a predictive index for OS and PFS of ESCC patients. In the present study, we discovered the median DFS and OS were 18.3 and 25.4 months in the low IKBKE group, while 50.0 and 59.8 months in the high group (p<0.001).
There were some shortcomings in our study. First, it was a retrospective cohort with a relatively small sample. Second, we could not confirm whether other cutoff values act as better predictors because we did not get esophageal specimens from healthy people. Third, we did not point out the causal relationship of IKBKE low expression and cancer development. In this study, we did not investigate the accurate mechanism underlying IKKε significant upregulation in human esophageal squamous carcinoma cells. Some of the future questions that should put a new premium on analyzing the role of IKBKE in tumor development are the following. Are there other substrates for these kinases that could explain their effect on cellular proliferation? The precise signal pathways are still needed.
Conclusion
In this study, we revealed that IKKε protein expression was significantly upregulated in esophageal squamous carcinoma and positively correlated with clinical stage and tumor grade. This study identified IKBKE as a mediator of ESCC OS. It could be used to diagnose ESCC with high specificity and sensitivity. Meanwhile, it may therefore represent a new opportunity for drug development.
Ethics approval and informed consent
Clinical protocols were approved by local regulatory authorities and the study was approved by the Ethics Committee of Qilu Hospital of Shandong University. All patients signed a study written informed consent before randomization, giving agreement for the trial to use biological material for research purposes. The clinical investigations have been conducted relying on the principles expressed in the Declaration of Helsinki.
Data availability
There are no additional unpublished data. Some original data are private. Please contact the authors with data requests if necessary.
Acknowledgments
The authors would like to thank Jian Zhao (professor in Department of Thoracic Surgery, Qilu Hospital of Shandong University, Jinan, People’s Republic of China) and Junhui Zhen (pathologist in Department of Pathology, Qilu Hospital of Shandong University) for their help in the realization of this work. The work was supported by the National Science Foundation of China (number 81602007) and the Science Foundation of Shandong Province (ZR2016HB68).
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381(9864):400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Lu CL, Lang HC, Luo JC, et al. Increasing trend of the incidence of esophageal squamous cell carcinoma, but not adenocarcinoma, in Taiwan. Cancer Causes Control. 2010;21(2):269–274. doi: 10.1007/s10552-009-9458-0. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Xie X, Zhou C, Peng S, Rao D, Fu J. Which factors are associated with actual 5-year survival of oesophageal squamous cell carcinoma? Eur J Cardiothorac Surg. 2012;41(3):e7–e11. doi: 10.1093/ejcts/ezr240. [DOI] [PubMed] [Google Scholar]
- 5.Sen R, Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappaB by a post translational mechanism. Cell. 1986;47(6):921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 7.Biswas DK, Shi Q, Baily S, et al. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc Natl Acad Sci U S A. 2004;101(27):10137–10142. doi: 10.1073/pnas.0403621101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkar FH, Li YW, Wang ZW, Kong DJ. NF-kappa B signaling pathway and its therapeutic implications in human diseases. Int Rev Immunol. 2008;27(5):293–319. doi: 10.1080/08830180802276179. [DOI] [PubMed] [Google Scholar]
- 9.Weaver KD, Yeyeodu S, Cusack JC, Jr, Baldwin AS, Jr, Ewend MG. Potentiation of chemotherapeutic agents following antagonism of nuclear factor kappa B in human gliomas. J Neurooncol. 2003;61(3):187–196. doi: 10.1023/a:1022554824129. [DOI] [PubMed] [Google Scholar]
- 10.Zheng M, Morgan-Lappe SE, Yang J, et al. Growth inhibition and radio-sensitization of glioblastoma and lung cancer cells by small interfering RNA silencing of tumor necrosis factor receptor-associated factor 2. Cancer Res. 2008;68(18):7570–7578. doi: 10.1158/0008-5472.CAN-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanarek N, Ben-Neriah Y. Regulation of NF-κB by ubiquitination and degradation of the IκBs. Immunol Rev. 2012;246(1):77–94. doi: 10.1111/j.1600-065X.2012.01098.x. [DOI] [PubMed] [Google Scholar]
- 12.Peters RT, Liao SM, Maniatis T. IKK epsilon is part of a novel PMA-inducible IkappaB kinase complex. Mol Cell. 2000;5(3):513–522. doi: 10.1016/s1097-2765(00)80445-1. [DOI] [PubMed] [Google Scholar]
- 13.Tenoever BR, Ng SL, Chua MA, et al. Multiple functions of the IKK-related kinase IKK epsilon in interferon-mediated antiviral immunity. Science. 2007;315(5816):1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 14.Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science. 1997;278(5339):866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 15.Shimada T, Kawai T, Takeda K, et al. IKK-i, a novel lipopolysaccharide-inducible kinase that is related to IkappaB kinases. Int Immunol. 1999;11(8):1357–1362. doi: 10.1093/intimm/11.8.1357. [DOI] [PubMed] [Google Scholar]
- 16.Schmitz ML, Bacher S. Novel molecular targets in the search for anti-inflammatory agents. Phytochem Rev. 2005;4:19–25. [Google Scholar]
- 17.Guo J, Kim D, Gao J, et al. IKBKE is induced by STAT3 and tobacco carcinogen and determines chemosensitivity in non-small cell lung cancer. Oncogene. 2013;32(2):151–159. doi: 10.1038/onc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Murphy LC, Seekallu SV, Watson PH. Clinical significance of estrogen receptor phosphorylation. Endocr Relat Cancer. 2011;18(1):R1–R14. doi: 10.1677/ERC-10-0070. [DOI] [PubMed] [Google Scholar]
- 19.Clément JF, Meloche S, Servant MJ. The IKK-related kinases: from innate immunity to oncogenesis. Cell Res. 2008;18(9):889–899. doi: 10.1038/cr.2008.273. [DOI] [PubMed] [Google Scholar]
- 20.Kravchenko VV, Mathison JC, Schwamborn K, Mercurio F, Ulevitch RJ. IKKi/IKKepsilon plays a key role in integrating signals induced by pro-inflammatory stimuli. J Biol Chem. 2003;278(29):26612–26619. doi: 10.1074/jbc.M303001200. [DOI] [PubMed] [Google Scholar]
- 21.Guo JP, Shu SK, He L, et al. Deregulation of IKBKE is associated with tumor progression, poor prognosis, and cisplatin resistance in ovarian cancer. Am J Pathol. 2009;175(1):324–333. doi: 10.2353/ajpath.2009.080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo JP, Tian W, Shu S, Xin Y, Shou C, Cheng JQ. IKBKE phosphorylation and inhibition of FOXO3a: a mechanism of IKBKE oncogenic function. PLoS One. 2013;8(5):e63636. doi: 10.1371/journal.pone.0063636. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Xie X, Zhang D, Zhao B, et al. IkappaB kinase epsilon and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proc Natl Acad Sci U S A. 2011;108(16):6474–6479. doi: 10.1073/pnas.1016132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natoli G, De Santa F. Shaping alternative NF-kappaB-dependent gene expression programs: new clues to specificity. Cell Death Differ. 2006;13(5):693–696. doi: 10.1038/sj.cdd.4401880. [DOI] [PubMed] [Google Scholar]
- 25.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8(1):49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 26.Silverman N, Maniatis T. NF-kappa B signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15(18):2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 27.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of Ikap-paBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84(6):853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 28.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-kappaB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 29.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 30.Chen WQ, Lin J, Cao H, Qian WB. Local and systemic IKKε and NF-κB signaling associated with Sjögren’s syndrome immunopathogenesis. J Immunol Res. 2015;2015:534648. doi: 10.1155/2015/534648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verhelst K, Verstrepen L, Carpentier I, Beyaert R. IκB kinase ε (IKKε): a therapeutic target in inflammation and cancer. Biochem Pharmacol. 2013;85(7):873–880. doi: 10.1016/j.bcp.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattioli I, Geng H, Sebald A, et al. Inducible phosphorylation of NF-kappa B p65 at serine 468 by T cell costimulation is mediated by IKK epsilon. J Biol Chem. 2006;281(10):6175–6183. doi: 10.1074/jbc.M508045200. [DOI] [PubMed] [Google Scholar]
- 33.Chau TL, Gioia R, Gatot JS, et al. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem Sci. 2008;33(4):171–180. doi: 10.1016/j.tibs.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Hemmi H, Takeuchi O, Sato S, et al. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med. 2004;199(12):1641–1650. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adi M, Baldwin AS. IKK-i/IKKepsilon controls constitutive, cancer cell-associated NF-kappaB activity via regulation of Ser-536 p65/RelA phosphorylation. J Biol Chem. 2006;281(37):26976–26984. doi: 10.1074/jbc.M603133200. [DOI] [PubMed] [Google Scholar]
- 36.Koop A, Lepenies I, Braum O, et al. Novel splice variants of human IKKε negatively regulate IKKε-induced IRF3 and NF-kB activation. Eur J Immunol. 2011;41(1):224–234. doi: 10.1002/eji.201040814. [DOI] [PubMed] [Google Scholar]
- 37.Harris J, Olière S, Sharma S, et al. Nuclear accumulation of cRel following C-terminal phosphorylation by TBK1/IKK epsilon. J Immunol. 2006;177(4):2527–2535. doi: 10.4049/jimmunol.177.4.2527. [DOI] [PubMed] [Google Scholar]
- 38.Moreno R, Sobotzik JM, Schultz C, Schmitz ML. Specification of the NF-kappaB transcriptional response by p65 phosphorylation and TNF-induced nuclear translocation of IKK epsilon. Nucleic Acids Res. 2010;38(18):6029–6044. doi: 10.1093/nar/gkq439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boehm JS, Zhao JJ, Yao J, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129(6):1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 40.Guo JP, Shu SK, Esposito NN, Coppola D, Koomen JM, Cheng JQ. IKKepsilon phosphorylation of estrogen receptor alpha Ser-167 and contribution to tamoxifen resistance in breast cancer. J Biol Chem. 2010;285(6):3676–3684. doi: 10.1074/jbc.M109.078212. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Guan H, Zhang H, Cai J, et al. IKBKE is over-expressed in glioma and contributes to resistance of glioma cells to apoptosis via activating NF-κB. J Pathol. 2011;223(3):436–445. doi: 10.1002/path.2815. [DOI] [PubMed] [Google Scholar]
- 42.Colas E, Perez C, Cabrera S, et al. Molecular markers of endometrial carcinoma detected in uterine aspirates. Int J Cancer. 2011;129(10):2435–2444. doi: 10.1002/ijc.25901. [DOI] [PubMed] [Google Scholar]
- 43.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 44.Kim D, Guo J, Challa S, Coppola D, Cheng JQ. IKBKE is a key mediator of Ras activation of NF-κB and Ras oncogenic function. Cancer Res. 2014;74(19 Suppl) Abstract 4416. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no additional unpublished data. Some original data are private. Please contact the authors with data requests if necessary.